SUMMARY
Mutations in several genes encoding centrosomal proteins dramatically decrease the size of the human brain. We show that Aspm and Wdr62 interact genetically to control brain size, with mice lacking Wdr62, Aspm or both showing gene dose-related centriole duplication defects that parallel the severity of the microcephaly, and increased ectopic basal progenitors, suggesting premature delamination from the ventricular zone. Wdr62 and Aspm localize to the proximal end of the mother centriole and interact physically, with Wdr62 required for Aspm localization, and both proteins, as well as microcephaly protein Cep63, required to localize CENPJ/CPAP/Sas-4, a final common target. Unexpectedly, Aspm and Wdr62 are required for normal apical complex localization and apical epithelial structure, providing a plausible unifying mechanism for the premature delamination and precocious differentiation of progenitors. Together, our results reveal links between centrioles, apical proteins, and cell fate, and illuminate how alterations in these interactions can dynamically regulate brain size.
INTRODUCTION
The human cerebral cortex is dramatically larger compared to other mammals, and is responsible for many complex cognitive abilities. Microcephaly (MCPH) is a clinical condition in which head size is severely reduced, resulting in intellectual disability. Mutations in ASPM (Abnormal Spindle-like, Microcephaly-associated) and WDR62 (WD Repeat-containing protein 62) are the two most common genetic causes of primary microcephaly, together accounting for more than half of all cases (Manzini and Walsh, 2011). ASPM mutations cause severe microcephaly with relatively well-preserved gyral pattern and cortical architecture (Bond et al., 2002), while null WDR62 mutations typically cause additional cortical malformations as well (Bilguvar et al., 2010; Nicholas et al., 2010; Yu et al., 2010).
Although most known microcephaly genes, including WDR62 and ASPM, encode proteins that localize to the mitotic spindle poles, suggesting a common biological function (Megraw et al., 2011), the role of these proteins in the interphase centrosome is not clear. The centrosome consists of a pair of centrioles surrounded by pericentriolar material, which nucleates microtubules that allow the centrosome to serve as the major microtubule-organizing center in mammalian cells (Nigg and Stearns, 2011). While the older or ‘mother’ centriole gives rise to the primary cilium in G1, centriole duplication occurs at the proximal end of existing mother and daughter centrioles during S phase in conjunction with DNA replication (Nigg and Stearns, 2011). Mutations in CENPJ, CEP152 and CEP63 all cause MCPH (Bond et al., 2005; Guernsey et al., 2010; Sir et al., 2011). CENPJ regulates centriole duplication and elongation (Schmidt et al., 2009), and mice conditionally mutant for CENPJ/Sas-4 show that neurogenesis defects in microcephaly may arise from depletion of centrosomes and subsequent loss of primary cilia (Insolera et al., 2014). CEP63 and CEP152 co-localize in a ring-like pattern at the proximal end of the maternal centriole (Sir et al., 2011), and are required for centriole duplication (Brown et al., 2013). Some have proposed that primary microcephaly-associated (MCPH) proteins may be required in a step-wise manner for recruiting each other to the centrosome, thereby ensuring proper centriole duplication (Delattre et al., 2006; Kodani et al., 2015).
Whereas Aspm and Wdr62 have been implicated in proliferation of neocortical progenitors occupying the ventricular zone (VZ) lining the ventricles and the adjacent subventricular zone (SVZ), two Aspm gene-trap mice show surprisingly mild reduction in brain size (Pulvers et al., 2010). RNAi knockdown of Wdr62 in mice results in premature cell cycle exit (Bogoyevitch et al., 2012), while Wdr62 intron 14 gene-trap mice, which show decreased protein levels, have defects in mitotic progression of embryonic neural progenitors and also modestly reduced brain size (Chen et al., 2014). These very mild phenotypes of single mutants of Aspm and Wdr62 create a challenge to understanding the mechanism of microcephaly, and the nature of their genetic and physical interactions.
Here we show that Aspm and Wdr62 have close interactions both genetically and biochemically, and that they and other microcephaly proteins converge on CENPJ/CPAP/Sas-4 as a final common target. Mice lacking both Wdr62 and Aspm are embryonically lethal, whereas heterozygous mutation of either gene enhances the phenotype of mutations in the other. We show for the first time that WDR62 and ASPM share common localization during interphase at mother centrioles, where they physically interact. Mouse embryonic fibroblasts (MEFs) deficient in Wdr62, Aspm or both show centriole duplication defects, with the severity of the cellular defect proportional to the severity of the microcephaly. Finally, we find that Wdr62 and Aspm are required for the organization of the apical complex proteins that function as cell fate determinants at the ventricular surface (Kim et al., 2010; Paridaen et al., 2013), providing a mechanism for their dynamic control of brain size.
RESULTS
Wdr62 and Aspm interact genetically in mice
We prepared new Wdr62 gene-trap mice and Aspm knockout mice that both express no detectable protein (Figure S1). The Wdr62 gene-trap cassette inserted between exons 21 and 22, upstream of several human patient mutations in WDR62 associated with severe phenotypes (Figures S1A and S1B). We confirmed by RT-PCR that Wdr62 exon 21 spliced to the Engrailed-2 splice acceptor of the gene-trap vector and that no wild-type Wdr62 mRNA (Figures S1C and S1D) or protein (Figure S1E) could be detected in homozygous gene-trap mutants. In the Aspm homozygous mutant mouse, a neomycin cassette replaced exons 1–3 (Figure S1F and S1G), and no full-length Aspm protein was detectable by Western blot (Figures S1H).
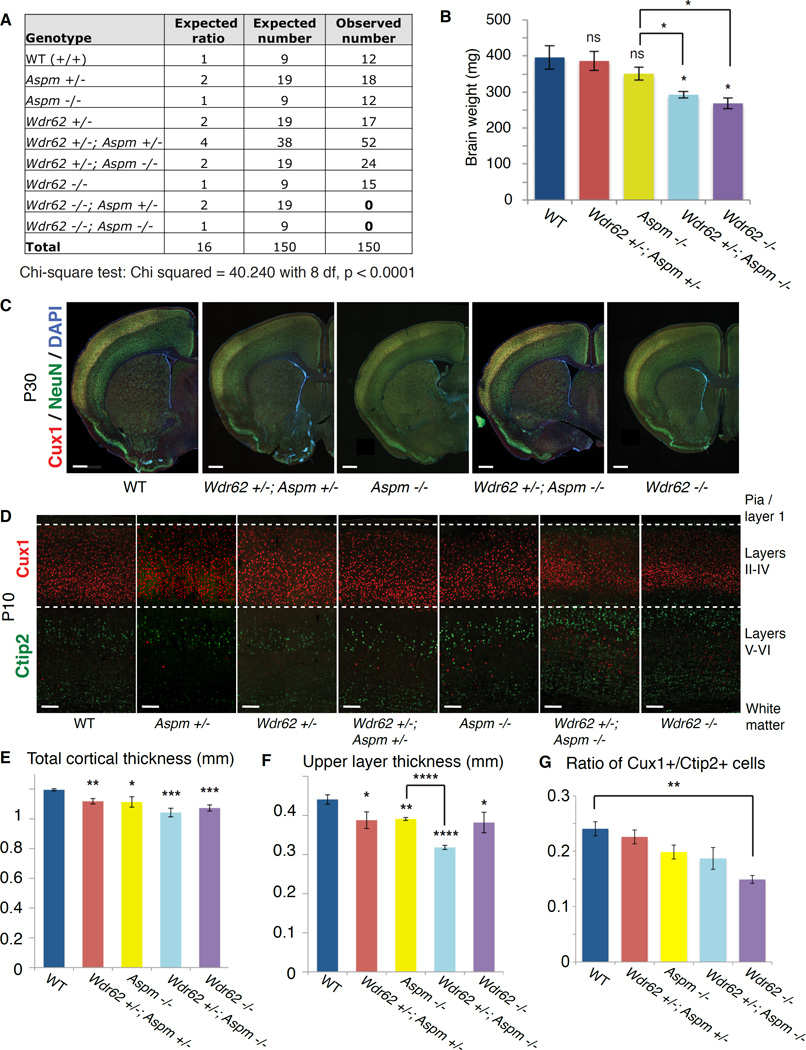
Wdr62-null mice show more severe brain phenotypes than Aspm-null mice, with severely reduced brain weight at postnatal day 30 (P30) compared to wild-type (WT) or heterozygous (het) controls (unpaired t-test, p = 0.03), as well as Aspm −/− mice (Figures S1I, S1J and 1B; p = 0.02). The reduced brain size is demonstrated by immunostaining for Cux1 and NeuN, which label upper cortical layer neurons (II–IV) and all neurons, respectively (Figure 1C). P10 Wdr62 −/− brain shows severely reduced cortical thickness (p = 0.0002), upper/superficial neuronal layer thickness (p = 0.03) and ratio of Cux1+ (upper layer) to Ctip2+ (lower layer) neurons (p = 0.003) compared to controls (Figures 1D–1G). P10 Aspm −/− brain shows a milder reduction in overall cortical thickness (p = 0.02), with a preferential reduction in the superficial cortical layers (p = 0.003; Figures 1D–1F).
Figure 1. Wdr62 and Aspm interact genetically to control brain size in mice.
(A) Table of expected genotypic classes, Mendelian ratios, and predicted vs. observed numbers of progeny resulting from crossing Wdr62 +/−; Aspm +/− × Wdr62 +/−; Aspm +/− mice (trans het × trans het). Chi square test, p < 0.0001.
(B) Brain weights measured at P30 show a declining trend, with Wdr62 +/−; Aspm −/− and Wdr62 −/− mice showing a significant reduction in brain weight compared to WT and het controls, as well as Aspm −/− mice. Error bars indicate mean ± SEM. “ns” = not significant. * = p < 0.05.
(C) P30 coronal brain sections of various genotypes (from left to right: WT, Wdr62 +/−; Aspm +/−, Aspm −/−, Wdr62 +/−; Aspm −/− and Wdr62 −/−) stained for Cux1 (to label upper layers) and NeuN (to label all neurons) show a severe reduction in overall brain size in the Wdr62 +/−; Aspm −/− and the Wdr62 −/−. Scale bar = 500 µm.
