Abstract
The recognition of β cell dedifferentiation in type 2 diabetes raises the translational relevance of mechanisms that direct and maintain β cell identity. LIM domain–binding protein 1 (LDB1) nucleates multimeric transcriptional complexes and establishes promoter-enhancer looping, thereby directing fate assignment and maturation of progenitor populations. Many terminally differentiated endocrine cell types, however, remain enriched for LDB1, but its role is unknown. Here, we have demonstrated a requirement for LDB1 in maintaining the terminally differentiated status of pancreatic β cells. Inducible ablation of LDB1 in mature β cells impaired insulin secretion and glucose homeostasis. Transcriptomic analysis of LDB1-depleted β cells revealed the collapse of the terminally differentiated gene program, indicated by a loss of β cell identity genes and induction of the endocrine progenitor factor neurogenin 3 (NEUROG3). Lineage tracing confirmed that LDB1-depleted, insulin-negative β cells express NEUROG3 but do not adopt alternate endocrine cell fates. In primary mouse islets, LDB1 and its LIM homeodomain–binding partner islet 1 (ISL1) were coenriched at chromatin sites occupied by pancreatic and duodenal homeobox 1 (PDX1), NK6 homeobox 1 (NKX6.1), forkhead box A2 (FOXA2), and NK2 homeobox 2 (NKX2.2) — factors that co-occupy active enhancers in 3D chromatin domains in human islets. Indeed, LDB1 was enriched at active enhancers in human islets. Thus, LDB1 maintains the terminally differentiated state of β cells and is a component of active enhancers in both murine and human islets.
Introduction
All forms of diabetes are characterized by reduced numbers and/or dysfunction of insulin-producing pancreatic islet β cells (1, 2). The β cell is unique in its ability to synthesize and secrete insulin and is 1 of 5 endocrine lineages that arise from neurogenin 3–expressing (NEUROG3-expressing) progenitors (3). As these lineages differentiate and mature, orchestrated mechanisms establish the epigenetic landscape and transcriptional networks that reinforce the mature, functional identity of each lineage (4, 5). The transcription factors pancreatic and duodenal homeobox 1 (PDX1) and paired box 4 (PAX4) specify fate, and MAF bZIP transcription factor A (MAFA) and teashirt zinc finger family member 1 (TSHZ1) drive the functional maturation of β cells (6–9). Ablation of Pdx1 in the mature β cell is sufficient to activate ectopic α cell features, illustrating the critical role PDX1 plays in maintaining β cell identity (10). Likewise, maintenance of the pancreatic endocrine cell epigenetic landscape is critical; inhibition of histone methyltransferases in human and murine islets leads α cells to ectopically express insulin and PDX1 (11).
LIM domain–binding protein 1 (LDB1), a nuclear protein lacking DNA-binding capacity and enzymatic activity (12), is also required for the maturation of pancreatic endocrine precursors (13). LDB1 homodimers dictate cell fate during fetal development in various progenitor populations (14). Two distinct LDB1 functional paradigms have been described and are distinguished by the high-affinity protein-protein interactions between LDB1 and LIM homeodomain (LIM-HD) transcription factors or LIM-only (LMO) scaffolding proteins (15). The LIM-HD–based paradigm involves LDB1-nucleating tetrameric and hexameric LIM-HD complexes and is exemplified by V2 interneuron and somatic motor neuron fate determination (16, 17). The LMO-based paradigm is implemented during erythropoiesis and hematopoiesis and involves the formation of a pentameric complex, in which LDB1 binds an LMO factor that in turn bridges a GATA factor and an E-box heterodimer (18–20). These LDB1-mediated complexes recruit chromatin-remodeling complexes and enlist transcriptional machinery (21–24). There is also growing evidence that LDB1 homodimerization is critical for establishing 3D chromatin architecture (25–27).
Roles for LDB1 have been primarily characterized in developmental and progenitor contexts (16, 25, 27–29), whereas the in vivo role of LDB1 in mature, terminally differentiated cell types, like the pancreatic β cell, has yet to be investigated. LDB1 and its direct binding partner, islet 1 (ISL1), remain enriched in terminally differentiated β cells (13, 30, 31). To determine the functional requirement for LDB1 with respect to ISL1 in terminally differentiated β cells, we generated and characterized inducible, β cell–specific loss-of-function mice for each factor. Using primary mouse islets, RNA-sequencing (RNA-seq) of FACS-enriched, loss-of-function β cells was integrated with cistromic analysis of LDB1 and ISL1. Our findings reveal that LDB1 and ISL1 function in a complex to maintain the terminal differentiation program of pancreatic β cells. Moreover, we demonstrate that LDB1-mediated complexes are incorporated into the transcriptional complexes occupying active enhancers in murine and human islets.
Results
Ablation of Ldb1 in mature β cells causes glucose intolerance by depleting pancreatic insulin.
