Abstract
We engineered covalently circularized nanodiscs (cNDs) with enhanced stability, defined diameter sizes and tunable shapes. Reconstitution into cNDs enhanced the quality of NMR spectra for both VDAC-1, a beta-barrel membrane protein, and the G protein-coupled receptor NTR1, an alpha-helical membrane protein. In addition, we utilized cNDs to visualize how simple, non-enveloped viruses translocate their genomes across membranes to initiate infection.
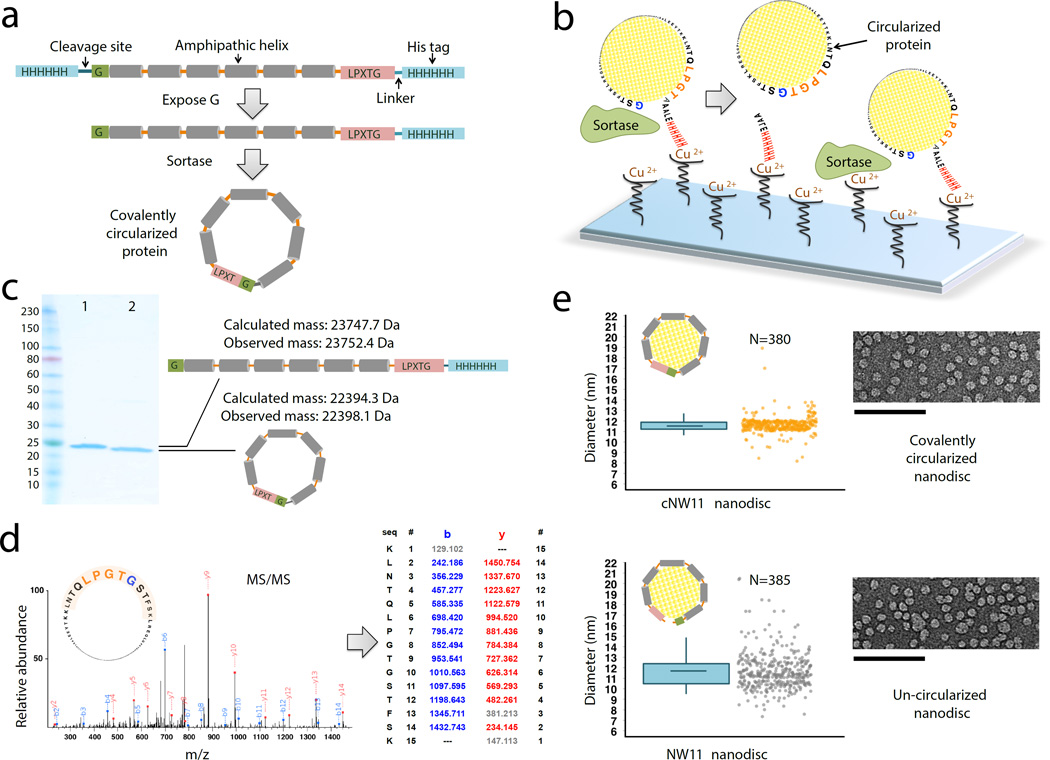
Phospholipid bilayer nanodiscs provide a detergent-free lipid bilayer model, enabling biochemical and biophysical characterization of membrane proteins in a physiologically relevant environment1. A traditional nanodisc is composed of a nanometer-sized phospholipid bilayer patch encircled by two copies of an α-helical, amphipathic membrane scaffold protein (MSP)2, 3. MSPs are truncated forms of apolipoprotein A1 (apoA1), which is the major protein component of high-density lipoprotein. To date, however, the utility of this system for structural studies has been limited by the heterogeneity in size and the number of membrane proteins enclosed, and only small nanodiscs could be constructed with the currently available protein scaffolds4–7. To resolve these problems we developed three different methods to covalently link the N and C termini of newly engineered variants based on apoA1, and produced nanodiscs with a large range of discrete sizes and defined geometric shapes. The protein constructs we used contain the consensus sequence recognized by sortase A (LPGTG) near the C terminus and a single glycine residue at the N terminus (Fig. 1a and supplementary Note 1). These two sites are sufficient to ensure covalent linkage between the N and C termini of a protein8 while still conserving the function to form nanodiscs.
Figure 1.
