Abstract
Potent and selective recognition and modulation of disease-relevant RNAs remains a daunting challenge. We previously examined the utility of the U1A N-terminal RNA recognition motif as a scaffold for tailoring new RNA hairpin recognition, and showed that as few as one or two mutations can result in moderate affinity (low μM dissociation constant) for the human immunodeficiency virus (HIV) Trans-Activation Response element (TAR) RNA, an RNA hairpin controlling transcription of the human immunodeficiency virus (HIV) genome. Here we use yeast display and saturation mutagenesis of established RNA binding regions in U1A to identify new synthetic proteins that potently and selectively bind TAR RNA. Our best candidate has truly altered, not simply broadened, RNA binding selectivity; it binds TAR with sub-nanomolar affinity (apparent dissociation constant ~0.5 nM), but does not appreciably bind the original U1A RNA target (U1hpII). It specifically recognizes the TAR RNA hairpin in the context of the HIV-1 5′-untranslated region, inhibits the interaction between TAR RNA and an HIV Trans-activator of transcription (Tat)-derived peptide, and suppresses Tat/TAR-dependent transcription. Proteins described in this work are among the tightest TAR RNA-binding reagents – small molecule, nucleic acid, or protein - reported to date, and thus have potential utility as therapeutics and basic research tools. Moreover, our findings demonstrate how a naturally occurring RNA recognition motif can be dramatically resurfaced through mutation, leading to potent and selective recognition—and modulation—of a disease-relevant RNA.
Graphical Abstract

Ribonucleic acids (RNAs) play a critical role in functionally diverse cellular processes, including many that are disease-relevant.(1–3) Ligands that bind these RNAs have value as basic research tools, possible therapeutic application, and have the potential to illuminate the molecular requirements for their selective recognition. Traditionally, research has focused on developing new small organic molecules (MW <800 Da) that bind RNAs of therapeutic relevance.(4–6) However, structural dynamics and electronic features associated with RNA makes identification of such reagents a daunting challenge. To date, only a relatively modest number of synthetic small (<1 kDa) to medium-sized (~1–2 kDa) RNA-binding molecules have been reported.(7–20) Of these, many are multimers of general nucleic acid binding molecules and only a fraction exhibit even moderate to good sequence selectivity.
By virtue of their size and molecular complexity, proteins can often bind macromolecules (including nucleic acids) that frustrate small-molecule drug discovery. Various screening and evolution-based techniques also permit relatively rapid analysis of large (≥106) protein libraries for binding to a target, and this effort is typically much higher-throughput and simpler than the analogous small-molecule discovery process. Finally, multiple technologies now exist – some developed in our own lab(21–23) - which enable functional protein delivery to the interior of mammalian cells and even specific subcellular environments(24). to the extent that multiple researchers have used exogenous natural or synthetic proteins as basic research tools or drug leads that act on intracellular targets.(25–30) Relatively few successful attempts at de novo protein design have been reported and de novo protein design remains a significant challenge.(31) Perhaps a most sensible solution is one of semi-design, i.e. start with a stable protein known to perform a related task, such as recognition of a particular macromolecular target, and modify it to selectively recognize a new disease-relevant target that shares features with the native binding partner.
Nature has evolved a portfolio of RNA-binding proteins, many of which recognize their target with excellent potency and selectivity profiles. Researchers have previously demonstrated that mutating residues within the putative RNA-binding face of the pumilio homology domain, which recognizes single-stranded RNA (ssRNA), created new variants with tailored ssRNA recognition.(32) While these studies establish that mutation of a naturally occurring RNA-binding protein can lead to altered (as opposed to merely broadened) binding selectivity, recognition is limited to ssRNA, while the majority of RNA exists as a globular-like structure, forming various hydrogen-bond mediated structures, including the RNA hairpin.
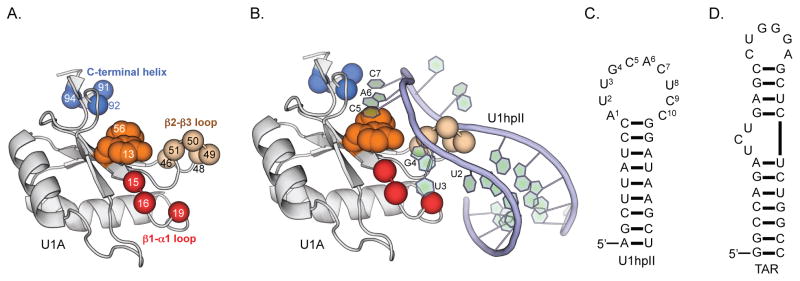
The RNA Recognition Motif (RRM) is among the best-studied RNA-binding protein scaffolds(33, 34), and the N-terminal U1A RRM (Figure 1A) is perhaps the best-studied RRM. The native U1A protein binds the RNA hairpin U1hpII (Figure 1B and Figure 1C) with exceptional affinity (KD ~40 pM) and exquisite selectivity, and the molecular dictates of this interaction are well characterized.(35) U1hpII recognition is stabilized across 4 putative RNA-binding regions of U1A (highlighted in Figure 1A and Figure 1B).(35) In addition to general recruitment of polyanionic RNA by cationic residues (lysines and arginines) on the RNA-binding face(36), solvent exposed nucleotides C5 and A6 π-stack with aromatic U1A residues F13 and Y56(37), in a manner that is not particularly sequence-selective. Additionally, the β2-β3 loop protrudes into the U1hpII RNA loop. While no residue in the β2-β3 loop directly engages any nucleotide in U1hpII, we have previously shown that this loop acts as a ‘steric ruler’ – compensatory mutations or deletions in this loop lead to U1A-derived proteins that bind U1hpII-derived RNA hairpins with smaller loops.(38, 39) Sequence-selectivity in this protein-RNA interaction is largely achieved through highly defined hydrogen bonding networks involving U1A residues N15, N16, and E19 (in the β1-α1 loop) and U1hpII nucleotides U2, U3, and G4, and residues S91 and D92 in the C-terminal helix and nucleotides A6 and C7.(35) The structural importance of I94 in the C-terminal helix has been inferred from observations that its substitution affected structural dynamics of the helix, which dramatically changes conformation upon recognition of U1hpII.(40)
Figure 1.
The U1A scaffold, and the molecular dictates for its interaction with U1hpII. (A) U1A N-terminal RRM, with putative RNA binding regions highlighted. (B) The U1A/U1hpII complex. Nucleotides that are directly engaged by U1A are highlighted. (C) Sequence and secondary structure of the U1hpII RNA hairpin used in this work, a binding partner of U1A. (D) Sequence and secondary structure of the TAR RNA hairpin used in this work.