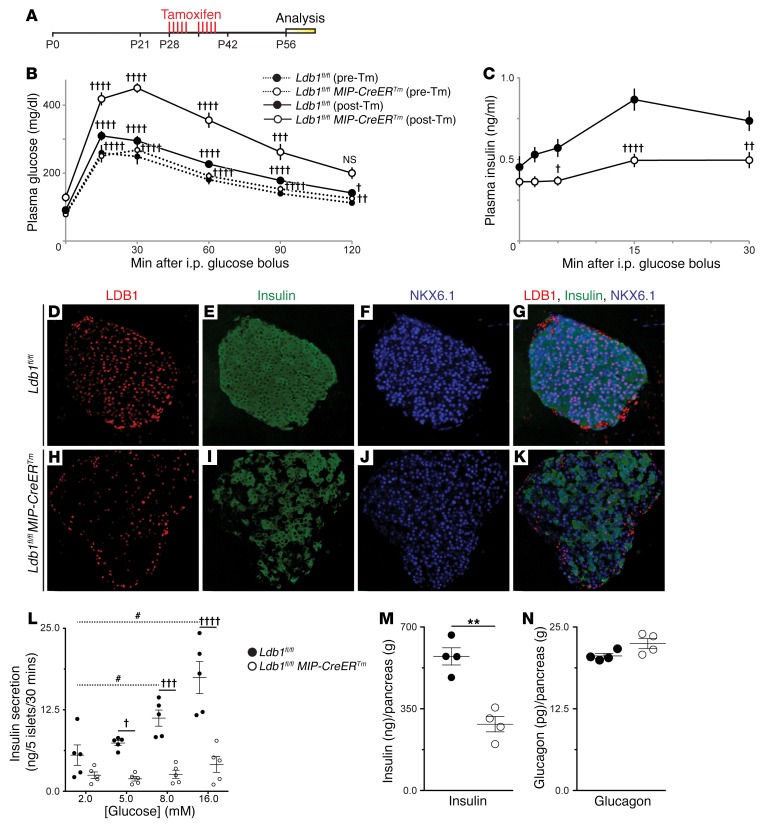
To determine the functional requirement for LDB1 in mature β cells, we crossed a floxed Ldb1 allele (Ldb1fl) with the β cell–specific, tamoxifen-inducible (Tm-inducible) MIP-CreERTm deleter strain to create Lbd1fl/fl MIP-CreERTm mice (32–34). At P28, Tm was orally administered over a 2-week period, followed by a 2-week washout (Figure 1A). Prior to Tm administration, the baseline glucose tolerance of Lbd1fl/fl MIP-CreERTm mice was indistinguishable from that of Ldb1fl/fl control mice (Figure 1B). During Tm administration and washout, both genotypes displayed similar weight gains and random blood glucose levels (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI88016DS1). After Tm washout, LDB1 was depleted from 40% to 60% of β cells (Figure 1, D–K). Induced Lbd1fl/fl MIP-CreERTm mice were glucose intolerant (Figure 1B) and showed an impaired glucose-stimulated insulin secretion response (Figure 1C). As expected, the insulin tolerance of Lbd1fl/fl MIP-CreERTm mice was unaffected (Supplemental Figure 1C).
Figure 1. Ablation of Ldb1 in the mature β cell results in impaired glucose homeostasis and insulin secretion.
(A) Tm administration schedule for Ldb1fl/fl and Ldb1fl/fl MIP-CreERTm mice. Red vertical hash marks indicate Tm gavages; yellow box indicates the experimental analysis window. (B) Intraperitoneal glucose tolerance tests (IPGTTs) performed at P28, prior to Tm administration (pre-Tm) (n = 9 Ldb1fl/fl mice; n = 11 Ldb1fl/fl MIP-CreERTm mice), and at P56 following completion of a Tm pulse-chase (post-Tm) (n = 14 Ldb1fl/fl mice; n = 15 Ldb1fl/fl MIP-CreERTm mice). Statistical annotation reflects the post-hoc comparisons at each time point in following order: post-Tm Ldb1fl/fl MIP-CreERTm mice compared to 1), post-Tm Ldb1fl/fl, 2) pre-Tm Ldb1fl/fl MIP-CreERTm, and 3) pre-Tm Ldb1fl/fl mice, respectively. (C) P56 glucose-stimulated insulin secretion (GSIS) assay (n = 9 Ldb1fl/fl mice; n = 10 Ldb1fl/fl MIP-CreERTm mice). (D–K) Co-IF for LDB1, insulin, and NKX6.1 at P56. (L) Static islet incubations: absolute insulin secretion. (M) Total pancreatic insulin content. (N) Total pancreatic glucagon content. (D–L) n = 5 Ldb1fl/fl mice; n = 5 Ldb1fl/fl MIP-CreERTm mice. (M and N) n = 4 Ldb1fl/fl mice; n = 4 Ldb1fl/fl MIP-CreERTm mice. †P < 0.05, ††P < 0.01, †††P < 0.001, and ††††P < 0.0001, by repeated-measures, 2-way ANOVA with (B) Holm-Sidak correction and (C and L) Bonferroni’s correction, and (L) #P < 0.05, by repeated-measures, 1-way ANOVA with Holm-Sidak correction. (L) Only the post-hoc comparisons to the respective 2.0 mM glucose treatments were analyzed. (M and N) **P < 0.01, by Student’s 2-tailed t test. NS in B and absent statistical annotation in C, L, and N indicates comparisons were not significant. Original magnification, ×20. Pooled data represent the mean ± SEM. See also Supplemental Figures 1 and 2.