Producing covalently circularized NW11 and nanodiscs. (a) A general outline of the constructs that are used for making covalently circularized nanodiscs. (b) Outline of the procedure for creating circularized proteins over a Cu2+ chip. Immobilizing NW11 on the Cu2+ chip for circularization reduces the chances for head-to-tail linkage of two neighboring NW11 molecules and also offers a quick reaction time. (c) SDS-PAGE analysis of NW11 before (lane 1) and after (lane 2) circularization. (d) MS/MS spectrum of a tryptic peptide of cNW11 confirming the ligation of the N-terminal residues (GSTFSK) to the C-terminal LPGTG motif. The b and y ions that were identified in the MS/MS spectrum are highlighted in blue and red. MS/MS and intact mass data for other NWs constructs (cNW9, cNW30, cNW50) are provided in Supplementary Table 1 and Supplementary Figs. 1–3. (e) Diameter distribution for nanodiscs made using circularized NW11 (top) and non-circularized NW11 (bottom) and representative negative-stain EM images. In the box-and-whisker plots, center lines show the means; box limits indicate the 25th and 75th percentiles; whiskers g o down to 5 percentile and up to 95 percentile. Raw data (jittered along x for clarity) are shown next to its representative plot. There is less variance in the lengths of cNW11 compared to NW11 nanodiscs (p < 0.001). Diameter distributions for NW30, cNW30 and cNW50 nanodiscs are provided in Supplementary Fig. 4. Scale bars, 100 nm (e).
First, we used a NW11 construct, which assembles an 11 nm nanodisc, to optimize the circularization over a Cu2+ chip (Fig. 1b). In this scheme, the Cu2+ is saturated with un-circularized NW11 protein prior to evolved sortase9 addition. Upon successful completion, the circularized NW11 (cNW11) is liberated to the solution and can be further purified via reverse nickel affinity chromatography. Reaction completion was confirmed by SDS-PAGE and tandem mass spectrometry (MS/MS) (Fig. 1c, d). Next, we tested whether the final circularized product was still capable of assembling nanodiscs. Indeed, cNW11 assembled nanodiscs, and the acquired electron microscopy (EM) images revealed more uniformly sized nanodiscs as compared to nanodiscs assembled with the linear counterpart (Fig. 1e).
Even though circularization over the surface of a Cu2+ chip usually results in a very clean final product, the approach is limited to small-scale production of circularized protein. In order to scale up the production of cNW11, we developed a modified approach by performing the circularization reaction over nickel beads (Supplementary Fig. 5a). To further increase the yield of cNW11, we developed a third method of performing the circularization reaction in solution (Supplementary Fig. 5b and Supplementary Protocol), and produced milligram quantities of cNW11 that is > 95% monomeric. Moreover, we created an array of higher molecular weight circularized species and used them to assemble larger nanodiscs (up to 80 nm in diameter) of well-defined, circular and polygonal shapes (Supplementary Figs 5–7 and Supplementary Note 2).
Comparisons of proton relaxation rates of lipid resonances in empty cNW9 and MSP1D1ΔH5 nanodiscs4 indicate more restriction imposed on the lipids inside cNW9 nanodiscs by the covalently circularized belt protein (Supplementary Fig. 8). This could partially explain the enhanced thermal stability of circularized nanodiscs as compared to the conventional ones (Supplementary Fig. 9). In addition to the improved thermal stability, we show that covalent circularization enhances the proteolytic stability of nanodiscs (Supplementary Fig. 10).