(D) P10 coronal brain sections (from left to right: WT, Aspm +/−, Wdr62 +/−, Wdr62 +/−; Aspm +/−, Aspm −/−, Wdr62 +/−; Aspm −/− and Wdr62 −/−) showing primary somatosensory cortex (barrel region) stained for Cux1 and Ctip2 to label upper layers (II–IV) and lower layers (V–VI), respectively. Overall cortical thickness and upper layer thickness are severely reduced in the Wdr62 −/− brain and the Wdr62 +/−; Aspm −/− brain, with a mild reduction in thickness of the upper layers in the Aspm −/− cortex. Scale bar = 100 µm.
(E–G) * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001 and “ns” = not significant. Wdr62 +/− and Aspm +/− are not significantly different from WT (not shown). Error bars indicate mean ± SEM.
(E) Quantification of overall cortical thickness.
(F) Quantification of thickness of upper layers. Wdr62 +/− and Aspm +/− are not significantly different from WT in upper layer thickness (not shown).
(G) Quantification of ratio of Cux1+ cells to Ctip2+ cells. See also Supplemental Figure S1.
Wdr62 and Aspm show extensive genetic interactions even in the heterozygous state (Figure 1). Double knockout (Wdr62 −/−; Aspm −/−) progeny were never recovered (chi-square test, p < 0.0001), even as early as E9.5, suggesting that knockout of both Wdr62 and Aspm in combination causes embryonic lethality (Figure 1A). Although Wdr62 −/− mice are viable, Wdr62 −/−; Aspm +/− progeny die before birth (Figure 1A), and are rarely recovered at E12.5 or earlier (data not shown). While Wdr62 +/−; Aspm −/− mice are viable (Figure 1A), the reduction in brain weight (Figure 1B, p = 0.02) and thinning of the upper layers (Figures 1D and 1F; p < 0.0001) are enhanced relative to Aspm −/− mice, suggesting preferential effects on late neurogenesis. P30 brains showed marked overall size reduction in Wdr62 +/−; Aspm −/− mice compared to controls (Figure 1C). Remarkably, even Wdr62 +/−; Aspm +/− mice show modest but statistically significant reduction in both overall cortical thickness (p = 0.002) and upper layer thickness (p = 0.03) relative to control (Figures 1E and 1F), demonstrating nonallelic noncomplementation, in which recessive alleles at two unlinked genetic loci fail to complement. Nonallelic noncomplementation typically implies that two genes share common cellular functions and often encode proteins that physically interact (Yook et al., 2001).
Loss of Wdr62 and Aspm results in expansion of basal progenitors beyond the germinal zones
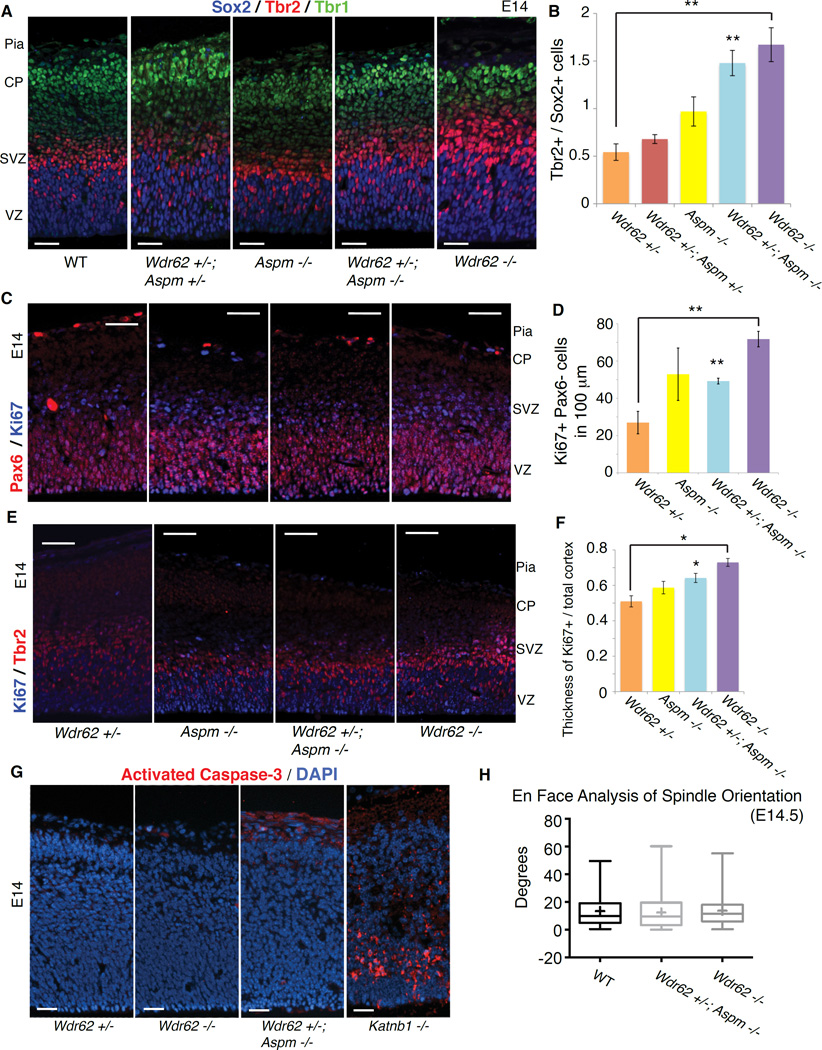
Disrupting Wdr62 and Aspm dramatically increases basal progenitor cells at mid-neurogenesis, suggesting regulation of cell fate rather than merely mitotic progression. Triple staining at E14.5 for Sox2, Tbr2 and Tbr1 shows dramatically increased Tbr2+ basal progenitors relative to Sox2+ apical progenitors in Wdr62 −/− (p = 0.005) and Wdr62 +/−; Aspm −/− (p = 0.004) animals (Figures 2A, 2B and S2A). Immunostaining of E14.5 embryos for Ki67 and Pax6 (which labels apical progenitors) revealed significantly increased numbers of Ki67-positive/Pax6-negative cells in the Wdr62 +/−; Aspm −/− (p = 0.007) and most notably in the Wdr62 −/− brain (p = 0.008), consistent with a relative reduction in apical progenitors (Figures 2C and 2D). Immunostaining for Ki67 and Tbr2 confirmed an increase in Tbr2+ cells in the mutants (Figures 2E and 2F). Together, these findings support the hypothesis of a fate switch from Pax6+ apical progenitors to Tbr2+ basal progenitors, leading to precocious formation of Tbr2+ cells.
Figure 2. Loss of Wdr62 and Aspm results in apical to basal progenitor fate switch with expansion beyond the germinal zones in developing neocortex.
(A) Loss of Wdr62 and Aspm results in expansion of the Tbr2+ basal progenitor pool at E14.5. Left to right: WT, Wdr62 +/−; Aspm +/−, Aspm −/−, Wdr62 +/−; Aspm −/− and Wdr62 −/− cortex stained for Sox2 (blue), Tbr2 (red) and Tbr1 (green) to label apical progenitors (APs) in the ventricular zone (VZ), basal progenitors (BPs) in the subventricular zone (SVZ) and neurons in layer VI of the cortical plate (CP), respectively. Scale bar = 25 µm.
(B) Quantification of ratio of Tbr2+ / Sox2+ cells in (A).
(C) Relative decrease in Pax6+ APs in Wdr62 −/− cortex at E14.5. Left to right: WT, Aspm −/−, Wdr62 +/−; Aspm −/− and Wdr62 −/− cortex stained for Pax6 (red) and Ki67 (blue) reveals an increase in the proportion of Ki67+ cycling cells that are Pax6− in the mutants, esp. Wdr62 −/−. Scale bar = 50 µm.
(D) Quantification of Ki67+Pax6− cells per 100 µm of ventricular surface in (C).
(E) Ectopic Tbr2+ basal progenitors outside the SVZ in Wdr62 −/− cortex at E14. Left to right: WT, Wdr62 +/−, Aspm −/−, Wdr62 +/−; Aspm −/− and Wdr62 −/− cortex stained for Ki67 (blue) and Tbr2 (red) to label proliferating cells and basal progenitors, respectively. The thickness of the region containing Ki67+ proliferating cells is marked by the bottom and middle hash lines, while the thickness of the entire cortex is marked by the bottom and top hash lines. Scale bar = 50 µm.
(F) Quantification of thickness of Ki67+ cells / overall cortical thickness shows that Ki67+ cells are expanded beyond the germinal zones (VZ/SVZ) in the mutants relative to the controls, suggesting that the neural progenitors are delocalized from the VZ/SVZ.
(G) Minimal apoptosis in single and double mutant cortex at E14. Left to right: Wdr62 +/− (negative control), Wdr62 −/−, Wdr62 +/−; Aspm −/− and Katnb1 homozygous gene-trap (positive control) mouse brain sections, stained for activated caspase-3 (red) and counterstained with DAPI. Scale bar = 25 µm.
(H) 3D en face analysis of spindle orientation shows no significant difference in mitotic spindle orientation of neural progenitors between WT, Wdr62 +/−; Aspm −/− and Wdr62 −/− embryos. Shown are box and whisker plots quantifying the spindle orientation of VZ progenitors in anaphase at E14.5 in WT (n = 60 cells, 4 animals), Wdr62 +/−; Aspm −/− (n = 40 cells, 3 animals) and Wdr62 −/− (n = 49 cells, 3 animals). Box boundaries extend from 25th to 75th percentiles; the line in the middle and the cross represent the median and mean, respectively.
The whiskers denote the range of values from smallest to largest. See also Supplemental Figure S2.
Immunostaining of E14.5 embryos for Ki67 also revealed that cycling progenitors are displaced from the germinal zones in Wdr62 −/− and Wdr62 +/−; Aspm −/− mutant cortex. As the cortex of all three mutants (including Aspm −/−) was reduced, we measured the thickness of the Ki67+ zone and normalized it to the thickness of the entire cortex. This ratio of Ki67+ to overall cortical thickness in the Wdr62 −/− and Wdr62 +/−; Aspm −/− mutants is expanded relative to the controls (Figures 2E and 2F), suggesting that some progenitors are leaving the VZ and SVZ, similar to the delamination of progenitors out of the VZ observed in Sas-4 −/− mice (Insolera et al., 2014). Staining for activated caspase-3, a marker of apoptosis, at E14.5 revealed little-to-no apoptosis in Wdr62 −/− and Wdr62 +/−; Aspm −/− mouse brains (Figure 2G), especially when compared to Katnb1 −/− (Hu et al., 2014) or Sas-4 −/− mice (Insolera et al., 2014) mice, which show robust apoptosis in developing brain. Together, these results suggest that loss of Wdr62 causes a cell fate change without significant apoptosis.