To directly interrogate the physiological capacity of Lbd1fl/fl MIP-CreERTm islets, we performed ex vivo static incubations using isolated islets challenged with increasing glucose concentrations in the presence and absence of exendin 4 (Ex-4), a GLP1 receptor (GLP1R) agonist (35). The absolute level of insulin secretion was decreased under all conditions, and the stimulatory responses to glucose and Ex-4 were blunted (Figure 1L and Supplemental Figure 1, D–F). Consistent with reduced insulin secretion, total pancreatic insulin content in Lbd1fl/fl MIP-CreERTm mice was significantly reduced compared with the content detected in Tm-treated littermates (Figure 1M). In contrast, total pancreatic glucagon content was unchanged (Figure 1N). Taken together, our in vivo and ex vivo physiologic analyses demonstrate that LDB1 expression in mature β cells maintains insulin content levels and a normal glucose-stimulated insulin secretion response, both of which are required for normal glucose homeostasis.
Ldb1-depleted β cells lose insulin but retain chromogranin A expression.
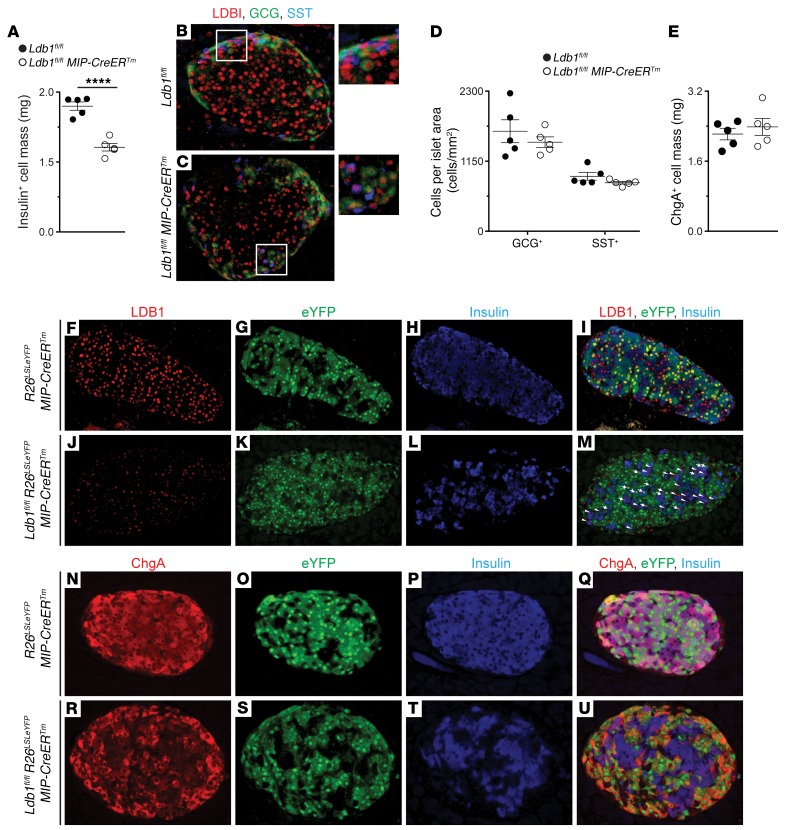
The decreased pancreatic insulin content of Lbd1fl/fl MIP-CreERTm mice was clearly evident from the disrupted insulin-staining pattern (Figure 1, E and I). Areas of insulin paucity corresponded to the population of LDB1– cells (Figure 1, H–K) and accounted for a 33% reduction in insulin+ cell mass (Figure 2A). In contrast, the somatostatin+ and glucagon+ cell populations within Lbd1fl/fl MIP-CreERTm pancreata were unaffected and retained LDB1 expression (Figure 2, B–D). These LDB1–, insulin– cells remained positive, albeit at a diminished intensity, for the β cell–specific transcription factor NK6 homeobox 1 (NKX6.1) (Figure 1, D–K), suggesting that β cells persisted following Ldb1 depletion. This conclusion was further supported by the observed mass of chromogranin A+ (ChgA+) endocrine cells as well as unchanged islet size and density in Lbd1fl/fl MIP-CreERTm pancreata (Figure 2E, and Supplemental Figure 1, G and H).
Figure 2. Ldb1-depleted β cells persist, retain ChgA, and are insulin depleted.