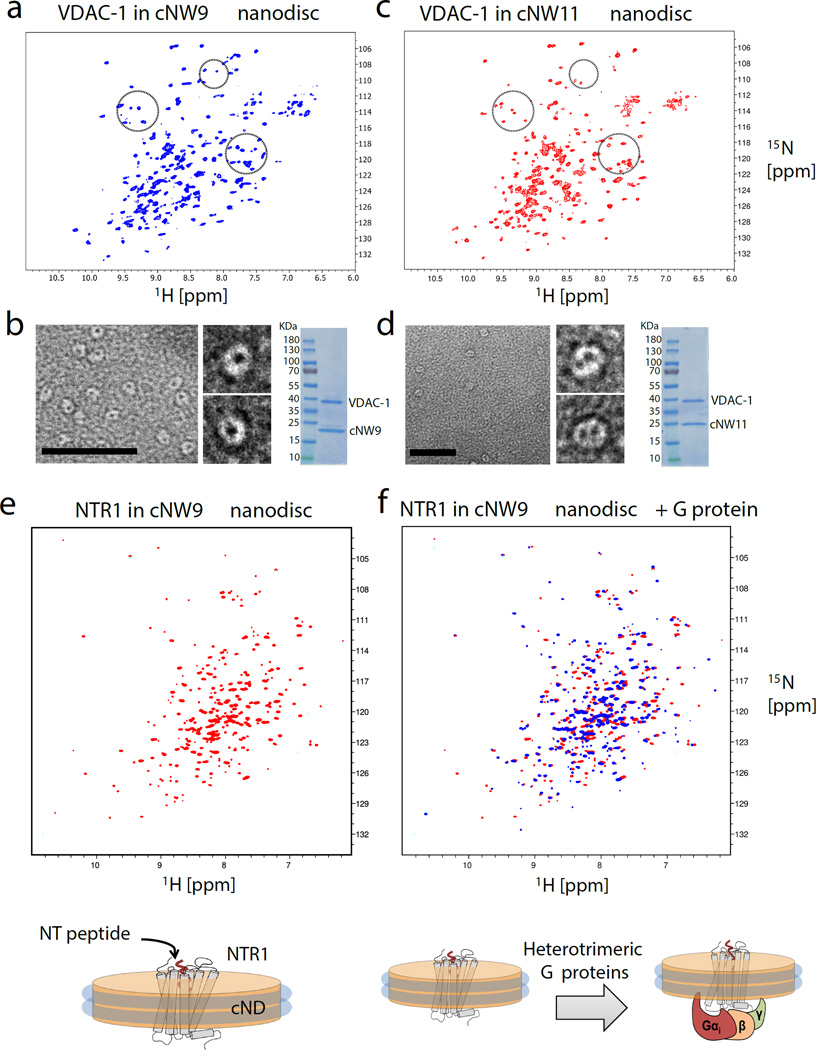
With the characterization of the circularized nanodiscs (cNDs) in place, we prepared [U-2H,15N] labeled VDAC-1 in cNW9 and cNW11 nanodiscs in order to incorporate one or two copies of the channel, respectively, and recorded 2D 1H-15N TROSY HSQC spectra at 45°C (Fig. 2a–d). Earlier preparations with open nanodiscs resulted in undefined numbers of embedded VDAC-1 molecules, which yield inconsistent NMR spectra due to sample heterogeneity5. Our new data exhibit enhanced signal intensities and spectral resolution compared to previously reported NMR spectra of VDAC-1 in nanodiscs5. The 1H-15N-TROSY HSQC spectra recorded for [U-2H,15N] labeled VDAC-1 in cNW9 and cNW11 nanodiscs are significantly different, suggesting conformational differences and/or intermolecular contacts between the monomer and dimer forms of membrane-embedded VDAC-1. These experiments demonstrate the power of the cND approach to control the oligomeric state of dynamically interacting proteins for structural studies. Moreover, the observed stability and quality of NMR spectra of VDAC-1 in cNDs at 45°C are dramatically better than those obtained with nanodiscs formed with linear MSPs. (Supplementary Fig. 12), and will greatly facilitate NMR assignments of VDAC-1. We next acquired a 1H-15N-TROSY HSQC spectrum of a 15N-labeled, signaling-competent variant of rat neurotensin receptor 1 (NTR1) in cNW9 nanodiscs (Fig. 2e,f). Again, the cND sample was stable at 45°C for longer than 10 days, which enabled us to test binding of a heterotrimeric G protein composed of Gαi1, Gβ1, and Gγ1 to cND-embedded NTR1, which resulted in numerous spectral changes. We believe that the cNDs will greatly facilitate the study of different dynamic events upon ligand binding in a near-native membrane environment, which may be critical for understanding the GPCR-mediated signal transduction process.
Figure 2.
Analysis of membrane proteins in different size nanodiscs. (a) 2D 1H-15N TROSY HSQC spectrum of monomeric [U-2H,15N] labeled VDAC-1 in cNW9 nanodiscs. (b) Representative image of negatively stained cNW9 nanodiscs containing single VDAC-1 channel (see also supplementary Fig. 11). The stain-filled channels appear as dark spot inside the nanodisc. Right: SDS-PAGE analysis of cNW9 nanodiscs containing VDAC-1. (c) 2D 1H-15N TROSY HSQC spectrum of dimeric [U-2H,15N] labeled VDAC-1 in cNW11 nanodiscs. (d) Representative image of negatively stained cNW11 nanodiscs containing two VDAC-1 channels. Right: SDS-PAGE analysis of cNW11 nanodiscs containing VDAC-1. (e) 1H-15N-TROSY HSQC spectrum of 40uM 15N-labeled NTR1 in cNW9 nanodisc acquired at 45°C on a Bruker 800MHz spectrometer. (f) Superimposed 1H-15N-TROSY HSQC spectra of 40uM 15N-labeled NTR1 in cNW9 nanodisc before (red) and after (blue) addition of purified heterotrimeric G protein composed of Gαi1, Gβ1, and Gγ1. The integrity of the cNW9 nanodiscs was not affected even though the G protein was added with a small amount of DDM (the final DDM concentration was less than 10% of its critical micellar concentration (CMC)). Scale bars, 50 nm (b, d).