Wdr62 −/− and Wdr62 −/−; Aspm +/− mice analyzed at E12.5 for mitotic spindle orientation using conventional 2D microscopy as well as a 3D en face technique in flat-mount cortex (Juschke et al., 2014) did not show consistent changes in mutants. We found that spindle orientation in the mutants was not significantly different from controls in anaphase (Figure 2H), indicating that spindle orientation changes are unlikely to explain the microcephaly in these mutants.
Wdr62 and Aspm genetically interact to control centriole duplication
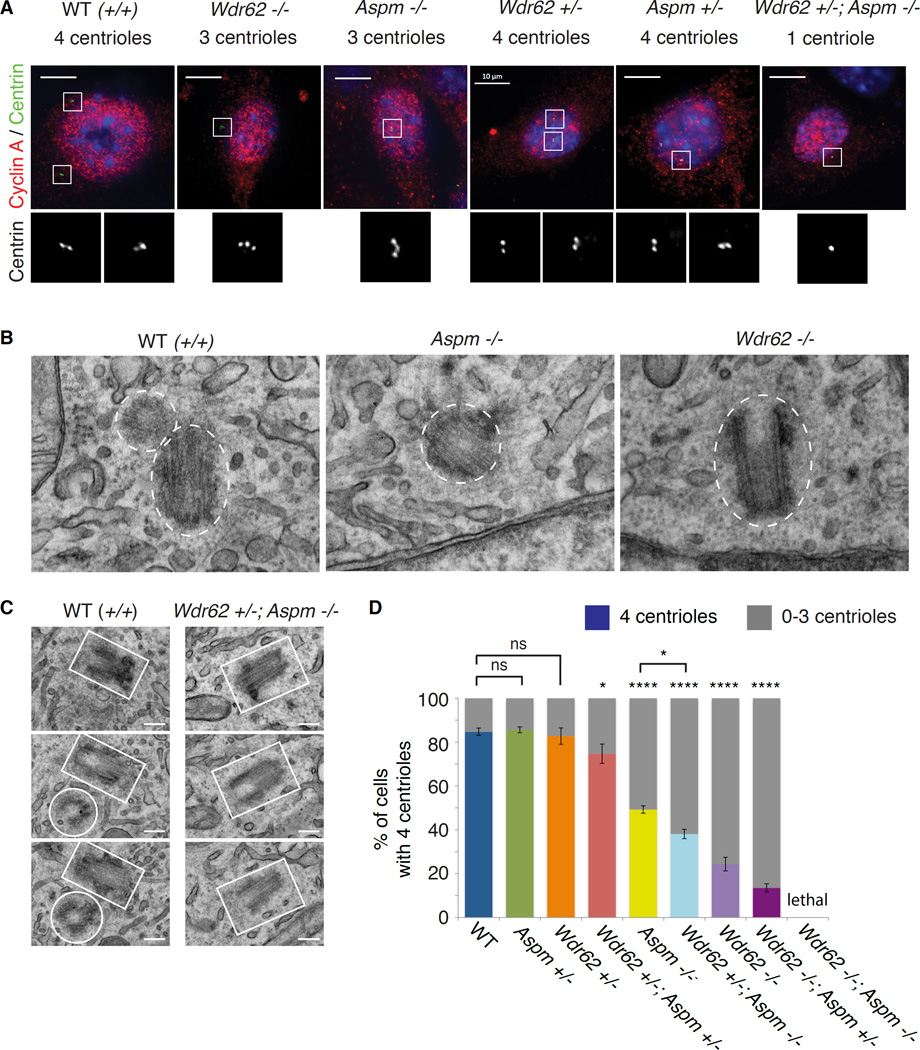
As sensitive genetic interactions are often observed between proteins that act closely together in a common molecular pathway, we explored potential cellular functions that Wdr62 and Aspm may share and identified a shared role for the two proteins in centriole duplication. As centriole duplication commences with procentriole formation at the G1-S transition (Nigg and Stearns, 2011), we examined centriole number in WT, Wdr62-null and Aspm-null MEFs stained for Centrin and Cyclin A to label centrioles and S-phase/G2 cells, respectively. Wdr62 −/− and Aspm −/− MEFs show reduced centriole numbers (from 0–4 in S-phase/G2), indicating defective duplication (Figure 3A). To confirm these results, we performed electron microscopy (EM) of WT, Aspm −/− and Wdr62 −/− MEFs (including serial EM of up to 10 consecutive 80 nm sections for Wdr62 −/− mutants). EM in WT cells revealed centrioles generally in pairs, positioned orthogonally and in close proximity, whereas some Aspm −/− and Wdr62 −/− cells contained single, unpaired centrioles, confirming defective centriole duplication (Figures 3B and 3C). Several mutant cells were acentriolar (not shown), consistent with immunofluorescence of the MEFs (Figure S3B). Whereas nearly 85% of Cyclin A-positive WT MEFs had 4 centrioles, < 25% of Wdr62 −/− MEFs (p < 0.0001) and < 50% of Aspm −/− MEFs (p < 0.0001) had 4 centrioles (Figure 3D). MEFs from Wdr62 +/−; Aspm +/− (‘trans het’) embryos had mild but statistically significant centriole duplication defects (Figures S3A and 3D; p = 0.01), while Wdr62 +/−; Aspm −/− MEFs showed severe, highly significant effects (Figures 3A and 3D; p < 0.0001) confirmed by serial EM (Figure 3C). The more severe centriole duplication defect observed in Wdr62 +/−; Aspm −/− MEFs compared to Aspm −/− MEFs (Fisher’s exact text, p = 0.02) further demonstrates the genetic interactions between Wdr62 and Aspm.
Figure 3. Wdr62 and Aspm genetically interact to control centriole duplication.
(A) Mouse embryonic fibroblasts (MEFs) stained with Centrin (green) and Cyclin A (red) to mark centrioles and S-phase/G2 cells, respectively, showing that loss of Wdr62, Aspm or both leads to underduplication of centrioles. Left to right: Wild-type (WT), Wdr62 −/−, Aspm −/−, Wdr62 +/−, Aspm +/−, Wdr62 +/−; Aspm −/−. Insets (in grayscale) show Centrin staining at higher magnification. Scale bar = 10 µm.
(B) EM of representative WT (left), Aspm −/− (middle) and Wdr62 −/− (right) MEFs, with white dotted ovals or circles outlining the individual centrioles. Pairs of centrioles are seen in WT MEFs, while only one unpaired centriole is visible in Aspm −/− and Wdr62 −/− MEFs. Scale bar = 100 nm.
(C) Serial EM confirms centriole duplication defect in Wdr62 +/−; Aspm −/− mouse embryonic fibroblasts (MEFs). Left: White rectangles and circles outline the pair of centrioles in serial sections of a representative WT cell. Right: Only one centriole (outlined in white) is visible through several serial sections of a representative Wdr62 +/−; Aspm −/− cell. Scale bar = 20 nm.
(D) Quantification of centriole number in S-phase cells of various genotypes indicates that the severity of the centriole duplication defect is proportional to the severity of the microcephaly phenotype. N = 200–300 cells per genotype. Fisher’s exact test, “ns” = not significant, * = p < 0.05 and **** = p < 0.0001 (compared to WT unless otherwise indicated). See also Supplemental Figure S3.
The severity of the centriole duplication defect in different Wdr62 and Aspm backgrounds parallels the severity of the microcephaly (Figure S3C). When the percentage of cells with 4 centrioles was plotted as a function of genotype, the most severe centriole reduction was in MEFs from Wdr62 −/−; Aspm +/− mice, which do not survive to birth (Figures 3D and 1B). Wdr62 −/− MEFs were slightly less severe, followed by Wdr62 +/−; Aspm −/−, while Aspm −/− S-phase MEFs showed a more moderate phenotype (Figure 3D), closely tracking cortical thickness and brain weights (Figures 3D and 1B). The mildest centriole defect was seen in the Wdr62 +/−; Aspm +/− (trans het) MEFs, consistent with the mild microcephaly (Figure 3D). Together, these data suggest that Wdr62 and Aspm may function together in a single pathway, likely with a common output, to control centriole duplication.
WDR62 interacts with ASPM and recruits it to the mother centriole
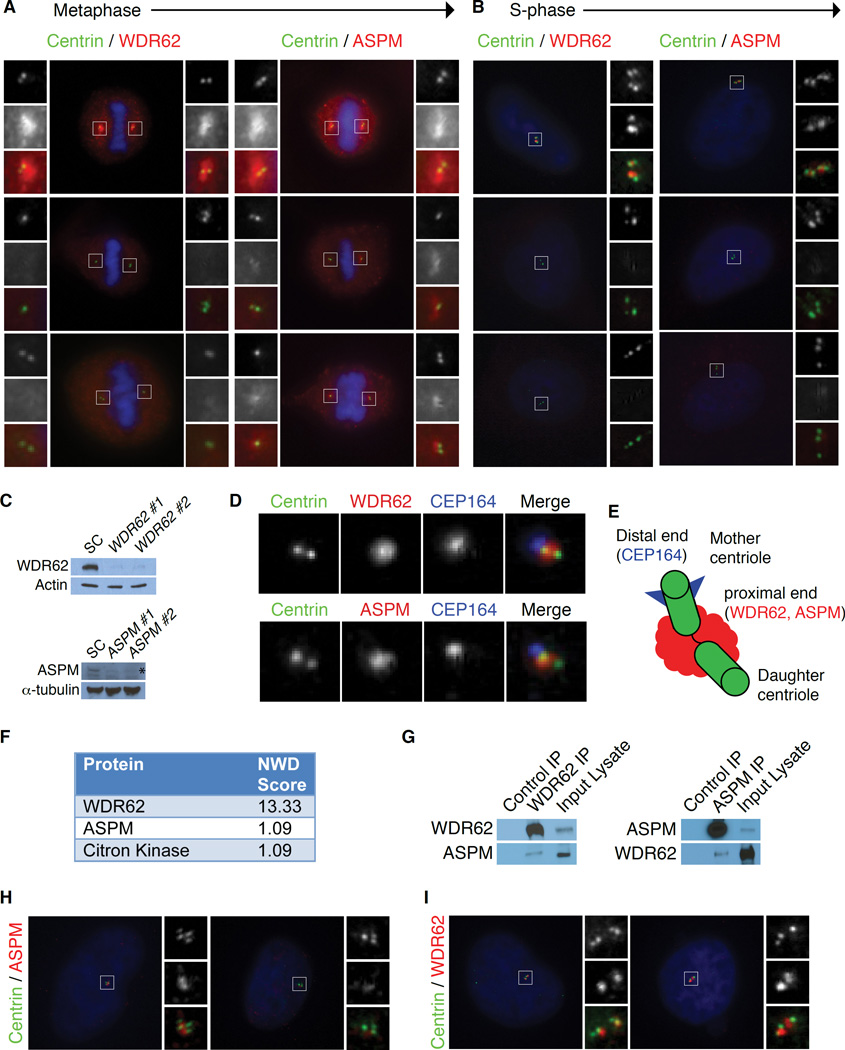
WDR62 and ASPM proteins share similar subcellular localization at the spindle poles in mitotic cells (Figure 4A)(Higgins et al., 2010; Yu et al., 2010), but this localization does not explain their role in centriole duplication, which occurs during interphase. In S-phase, WDR62 and ASPM were detected between each pair of centrioles in WT cells transfected with a control siRNA (Figure 4B). Transfection of two distinct siRNAs each against human WDR62 and ASPM abolished the centrosomal signal in metaphase and S-phase (Figures 4A and 4B), and produced efficient protein knockdown by Western analysis (Figure 4C), confirming the specificity of both antibodies. Most WDR62- and ASPM-depleted S-phase cells had 2–3 centrioles, confirming that both are required for centriole duplication in human cells (Figures 4A and 4B). WDR62 and ASPM localize asymmetrically to the proximal end of the mother centriole by late G1 (Figure S4A and Kodani et al., 2015), confirmed by co-staining with an antibody to the distal appendage marker CEP164 (Figure 4D). This colocalization hints at a physical complex of WDR62 and ASPM at the mother centriole in interphase (Figure 4E).