Previously, high-throughput screening has revealed U1A-derived proteins that recognize U1hpII-derived RNA hairpins.(41, 42) One goal of our lab has been to develop synthetic RRMs that potently and selectively bind and modulate a disease-relevant RNA hairpin. We previously showed how mutating the N-terminal U1A RRM (U1A, Figure 1A) alters its target RNA sequence selectivity.(39, 43) In particular, we probed how one or two mutations to putative RNA-binding residues within the β1-α1 and β2-β3 loops lower affinity for U1hpII and increase affinity for Trans-Activation Reporter (TAR) element RNA(44) (Figure 1D). TAR RNA has an extended stem-loop structure that is crucial for efficient transcription of the integrated HIV genome, and is an established therapeutic target.(45) This RNA also protects against apoptosis in infected cells by altering cellular gene expression(46), and is sequence and structurally non-homologous to U1hpII. From our previous mutagenesis studies we identified U1A E19S, a single amino acid mutant that binds TAR RNA with moderate affinity (dissociation constant, KD ~4 μM) and ~3-fold selectivity over U1hpII.(43) In this work, using U1A E19S as a starting point, we employ yeast display and sequential random mutagenesis of the β2-β3 loop and C-terminal helix to identify new U1A-derived proteins that are tailored for recognition of TAR RNA.
RESULTS
Yeast Display Maturation of U1A for TAR RNA Recognition
As stated above, in the native U1A/U1hpII interaction the β2-β3 loop acts as a ‘steric ruler’ for RNA recognition. Based on previous studies(38), including our own(39), we hypothesized that high-throughput screening would yield a privileged β2-β3 loop which selectively recognizes TAR RNA. Based on this hypothesis, we hypothesized that high-throughput screening would yield a privileged β2-β3 loop that is selective for TAR RNA recognition. Based on this hypothesis, we used saturation mutagenesis to prepare a ~3x106 member library of U1A-derived proteins with randomized β2-β3 loop residues 46, 48, 49, 50, and 51. Since it plays a role in general recruitment of anionic RNA(36), R47 in the β2-β3 loop was retained.
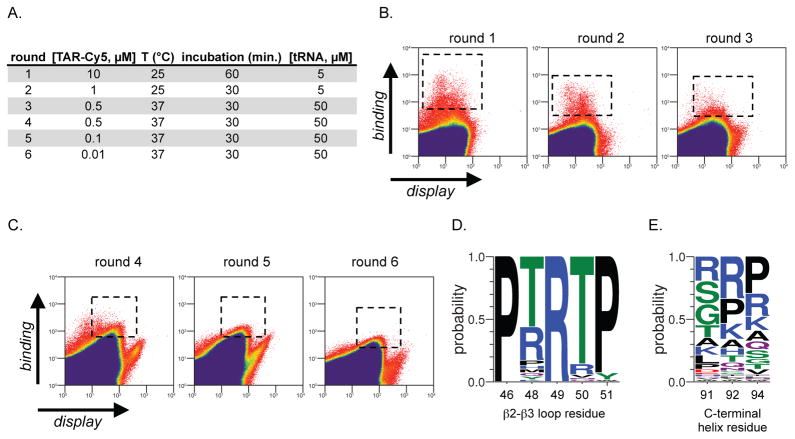
This U1A β2-β3 loop protein library was screened for TAR recognition by yeast display(47, 48) over the course of three rounds. Briefly, a library of U1A E19S-derived proteins with a randomized β2-β3 loop was displayed on EBY100 Saccharomyces cerevisiae with a C-terminal myc tag. The display of the protein library on yeast was characterized by treatment with a FITC-labeled anti-myc antibody and flow cytometry. The U1A yeast display library was concomitantly incubated with a TAR RNA-cyanine-5 conjugate (TAR-Cy5). Since the excitation/emission profiles of FITC and Cy5 are orthogonal, relative FITC:Cy5 fluorescence was used to identify yeast with the highest levels of displayed protein (FITC) and bound TAR RNA Cy5), and these yeast cells were isolated by fluorescence activated cell sorting (FACS). Following the first round of yeast display and FACS, enriched yeast was grown to confluence over 3 days and re-induced for another round of screening. This process (induce U1A protein library expression, incubate with FITC-labelled anti-myc antibody and TAR-Cy5, FACS to isolate best binders, grow yeast to confluence) was conducted over three rounds, and the stringency of screening was increased with each round (Figure 2A). Rounds one and two were performed at 25 °C with 5 μM unlabeled tRNAs from E. coli, while round three was performed at physiological temperature (37 °C), with 50 μM unlabeled tRNAs from E. coli. The concentration of TAR-Cy5 (round 1: 10 μM; round 2: 1 μM; round 3: 0.5 μM) and incubation time (round 1: 60 minutes; round 2: 30 minutes; round 3: 30 minutes) were also decreased as rounds increased, to enrich the highest affinity interactions. Flow cytometry data from rounds 1–3 is shown in Figure 2B (dashed boxes indicate library members that were enriched by FACS).
Figure 2.
Yeast display screening identifies new TAR-binding proteins. (A) Yeast display screening conditions for rounds 1–6. (B) Flow cytometry data of the yeast display library screening for rounds 1–3, which focused on maturation of the β2-β3 loop. Boxes with dashed lines represent the portion of the library that is sorted in each round. (C) Flow cytometry data of the yeast display library screening for rounds 4–6, which focused on maturation of the C-terminal helix. Boxes with dashed lines represent the portion of the library that is sorted in each round. (D) Consensus sequence of the β2-β3 loop found in library members following the 6th round of yeast display screening. (E) Consensus sequence of the C-terminal helix found in library members following the 6th round of yeast display screening.
We next set out to mature the C-terminal helix for TAR recognition. In the native U1A/U1hpII complex, the C-terminal helix undergoes a dramatic conformational rearrangement to directly engage U1hpII.(40) In particular, residues S91 and D92 form a sequence-selective hydrogen bond network with U1hpII loop nucleotides A6 and C7. This strutural rearrangement is facilitated by residue I94. Given the role of these residues in the U1A/U1hpII complex, we hypothesized that randomization of these residues, and selection for those that bind TAR should lead to a new C-helix that is selective for TAR recognition. To test this hypothesis the enriched β2-β3 loop library was randomized to all proteinogenic amino acids at positions 91, 92, and 94, and subjected to three additional rounds of yeast display screening as described above. The conditions for screening rounds 4–6 are described in Figure 2A and flow cytometry data from screening rounds 4–6 is shown in Figure 2C (dashed boxes indicate library members that were enriched by FACS). Again, screening stringency was increased by addition of 50 μM tRNA and decreasing the concentration of TAR-Cy5 in each round.
Following enrichment in round 6, we sequenced 70 clones. Interestingly, a strong consensus sequence emerged in the β2-β3 loop (Figure 2D), suggesting that we had evolved a privileged sequence for TAR recognition. Positions 46 and 51 show a very strong (100% and 93%, respectively) preference for proline, in contrast to the serine and lysine present at these positions in the native U1A protein. Position 48 is largely populated by threonine and arginine (60% and 27%, respectively), and the most popular mutation is chemically similar to the serine found at that position in the native U1A protein. However, the most avid TAR-binding protein (TBP) we characterized (TBP 6.7, see below) contains a glutamine at position 48, while glutamine is present in only ~3% of sequenced clones. Residue 49 gains a cationic charge by virtue of undergoing a leucine to arginine mutation in all sequenced library members. In contrast, position 50 loses positive charge in most of the clones; a lysine to threonine mutation is the most common among the group (87%). Although the consensus sequence in the C-terminal helix (Figure 2E) is considerably more heterogeneous than that of the β2-β3 loop, themes emerge. In native U1A residues 91 and 92 are serine and aspartic acid, respectively. Interestingly, a significant portion of the TAR-binding RRMs retain serine at position 91, although arginine, glycine, and threonine are also prominent. Position 92 and 94 undergo more dramatic changes. In particular, cationic residues arginine and lysine are found in a significant number of TAR-binding RRMs, as is proline. Proline, arginine, and lysine are also prominent at position 94, which, in the native protein is known to modulate the conformation of the C-terminal helix.(40) The prominence of proline at positions 92 and 94 within the C-terminal helix is interesting and suggests that conformational rigidity might be favorable.