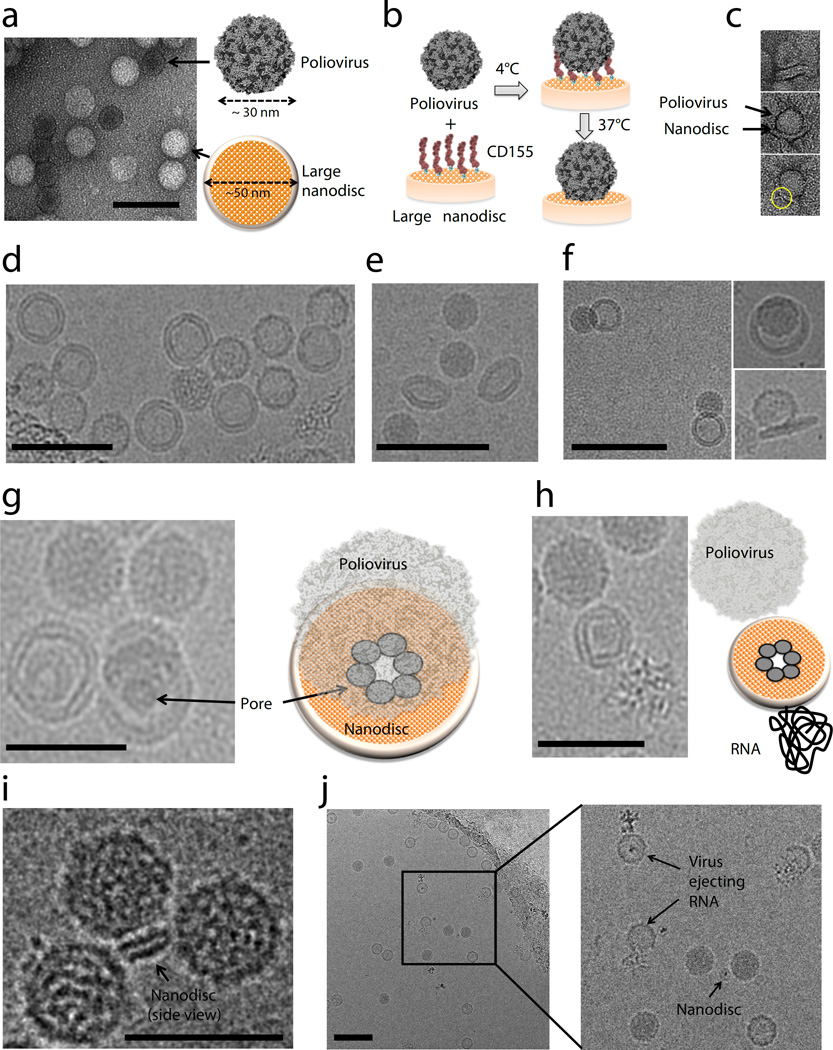
Next, we utilized the cNW50 nanodiscs as a model membrane to study the question of how simple non-enveloped viruses transfer their genomes across membranes to initiate infection. Unlike enveloped viruses, non-enveloped viruses lack an external membrane, and the delivery of their genome into cells requires translocation across a membrane to gain access to the inside of the host cell10, 11. Although there are now several model systems being used to study this process, the mechanism of genome translocation remains poorly understood12, and a more detailed structural analysis of the membrane-associated forms of the cell-entry intermediates is required. So far, mechanistic insights have been limited, due in part to technical difficulties involved in direct visualization of viral gene delivery and size heterogeneity of liposomes. The availability of large nanodiscs of a defined size encouraged us to pursue studies that could provide structural information at a resolution sufficient to gain insights into the mechanism of RNA translocation. As a proof of principal we have used the cNW50 nanodiscs to visualize the RNA-translocation pore of poliovirus.
Poliovirus (~30 nm diameter) is the prototype member of the enterovirus genus of the picornavirus family, which are positive-sense, single-stranded RNA viruses with ~7500b genomes enclosed by an icosahedral capsid, and lacking an envelope13. Viral infection is mediated by a specific receptor, CD155 (also known as the poliovirus receptor, PVR)14. Upon raising the temperature from 4°C to 37°C the receptor catalyzes a conformational rearrangement and expansion of the virus particle. The expanded virus is then endocytosed by a non-canonical, actin-independent pathway15, and the RNA is released across the endosomal membrane. A 50-nm nanodisc is sufficiently large to accommodate multiple CD155 copies and has enough surface area to act as a surrogate membrane for the RNA-translocation complex during viral uncoating (Fig. 3a). Similar to studies that used liposomes16–18, 50-nm nanodiscs containing lipids derivatized with a NTA nickel-chelating head group were generated and functionalized with the His-tagged CD155 ectodomain. The receptor-decorated nanodiscs were incubated with poliovirus for 5 minutes at 4°C. The complex was then heated to 37°C for 15 minutes to initiate receptor-mediated viral uncoating (Fig. 3b). Negative-stain EM confirmed virus binding to the CD155-decorated nanodiscs and subsequent insertion of viral components into and across the membrane (Fig. 3c). Additionally, the negative-stain EM images indicated that the virus started to form a pore in the nanodisc. To obtain a view of the molecular interactions involved in the RNA-translocation complex as well as to elucidate in more detail the formation of a pore, we conducted cryo-EM studies using an FEI Polara electron microscope. Figure 3d and Supplementary Figure 13 show the dark RNA-filled virus next to the slightly larger 50 nm nanodisc. The nanodisc is tilted in Figure 3e and tethered to the virus in Figure 3f. We were able to visualize the formation of a putative pore inside the nanodiscs (Fig. 3g, h), through which the virus ejects its RNA. The identities of the pore forming proteins are currently being determined. The availability of the circularized 50-nm nanodiscs greatly facilitates imaging as compared to using liposomes because the nanodiscs are more homogenous in size and shape, and allow for the use of a thinner ice layer. Also, RNA can be visualized more easily in the absence of large liposomal membranes. In order to reduce complexity even further, we used cNW30 nanodiscs (~15 nm) decorated with CD155. Surprisingly, the virus also tethers to this smaller nanodisc (Fig. 3i), and ejects RNA, leaving an empty viral capsid behind (Fig. 3j and Supplementary Fig. 14). Three-dimensional reconstruction of these complexes will be challenging and is beyond the scope of this study. However, the quality of the data collected on the virus-nanodisc complexes represents a vast improvement over data previously used to obtain low-resolution structural models of the translocation complex using the receptor-decorated liposome model19, and we are confident that the nanodisc model will allow determination of structures with greatly improved resolution and quality.