Figure 4. WDR62 recruits ASPM to form a physical complex at the mother centriole.
(A–B) WDR62 and ASPM are proximal centriole proteins required for centriole duplication in human cells. Using two distinct siRNAs against human WDR62 (WDR62 #1 and WDR62 #2) and ASPM (ASPM #1 and ASPM #2), the centrosomal signal can be abolished in metaphase and S-phase transfected cells. The majority of WDR62- and ASPM-depleted cells had two or three centrioles. Insets: c = Centrin; w = WDR62 and a = ASPM.
(A) WDR62 and ASPM localize to the mitotic spindle poles during metaphase.
(B) During S-phase, WDR62 and ASPM can be detected between each centriole pair in scrambled control (SC) transfected cells.
(C) Top: WDR62 antibody specifically detects endogenous WDR62 by Western in scrambled control (SC) transfected cells; signal decreases upon knockdown with siRNAs WDR62 #1 and WDR62 #2. Bottom: VTKR peptide antibody specifically detects endogenous ASPM by Western in scrambled control (SC) transfected cells; signal decreases upon knockdown with siRNAs ASPM #1 and ASPM #2. The asterisk represents the specific band.
(D) Both WDR62 and ASPM localize to the mother centriole in interphase cells. Top: HeLa cell co-stained for Centrin (to label centrioles), endogenous WDR62 and CEP164 (to label the mother centriole). Bottom: HeLa cell co-stained for Centrin (to label centrioles), endogenous ASPM and CEP164 (to label the mother centriole).
(E) Schematic of the centrosome composed of two centrioles (green), including the mother centriole marked with distal appendages (blue) and the daughter centriole (unmarked), connected by a linker (black). WDR62 and ASPM both localize to the proximal end of the mother centriole (red cloud).
(F) Table showing a selected list of hits from the IP-mass-spectrometry screen that meet the threshold for significance (NWD-score > 1) includes ASPM.
(G) Co-immunoprecipitation of ASPM with an antibody to endogenous WDR62 confirms the interaction. Left: Lane 1: IgG control IP with c-Myc antibody (SCBT) as a negative control for “stickiness.” Lane 2: IP with antibody to endogenous WDR62. Lane 3: Input (lysate). Right, lane 2: Immunoprecipitation of ASPM confirmed the reciprocal co-precipitation of WDR62.
(H) Centrosomal localization of ASPM is dependent on WDR62. ASPM localizes to centrosomes in scrambled control (SC) transfected cells but is absent from centrosomes in cells depleted of WDR62. Insets: c = Centrin and a = ASPM.
(I) Centrosomal localization of WDR62 is not dependent on ASPM. WDR62 levels at the centrosome were unchanged in cells treated with scrambled control (SC) or ASPM siRNA (ASPM #2). Insets: c = Centrin; w = WDR62.
See also Supplemental Figure S4.
Liquid chromatography-mass spectrometry (LC-MS/MS) analysis identified interacting partners of WDR62 that included ASPM. HeLa S3 cells stably overexpressing C-terminal HA tagged WDR62 were lysed and immunoprecipitated with anti-HA resin as previously described (Sowa et al., 2009), with LC-MS/MS identifying ASPM as well as Citron kinase, a known interactor of ASPM (Paramasivam et al., 2007), as high-confidence candidate interacting partners (Figures 4F and S4B). Immunoprecipitation of endogenous WDR62 confirmed ASPM as a binding partner (Figure 4G).
Given our findings that WDR62 and ASPM interact and function in centriole biogenesis, we tested whether they depend on each other for their localization and found that WDR62 is required to localize ASPM to the centrosome. ASPM was absent from centrosomes in HeLa cells depleted of WDR62, whereas WDR62 localized normally to centrosomes after siRNA knockdown of ASPM (Figures 4H and 4I). Cep152, which localizes Wdr62 to the centrosome (Kodani et al., 2015), was unperturbed in Wdr62 −/− MEFs (Figures S4C and S4D). The dependence of ASPM localization on WDR62 may potentially explain the more severe phenotype of WDR62 mutation in mice and humans.
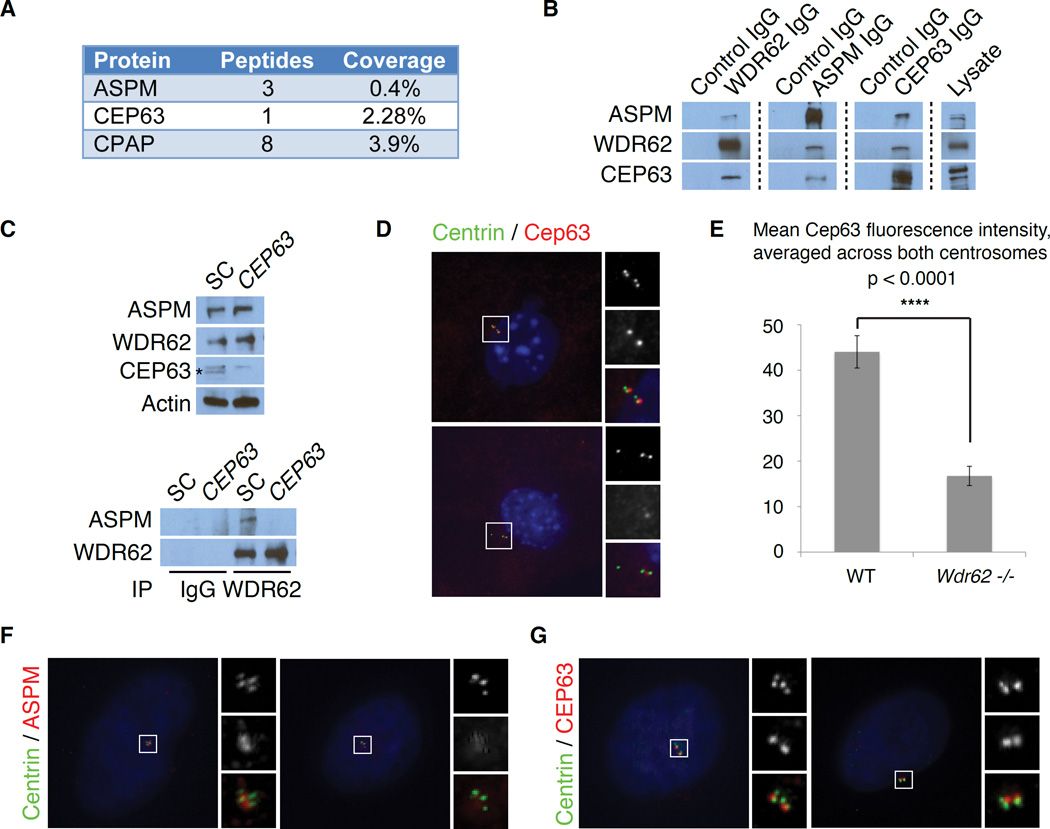
WDR62, CEP63 and ASPM interact and assemble at the centrosome in a hierarchy
WDR62 and ASPM form a complex with CEP63, which is also disrupted in human microcephaly and localizes to the maternal centriole (Sir et al., 2011). Similar to WDR62, CEP63 localizes in a ring-shaped pattern around the mother centriole in G1 and between each pair of centrioles in S-phase (Sir et al., 2011). WDR62 interacts with CEP63 (Kodani et al., 2015), while MS of endogenous ASPM detected an interaction between ASPM and CEP63 (Figure 5A). Endogenous WDR62 and ASPM both reciprocally interacted with CEP63 and with each other, suggesting a three-protein complex (Figure 5B). Knockdown of CEP63 with siRNA does not alter ASPM and WDR62 protein levels but abolishes their interaction (Figure 5C), suggesting that this interaction is indirect and mediated by CEP63. RNAi knockdown of CEP63 in HeLa cells causes a profound centriole duplication defect, with <20% of cells carrying 4 centrioles (Figures S5A–S5C), consistent with previously published findings (Brown et al., 2013; Kodani et al., 2015; Sir et al., 2011).
Figure 5. WDR62 and ASPM assemble at the centrosome in a sequential hierarchy mediated by CEP63.
(A) Table showing a selected list of interactors from an IP-mass-spectrometry screen, with the number of peptides and percent coverage, suggests that CPAP and CEP63 interact with ASPM.
(B) Reciprocal co-immunoprecipitation (IP) of endogenous ASPM, WDR62 and CEP63 demonstrates that they form a 3-protein complex. Lanes 1, 3 and 5: IgG control IP with c-Myc antibody (SCBT) as a negative control. Lanes 2, 4 and 6: IP with antibodies to endogenous CEP63, WDR62 and ASPM, respectively. Rows (top to bottom): Western blot for ASPM, WDR62 and CEP63.