Characterization of New Synthetic Protein/TAR RNA Complexes
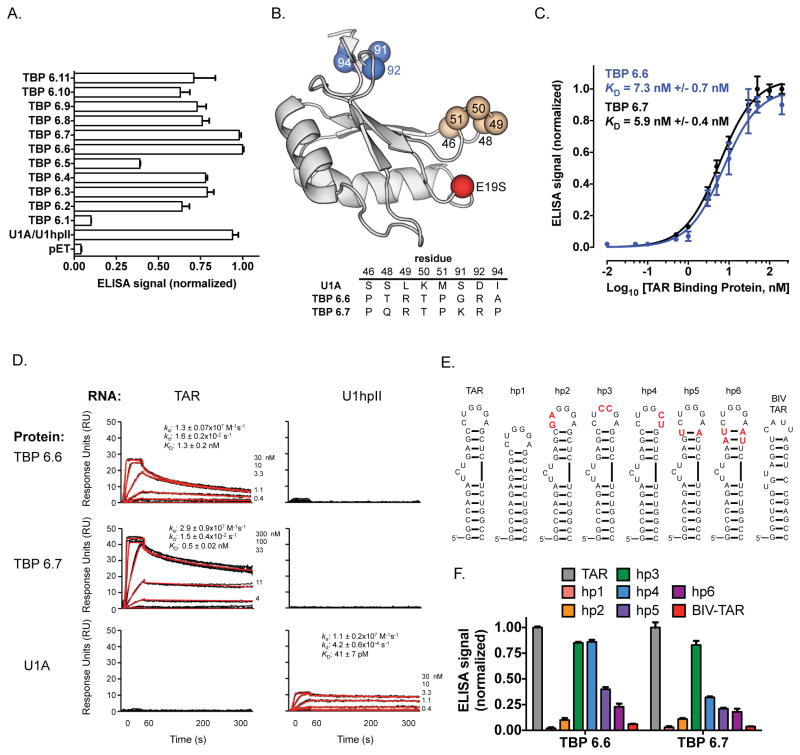
We next assessed the ability of enriched library members to bind TAR RNA by an enzyme linked immunosorbent assay (ELISA). Briefly, 5′-biotinylated TAR RNA was immobilized onto streptavidin-coated plates. TAR RNA-displaying wells were then incubated with a 20 nM solution containing the one of the TAR-Binding Proteins (TBP 6.1–6.11), with a C-terminal His6 spacer and FLAG tag. Following washing steps, incubation with horseradish peroxidase (HRP) conjugated anti-FLAG antibody and HRP substrate, relative levels of the protein-RNA complex were measured with a plate reader. With the exception of TBP 6.1, which did not tightly bind TAR RNA, all other proteins appreciably bound the target RNA (Figure 3A). Among the group, two TAR-Binding Proteins (TBPs) stood out as superior: TBP 6.6 and TBP 6.7, which compares similarly to U1A/U1hpII positive control. In contrast, a negative control featuring the output of a protein purification performed on E. coli transformed with a pETDuet plasmid (without an encoded RRM) did not generate appreciable signal. The β2-β3 loop and C-terminal helix mutations observed in the two best performing proteins are shown in Figure 3B. Based on this initial characterization of TAR RNA binding affinity, these two proteins were investigated further.
Figure 3.
Characterization of TAR RNA binding affinity and selectivity. (A) ELISA analysis of selected 6th generation TAR-Binding Proteins (TPBs). (B) U1A scaffold highlighting mutations identified in the tightest TBPs: TBP 6.8 and TBP 6.9. (C) ELISA data showing binding between TAR RNA and TBP 6.6 (blue) or TBP 6.7 (black). (D) SPR data for the binding interactions between TBP 6.6 and TAR RNA (left column, top row); TBP 6.7 and TAR RNA (left column, middle row); U1A and TAR RNA (left column, bottom row); TBP 6.6 and U1hpII RNA (right column, top row); TBP 6.7 and U1hpII RNA (right column, middle row); U1A and U1hpII RNA (right column, bottom row). (E) Sequence and secondary structures of TAR-derived mutants used to characterize the requirements for TAR recognition by TBP 6.6 and TBP 6.7. (F) ELISA data showing binding between TBP 6.6 or TBP 6.7 and TAR mutants hp1-6 and BIV TAR.
Quantitative evaluation of these new protein-RNA binding interactions was first measured by ELISA. Similar to the experiment described above, biotinylated RNA was immobilized onto a streptavidin-coated plate, then incubated with varied concentrations (0.01 – 200 nM) of either TBP 6.6 or 6.7 with a C-terminal FLAG tag. Following incubation with an HRP-conjugated anti-FLAG antibody and HRP substrate, relative levels of the protein-RNA complex was measured with a plate reader. Both proteins bound TAR RNA with single digit nanomolar apparent dissociation constants (KD ~6 nM for TBP 6.6; KD ~7 nM for TBP 6.7, Figure 3C).
Surface Plasmon Resonance (SPR) Analysis of New Protein/RNA Complexes
We next characterized the binding interaction, kinetics, and selectivity by surface plasmon resonance (SPR), as described.(38, 49) A streptavidin-coated sensorchip (SA, GE Healthcare) was used to coat 25 response units (RU) of 5′-biotinylated U1hpII (5′-AGCUUAUCCAUUGCACUCCGGAUGAGCU-3′) on flow cell 1, and 25 RU of TAR (5′-GGCAGAUCUGAGCCUGGGAGCUCUCUGCC-3′) on flow cell 3, leaving flow cells 2 and 4 blank for background correction. Proteins were serially diluted in running buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5% glycerol, 62.6 mg mL−1 bovine serum albumin, 1 mM dithiothreitol, 0.05% surfactant P20, and 125 μg mL−1 yeast tRNA) to the concentrations indicated (Figure 3D) and injected at 20 °C with a flow rate of 50 μL/min over all surfaces consecutively. In each for three independent experiments, triplicate injections were fully randomized and interspersed with buffer injections to allow double referencing.(50) After each protein injection the surface was regenerated with a 1-minute 2 M NaCl injection, followed by a buffer injection. Data was processed using Scrubber and analyzed using CLAMP XP(51) and a simple 1:1 Langmuir interaction model with a correction for mass transport(52). TBP 6.6 and 6.7 show similar affinities for the TAR hairpin (apparent KD = 1.3 ± 0.2 nM and 0.5 ± 0.1 nM, respectively, Figure 3D), which is consistent with our ELISA data (Figure 3C). Gratifyingly, both TBP 6.6 and 6.7 show negligible binding to the U1hpII surface. These results confirm the evolution of two very high affinity TAR-binding proteins that have essentially lost their ability to bind U1hpII, their original RNA target. Native U1A, injected for comparison, did not bind to TAR, but did bind to the U1hpII surface with the expected affinity of ~40 pM.