Figure 3.
Poliovirus caught in the act. (a) Negative-stain EM of 50-nm circularized nanodiscs plus poliovirus. A control (nanodiscs without CD155) is shown to illustrate the relative dimensions of the 30-nm poliovirus and the 50-nm nanodisc. (b) Outline of the procedure used to initiate poliovirus bridging and fusion with nanodiscs decorated with CD155. (c) Negative-stain EM images showing individual viruses tethered to nanodiscs. (d) Cryo-EM image of 50-nm nanodiscs plus poliovirus. (e) Cryo-EM image showing a tilted view of 50-nm nanodisc. (f) Cryo-EM images showing individual viruses tethered to nanodiscs. (g, h) Cryo-EM images showing the creation of a putative pore in the nanodisc by the poliovirus. (i) Cryo-EM image showing three viral particles around a 15-nm nanodisc. (j) Cryo-EM image showing individual viruses ejecting RNA after incubation with CD155-decorated 15-nm nanodiscs. Scale bars, 100 nm (a, d, e, f, j) and 50 nm (g, h, i).
In conclusion, we have demonstrated the construction of covalently circularized nanodiscs with a wide range of geometric shapes and sizes. The ability to make stable cNDs at multiple defined sizes up to 80 nm diameter provides tools to tightly embed much larger membrane proteins or their intra- and extra-membrane complexes than previous nanodisc systems have allowed. Moreover, we have shown that the newly engineered covalently circularizable scaffold proteins produce nanodiscs with high homogeneity in size and shape and with significantly improved stability compared to non-circularized forms, both of which would greatly facilitate their use for NMR and cryo-EM. We have demonstrated the utility of this model system to probe an outstanding question in the field of virology, and believe that the system will be similarly enabling for the structural and functional study of other large protein/membrane complexes.
ONLINE METHODS
We provide a Supplementary Protocol that includes detailed step-by-step instructions for the production of NWs proteins, circularization reaction and nanodisc assembly. Additionally, materials will be available upon request.
Expression of NW9, NW11, NW30 and NW50
NW9, NW11, NW30 and NW50 (all in pET-28a) containing a tobacco etch virus (TEV) protease-cleavable N-terminal His6 tag and a C-terminal sortase-cleavable His6 tag were transformed into BL21-Gold (DE3) competent E. coli cells (Agilent). 3L cell cultures were grown at 37°C, with agitation at 200 rpm in Luria broth (LB) medium supplemented with 50 µg/ml Kanamycin. Expression was induced at an OD600 of 0.6 with 1 mM IPTG and cells were grown for another 3 hours at 37°C (NW9 and NW11) or 16 hours at 18°C (NW30 and NW50). Cells were harvested by centrifugation (7000×g, 15 minutes, 4°C) and cell pellets were stored at −80°C.
Purification of NW9 and NW11
Pellets of cells expressing NW9 or NW11 were resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1% Triton X-100) and lysed by sonication on ice. Lysate was centrifuged (35,000×g, 50 minutes, 4°C), and the supernatant was loaded onto a Ni2+-NTA column. The column was washed with lysis buffer then with buffer A (50 mM Tris, pH 8.0, 500 mM NaCl). To recover additional protein from the insoluble fractions, the pellets recovered from lysate centrifugation were dissolved in denaturing buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 6M guanidine hydrochloride), centrifuged (35,000×g, 50 minutes, 4°C), and the supernatant was applied to the same Ni2+-NTA column containing bound protein from the soluble fraction. The column was washed with denaturing buffer, and NW9 and NW11 were refolded on-column with 10 column volumes (CV) buffer A. Resin was then washed with 10 CV of the following buffers: buffer A + 1% Triton X-100, buffer A + 50 mM sodium cholate, buffer A, and buffer A + 20 mM imidazole. Proteins were eluted with buffer A + 500 mM imidazole, TEV (His6-tagged; produced in-house) was added to cleave the N-terminal His6 tag, and the samples were dialyzed against 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA, 2 mM DTT at 4°C for 16 hours. NW9 and NW11 (still containing a C-terminal His6 tag) were exchanged into nanodisc-assembly buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 0.02% NaN3) using Centricon concentrators (10 kDa MW cutoff, Millipore).