(C) RNAi knockdown of CEP63 blocks the ability of WDR62 to bind ASPM, without changes in protein levels of WDR62 or ASPM. Left: Lane 1 “SC” indicates scrambled control lysate. Lane 2 shows CEP63 RNAi knockdown lysate. Rows, from top to bottom: Western blot for ASPM, WDR62 and CEP63. Actin served as a loading control. The band marked with an asterisk (*) is specific to CEP63.
Right: Lanes 1 and 3 show scrambled control (SC) lysate, Lanes 2 and 4 show CEP63 RNAi knockdown lysate. Lanes 1–2: IgG control IP. Lanes 3–4: IP with WDR62 antibody. Row 1: Western blot for ASPM. Row 2: Western blot for WDR62.
(D) Wdr62 is required for centrosomal localization of Cep63. Wild-type (WT) and Wdr62 −/− mouse embryonic fibroblasts (MEFs) stained with Centrin (green), to mark centrioles, and Cep63 (red). Insets: c = Centrin and 3 = Cep63.
(E) Quantification of mean Cep63 immunofluorescence intensity across a fixed area, averaged across both centrosomes with background fluorescence subtracted, in WT and Wdr62 −/− MEFs. Mean ± SEM. **** p < 0.0001.
(F) Centrosomal localization of ASPM is dependent on CEP63. ASPM localizes to centrosomes in scrambled control (SC) transfected cells but is absent from centrosomes in cells depleted of CEP63. Insets: c = Centrin and a = ASPM.
(G) CEP63 localization does not depend on ASPM. CEP63 levels at the centrosome were unchanged in cells treated with scrambled control (SC) or ASPM siRNA (ASPM #2). Insets: c = Centrin; 3 = Cep63.
See also Supplemental Figure S5.
Wdr62 is not only required for Aspm localization but is also required to localize Cep63 to the centrosome. Centrosomal Cep63 was reduced in Wdr62 −/− MEFs relative to WT MEFs (Figures 5D and 5E; unpaired t-test, p < 0.0001), similar to the effects of Wdr62 siRNA (Kodani et al., 2015). CEP63 depletion in HeLa cells did not perturb centrosomal localization of WDR62 (Figure S5D and Kodani et al., 2015), indicating a unidirectional dependence of CEP63 on WDR62 for its localization.
ASPM localization depends on CEP63, as ASPM was absent from centrosomes in cells depleted of CEP63 and CEP152 (Figures 5F and S5E). The centrosomal localization of CEP63 and CEP152 was unchanged after siRNA knockdown of ASPM (Figures 5G and S5F). These findings, along with other studies (Kodani et al., 2015), support the existence of a more general hierarchy of primary microcephaly-associated proteins, which are required in a step-wise manner to recruit each other to the centrosome and ensure proper centriole duplication.
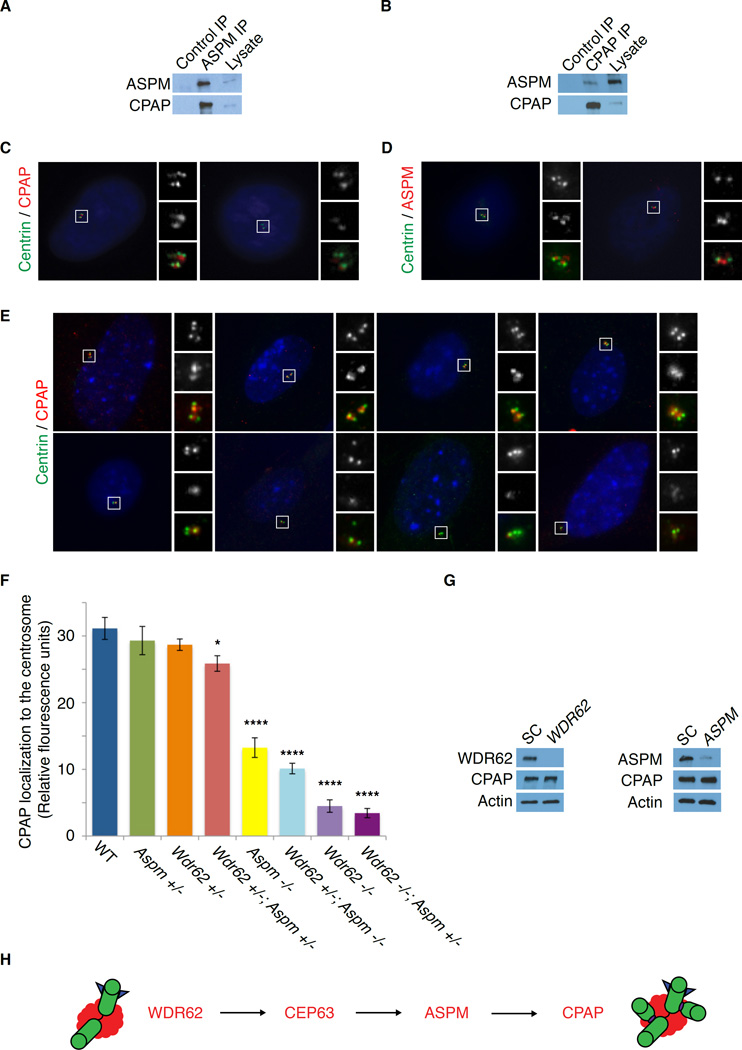
Microcephaly-associated proteins cooperate additively to recruit CPAP/Sas-4 to the centrosome
Mass spectrometry using endogenous ASPM as bait identified CPAP as an ASPM-interacting protein (Figure 5A). Endogenous ASPM can co-precipitate CPAP, and ASPM and CPAP reciprocally interact with each other (Figures 6A and 6B). ASPM is required for centrosomal localization of CPAP, whereas CPAP was virtually absent from centrosomes in ASPM-depleted cells (Figure 6C). In contrast, centrosomal ASPM localization was unperturbed in CPAP-depleted cells (Figure 6D). These findings suggest that ASPM lies between CEP63 and CPAP in the pathway of sequential centrosomal MCPH protein accumulation.
Figure 6. Microcephaly-associated proteins cooperate additively to recruit CPAP/Sas-4 to the centrosome.
(A) IP of endogenous ASPM followed by Western blot with an antibody to CPAP confirms that ASPM interacts with CPAP.
(B) IP of endogenous CPAP followed by Western blot with an antibody to ASPM confirms reciprocal interaction of CPAP and ASPM.
(C) ASPM is required to localize CPAP to the centrosome. Cells were treated with scrambled control (SC) or ASPM siRNAs (ASPM #2) and then stained for CPAP. CPAP was practically absent from the centrosomes in ASPM-depleted cells. Insets: c = Centrin; p = CPAP.
(D) ASPM localization does not depend on CPAP. Cells were treated with scrambled control (SC) or CPAP siRNA and then stained for ASPM. ASPM localization was unchanged in CPAP-depleted cells. Insets: c = Centrin; a = ASPM.
(E) Mouse embryonic fibroblasts (MEFs) stained with Centrin (green) to mark centrioles and CPAP (red), showing that Wdr62 and Aspm cooperate in a dose-dependent manner to localize CPAP to the centrosome. Top row, left to right: Wild-type (WT), Aspm +/−, Wdr62 +/−, Wdr62 +/−, Aspm +/−. Bottom row, left to right: Aspm −/−, Wdr62 +/−; Aspm −/−, Wdr62 −/−, Wdr62 −/−; Aspm +/−. Insets (in grayscale): c = Centrin; p = CPAP.
(F) Quantification of CPAP relative immunofluorescence intensity in MEFs of various genotypes: Wild-type (WT), Aspm +/−, Wdr62 +/−, Wdr62 +/−, Aspm +/−, Aspm −/−, Wdr62 +/−; Aspm −/−, Wdr62 −/− and Wdr62 −/−; Aspm +/−. Mean ± SEM. * p < 0.05 and **** = p < 0.0001 (vs. WT).
(G) RNAi knockdown of WDR62 (left) or ASPM (right) does not affect CPAP levels.
(H) Model of sequential recruitment of microcephaly-associated proteins to the centrosome: WDR62, followed by CEP63, ASPM and CPAP, respectively. All are required for centriole biogenesis.
Finally, we found that similar to the trends seen for brain size and centriole duplication, the localization of CPAP to the centrosome depends on the dosage of Wdr62 and Aspm. CPAP localization to the centrosome was intact in MEFs derived from WT, Aspm +/− or Wdr62 +/− embryos but was significantly reduced in Wdr62 +/−; Aspm +/− MEFs (p = 0.03), as well as Aspm −/− and Wdr62 −/− MEFs (Figures 6E and 6F, p < 0.0001). Similar to the trends for brain size and centriole duplication, Wdr62 −/−; Aspm +/− MEFs had the most severe reduction in CPAP localization; Wdr62 −/− MEFs were less severe, followed by Wdr62 +/−; Aspm −/−, whereas Aspm −/− MEFs had a more moderate phenotype and Wdr62 +/−; Aspm +/− MEFs had the mildest phenotype (Figure 6F). This progressive reduction in CPAP localization with decreasing dosage of Wdr62 and Aspm is not due to a reduction in CPAP protein levels (Figure 6G), further supporting the notion that the genetic interaction between Wdr62 and Aspm is critical for CPAP localization, correlating with centriole duplication and normal brain size. The additive effect of Wdr62 and Aspm on centrosomal recruitment of CPAP, which is required for centriole duplication, helps explain why the two genes are not wholly redundant and why the phenotype of mutations in one gene is enhanced by mutations in the other. Together, these findings support our model of sequential recruitment of WDR62, followed by CEP63, ASPM and CPAP, to the centrosome (Figure 6H).