Characterization of TAR Binding Selectivity
Binding selectivity, and the requirements for TAR RNA recognition, was further characterized by ELISA using a variety of TAR-derived RNAs (Figure 3E). We used a TAR-derived RNA hairpin, designated hairpin 1 (hp1) that lacks the UCU bulge (Figure 3E), and three TAR-derived RNAs designated hairpins 2, 3, and 4 (hp2, hp3, and hp4), which have two consecutive mutations in the apical loop (Figure 3E, mutations are highlighted in red). The most dramatic change in binding was observed when we removed the UCU bulge in TAR (hp1). When we performed an ELISA using this immobilized RNA, essentially no binding was observed with either TBP 6.6 or TBP 6.7 (Figure 3F, red bar). This finding is important, since the native TAR-binding protein (Tat) largely recognizes the UCU bulge. Thus, if our proteins occupy this space they should antagonize the Tat-TAR interaction (see below). Dramatically decreased binding was also observed when the first two loop nucleotides (5′-CU-3′) were mutated to 5′-GA-3′ (hp2). Less dramatic, but significant, changes in affinity were observed when we mutated other loop residues. When the last two loop nucleotides (5′-GA-3′) are mutated to 5′-CU-3′ in hp4, while no appreciable loss in affinity was observed for TBP 6.6, but significant lower binding was observed for the complex involving this RNA and TBP 6.7 (Figure 3F, blue bar). In contrast, mutating the central nucleotides (5′-GG-3′) to 5′-CC-3′ did not appreciably alter affinity for either protein (Figure 3F, green bar). The ability of synthetic RRMs to recognize specific loop nuceotides begs the question: are nucleotides in the top of the stem (which link the bulge and loop) recognized by U1A-derived proteins? To probe this, we assessed binding of mutants hp5 and hp6, which have mutated residues in the top of the stem. Both mutations significantly decreased affinity to TBP 6.6 and TBP 6.7, suggesting that the uppermost stem nucleotides directly participates in complex formation. Additionally, we also tested binding to the TAR sequence from the Bovine Immunodeficiency Virus (BIV TAR(53)). This RNA is structurally similar to HIV TAR, but differs in the sequence and size of the loop (4 bases in BIV TAR, versus 6 bases in HIV TAR) and in the nature of the stem-bulge. We found that neither TBP 6.6, nor TBP 6.7 had appreciable affinity for this RNA (Figure 3F).
SHAPE Analysis of Binding Within the HIV-1 5′-UTR
We next explored binding of TBP 6.6 and 6.7 to the TAR region of the HIV-I (NL4-3) 5′-UTR RNA, using Selective 2′-Hydroxyl Acylation analyzed by Primer Extension (SHAPE), which probes RNA conformational flexibility through accessibility of the 2′-OH group to acylation. TBP binding to TAR predicts this region would become less amenable to chemical modification. We studied binding in the context of the 362-nt 5′-UTR, since this RNA is highly structured, harboring the TAR and polyA hairpins, the primer binding site (PBS), packaging signal (Ψ), dimer linkage sequence (DLS), and the major 5′ splice site (5′ss).(54, 55) These multiple cis-acting elements bind different ligands and support long-range inter- and intramolecular interaction(s) that facilitate genome transcription, translation, RNA dimerization/packaging and splicing. Therefore, TBP binding to the 5′-UTR provides a direct and biologically relevant measure of potential off-target effects.
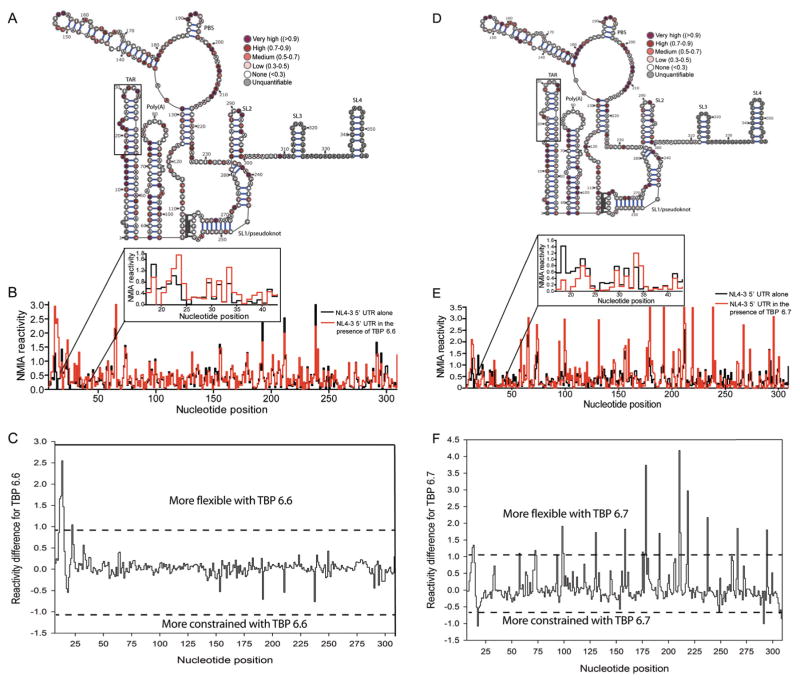
The 5′-UTR RNA exists in monomeric and dimeric forms (56–59), and since SHAPE measures ensemble-average reactivity, folding conditions were optimized to prepare a homogeneous monomeric RNA (Figure S1A). Averaged SHAPE reactivity values from 3 independent experiments were color-coded onto the proposed pseudoknot monomeric 5′-UTR structure (57, 58) as the algorithm of the software commonly used for RNA secondary structure prediction (RNAStructure) cannot be used to predict pseudoknot structures.(57) As shown in Figure S1B, data for the 5′-UTR RNA in the absence of protein are at slight variance with the proposed pseudoknot structure. Such discrepancy in HIV-I monomeric RNA secondary structure, which was previously reported (57), may reflect differences in folding conditions and tertiary interactions. SHAPE analysis was performed at different RNA:TBP ratios (1:1, 1:2, 1:4 and 1:8), and appreciable changes in acylation sensitivity were observed only when either protein was present in a 4-fold or 8-fold molar excess (Figure S3 and Figure S4, and Figure 4, respectively). Reactivity values were color coded (Figure 4A for TBP 6.6 and Figure 4C for TBP 6.7) and plotted as a function of nucleotide position (Figure 4B for TBP 6.6 and Figure 4D for TBP 6.7). To determine which nucleotides were conformationally flexible or constrained by TBP binding, reactivity values in the absence of the protein were substracted from those in its presence and the resulting difference values were plotted as a function of nucleotide position (Figure 4C for TBP 6.6 and Figure 4F for TBP 6.7). Following the previously-reported quantitative SHAPE difference cutoffs(60), nucleotides with a reactivity difference > +1 were designated conformationally more flexible and those with a reactivity difference < −1 were assigned conformationally more constrained.