Purification of NW30 and NW50
NW30 and NW50 were purified under denaturing conditions and refolded as follows. Pellets of cells expressing NW30 or NW50 were resuspended in denaturing lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 6M guanidine hydrochloride) and lysed by sonication on ice. Lysate was centrifuged (35000×g, 50 minutes, 4°C), and the supernatant was loaded onto a Ni2+-NTA column. Resin was washed with 10 column volumes (CV) of denaturing lysis buffer to remove unbound proteins, and NW30 and NW50 were refolded on-column with 10 CV buffer A (50 mM Tris HCl, pH 8.0, 500 mM NaCl). Resin was washed with 10 CV of the following buffers: buffer A + 1% Triton X-100, buffer A + 50 mM sodium cholate, buffer A, and buffer A + 20 mM imidazole. Proteins were eluted with buffer A + 500 mM imidazole, TEV protease was added to cleave the N-terminal His6 tag, and samples were dialyzed against 50 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, 2 mM DTT at 4°C for 16 hours. NW30 and NW50 (still containing a C-terminal His6 tag) were further purified by size exclusion chromatography (SEC; Superdex 200 16/60 [GE Healthcare] equilibrated in 20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 50 mM sodium cholate, 0.5 mM EDTA). SEC fractions containing NW30 and NW50 were further purified over Ni2+-NTA resin to remove truncation products (which lack a C-terminal His6 tag). Purified proteins were exchanged into nanodisc assembly buffer (50 mM Tris HCl, pH 8.0, 500 mM NaCl, 0.02% NaN3) using Centricon concentrators (30 kDa MW cutoff, Millipore).
MSP circularization
A 50 mL reaction was prepared with 10 µM NWs and 5 µM (final concentrations) freshly made evolved sortase (Addgene plasmid # 75144, a gift from David Liu) in 300 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 10 mM CaCl2. The reaction was incubated at 37°C for 3–4 hours or at 4°C overnight with gentle shaking on a rotating platform. A covalent sortase inhibitor AAEK220 was added to a concentration of 500 µM, and the solution was incubated for another 30 minutes at room temperature with gentle shaking. Proteins that did not undergo circularization were removed by binding to a Ni2+-NTA column. Circularized NWs (cNWs) were further purified by size-exclusion chromatography (Superdex 75 16/60) equilibrated in buffer containing 20 mM Tris-HCl, pH 7.5, 500 mM NaCl containing 50 mM sodium cholate or 1 mM dodecyl-β-D-maltoside (DDM). Purified protein was exchanged into buffer Aix (20 mM Tris, pH 8.2, 1 mM DDM) using centricon concentrators (10 kDa MW cutoff, Millipore) and was then applied to a Resource Q column equilibrated with the same buffer (buffer Aix). A linear salt gradient from 0–60 % buffer Bix (20 mM Tris, pH 8.2, 1 mM DDM, 1M NaCl) was applied. Circularized proteins were eluted around 150–200 mM NaCl.
Reconstitution of cNW11, cNW30 and cNW50 nanodiscs
cNWs:lipid ratios of 1:60, 1:75, 1:1000 and 1:4000 were used to assemble cNW9, cNW11, cNW30, cNW50 nanodiscs, respectively. Lipids (POPC:POPG 3:2, solubilized in sodium cholate) and cNWs were incubated on ice for 1 hour. After incubation, sodium cholate was removed by incubation with Bio-beads SM-2 (Bio-Rad) for 1 hour on ice followed by incubation overnight at 4°C. The nanodisc preparations were filtered through 0.22 µm nitrocellulose-filter tubes to remove the Bio-beads. The nanodisc preparations were further purified by size-exclusion chromatography while monitoring the absorbance at 280 nm on a Superdex 200 10×300 column (for cNW9 and cNW11 nanodiscs) or Superose 6 10/300 column (for cNW30 and cNW50 nanodiscs) equilibrated in 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.5 mM EDTA. Fractions corresponding to the size of each nanodisc were collected and concentrated. The purity of nanodisc preparations was assessed using SDS-PAGE.
In vitro reconstitution of VDAC-1 into POPC/POPG or DMPC/DMPG nanodiscs
To assemble VDAC-1 into POPC/POPG nanodiscs, 15 µM of VDAC-1, 150 µM of cNW11 and 8.5 mM lipids (POPC:POPG 3:2, solubilized in sodium cholate) were incubated over ice for 1 hour. To assemble monomeric VDAC-1 into DMPC/DMPG nanodiscs for NMR experiments, 25 µM of VDAC-1, 175 µM of cNW9, and 9.5 mM lipids (DMPC:DMPG 3:1) were incubated for 1 hour at room temperature. To assemble dimeric VDAC-1 into DMPC/DMPG nanodiscs for NMR experiments, 79 µM of VDAC-1, 75 µM of cNW11, and 3.75 mM lipids (DMPC:DMPG 3:1) were incubated for 1 hour at room temperature. After incubation, detergents were removed by the addition of Bio-beads SM-2 (Bio-Rad) and incubation for 1 hour on ice followed by overnight incubation at 4°C (POPC/POPG nanodiscs) or overnight incubation at room temperature (DMPC/DMPG nanodiscs). The nanodisc preparation was filtered through 0.22 µm nitrocellulose-filter tubes to remove the Bio-beads. To remove the VDAC-free nanodiscs, the sample was mixed with Ni2+-NTA resin for 1 hour at 4°C. The resin bed volume was equal to the assembly mixture. The resin was washed with buffer E (20 mM Tris-HCl, pH 8.0, 0.1 M NaCl, 20 mM imidazole). Nanodiscs containing VDAC were eluted with buffer E containing 0.5 M imidazole. The nanodisc preparation was further purified by size-exclusion chromatography while monitoring the absorbance at 280 nm on a Superdex 200 10 × 300 or a 16/60 Superdex 200 prep grade columns (GE Healthcare) equilibrated in buffer F (20 mM sodium phosphate, 50 mM NaCl, 5 mM DTT, 1 mM EDTA, pH 7.0). Fractions corresponding to the size of the VDAC-nanodisc complex were collected and concentrated. The purity of VDAC-containing nanodiscs was assessed using SDS-PAGE.