Loss of Wdr62 and Aspm disrupts apical complex proteins and leads to premature dissociation of ciliary remnants from centrosomes
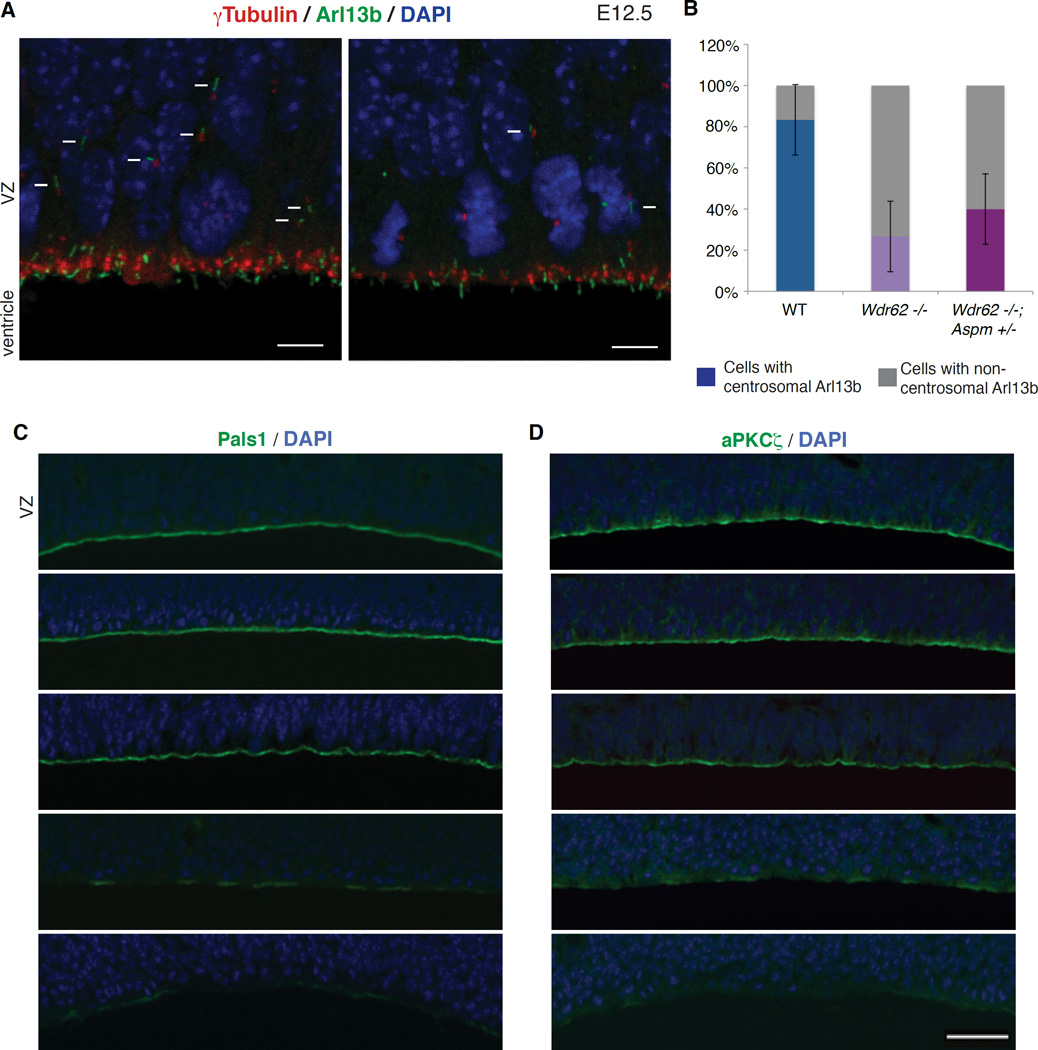
We confirmed the dependence of centriole duplication on Wdr62 and Aspm by crossing EGFP-Centrin transgenic mice (Higginbotham et al., 2004) to Wdr62 +/− mice, and then crossing the resulting Wdr62 +/−; EGFP-Centrin progeny to generate Wdr62 −/−; EGFP-Centrin mice and transgenic control littermates. Immunostaining for Arl13b-labeled cilia in Wdr62 +/−; EGFP-Centrin mice revealed an orderly association with pairs of centrioles along the surface of the lateral ventricle (Figure S6A). By contrast, Wdr62 −/−; EGFP-Centrin mice showed cilia associated with odd numbers of centrioles (1 or 3), as well as pairs of centrioles, suggesting that centriole duplication is impaired in mutant brain (Figure S6A). Consistent with the asymmetric localization of WDR62 and ASPM to the mother centriole, which is critical in organizing primary cilia, there was a loss of Arl13b staining in the brains of Wdr62 −/− and Wdr62 +/−; Aspm −/− mice, implicating Wdr62 and Aspm in maintenance of cilia in developing brain (Figure S6B). Co-immunostaining of E12.5 WT and Wdr62 −/− brains for γ-tubulin and Arl13b revealed a marked reduction in the intensity of γ-tubulin immunofluorescence at the centrosomes (Figure 7A), consistent with Wdr62 −/− mouse embryonic fibroblasts (MEFs), which show a 40% reduction in γ-tubulin staining at the centrosome (Figures S6C and S6D; p = 0.0001).
Figure 7. Loss of Wdr62 and Aspm disrupts apical complex proteins and leads to premature dissociation of ciliary remnants from centrosomes.
(A) Reduction in centrosomes and cilia in the Wdr62 −/− mouse brain, both at and away from the ventricular surface, along with an increase in dissociated ciliary remnants. Shown here are E12.5 WT (top) and Wdr62 −/− (bottom) brain sections immunostained for γ-tubulin (centrosomes) and Arl13b (cilia). Arrows denote cilia located away from the ventricular surface, while arrowheads mark non-centrosomal Arl13b (dissociated ciliary remnants). Scale bar = 5 µm.
(B) Quantification of centrosomal versus non-centrosomal Arl13b in WT, Wdr62 −/− and Wdr62 −/−; Aspm +/− brains. Fisher’s exact test, ** p = 0.002 for WT vs. Wdr62 −/− and ** p = 0.009 for WT vs. Wdr62 −/−; Aspm +/−.
(C–D) Loss-of-function of Wdr62, Aspm or both severely disrupts apical complex proteins Pals1 and aPKCζ in a dose-dependent manner. From top to bottom: WT, Wdr62 +/−, Aspm −/−, Wdr62 +/−; Aspm −/− and Wdr62 −/− brains at E14.5 stained for (C) Pals1 and (D) aPKCζ to label the apical complex, and counterstained with DAPI. Scale bar = 50 µm.
Recent work has suggested that primary cilia do not completely disassemble prior to mitosis, with centrosomal association of the remnants of ciliary membrane preferentially occurring in cells that maintain a stem cell fate; this association then decreases over the course of neurogenesis, coinciding with differentiation of cells from apical progenitors to basal progenitors and neurons (Paridaen et al., 2013). Co-immunostaining for γ-tubulin and Arl13b (Paridaen et al., 2013) confirmed this relationship in normal brain but showed a premature increase in non-centrosomal Arl13b staining in Wdr62 −/− mouse brains at E12, when most normal cell divisions show Arl13b staining at one of the two centrosomes (Figures 7A and 7B; Fisher’s exact test, p = 0.002). We concomitantly observed a pronounced loss of cilia away from the ventricular surface (Figure 7A). Similarly, co-immunostaining for γ-tubulin and Arl13b revealed a premature increase in non-centrosomal Arl13b staining in Wdr62 −/−; Aspm +/− mouse brains at E12 (Figure 7B; Fisher’s exact test, p = 0.009). Transmission EM from WT and Wdr62 −/−; Aspm +/− mice at E12.5 confirmed a dose-dependent decrease in centrioles and cilia in the Wdr62 −/− and Wdr62 −/−; Aspm +/− brain (Figure S6E). Interestingly, the presence of grossly normal cilia (albeit in smaller numbers) in the mutants suggests that some mutant cells can form cilia. Together, these results suggest that Aspm and Wdr62 control centriole number and the depletion of centrioles in Aspm and Wdr62 mutants leads to premature formation of ciliary remnants.
Given the importance of the apical complex in maintaining neural stem cell fate (Kim et al., 2010), we examined the apical complex in Wdr62 and Aspm mutant mice and found that loss-of-function of Wdr62, Aspm or both disrupts the apical complex in a dose-dependent manner. Immunostaining for the apical complex proteins Pals1 and aPKCζ (Kim et al., 2010) at E14.5 revealed that Aspm −/− embryos show a mild reduction in localization of both Pals1 and aPKCζ at the ventricular surface compared to WT or Wdr62 +/− embryos, whereas Wdr62 +/−; Aspm −/− embryos show an intermediate phenotype, and Wdr62 −/− embryos have the most dramatic reduction in Pals1 and aPKCζ immunoreactivity at the apical surface (Figures 7C and 7D), tracking the trends for brain weight, centriole duplication and CPAP levels (Figures 1B, 3D and 6F). In the most severely affected genotypes (Wdr62 −/− and Wdr62 +/−; Aspm −/−), the loss of apical complex proteins is associated with disruptions and irregularities in the ventricular lining, and in the ordered arrangement of M phase nuclei at the ventricular surface. These disruptions of the epithelial lining suggest a ready mechanism to explain the premature delamination of progenitors from the VZ, and the cell fate changes seen in these mutants, as an intact apical complex has been shown to be essential to maintain progenitors in the VZ (Kim et al., 2010), and to allow signaling by CSF signals to promote stem cell proliferation (Lehtinen et al., 2011). These findings suggest that Wdr62 and Aspm are essential not only for centriole biogenesis, but also for normal expression and localization of the apical complex, providing a link between centrioles, ciliary remnants and cell fate.
DISCUSSION
Here, we identify genetic interactions between Wdr62 and Aspm, physical interactions between the proteins that they encode, essential roles for both proteins in centriole biogenesis, and unexpected regulation by these proteins of apical epithelial organization. WDR62 and ASPM physically associate at the proximal end of the mother centriole, mediated by CEP63. Loss of Wdr62, Aspm or both impairs centriole duplication in a dose-dependent manner, correlating with the severity of microcephaly, and leads to loss of centrosomes and cilia in the developing brain. Wdr62 −/− and Aspm −/− mice resemble the Sas-4 conditional mutant (cKO) mouse in crucial ways, including centriole duplication defects and precocious delamination of progenitors from the germinal zones, consistent with our evidence that CPAP localization depends on Wdr62 and Aspm, suggesting that CPAP/Sas-4 is the common target of a hierarchy of microcephaly proteins. Most surprisingly, mice deficient in Aspm and Wdr62 show gene-dose dependent perturbation of apical complex components, and a fate switch from apical to basal progenitors. These data implicate microcephaly-associated proteins like WDR62 and ASPM not only in centriole biogenesis through their effect on CPAP/Sas-4 but also in regulating the interaction of centrioles and cell fate determinants, thereby maintaining stem cell character, and suggest a plausible mechanism for how microcephaly proteins can regulate cell fate in the normal brain.