Figure 4.
SHAPE analysis of the monomeric NL4-3 5′-UTR RNA in the presence of an 8-fold excess of TBP 6.6 and TBP 6.7. (A) SHAPE reactivity profile of the 5′-UTR obtained in the presence of TBP 6.6, color-coded on the proposed pseudo-knot 5′-UTR monomeric structure (57, 58) (B) Step plot comparing the NMIA reactivity of the 5′-UTR in the presence (red) or absence (black) of TBP 6.6. (C) SHAPE difference profile showing conformational changes induced by TBP 6.6. (D) SHAPE reactivity profile of the 5′-UTR obtained in the presence of TBP 6.7, color-coded on the proposed pseudo-knot 5′-UTR monomeric structure. (E) Step plot comparing the NMIA reactivity of the 5′-UTR in the presence (red) or absence (black) of TBP 6.7. (F) SHAPE difference profile showing conformational changes induced by TBP 6.6. In (A), (B), (C), and (D), the boxed region corresponds to the test TAR region (nucleotides 17–43) used in the other binding selectivity assays.
A common feature of TBP 6.6 and 6.7 complexes was increased acylation at several important positions of the TAR hairpin. Protein binding destabilized the local helix at nucleotides U12, U13, A14, G15 for TBP 6.6 and U13, and A14 for TBP 6.7. Nucleotide C23 of the UCU loop (which was deleted in the hp1 mutant) was also rendered more flexible in the presence of TBP 6.6. Interestingly, of all TAR mutants tested for reduced binding to TBP 6.6 and 6.7 by ELISA (Figure 3F), hp1 mutant most significantly disrupted the interaction. Therefore, the SHAPE and ELISA assays collectively implicate UCU loop nucleotide C23 in TBP 6.6 binding. TBP 6.7 significantly constrained nucleotide C18 located at the base stem implying C18 as an important contact site for the protein. Conversely, no TAR nucleotide was rendered more constrained by TBP 6.6 binding suggesting it may bind to an already constrained (base-paired) region, e.g. the upper stem, which was shown to be important for binding by ELISA (Figure 3F). Therefore, both TBP 6.6 and TBP 6.7 proteins induce significant, yet distinctly different, conformational changes in TAR, suggesting they interact differently.
No significant changes in acylation sensitivity were observed outside the TAR region for TBP 6.6, indicative of a local interaction. In contrast, TBP 6.7 significantly increased conformational flexibility at nucleotides C58, A73, A74, U94, G99, U100, U131, C159, U176, G178, C179, A192, A211, G212, C219, C238, C267, and U295. TBP 6.7 also significantly constrained nucleotide C292 in the SL2 loop (the major 5′-splice site). SL2 residues were shown to mediate long-range contact with residues at the base of SL1 and upstream of the U5 region(56) in the dimeric UTR. Therefore, by decreasing reactivity of the SL2 loop residue, TBP 6.7 could shift the equilibrium towards the dimeric UTR conformer. Furthermore, we rule out non-specific protein-RNA interactions driving the conformational flexibility observed outside the TAR hairpin of the 5′-UTR as TBP 6.7 strongly discriminates against (a) BIV TAR, a highly homologous relative of HIV-1 TAR (Figure 3F), and (b) 5000-fold excess of competitor tRNA (Figure 2A). Therefore, we propose that TBP 6.7 binding to TAR in the context of the 5′-UTR induces long-range alterations in overall topology that are more pronounced than those promoted by TBP 6.6. Stated differently, secondary consequences of TBP 6.7 binding to TAR on global 5′-UTR topology cannot be ruled out. This notion is supported by our recent work that identified small-molecule ligands specific for TAR(61) where we likewise observed changes in SHAPE reactivity profiles distal to the ligand binding site.
Thus, we believe that differences in the nature of the primary interaction between TPB 6.6-TAR and TPB 6.7-TAR, and the structural consequences thereof, might explain why TPB 6.7 selectively induces long range topological changes. These differences might also explain why despite having equal affinity towards TAR RNA, the two TAR binding proteins have different biological activity. As shown subsequently in the manuscript, TBP 6.7 prevents Tat-TAR interaction and inhibits transcription from TAR region while TBP 6.6 does not.
Inhibition of the Tat/TAR complex by a Synthetic TAR-Binding Protein
The synthetic proteins described in this work recognize TAR RNA with excellent affinity and exquisite selectivity. However, any potential therapeutic or basic research utility of these new proteins rests on their ability to inhibit a protein-RNA interaction involving the trans-activator of transcription (Tat) protein and TAR RNA. In binding to TAR, Tat alters the transcription complex, recruits the positive transcription elongation complex (PTEFb) of cellular CDK9 and cyclin T1, resulting in an increase in the production of full-length viral RNA.(62) Reagents that inhibit the Tat/TAR complex can suppress the transcription of full-length HIV RNA, leading to suppression or abrogation of HIV protein expression and production of virus.(63)
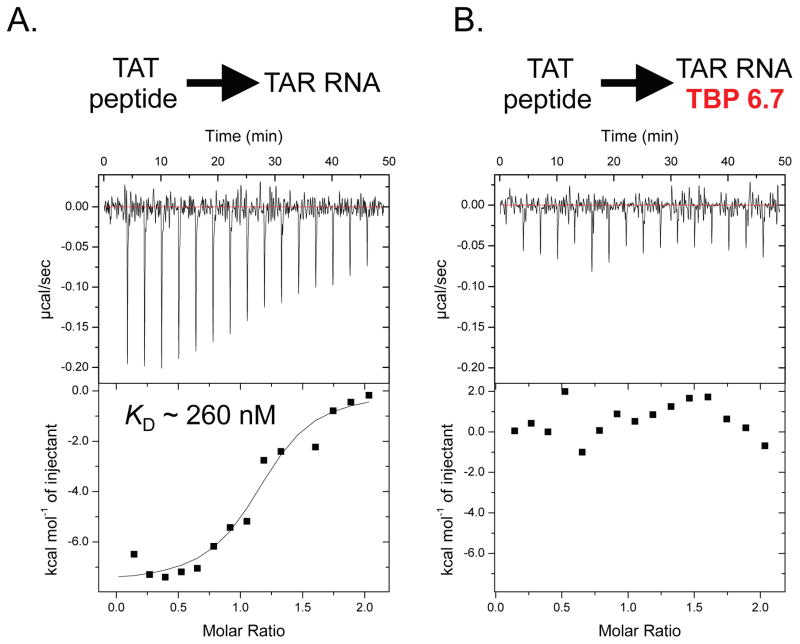
In order to determine if our new TAR-binding proteins inhibit an interaction with Tat, we utilized a previously described Tat peptide comprising the TAR-binding portion. This peptide, and variations on this theme have previously been used to characterize the Tat/TAR interaction. Initially we used ELISA to determine if our protein could disrupt or inhibit the Tat/TAR, complex; however, the high theoretical charge of this peptide (RKKRRQRRRPPQGSQTHQVSLSKQPTSQPRGDPTGPKE, theoretical charge = +9) complicated these experiments. When biotinylated TAR RNA is immobilized on streptavidin-coated ELISA plates, we found that Tat peptide absorbed onto the plates (presumably through non-selective interactions with streptavidin) and could not be easily removed. To overcome this challenge, we used a solution phase experiment (isothermal titration calorimetry, ITC) to characterize the interaction, and the effect our TAR-binding proteins have on complex formation. As seen in Figure 5A, the Tat peptide binds TAR with good affinity (KD ~ 260 nM). However, when the peptide is titrated into a preformed 1:1 TAR/TBP 6.7 complex, we observe no appreciable binding (Figure 5B), suggesting the protein developed in this work inhibits formation of the Tat/TAR complex. This inhibition is supported not only by the significantly tighter binding observed here between our protein and TAR, compared to the Tat peptide/TAR interaction (apparent KD ~0.5 nM versus ~260 nM, respectively), and by the observation in our RNA mutagenesis studies that TBP 6.7 binds the trinucleotide bulge of TAR, the same site that is recognized by Tat peptide.
