In Fig 2A, the color labels on the bar graph for WT and SLP-76 KO were incorrect. WT should be in red, and SLP-76 KO should be in blue. Please see the correct Fig 2 here.
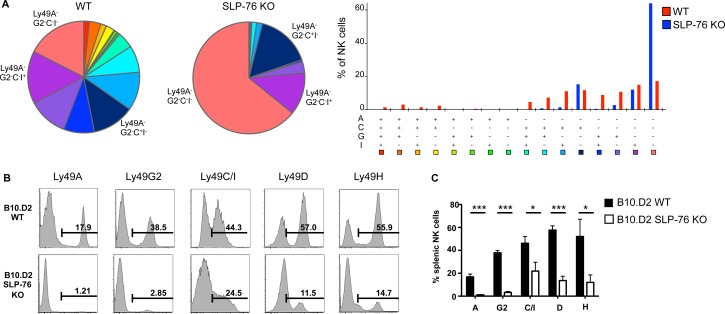
Fig 2. Ly49 receptor expression loss in SLP-76 KO NK cells is independent of MHC I haplotype.
(A) The coexpression pattern of inhibitory Ly49 receptor in WT vs SLP-76 KO splenic NK cells was assessed through SPICE analysis. (B) Representative histograms of Ly49 receptor expression by WT B10.D2 and B10D2.SLP-76 KO splenic NK cells are shown. (C) The proportion of Ly49 receptor-expressing NK cells from multiple WT B10.D2 (black bars) and B10D2.SLP-76 KO (white bars) mice is represented as mean percent positive ± SEM of n = 3 mice/group. *p < 0.05, ***p < 0.001 by student’s t test. See S1 Data for raw data.
Reference
- 1.Freund J, May RM, Yang E, Li H, McCullen M, Zhang B, et al. (2016) Activating Receptor Signals Drive Receptor Diversity in Developing Natural Killer Cells. PLoS Biol 14(8): e1002526 doi: 10.1371/journal.pbio.1002526 [DOI] [PMC free article] [PubMed] [Google Scholar]