Microcephaly as a ‘centriolopathy’
Our studies not only assign a novel cellular function to Aspm in centriole duplication but also place both Aspm and Wdr62 in the context of other microcephaly genes, in particular CENPJ/CPAP/Sas-4 and CEP63, whose gene-products also function in centriole biogenesis. Our studies confirm that Wdr62 is required for adequate centrosomal accumulation of Cep63 and, in turn, reveal a role for WDR62 and CEP63 in ensuring proper centrosomal accumulation of ASPM. Finally, we show that ASPM, in turn, helps localize CPAP to the centrosome. Given the critical role of CPAP in centriole biogenesis and elongation (Nigg and Stearns, 2011), the final common pathway of these several microcephaly-associated proteins may be to bring CPAP to the centrosome, enabling centriole biogenesis (Figure 7H).
Our findings in Wdr62 and Aspm mutant mice are consistent with studies in the Sas-4 mutant mouse suggesting that neurogenesis defects in microcephaly may be due to loss of centrioles and a secondary reduction in cilia (Insolera et al., 2014). Consistent with the requirement of CPAP/Sas-4 for centriole/centrosome duplication, the authors observed a progressive depletion of centrosomes, as well as of primary cilia that grow from the few functional centrioles remaining in the Sas-4 −/− cortex (Insolera et al., 2014). Wdr62 −/− mice on a GFP-Centrin transgenic background confirmed the centriole duplication defect in vivo, as cilia were often associated with odd numbers of centrioles. Another phenotypic similarity with Sas-4 cKO mice is the displacement of cycling progenitors from the germinal zones in Wdr62 −/− and Wdr62 +/−; Aspm −/− mutant cortex. Ectopic Pax6+ progenitors with almost no cilia (unlike apical progenitors at the ventricular surface) were observed in the Sas-4 −/− cortex (Insolera et al., 2014), whereas we observed an expansion of the Ki67+ population of cycling progenitors beyond the VZ and SVZ, an increase in Pax6− Ki67+ cells and an expansion of Tbr2+ basal progenitors in Wdr62 −/− and Wdr62 +/−; Aspm −/− mutant cortex (Figure 2), consistent with delamination of apical progenitors and a fate switch to basal progenitors in the absence of centrioles and cilia. This delamination of progenitors can be explained by the dose-dependent loss of apical complex localization seen with loss of Wdr62 and Aspm, as the apical complex is critical for maintenance of stem cell fate.
Our 3D en face analysis (Juschke et al., 2014) excludes mitotic spindle orientation as a major mechanism in Wdr62 and Aspm mutant mice, as we detected no difference in neural progenitor spindle orientation between WT, Wdr62 +/−; Aspm −/− and Wdr62 −/− embryos, suggesting that other mechanisms contribute to the microcephaly observed in these mutants. Our finding that Aspm and Wdr62 function together to promote centriole duplication provides a novel insight into how defects in centriole biogenesis may underlie the pathogenesis of MCPH. It seems likely that centriole number and organization are critical for progenitor attachment to the ventricular surface and the maintenance of neural progenitors, but how centrioles associate and control the organization of the apical surface remains unclear. Increased phosphohistone H3 (pHH3) immunoreactivity suggests a delay in mitotic progression in Cdk5rap2 −/− mice, Nde1 −/− mice and Sas-4 mice (Feng and Walsh, 2004; Insolera et al., 2014; Lizarraga et al., 2010), as well as a Wdr62 hypomorphic mouse (Chen et al., 2014), and may contribute to premature neuronal formation and depletion of progenitor cells (Pilaz et al., 2016).
Primary autosomal recessive microcephaly (MCPH) has been described as a centrosomal disease (Megraw et al., 2011), but based on our findings in Wdr62 and Aspm mutant mice, we propose that MCPH is more properly a ‘centriolopathy’, with a secondary reduction in cilia. Given the role of Wdr62 and Aspm in localizing CPAP, we propose that loss of these microcephaly proteins results in a progressive dilution of centrioles and centrosomes over the course of several rapid cell cycles in neural progenitors, leading to the loss of mother centrioles with which ciliary remnants can associate, while also causing perturbation of the apical complex and delamination of progenitors from the germinal zones; the eventual depletion of the neural stem cell pool results in a smaller brain.
Microcephaly proteins, the apical complex, and the ciliary remnant
Although our studies provide the first direct test of the recently proposed model of asymmetric ciliary membrane inheritance in regulating cell fate, the role of this mechanism is not yet clear. During normal progenitor cell division around E12.5 in mice, a remnant of the ciliary membrane is linked to the older mother (‘grandmother’) centriole and is preferentially inherited by the daughter cell that is destined to remain a neuroepithelial or apical progenitor cell (Paridaen et al., 2013). The degree of centrosomal association with ciliary remnants correlates with the maintenance of stem cell character and decreases over the course of neurogenesis, coinciding with the shift from symmetric proliferative divisions to asymmetric or neurogenic divisions, and with differentiation from apical progenitors to basal progenitors or neurons. Our data show premature dissociation of ciliary remnants from centrosomes at E12.5 in Wdr62 −/− mice (two days earlier than WT), followed by precocious generation of basal progenitors relative to apical progenitors by E14.5 consistent with, but not yet proving, the model that ciliary remnant/centriole association may maintain apical stem cell fate.
However, our data suggest an alternative potential mechanism for the cell fate changes seen in Wdr62 and Aspm mutants in the dramatic disruption of apical complex proteins, which is modest and patchy in Aspm −/− mutants, striking in Wdr62 +/−; Aspm −/−, and virtually complete in Wdr62 −/− mutants, associated with loss of epithelial organization. Apical complex proteins such as Pals1, and basolateral proteins such as β-catenin show dosage-sensitive effects on proliferation and cell fate, with overexpression inducing proliferation (Chenn and Walsh, 2002), while depletion (Kim et al., 2010) or mislocalization (Chae et al., 2004) cause premature differentiation into neurons and subsequent microcephaly. Loss of apical complex proteins may influence proliferation directly, or may regulate proliferation indirectly by maintaining the apical surface of neuroepithelial progenitors in the ventricular zone, where the apical complex is bathed in growth factors present in the cerebrospinal fluid (Lehtinen et al., 2011). Hence, premature withdrawal of progenitors from the ventricular zone would remove this proliferative effect and result in premature differentiation.
The defects of epithelial structure and apical proteins suggest a likely mechanism for the structural defects such as polymicrogyria, schizencephaly, and periventricular heterotopia that are commonly observed in WDR62 mutations in humans (Bilguvar et al., 2010; Yu et al., 2010), and occasionally reported in ASPM mutations as well (Passemard et al., 2009). These structural defects have not been well explained before by a centriolar model, but several of these malformations (especially periventricular heterotopia and schizencephaly) reflect abnormal neuroepithelial structure.
The mechanisms by which centriolar proteins would regulate the apical protein complex are obscure, and bear further study. The apical complex components Par3 (Inaba et al., 2015), Par6 (Kodani et al., 2010), and aPKC (Atwood et al., 2013) associate with centrosomes, and Arl13b itself is essential for the formation of polarized apical progenitors and normal localization of Igf1R apical complex receptors (Higginbotham et al., 2013). In C. elegans, ASPM-1 binds LIN-5, a homolog of mammalian NuMA that is part of the apical complex (van der Voet et al., 2009). Centrosomes at M phase accumulate vesicular structures that appear to localize a variety of plasma membrane proteins including Arl13b and other apical proteins (Paridaen et al., 2013). Perhaps the cycling of apical complex proteins through the centrosome at M phase is an essential component of maintaining their normal polarized expression, with the loss of that centriolar association resulting in their eventual depletion from the apical membrane.
EXPERIMENTAL PROCEDURES
Animals
Targeted Wdr62 gene-trap ES cells (MMRRC Cat# 021135-UCD, RRID: MMRRC_021135-UCD) from the Sanger Institute Gene Trap Resource (SIGTR) were injected into 129Sv blastocysts to generate chimeric mice. Male chimeras were bred to WT 129Sv background females (Charles River) to transmit the gene-trap allele. The mice were later bred to C57Bl/6 background females (Charles River) and are maintained separately on 129Sv and C57Bl/6 backgrounds.
Aspm gene targeting was carried out by InGenious Targeting Laboratory, Inc. using a targeting vector, in which a Neo cassette with 1.5-kb and 8.0-kb homology arms replaced exons 1–3 of Aspm. This targeting vector, which was generated by the authors, was first electroporated into 129/Sv embryonic stem (ES) cells, followed by selection in G418. Surviving clones were expanded for PCR analysis to identify recombinant ES clones, which were then injected into C57Bl/6 blastocysts. Male chimeras were then bred with wild-type C57Bl/6 females to ensure germline transmission.
Transgenic EGFP-Centrin2 mice, referred to here as EGFP-Centrin, were obtained from Jackson Laboratory (JAX strain name CB6-Tg(CAG-EGFP/CETN2)3-4Jgg/J; IMSR Cat# JAX:008234, RRID:IMSR_JAX:008234), by rederivation from cryopreserved sperm. The transgenic mice express Enhanced Green Fluorescent Protein-labeled human Centrin-2 driven by a chicken β-actin promoter with CMV immediate early enhancer (Higginbotham et al., 2004). Additional details are available in Supplemental Experimental Procedures.
Cell Culture
Primary mouse embryonic fibroblasts (MEFs) were isolated from E14.5 embryos and dissociated by trypsinization. To establish immortalized cell lines, MEFs were derived from Wdr62 and Aspm single- and double-mutant embryos at E9.5 or E14.5 and transformed with SV-40 retrovirus using the Phoenix packaging system (RRID:SCR_003163). Both primary and immortalized MEFs were maintained in Dulbecco’s Modified Eagle Medium, High Glucose (HyClone) with 15% fetal bovine serum (FBS, Gibco) and 1mM penicillin, streptomycin and L-glutamine. Additional details are in Supplemental Experimental Procedures.
Immunoprecipitation, Western blotting and Mass spectrometry
Data on the WDR62-interactome were deposited in the PeptideAtlas database (PeptideAtlas: PASS00927, http://www.peptideatlas.org/PASS/PASS00927). See Supplemental Experimental Procedures for additional details.