(A) Quantified insulin+ cell mass. (B and C) Co-IF for LDB1, glucagon (GCG), and somatostatin (SST) at P56. (D) Frequency of glucagon+ and somatostatin+ cells within ChgA+ cell mass. (E) Quantified ChgA+ cell mass. (F–M) Co-IF for LDB1, insulin, and eYFP at P56. White arrows indicate eYFPlo, LDB1lo, and insulin+ cell population; white arrowheads indicate eYFP–, LDB1+, and insulin+ cell population. (N–U) Co-IF for ChgA+, insulin, and eYFP at P56. (A–U) n = 5 Ldb1fl/fl mice; n = 5 Ldb1fl/fl MIP-CreERTm mice. (D) Repeated-measures, 2-way ANOVA with Bonferroni’s correction. (A and E) ****P < 0.0001, by 2-tailed Student’s t test. Absent statistical annotation in D and E indicates that the comparisons that were not significant. Original magnification, ×20 and ×40 (insets in B and C). Pooled data represent the mean ± SEM. See also Supplemental Figure 3.
To conclusively determine the fate of Ldb1-depleted β cells, we crossed the R26LSLeYFP lineage-tracing allele with Lbd1fl/fl MIP-CreERTm mice (36). Following Tm administration, enhanced yellow fluorescent protein (eYFP) signal in the control R26LSLeYFP MIP-CreERTm strain was exclusively detected in LDB1+, insulin+ cells (Figure 2, F–I). In contrast, eYFP signal in Ldb1fl/fl R26LSLeYFP MIP-CreERTm islets almost exclusively labeled the LDB1–, insulin– cells (Figure 2, J–M), indicating that a) eYFP reliably traced Ldb1 ablation, and b) LDB1 maintained insulin expression in a cell-autonomous manner. Additionally, ChgA expression was replete throughout Ldb1fl/fl R26LSLeYFP MIP-CreERTm islets including the population of eYFP+, insulin– cells (Figure 2, N–U). Thus, ablation of Ldb1 in mature β cells yields a population of insulin– cells that continue to express the endocrine marker ChgA and the β cell factor NKX6.1.
Ablation of Isl1 in mature β cells phenocopies Ldb1 loss of function.
To determine whether LDB1 functions in a complex with ISL1 in mature β cells, we generated an Isl1 loss-of-function strain (Isl1fl/fl R26LSLeYFP MIP-CreERTm) to compare the transcriptomes of Ldb1-depleted β cells and Isl1-depleted β cells (37). Prior to Tm administration, the baseline glucose tolerances of Isl1fl/fl and Isl1fl/fl MIP-CreERTm mice were indistinguishable (Supplemental Figure 2F). As with the Ldb1fl/fl MIP-CreERTm model, we administered Tm starting at P28; however, fewer doses of Tm were required to achieve maximal ablation of Isl1 (Supplemental Figure 2, A–C). Ablation of Isl1 in mature β cells closely recapitulated the glucose homeostasis defects observed in the Ldb1fl/fl MIP-CreERTm mice and were similar to previous findings in Isl1fl/fl Pdx1-CreERTm and Isl1mER-Cre-mER/L mice (30, 38). Weight gain and insulin tolerance were similar between Isl1fl/fl MIP-CreERTm mice and their littermate controls (Supplemental Figure 2, D and H), but the random blood glucose levels of Isl1fl/fl MIP-CreERTm mice became significantly elevated (Supplemental Figure 2E). Tm-induced Isl1fl/fl MIP-CreERTm mice were glucose intolerant and displayed negligible in vivo glucose-stimulated insulin secretion (Supplemental Figure 2, F and G). Likewise, static incubations of Isl1fl/fl MIP-CreERTm islets revealed reduced absolute insulin secretion and nearly absent glucose-stimulated insulin secretion and Ex-4 potentiation (Supplemental Figure 2, I–L). Consistent with these observations, total pancreatic insulin content and insulin+ cell mass were decreased in Isl1fl/fl MIP-CreERTm mice (Supplemental Figure 2M and Supplemental Figure 3, A–I). There were no changes in pancreatic glucagon content, ChgA+ cell mass, average islet size, pancreatic islet density, or the number of somatostatin+ and glucagon+ cells per islet (Supplemental Figure 2N and Supplemental Figure 3, J–O). Overall, the physiological characterization of Isl1fl/fl MIP-CreERTm mice closely phenocopied that of Ldb1fl/fl MIP-CreERTm mice, suggesting that LDB1 indeed functionally interacts with ISL1 in the mature β cell.
To comprehensively interrogate the fates of Ldb1- and Isl1-depleted β cells, we performed RNA-seq on the FACS-enriched eYFP+ cell populations collected from the respective WT and loss-of-function lineage-tracing strains. Following Tm administration, we detected an eYFP+ cell population in the dispersed islets of all 3 lineage-tracing strains (Figure 3A) that was absent in the respective Tm-treated, Cre-negative littermates (Supplemental Figure 4). Notably, side scatter (SSC), an indicator of cellular granularity, was significantly reduced in eYFP+ cells collected from both loss-of-function strains (Figure 3, A and B), an observation that may be attributable to the reduced insulin content in both models (Figure 1M and Supplemental Figure 2M). Using the transcriptome of the FACS-enriched WT β cells as the common baseline, the FACS-enriched Ldb1– and Isl1– β cell transcriptomes were highly correlated (r = 0.74), with 1,876 and 1,234 genes identified as differentially regulated at an FDR of 0.01 or lower, respectively (Figure 3C). As would be expected if LDB1 and ISL1 were playing concerted transcriptional roles, the majority of these differentially regulated genes showed correlated misexpression in both loss-of-function transcriptomes (Figure 3D). The similarity between the 2 loss-of-function transcriptomes strongly supports the idea of overlapping functional roles for LDB1 and ISL1 in the mature β cell.
