Abstract
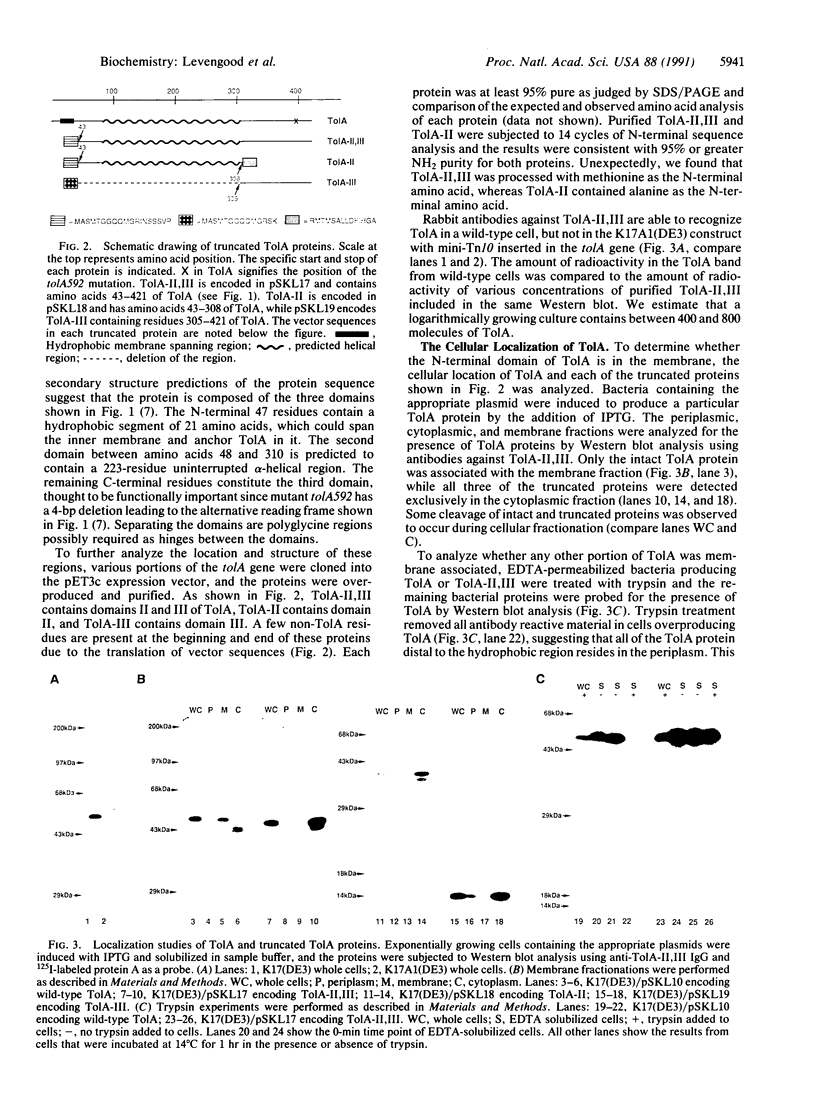
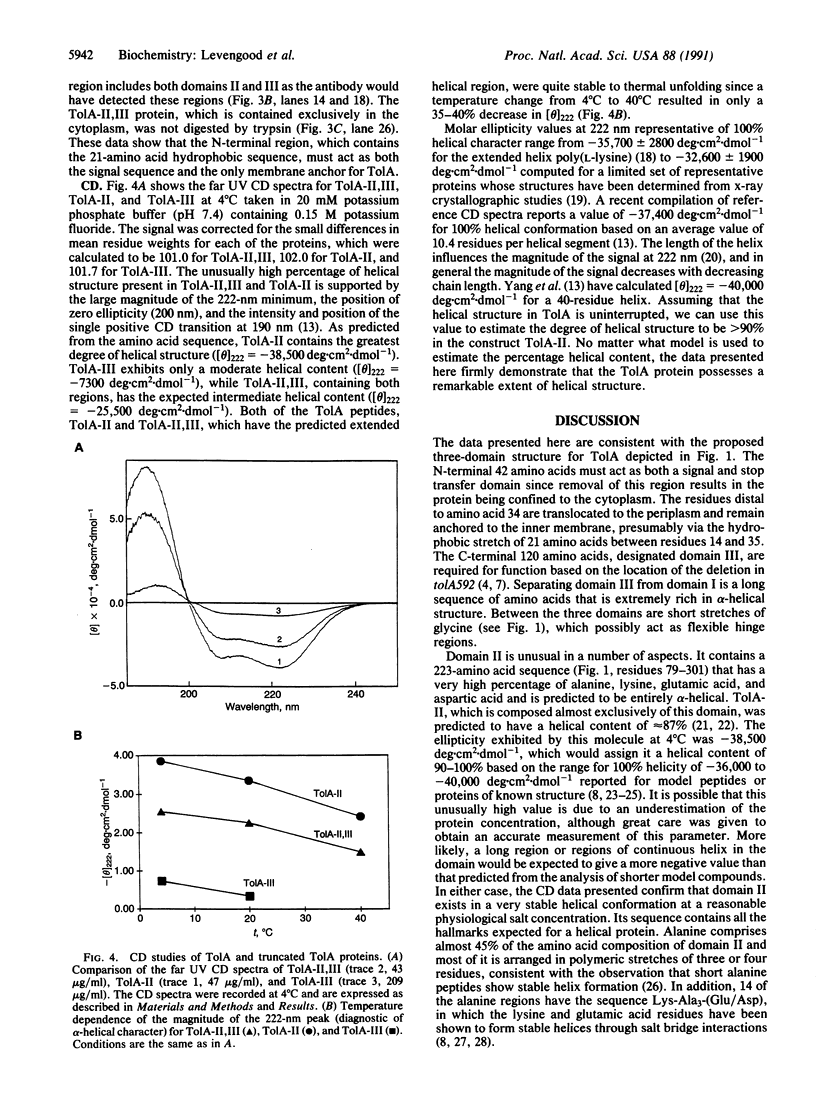
The group A colicins and the DNA of many single-stranded filamentous bacteriophage are able to use combinations of the Tol proteins to gain entrance into or across the membrane of Escherichia coli. The TolA protein is a 421-amino acid residue integral membrane protein composed of three domains. Domain I, consisting of the amino-terminal 47 amino acids, contains a 21-residue hydrophobic segment that anchors the protein in the inner membrane. The remaining 374 amino acids, containing the other two domains, reside in the periplasmic space. Domain III, consisting of the carboxyl-terminal 120 residues, is considered to be the functional domain based on the location of the tolA592 deletion mutation. The internal 262 amino acids comprise domain II, which connects domains I and III together via short regions of polyglycine. It contains a large number of 3- to 5-residue polyalanine stretches, many of which have a repeat of the sequence Lys-Ala-Ala-Ala-(Glu/Asp). Circular dichroism analysis of different portions of TolA show domain II to be predominantly alpha-helical in structure while domain III contains approximately 10% helical structure.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayer M. E. Areas of adhesion between wall and membrane of Escherichia coli. J Gen Microbiol. 1968 Oct;53(3):395–404. doi: 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- Chang C. T., Wu C. S., Yang J. T. Circular dichroic analysis of protein conformation: inclusion of the beta-turns. Anal Biochem. 1978 Nov;91(1):13–31. doi: 10.1016/0003-2697(78)90812-6. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T. A new approach to the calculation of secondary structures of globular proteins by optical rotatory dispersion and circular dichroism. Biochem Biophys Res Commun. 1971 Sep 17;44(6):1285–1291. doi: 10.1016/s0006-291x(71)80225-5. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974 Jul 30;13(16):3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Hill C., Holland I. B. Genetic basis of colicin E susceptibility in Escherichia coli. I. Isolation and properties of refractory mutants and the preliminary mapping of their mutations. J Bacteriol. 1967 Sep;94(3):677–686. doi: 10.1128/jb.94.3.677-686.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ITZHAKI R. F., GILL D. M. A MICRO-BIURET METHOD FOR ESTIMATING PROTEINS. Anal Biochem. 1964 Dec;9:401–410. doi: 10.1016/0003-2697(64)90200-3. [DOI] [PubMed] [Google Scholar]