Production of NTR1 and reconstitution into DMPC/DMPG nanodiscs
Expression and the purification of a signaling-competent, thermo-stabilized variant of rat neurotensin receptor 1 (termed HTGH4) were performed as described previously21,22 with some modifications. Briefly, the full-length fusion protein consisting of maltose-binding protein (MBP), followed by a His tag, a 3C protease recognition site, NTR1, a second 3C protease recognition site, thioredoxin (TrxA), and a His tag at the C terminus was purified by Ni2+ affinity chromatography. The purified full-length fusion protein was immediately incorporated into DMPC:DMPG (3:1) cNW9 nanodiscs. The assembled nanodiscs were subjected to Ni2+ affinity chromatography to remove empty nanodiscs from the receptor-containing nanodiscs utilizing the His tag of the fusion protein. The receptor-containing nanodiscs were further purified by size-exclusion chromatography followed by incubation with resin decorated with 3C protease-cleavable neurotensin peptide (pD-NT) to enrich for nanodiscs containing correctly folded receptors. Next, the pD-NT column was incubated for 2 hours with His-tagged 3C protease (produced in house). The eluted proteins (neurotensin-bound NTR1 in nanodiscs, MBP, TrxA and 3C protease) were subjected to another Ni affinity chromatography followed by size-exclusion chromatography using a Superdex-200 10/300 column that was pre-equilibrated with running buffer containing 20 mM sodium phosphate, pH 6.9, 50 mM NaCl, 5 mM DTT, 1 mM EDTA.
Heterotrimeric G protein (αi1β1γ1) was expressed in Sf9 cells using a single baculovirus encoding all three subunits as previously described22 and purified following a published procedure23.
Negative-stain electron microscopy
Samples were prepared by conventional negative staining as described previously24. Briefly, 3.5 µl of nanodisc samples were adsorbed to glow-discharged, carbon-coated copper grids and stained with 0.75% (w/v) uranyl formate. All EM images (except images for VDAC-1 in cNW9 nanodiscs in Supplementary Figure 11) were collected with a Philips CM10 electron microscope (FEI) equipped with a tungsten filament and operated at an acceleration voltage of 100 kV. Images were recorded with a Gatan 1 K × 1 K CCD camera (Gatan, Inc., Pleasanton, CA, USA). Using EMAN software package25, 9258 particles were selected from 69 images of VDAC-1 in cNW9 nanodiscs (recorded at a magnification of 42,000×, defocus value of −1.5 µm) and classified into 150 classes.
NMR spectroscopy
[U-2H,15N] labeled VDAC-1 in DMPC:DMPG (3:1) nanodiscs (611 µM and 500 µM of VDAC1 in cNW9 and cNW11 nanodiscs, respectively) were prepared as described above in NMR buffer (20 mM NaPO4, 50 mM NaCl, 5 mM DTT, 1 mM EDTA, pH 7.0, 6% D2O). 15N-TROSY HSQC data were collected at 45°C on a Bruker 800-MHz spectrometer equipped with a TXO cryogenic probe. Data for VDAC-1 in cNW9 and cNW11 nanodiscs were acquired with 48 and 96 scans, respectively, and 128 complex points in the 15N-indrect dimension.
The 15N-TROSY HSQC spectrum of 40 µM 15N-labeled NTR1 in cNW9 nanodiscs was acquired at 45°C on a Bruker 800-MHz spectrometer. Data were collected with 2048 scans per FID and 36 non-uniformly sampled 15N-dimension complex points (maximum point of 64). The 15N-dimension time domain data were reconstructed with hmsIST26 and processed by NMRPipe software programs27.
Cryo-electron microscopy
A 3.5 µl droplet of the poliovirus-nanodisc complex was loaded onto a glow-discharged holey carbon grid (Protochips, Morrisville, NC). Excess liquid was removed from the grid surface before it was rapidly plunged into liquid ethane. Grids were transferred into an FEI Polara electron microscope operating at an acceleration voltage of 300 kV. Micrographs were acquired on a K2 Summit camera (Gatan, Pleasanton, CA) in super-resolution mode using SerialEM28, whereby 25 frames were collected for a total dose of 30 electrons per square Ångstrom. These frames were aligned and averaged using motioncorr29.