Figure 3. Ldb1 and Isl1 β cell transcriptomes are highly correlated.
(A) Sorting strategy for FACS-enriched, eYFP+ WT, Isl1– β cell, and Ldb1– β cell populations. (B) SSC area mode of the FACS-enriched WT (n = 5), Isl1– β cell (n = 6), and Ldb1– β cell (n = 8) populations. (C) Scatter plot comparing the average expression change between the FACS-enriched Isl1– β cell (n = 5) and Ldb1– β cell (n = 8) populations across all genes. Gray circles denote unchanged genes; red circles denote misexpressed genes (FDR ≤0.01, no fold-change restriction). Pearson’s product-moment correlation coefficient: r = 0.74. (D) Heatmap of the union set of differentially regulated genes from FACS-enriched Ldb1– β cells (n = 8) and Isl1– β cells (n = 5), ordered on the basis of the average expression change in the Ldb1– β cell populations. (C and D) Fold-change (FC) in gene expression determined with respect to the average expression level in the FACS-enriched WT β cell (n = 5) populations and displayed as log2(FC)/log2(FC). ####P < 0.001, by 1-way ANOVA with Holm-Sidak correction. Absent statistical annotation in B indicates comparisons that were not significant. Pooled data represent the mean ± SEM. See also Supplemental Figure 4.
Collapse of the terminally differentiated β cell transcriptional program after depletion of Ldb1 or Isl1.
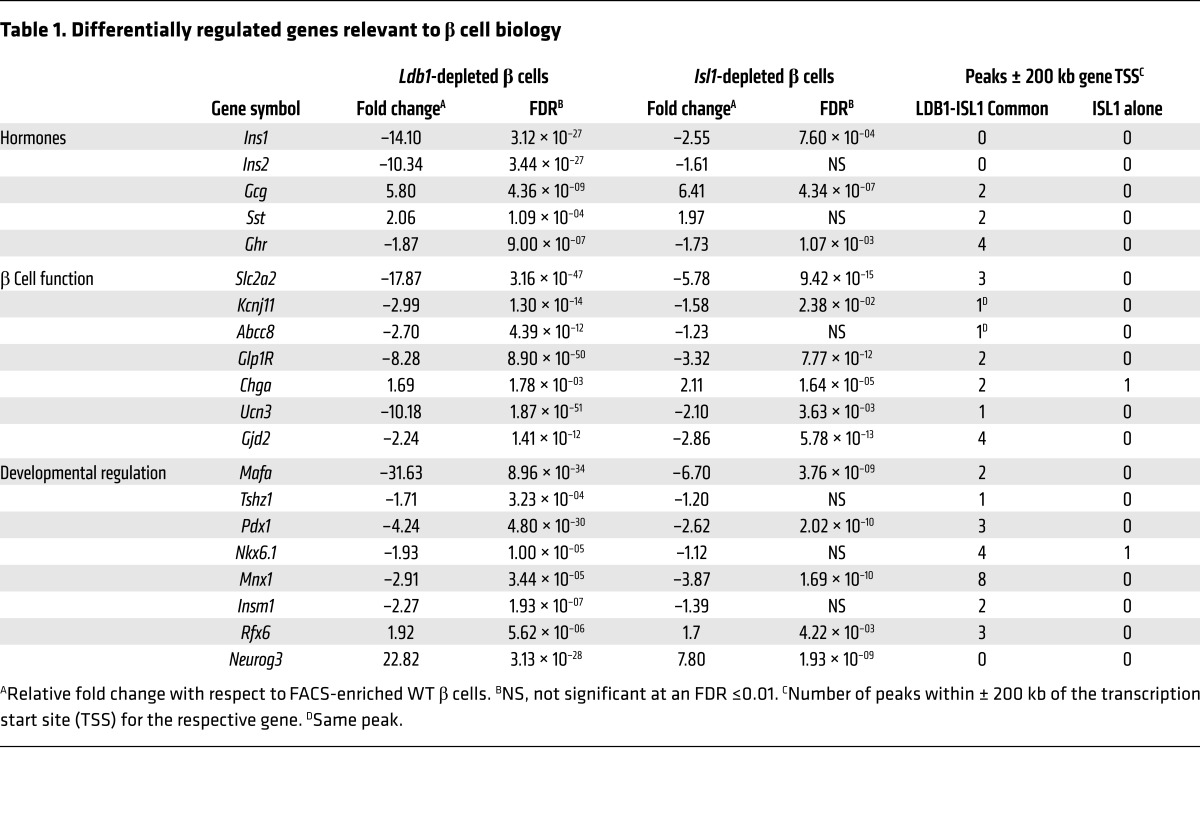
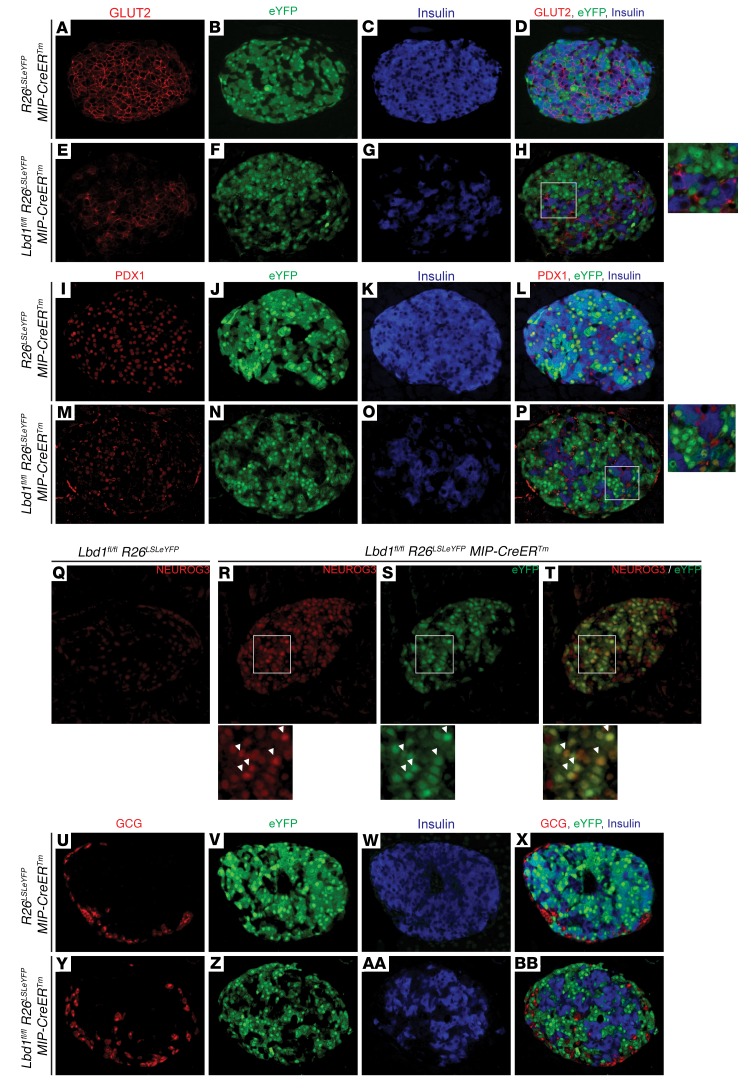
Targeted analysis of the RNA-seq data set revealed that genes critical to β cell function and/or development were misexpressed in one or both loss-of-function models (Table 1). Consistent with the in vivo and ex vivo physiological analyses, expression of Ins1 and Ins2 was significantly reduced, as was the expression of genes encoding other components critical to