Figure 5.
Inhibition of the Tat/TAR complex by TAR-binding proteins. (A) Isothermal Titration Calorimetry (ITC) data for a titration of 66 μM Tat peptide into 6.6 μM TAR RNA. (B) ITC data for a titration of 66 μM Tat peptide into 6.6 μM pre-formed TAR RNA/TBP 6.7 complex. All ITC experiments were performed at 25 °C.
Interestingly, despite the fact that TBP 6.6 also binds TAR with exceptional affinity, and binds the trinucleotide bulge, we observe no appreciable change in the binding isotherm for ITC experiments involving titration of Tat peptide into a solution of TAR RNA, or a pre-formed TAR RNA/TBP 6.6 complex. This result suggests that that despite their virtually identical affinity for TAR RNA, only TBP 6.7 inhibits the interaction between Tat peptide and TAR RNA.
Suppression of Tat-TAR-Dependent Transcription by a Synthetic TAR-Binding Protein
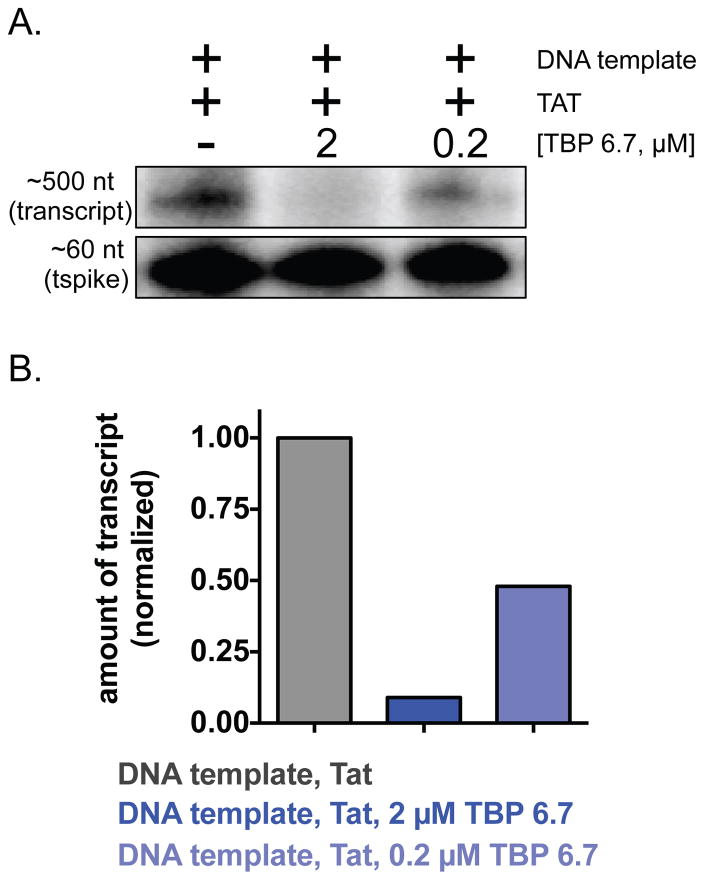
A previously described function-based assay(63–65) was performed to measure the ability of TBP 6.7 to suppress Tat/TAR-dependent transcription of a portion of the HIV-1 genomic DNA that includes the TAR element. We performed in vitro transcription in HeLa cell nuclear extract in the presence or absence 2 μM or 0.2 μM TBP 6.7. Gratifyingly, we observed a concentration-dependent suppression of Tat/TAR-dependent transcription in the presence of TBP 6.7. At the highest concentration of TBP 6.7 tested, we observe virtually complete suppression of transcription (Figure 6).
Figure 6.
Suppression of Tat/TAR-dependent transcription by TBP 6.7. A plasmid encoding a portion of the HIV-1 genome with an upstream TAR element was in vitro transcribed in HeLa cell extract in the presence or absence of Tat or TBP 6.7. 32P-radiolabelled RNA transcript yield was measured for each condition; those transcripts are shown in (A) and densitometry values are plotted in (B).
Conclusion
Folded RNAs, including a number of sequence and structurally diverse RNA hairpins, have been identified as therapeutic targets. However, relatively little is known about how to develop potent and sequence-selective RNA-binding reagents. Nature has evolved a number of RNA-binding proteins, many of which recognize their cognate RNAs with exceptional affinity and selectivity. Recently, we have shown that as few as one or two mutations to the RNA Recognition Motif U1A can turn ‘on’ affinity for TAR RNA, while turning ‘off’ affinity for U1hpII – a physiological binding partner of U1A. Thus, U1A may represent a privileged scaffold from which new RNA-binding proteins can be developed.
In this work, starting from U1A E19S—a mutant we recently described(43) we used yeast display high-throughput screening and saturation mutagenesis of putative RNA-binding regions to develop new proteins with potent TAR RNA recognition. This effort resulted in new synthetic RRMs with good affinity for TAR RNA (apparent KD ~0.5 nM). Not only were we able to generate excellent affinity for TAR, these new proteins also exhibit exceptional selectivity for TAR. TAR-binding proteins recognize this target RNA the presence of ~5000-fold molar excess tRNAs (Figure 3C), and show virtually no affinity for U1hpII (Figure 4D), a physiological binding partner of U1A. The proteins described in this work selectively recognize TAR RNA in the context of the HIV-1 5′-UTR, and the best performing protein inhibits a complex between the Tat peptide and TAR RNA, and suppresses Tat-TAR-dependent transcription in vitro. Collectively, the proteins reported here (TBP 6.6 and TBP 6.7) are among the tightest TAR RNA binding reagents reported to date and represent the first RRMs that potently and selectively recognize TAR RNA (as opposed to U1hpII-derived RNA hairpins). Moreover, this work shows that a naturally occurring RNA-binding protein (U1A) can be dramatically mutated and evolved for a new function: potent and selective recognition—and modulation—of a disease-relevant RNA.