Statistics
We graphically display the distribution of diameters of the circularized and uncircularized nanodiscs using box and whisker plots (Figure 1 and Supplementary Figure 4) and compared the variances using a variance ratio test.
Availability of data
The data supporting the results of this paper are included within the paper (and its supplementary files).
Supplementary Material
Acknowledgments
The authors would like to thank T. Raschle for his early contribution to this work and H. Chou, H. Arthanari, J. Ziarek for helpful discussions. The authors gratefully acknowledge the use of the services of the Taplin Mass Spectrometry Facility and the Center of Macromolecular Interactions (CMI). This work was supported by NIH grant F32GM113406 to M.L.N, by grant AI20566 to J.M.H., and grants GM047467, GM075879 and AI037581 to G.W. G.W., A.P. and F.H. are supported by the Human Frontiers Science program. D.B. acknowledges support from the HHMI as a Gilliam Fellow. F.H. acknowledges support by the TUM Institute for Advanced Study co-funded by the European Commission, as well as the CIPSM excellence cluster and the DFG (SFB1035).
Footnotes
Note: Supplementary Information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
The project was conceived by M.L.N. and G.W. Experiments were designed by M.L.N., D.B., G.W., M.S. and J.H. Experiments were carried out by M.L.N. D.B., M.S., Z.J.S., S.G. and S.H. Manuscript was written by M.L.N. and G.W. T.W., J.H., D.B., F.H., A.P., S.G. contributed to discussions and editing of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors applied for a patent on cND technology but declare no other competing financial interests.
REFERENCES
- 1.Bayburt TH, Carlson JW, Sligar SG. J Struct Biol. 1998;123:37–44. doi: 10.1006/jsbi.1998.4007. [DOI] [PubMed] [Google Scholar]
- 2.Bayburt TH, Grinkova YV, Sligar SG. Nano Letters. 2002;2:853–856. [Google Scholar]
- 3.Ritchie TK, et al. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagn F, Etzkorn M, Raschle T, Wagner G. J Am Chem Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raschle T, et al. J Am Chem Soc. 2009;131:17777–17779. doi: 10.1021/ja907918r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grinkova YV, Denisov IG, Sligar SG. Protein Eng Des Sel. 2010;23:843–848. doi: 10.1093/protein/gzq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raschle T, Hiller S, Etzkorn M, Wagner G. Current opinion in structural biology. 2010;20:471–479. doi: 10.1016/j.sbi.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antos JM, et al. J Biol Chem. 2009;284:16028–16036. doi: 10.1074/jbc.M901752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen I, Dorr BM, Liu DR. Proc Natl Acad Sci U S A. 2011;108:11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogle JM. Annual Review of Microbiology. 2002;56:677–702. doi: 10.1146/annurev.micro.56.012302.160757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuthill TJ, Groppelli E, Hogle JM, Rowlands DJ. Viruses. 2010;343:43–89. doi: 10.1007/82_2010_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh M, Helenius A. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuthill TJ, Groppelli E, Hogle JM, Rowlands DJ. Curr Top Microbiol Immunol. 2010;343:43–89. doi: 10.1007/82_2010_37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendelsohn CL, Wimmer E, Racaniello VR. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 15.Brandenburg B, et al. PLoS Biol. 2007;5:e183. doi: 10.1371/journal.pbio.0050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuthill TJ, Bubeck D, Rowlands DJ, Hogle JM. J Virol. 2006;80:172–180. doi: 10.1128/JVI.80.1.172-180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bubeck D, Filman DJ, Hogle JM. Nat Struct Mol Biol. 2005;12:615–618. doi: 10.1038/nsmb955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. J Virol. 2013;87:3903–3914. doi: 10.1128/JVI.03209-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strauss M, et al. J Virol. 2015;89:4143–4157. doi: 10.1128/JVI.03101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 20.Maresso AW, et al. J Biol Chem. 2007;282:23129–23139. doi: 10.1074/jbc.M701857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egloff P, et al. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E655–E662. doi: 10.1073/pnas.1317903111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egloff P, Deluigi M, Heine P, Balada S, Pluckthun A. Protein Expr Purif. 2015;108:106–114. doi: 10.1016/j.pep.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Rasmussen SG, et al. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohi M, Li Y, Cheng Y, Walz T. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang G, et al. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Hyberts SG, Milbradt AG, Wagner AB, Arthanari H, Wagner G. J Biomol NMR. 52:315–327. doi: 10.1007/s10858-012-9611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delaglio F, et al. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 28.Mastronarde DN. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Grigorieff N, Cheng Y. J Struct Biol. 2010;172:407–412. doi: 10.1016/j.jsb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schneider CA, Rasband WS, Eliceiri KW. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.