mature murine β cell function, including the GLUT2 glucose transporter, subunits of the ATP-dependent potassium channel (SUR1 and KIR6.2), the GLP1 receptor, and the paracrine hormone urocortin 3 (Table 1). We confirmed by coimmunofluorescence (co-IF) that GLUT2 downregulation, like insulin, was cell autonomous (Figure 4, A–H). Within Ldb1fl/fl R26LSLeYFP MIP-CreERTm islets, GLUT2 expression was restricted to eYFP–, insulin+ cells and depleted in eYFP+, insulin– cells (Figure 4H).
Table 1. Differentially regulated genes relevant to β cell biology.
Figure 4. Loss of β cell identity gene expression and acquisition of NEUROG3 in Ldb1-depleted β cells.
Co-IF at P56 for (A–H) GLUT2, insulin, and eYFP; (I–P) PDX1, insulin, and eYFP; (Q–T) NEUROG3 and eYFP; and (U–BB) glucagon, insulin, and eYFP. (A–BB) n = 5 Ldb1fl/fl mice; n = 5 Ldb1fl/fl MIP-CreERTm mice. All images were captured at ×20 zoom (insets, ×40 zoom). See also Supplemental Figure 5.
The transcription factor network that establishes and directs β cell fate was also remarkably altered in Ldb1– β cells (Table 1). Expression of genes encoding factors that regulate β cell specification (PDX1 and NKX6.1) and maturation (MAFA and TSHZ1) was decreased (Table 1) (6, 8, 9, 39, 40). The genes encoding transcription factors required for endocrine precursor differentiation (Insulinoma-associated protein 1 [INSM1], motor neuron and pancreas homeobox 1 [MNX1], and Regulatory factor X6 [RFX6]) were also misexpressed (Table 1) (41–44). As seen with GLUT2, MAFA and PDX1 were exclusively depleted in eYFP+, insulin– cells in Ldb1fl/fl R26LSLeYFP MIP-CreERTm islets (Figure 4, I–P, and Supplemental Figure 5, A–H), indicating that Ldb1 ablation impacted the β cell transcription factor network in a cell-autonomous manner.