Supplementary Material
Acknowledgments
We thank Paul Chang for pointing out the maternal centriolar localization of ASPM, Victor Larionov for reagents, Kutay Atabay and Cameron Sadegh for technical expertise, and Garrett Kingman and Alberta Yen for help with MS and the Aspm mutant mouse. We thank the Boston Children’s Hospital (BCH) Mouse Gene Manipulation Core and BCH Cellular Imaging Core (BCH IDDRC, P30 HD18655), the Beth Israel Deaconess Medical Center (BIDMC) Histology Core Facility, the Harvard Medical School Electron Microscopy Facility, and the BIDMC Confocal Imaging Core for technical support. This research was supported by the National Institutes of Health (NIH) MSTP training grant NIGMS T32GM007753 (D.J.); NIH / NHLBI (T32HL007731), the UCSF Program for Breakthrough Biomedical Research, and the Sandler Foundation (A.K.); NIH / NINDS 1 R21 NS091865-01 (B.-I.B.); American Society of Radiation Oncology Junior Faculty Career Research Training Award (JF2013-2), KL2/Catalyst Medical Research Investigator Training award (TR001100), and Burroughs-Wellcome Career Award for Medical Scientists (J.D.M.); Nancy Lurie Marks Junior Faculty MeRIT Fellowship (T.W.Y.); Manton Center for Orphan Disease Research, and F. Hoffman-La Roche Ltd (G.H.M.); NIH GM070565 and GM095567 (J.W.H.); NIAMS R01AR054396, NIGMS R01GM095941, the Burroughs Wellcome Fund, the Packard Foundation, and the Sandler Family Supporting Foundation (J.F.R.); NINDS (R01 NS035129 and R01 NS32457) and the Manton Center for Orphan Disease Research (C.A.W.). C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, D.J., A.K., T.W.Y, B.-I.B., and C.A.W.; Methodology, D.J., A.K., G.H.M., and C.V.; Validation, D.J., A.K., and D.M.G.; Investigation, D.J., A.K., D.M.G., J.D.M., C.V., J.J., and N.K.; Resources, J.W.H., J.F.R., and C.A.W. Writing – Original Draft, D.J., A.K., and C.A.W.; Writing – Review & Editing, D.J., A.K., T.W.Y, B.-I.B., and C.A.W.; Visualization, D.J., A.K., D.M.G., J.D.M., and B.-I.B.; Supervision, J.W.H., J.F.R., T.W.Y, B.-I.B., and C.A.W.; Project Administration, C.A.W.; Funding Acquisition, D.J., A.K., J.D.M., G.H.M., J.W.H., J.F.R., T.W.Y., B.-I.B., and C.A.W.
References
- Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature. 2013;494:484–488. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilguvar K, Ozturk AK, Louvi A, Kwan KY, Choi M, Tatli B, Yalnizoglu D, Tuysuz B, Caglayan AO, Gokben S, et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature. 2010;467:207–210. doi: 10.1038/nature09327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoyevitch MA, Yeap YY, Qu Z, Ngoei KR, Yip YY, Zhao TT, Heng JI, Ng DC. WD40-repeat protein 62 is a JNK-phosphorylated spindle pole protein required for spindle maintenance and timely mitotic progression. J Cell Sci. 2012;125:5096–5109. doi: 10.1242/jcs.107326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J, Roberts E, Mochida GH, Hampshire DJ, Scott S, Askham JM, Springell K, Mahadevan M, Crow YJ, Markham AF, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Brown NJ, Marjanovic M, Luders J, Stracker TH, Costanzo V. Cep63 and Cep152 cooperate to ensure centriole duplication. PLoS One. 2013;8:e69986. doi: 10.1371/journal.pone.0069986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae TH, Kim S, Marz KE, Hanson PI, Walsh CA. The hyh mutation uncovers roles for alpha Snap in apical protein localization and control of neural cell fate. Nat Genet. 2004;36:264–270. doi: 10.1038/ng1302. [DOI] [PubMed] [Google Scholar]
- Chen JF, Zhang Y, Wilde J, Hansen KC, Lai F, Niswander L. Microcephaly disease gene Wdr62 regulates mitotic progression of embryonic neural stem cells and brain size. Nat Commun. 2014;5:3885. doi: 10.1038/ncomms4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, et al. Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet. 2010;87:40–51. doi: 10.1016/j.ajhg.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Bielas S, Tanaka T, Gleeson JG. Transgenic mouse line with green-fluorescent protein-labeled Centrin 2 allows visualization of the centrosome in living cells. Transgenic Res. 2004;13:155–164. doi: 10.1023/b:trag.0000026071.41735.8e. [DOI] [PubMed] [Google Scholar]
- Higginbotham H, Guo J, Yokota Y, Umberger NL, Su CY, Li J, Verma N, Hirt J, Ghukasyan V, Caspary T, Anton ES. Arl13b-regulated cilia activities are essential for polarized radial glial scaffold formation. Nat Neurosci. 2013;16:1000–1007. doi: 10.1038/nn.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J, Midgley C, Bergh AM, Bell SM, Askham JM, Roberts E, Binns RK, Sharif SM, Bennett C, Glover DM, et al. Human ASPM participates in spindle organisation, spindle orientation and cytokinesis. BMC Cell Biol. 2010;11:85. doi: 10.1186/1471-2121-11-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WF, Pomp O, Ben-Omran T, Kodani A, Henke K, Mochida GH, Yu TW, Woodworth MB, Bonnard C, Raj GS, et al. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 2014;84:1240–1257. doi: 10.1016/j.neuron.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Venkei ZG, Yamashita YM. The polarity protein Baz forms a platform for the centrosome orientation during asymmetric stem cell division in the Drosophila male germline. Elife. 2015:4. doi: 10.7554/eLife.04960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insolera R, Bazzi H, Shao W, Anderson KV, Shi SH. Cortical neurogenesis in the absence of centrioles. Nat Neurosci. 2014;17:1528–1535. doi: 10.1038/nn.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juschke C, Xie Y, Postiglione MP, Knoblich JA. Analysis and modeling of mitotic spindle orientations in three dimensions. Proc Natl Acad Sci U S A. 2014;111:1014–1019. doi: 10.1073/pnas.1314984111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lehtinen MK, Sessa A, Zappaterra MW, Cho SH, Gonzalez D, Boggan B, Austin CA, Wijnholds J, Gambello MJ, et al. The apical complex couples cell fate and cell survival to cerebral cortical development. Neuron. 2010;66:69–84. doi: 10.1016/j.neuron.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Tonthat V, Wu B, Sutterlin C. Par6 alpha interacts with the dynactin subunit p150 Glued and is a critical regulator of centrosomal protein recruitment. Mol Biol Cell. 2010;21:3376–3385. doi: 10.1091/mbc.E10-05-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani A, Yu TW, Johnson JR, Jayaraman D, Johnson TL, Al-Gazali L, Sztriha L, Partlow JN, Kim H, Krup AL, et al. Centriolar satellites assemble centrosomal microcephaly proteins to recruit CDK2 and promote centriole duplication. Elife. 2015:4. doi: 10.7554/eLife.07519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Zappaterra MW, Chen X, Yang YJ, Hill AD, Lun M, Maynard T, Gonzalez D, Kim S, Ye P, et al. The cerebrospinal fluid provides a proliferative niche for neural progenitor cells. Neuron. 2011;69:893–905. doi: 10.1016/j.neuron.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarraga SB, Margossian SP, Harris MH, Campagna DR, Han AP, Blevins S, Mudbhary R, Barker JE, Walsh CA, Fleming MD. Cdk5rap2 regulates centrosome function and chromosome segregation in neuronal progenitors. Development. 2010;137:1907–1917. doi: 10.1242/dev.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzini MC, Walsh CA. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev. 2011;21:333–339. doi: 10.1016/j.gde.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Sharkey JT, Nowakowski RS. Cdk5rap2 exposes the centrosomal root of microcephaly syndromes. Trends Cell Biol. 2011;21:470–480. doi: 10.1016/j.tcb.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas AK, Khurshid M, Desir J, Carvalho OP, Cox JJ, Thornton G, Kausar R, Ansar M, Ahmad W, Verloes A, et al. WDR62 is associated with the spindle pole and is mutated in human microcephaly. Nat Genet. 2010;42:1010–1014. doi: 10.1038/ng.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam M, Chang YJ, LoTurco JJ. ASPM and citron kinase co-localize to the midbody ring during cytokinesis. Cell Cycle. 2007;6:1605–1612. doi: 10.4161/cc.6.13.4356. [DOI] [PubMed] [Google Scholar]
- Paridaen JT, Wilsch-Brauninger M, Huttner WB. Asymmetric inheritance of centrosome-associated primary cilium membrane directs ciliogenesis after cell division. Cell. 2013;155:333–344. doi: 10.1016/j.cell.2013.08.060. [DOI] [PubMed] [Google Scholar]
- Passemard S, Titomanlio L, Elmaleh M, Afenjar A, Alessandri JL, Andria G, de Villemeur TB, Boespflug-Tanguy O, Burglen L, Del Giudice E, et al. Expanding the clinical and neuroradiologic phenotype of primary microcephaly due to ASPM mutations. Neurology. 2009;73:962–969. doi: 10.1212/WNL.0b013e3181b8799a. [DOI] [PubMed] [Google Scholar]
- Pilaz LJ, McMahon JJ, Miller EE, Lennox AL, Suzuki A, Salmon E, Silver DL. Prolonged Mitosis of Neural Progenitors Alters Cell Fate in the Developing Brain. Neuron. 2016;89:83–99. doi: 10.1016/j.neuron.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvers JN, Bryk J, Fish JL, Wilsch-Brauninger M, Arai Y, Schreier D, Naumann R, Helppi J, Habermann B, Vogt J, et al. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A. 2010;107:16595–16600. doi: 10.1073/pnas.1010494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Sir JH, Barr AR, Nicholas AK, Carvalho OP, Khurshid M, Sossick A, Reichelt S, D'Santos C, Woods CG, Gergely F. A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet. 2011;43:1147–1153. doi: 10.1038/ng.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, Boxem M, van den Heuvel S. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- Yook KJ, Proulx SR, Jorgensen EM. Rules of nonallelic noncomplementation at the synapse in Caenorhabditis elegans. Genetics. 2001;158:209–220. doi: 10.1093/genetics/158.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, Sergi CM, Topcu M, McDonald MT, Barry BJ, Felie JM, et al. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Genet. 2010;42:1015–1020. doi: 10.1038/ng.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.