Supplementary Material
Acknowledgments
This work is supported by the NIH/NIGMS (R01GM107520, B.R.M.) and NIH/NCI P30 CA014089 (I.A.L-O.). We thank A. Chapman for assistance with isothermal titration calorimetry experiments, as well as valuable discussions. The authors are grateful to J.T. Sczepanski (Texas A&M) for supplying plasmid pLAI-BS used in transcription assays, and for helpful discussions. S.L.G. and C.S. are supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health, Department of Health and Social Services.
Footnotes
Supporting Information. Detailed experimental methods, supplementary data, characterization and sequence of proteins used in this work.
References
- 1.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Non-coding RNAs in human disease. Nature reviews Genetics. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 3.Osborne RJ, Thornton CA. RNA-dominant diseases. Human molecular genetics. 2006;15(Spec No 2):R162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 4.Gallego J, Varani G. Targeting RNA with small-molecule drugs: therapeutic promise and chemical challenges. Accounts of chemical research. 2001;34:836–843. doi: 10.1021/ar000118k. [DOI] [PubMed] [Google Scholar]
- 5.Hermann T, Tor Y. RNA as a target for small-molecule therapeutics. Expert Opin Ther Pat. 2005;15:49–62. [Google Scholar]
- 6.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chemical reviews. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 7.Gareiss PC, Sobczak K, McNaughton BR, Palde PB, Thornton CA, Miller BL. Dynamic Combinatorial Selection of Molecules Capable of Inhibiting the (CUG) Repeat RNA-MBNL1 Interaction In Vitro: Discovery of Lead Compounds Targeting Myotonic Dystrophy (DM1) J Am Chem Soc. 2008;130:16254–16261. doi: 10.1021/ja804398y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNaughton BR, Gareiss PC, Miller BL. Identification of a selective small-molecule ligand for HIV-1 frameshift-inducing stem-loop RNA from an 11,325 member resin bound dynamic combinatorial library. J Am Chem Soc. 2007;129:11306–+. doi: 10.1021/ja072114h. [DOI] [PubMed] [Google Scholar]
- 9.Childs-Disney JL, Hoskins J, Rzuczek SG, Thornton CA, Disney MD. Rationally Designed Small Molecules Targeting the RNA That Causes Myotonic Dystrophy Type 1 Are Potently Bioactive. Acs Chem Biol. 2012;7:856–862. doi: 10.1021/cb200408a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Childs-Disney JL, Parkesh R, Nakamori M, Thornton CA, Disney MD. Rational Design of Bioactive, Modularly Assembled Aminoglycosides Targeting the RNA that Causes Myotonic Dystrophy Type 1. Acs Chem Biol. 2012;7:1984–1993. doi: 10.1021/cb3001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karan C, Miller BL. RNA-selective coordination complexes identified via dynamic combinatorial chemistry. J Am Chem Soc. 2001;123:7455–7456. doi: 10.1021/ja010325v. [DOI] [PubMed] [Google Scholar]
- 12.Bryson DI, Zhang WY, McLendon PM, Reineke TM, Santos WL. Toward Targeting RNA Structure: Branched Peptides as Cell-Permeable Ligands to TAR RNA. Acs Chem Biol. 2012;7:210–217. doi: 10.1021/cb200181v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XJ, Thomas JR, Hergenrother PJ. Deoxystreptamine dimers bind to RNA hairpin loops. J Am Chem Soc. 2004;126:9196–9197. doi: 10.1021/ja048936l. [DOI] [PubMed] [Google Scholar]
- 14.Meyer ST, Hergenrother PJ. Synthetic small molecule ligands for bulged RNA secondary structure. Abstr Pap Am Chem S. 2007;233:782–782. doi: 10.1021/ol901478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer ST, Hergenrother PJ. Small Molecule Ligands for Bulged RNA Secondary Structures. Org Lett. 2009;11:4052–4055. doi: 10.1021/ol901478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas JR, Liu XJ, Hergenrother PJ. Size-specific ligands for RNA hairpin loops. J Am Chem Soc. 2005;127:12434–12435. doi: 10.1021/ja051685b. [DOI] [PubMed] [Google Scholar]
- 17.Blount KF, Tor Y. A tale of two targets: Differential RNA selectivity of nucleobase-aminoglycoside conjugates. Chembiochem. 2006;7:1612–1621. doi: 10.1002/cbic.200600109. [DOI] [PubMed] [Google Scholar]
- 18.Bonnard V, Pascale L, Azoulay S, Di Giorgio A, Rogez-Kreuz C, Storck K, Clayette P, Patino N. Polyamide Amino Acids trimers as TAR RNA ligands and anti-HIV agents. Bioorgan Med Chem. 2010;18:7432–7438. doi: 10.1016/j.bmc.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Arambula JF, Ramisetty SR, Baranger AM, Zimmerman SC. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. P Natl Acad Sci USA. 2009;106:16068–16073. doi: 10.1073/pnas.0901824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nature chemical biology. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePorter SM, McNaughton BR. Engineered M13 bacteriophage nanocarriers for intracellular delivery of exogenous proteins to human prostate cancer cells. Bioconjugate chemistry. 2014;25:1620–1625. doi: 10.1021/bc500339k. [DOI] [PubMed] [Google Scholar]
- 22.DePorter SM, Lui I, Bruce VJ, Gray MA, Lopez-Islas M, McNaughton BR. Mutagenesis modulates the uptake efficiency, cell-selectivity, and functional enzyme delivery of a protein transduction domain. Molecular bioSystems. 2014;10:18–23. doi: 10.1039/c3mb70429g. [DOI] [PubMed] [Google Scholar]
- 23.DePorter SM, Lui I, Mohan U, McNaughton BR. A protein transduction domain with cell uptake and selectivity profiles that are controlled by multivalency effects. Chem Biol. 2013;20:434–444. doi: 10.1016/j.chembiol.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 24.Horton KL, Stewart KM, Fonseca SB, Guo Q, Kelley SO. Mitochondria-penetrating peptides. Chem Biol. 2008;15:375–382. doi: 10.1016/j.chembiol.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs SM, Raines RT. Arginine grafting to endow cell permeability. Acs Chem Biol. 2007;2:167–170. doi: 10.1021/cb600429k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson DB, Cronican JJ, Liu DR. Engineering and identifying supercharged proteins for macromolecule delivery into mammalian cells. Methods in enzymology. 2012;503:293–319. doi: 10.1016/B978-0-12-396962-0.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cronican JJ, Beier KT, Davis TN, Tseng JC, Li W, Thompson DB, Shih AF, May EM, Cepko CL, Kung AL, Zhou Q, Liu DR. A class of human proteins that deliver functional proteins into mammalian cells in vitro and in vivo. Chem Biol. 2011;18:833–838. doi: 10.1016/j.chembiol.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cronican JJ, Thompson DB, Beier KT, McNaughton BR, Cepko CL, Liu DR. Potent delivery of functional proteins into Mammalian cells in vitro and in vivo using a supercharged protein. Acs Chem Biol. 2010;5:747–752. doi: 10.1021/cb1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sundlass NK, Raines RT. Arginine residues are more effective than lysine residues in eliciting the cellular uptake of onconase. Biochemistry. 