In contrast to MAFA and PDX1, the gene encoding the endocrine progenitor transcription factor NEUROG3 was upregulated 22.8-fold in FACS-enriched Ldb1– β cells (Table 1). While low levels of Neurog3 transcript are recognized as being expressed in mature murine β cells (45), translation of the Neurog3 transcript is more controversial (46–48). As has been previously described, we observed a faint nuclear signal for NEUROG3 in a fraction of cells populating the islets in Ldb1fl/fl R26LSLeYFP animals (Figure 4Q). In contrast, the NEUROG3 signal in Ldb1fl/fl R26LSLeYFP MIP-CreERTm islets was noticeably more intense and expressed in a larger proportion of islet cells (Figure 4R). Critically, the eYFP lineage tracer marked the majority of NEUROG3+ cells (Figure 4, R–T). Taken together, these results suggest that ablation of Ldb1 in mature β cell leads to a collapse of the terminally differentiated gene program, concomitant with a reversion to a dedifferentiated, possibly more progenitor-like state.
Last, a modest but significant increase in the levels of both Sst and Gcg was detected in the Ldb1– β cell transcriptome (Table 1), raising the possibility that ablation of Ldb1 in mature β cells also resulted in the adoption of alternate endocrine cell fates, as has been reported in other models of murine β cell dedifferentiation (49). However, neither glucagon nor somatostatin colocalized with eYFP or insulin in Ldb1fl/fl R26LSLeYFP MIP-CreERTm pancreata (Figure 4, U–BB, and Supplemental Figure 5, I–P), indicating that reprogramming of β cells to α or δ cells was not occurring. We believe that the Gcg and Sst transcripts detected by RNA-seq are derived from a minor population of non–β-eYFP– cells contaminating the FACS-enriched eYFP+ population. Technical limitations associated with sorting small, low-yield cell populations precluded the implementation of purifying protocols for this experiment.
Highly correlated LDB1 and ISL1 enrichment across the β cell genome.
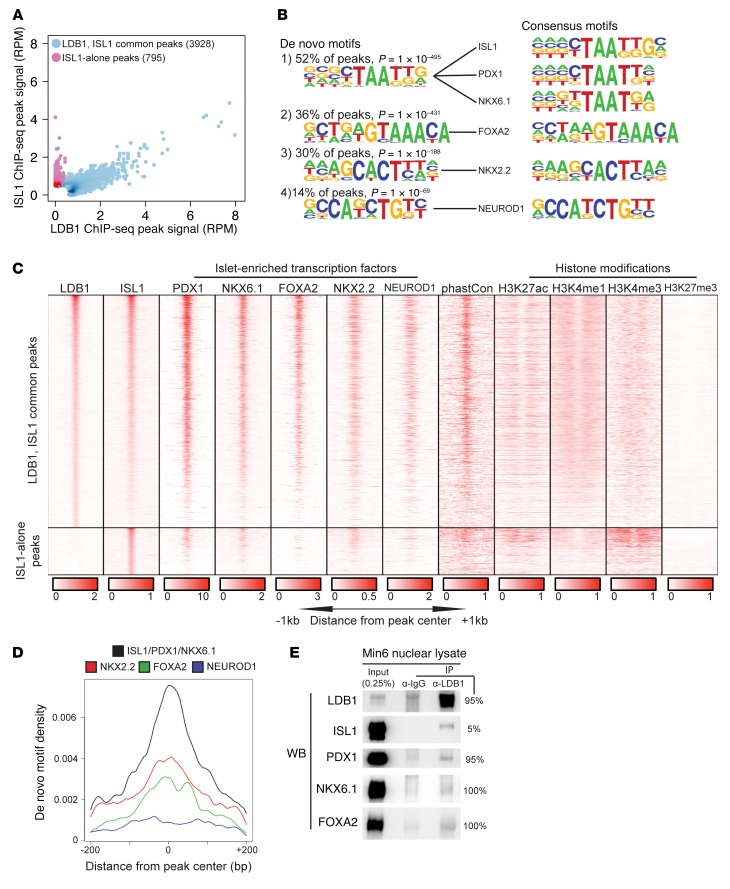
Our physiological and transcriptomic analyses implicate ISL1 as a key partner of LDB1 in the mature β cell. In support of this hypothesis, Isl1 expression was considerably higher than that of any other LIM-HD or LMO transcript detected in the FACS-enriched WT β cell transcriptome (Supplemental Figure 6). Nonetheless, even minimal expression of other LIM-HD and LMO species raises the possibility that LDB1 acts independently of ISL1. To determine the extent to which LDB1 and ISL1 partner in the mature endocrine pancreas, ChIP-seq for both LDB1 and ISL1 was performed using primary mouse islets.