- Kellenberger E. The 'Bayer bridges' confronted with results from improved electron microscopy methods. Mol Microbiol. 1990 May;4(5):697–705. doi: 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levengood S. K., Webster R. E. Nucleotide sequences of the tolA and tolB genes and localization of their products, components of a multistep translocation system in Escherichia coli. J Bacteriol. 1989 Dec;171(12):6600–6609. doi: 10.1128/jb.171.12.6600-6609.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqusee S., Baldwin R. L. Helix stabilization by Glu-...Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqusee S., Robbins V. H., Baldwin R. L. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merutka G., Stellwagen E. Positional independence and additivity of amino acid replacements on helix stability in monomeric peptides. Biochemistry. 1990 Jan 30;29(4):894–898. doi: 10.1021/bi00456a007. [DOI] [PubMed] [Google Scholar]
- Nagel de Zwaig R., Luria S. E. Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J Bacteriol. 1967 Oct;94(4):1112–1123. doi: 10.1128/jb.94.4.1112-1123.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu H. C., Heppel L. A. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J Biol Chem. 1965 Sep;240(9):3685–3692. [PubMed] [Google Scholar]
- Nomura M., Witten C. Interaction of colicins with bacterial cells. 3. Colicin-tolerant mutations in Escherichia coli. J Bacteriol. 1967 Oct;94(4):1093–1111. doi: 10.1128/jb.94.4.1093-1111.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki Y. Determination of the concentration of protein by dry weight--a comparison with spectrophotometric methods. Arch Biochem Biophys. 1986 Sep;249(2):437–446. doi: 10.1016/0003-9861(86)90020-2. [DOI] [PubMed] [Google Scholar]
- Ozols J. Amino acid analysis. Methods Enzymol. 1990;182:587–601. doi: 10.1016/0076-6879(90)82046-5. [DOI] [PubMed] [Google Scholar]
- Rosenberg A. H., Lade B. N., Chui D. S., Lin S. W., Dunn J. J., Studier F. W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56(1):125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Sun T. P., Webster R. E. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J Bacteriol. 1987 Jun;169(6):2667–2674. doi: 10.1128/jb.169.6.2667-2674.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T. P., Webster R. E. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J Bacteriol. 1986 Jan;165(1):107–115. doi: 10.1128/jb.165.1.107-115.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaralingam M., Sekharudu Y. C., Yathindra N., Ravichandran V. Ion pairs in alpha helices. Proteins. 1987;2(1):64–71. doi: 10.1002/prot.340020108. [DOI] [PubMed] [Google Scholar]
- Yang J. T., Wu C. S., Martinez H. M. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]