2011;50:10293–10299. doi: 10.1021/bi200979k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanzl EG, Trantow BM, Vargas JR, Wender PA. Fifteen years of cell-penetrating, guanidinium-rich molecular transporters: basic science, research tools, and clinical applications. Accounts of chemical research. 2013;46:2944–2954. doi: 10.1021/ar4000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khoury GA, Smadbeck J, Kieslich CA, Floudas CA. Protein folding and de novo protein design for biotechnological applications. Trends Biotechnol. 2014;32:99–109. doi: 10.1016/j.tibtech.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clery A, Blatter M, Allain FHT. RNA recognition motifs: boring? Not quite. Curr Opin Struc Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Maris C, Dominguez C, Allain FHT. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 35.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal-Structure at 1.92 Angstrom Resolution of the Rna-Binding Domain of the U1a Spliceosomal Protein Complexed with an Rna Hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 36.Law MJ, Linde ME, Chambers EJ, Oubridge C, Katsamba PS, Nilsson L, Haworth IS, Laird-Offringa IA. The role of positively charged amino acids and electrostatic interactions in the complex of U1A protein and U1 hairpin II RNA. Nucleic Acids Res. 2006;34:275–285. doi: 10.1093/nar/gkj436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Law MJ, Chambers EJ, Katsamba PS, Haworth IS, Laird-Offringa IA. Kinetic analysis of the role of the tyrosine 13, phenylalanine 56 and glutamine 54 network in the U1A/U1 hairpin II interaction. Nucleic Acids Res. 2005;33:2917–2928. doi: 10.1093/nar/gki602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katsamba PS, Bayramyan M, Haworth IS, Myszka DG, Laird-Offringa IA. Complex role of the beta(2)-beta(3) loop in the interaction of U1A with U1 hairpin II RNA. J Biol Chem. 2002;277:33267–33274. doi: 10.1074/jbc.M200304200. [DOI] [PubMed] [Google Scholar]
- 39.Blakeley BD, Shattuck J, Coates MB, Tran E, Laird-Offringa IA, McNaughton BR. Analysis of protein-RNA complexes involving a RNA recognition motif engineered to bind hairpins with seven- and eight-nucleotide loops. Biochemistry. 2013;52:4745–4747. doi: 10.1021/bi400801q. [DOI] [PubMed] [Google Scholar]
- 40.Law MJ, Lee DS, Lee CS, Anglim PP, Haworth IS, Laird-Offringa IA. The role of the C-terminal helix of U1A protein in the interaction with U1hpII RNA. Nucleic Acids Res. 2013;41:7092–7100. doi: 10.1093/nar/gkt326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laird-Offringa IA. In vitro genetic analysis of RNA-binding proteins using phage display. Methods in molecular biology. 1999;118:189–216. doi: 10.1385/1-59259-676-2:189. [DOI] [PubMed] [Google Scholar]
- 42.Fan Y. PhD. University of Illinois; 2009. Exploring Protein-RNA Interactions with Site-Directed Mutagenesis and Phage Display. [Google Scholar]
- 43.Blakeley BD, McNaughton BR. Synthetic RNA recognition motifs that selectively recognize HIV-1 trans-activation response element hairpin RNA. Acs Chem Biol. 2014;9:1320–1329. doi: 10.1021/cb500138h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bannwarth S, Gatignol A. HIV-1 TAR RNA: the target of molecular interactions between the virus and its host. Current HIV research. 2005;3:61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 45.Richter S, Ping YH, Rana TM. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7928–7933. doi: 10.1073/pnas.122119999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klase Z, Winograd R, Davis J, Carpio L, Hildreth R, Heydarian M, Fu S, McCaffrey T, Meiri E, Ayash-Rashkovsky M, Gilad S, Bentwich Z, Kashanchi F. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology. 2009;6:18. doi: 10.1186/1742-4690-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepper LR, Cho YK, Boder ET, Shusta EV. A decade of yeast surface display technology: where are we now? Combinatorial chemistry & high throughput screening. 2008;11:127–134. doi: 10.2174/138620708783744516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nature biotechnology. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 49.Katsamba PS, Myszka DG, Laird-Offringa IA. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J Biol Chem. 2001;276:21476–21481. doi: 10.1074/jbc.M101624200. [DOI] [PubMed] [Google Scholar]
- 50.Myszka DG. Improving biosensor analysis. Journal of molecular recognition: JMR. 1999;12:279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 51.Myszka DG, Morton TA. CLAMP: a biosensor kinetic data analysis program. Trends in biochemical sciences. 1998;23:149–150. doi: 10.1016/s0968-0004(98)01183-9. [DOI] [PubMed] [Google Scholar]
- 52.Myszka DG, Jonsen MD, Graves BJ. Equilibrium analysis of high affinity interactions using BIACORE. Analytical biochemistry. 1998;265:326–330. doi: 10.1006/abio.1998.2937. [DOI] [PubMed] [Google Scholar]
- 53.Goel T, Kumar S, Maiti S. Thermodynamics and solvation dynamics of BIV TAR RNA-Tat peptide interaction. Molecular bioSystems. 2013;9:88–98. doi: 10.1039/c2mb25357g. [DOI] [PubMed] [Google Scholar]
- 54.Berkhout B, van Wamel JL. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA. 2000;6:282–295. doi: 10.1017/s1355838200991684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clever J, Sassetti C, Parslow TG. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. RNA structure. Structure of the HIV-1 RNA packaging signal. Science. 2015;348:917–921. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kenyon JC, Prestwood LJ, Le Grice SF, Lever AM. In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res. 2013;41:e174. doi: 10.1093/nar/gkt690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stephenson JD, Li H, Kenyon JC, Symmons M, Klenerman D, Lever AM. Three-dimensional RNA structure of the major HIV-1 packaging signal region. Structure. 2013;21:951–962. doi: 10.1016/j.str.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nature methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sztuba-Solinska J, Shenoy SR, Gareiss P, Krumpe LR, Le Grice SF, O’Keefe BR, Schneekloth JS., Jr Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J Am Chem Soc. 2014;136:8402–8410. doi: 10.1021/ja502754f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeang KT, Xiao H, Rich EA. Multifaceted activities of the HIV-1 transactivator of transcription, Tat. The Journal of biological chemistry. 1999;274:28837–28840. doi: 10.1074/jbc.274.41.28837. [DOI] [PubMed] [Google Scholar]
- 63.Sczepanski JT, Joyce GF. Binding of a structured D-RNA molecule by an L-RNA aptamer. J Am Chem Soc. 2013;135:13290–13293. doi: 10.1021/ja406634g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marciniak RA, Calnan BJ, Frankel AD, Sharp PA. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990;63:791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- 65.Arzumanov A, Walsh AP, Liu X, Rajwanshi VK, Wengel J, Gait MJ. Oligonucleotide analogue interference with the HIV-1 Tat protein-TAR RNA interaction. Nucleosides Nucleotides Nucleic Acids. 2001;20:471–480. doi: 10.1081/NCN-100002321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.