While there was no clear evidence for the existence of LDB1-alone peaks, a small group of 795 ISL1-alone peaks was observed (Figure 5A). De novo motif analysis of this peak set failed to identify any significantly enriched motifs (data not shown). Further subdivision of this peak set was performed on the basis of histone h3 modifications indicative of inactive chromatin (H3K27me3), active chromatin (H3K27ac and H3K4me1), and active promoters (H3K4me3) (Figure 5C) (50, 51). One set of 472 ISL1-alone peaks were devoid of any histone modifications and deemed nonfunctional artifacts. The remaining 323 peaks were enriched at active promoters; however, the de novo motif analysis was, again, unsuccessful in identifying enriched motifs with clear biological relevance (Supplemental Figure 7). These 323 peaks may represent “phantom” peaks that have been described as occurring within the open chromatin of active promoters (52).
Figure 5. LDB1 and ISL1 are coenriched throughout the β cell genome.
(A) Scatter plot comparing signal strength in reads per million (RPM) of ISL1 and LDB1 primary islet ChIP-seqs at the combined set of called peaks from both ChIP-seqs. Blue dots represent peaks in the LDB1, ISL1 common set; red-purple dots represent peaks in the ISL1-alone set. log2(ISL1 ChIP-seq RPM/LDB1 ChIP-seq RPM) = 2.0 delineates the 2 sets. (B) De novo motif analysis of the LDB1, ISL1 common peak set. The de novo motif is presented with the percentage of LDB1-ISL1 common peaks containing the motif and the P value. (C) Heatmap of primary mouse islet LDB1, ISL1, PDX1, NKX6.1, FOXA2, NEUROD1, H3K27ac, H3K4me1, H3K4me3, and H3K27me3 ChIP-seq data, Min6 NKX2.2 ChIP-seq data, and evolutionary conservation (phastCon) for the LDB1, ISL1 common and ISL1-alone peak sets. LDB1, ISL1 common peaks are displayed from the highest to lowest average LDB1 and ISL1 ChIP-seq signal. (D) Density of motifs with respect to identified LDB1, ISL1 common peaks. Colors above the plot correspond to the respective curve in the overlay panel. (E) Western blot (WB) of LDB1 and IgG immunoprecipitates using Min6 nuclear lysates (n = 3). The loaded volume is displayed as the percentage of immunoprecipitate volume. See also Supplemental Figures 6 and 7.
The remaining 3,928 peaks (83.2% of all peaks) displayed a correlated enrichment for both LDB1 and ISL1, referred to herein as LDB1-ISL1 common peaks (Figure 5A). Intriguingly, the majority of genes in Table 1 had one or more LDB1-ISL1 common peaks within ± 200 kb of their respective transcription start sites, including the previously published direct targets of ISL1: Slc2a2, Glp1r, Mafa, and Pdx1 (13, 30, 53). Over half of the LDB1-ISL1 common peaks were enriched for a de novo motif matching the consensus binding sequence of ISL1 that is also highly similar to the consensus binding sequences of the homeodomain factors PDX1 and NKX6.1 (Figure 5B). Three other de novo motifs matching the consensus binding sequences of the pancreatic endocrine–enriched factors forkhead box A2 (FOXA2), NK2 homeobox 2 (NKX2.2), and neuronal differentiation 1 (NEUROD1) were also identified within the LDB1-ISL1 common peak set (Figure 5B).
To determine whether the de novo motifs accurately reflect PDX1, NKX6.1, FOXA2, NKX2.2, and NEUROD1 enrichment in proximity to sites of LDB1, ISL1 coenrichment, we incorporated primary mouse islet cistrome data for PDX1, NKX6.1, FOXA2, and NEUROD1 as well as Min6 insulinoma cistrome data for NKX2.2 into our analysis (54–57) (Dominguez et al., unpublished observations). Strikingly, LDB1 and ISL1 co-occupancy was highly correlated with PDX1, NKX6.1, and FOXA2 occupancy in terms of both location and signal intensity (Figure 5C). NKX2.2 and NEUROD1 enrichment at sites of LDB1 and ISL1 co-occupancy was also frequently but less consistently observed than the other 3 factors (Figure 5C). Remarkably, the de novo motif density analysis of the LDB1-ISL1 common peak set accurately predicted the enrichment pattern of each islet-enriched transcription factor in terms of relative motif frequency and positioning to peak centers (Figure 5D). Given the strong correlation among LDB1, ISL1, PDX1, NKX6.1, and FOXA2 chromatin occupancy, we assessed whether they occur in a complex by performing co-IPs of endogenous LDB1 using Min6 insulinoma nuclear lysates. Indeed, we detected endogenous ISL1, PDX1, NKX6.1, and FOXA2 in LDB1 immunoprecipitates (Figure 5E; see complete unedited blots in the supplemental material). Collectively, our analysis positions LDB1-ISL1 complexes as trans-acting components that interact with key islet-enriched transcription factors.
Ldb1 occupies active enhancers in normal human islets.
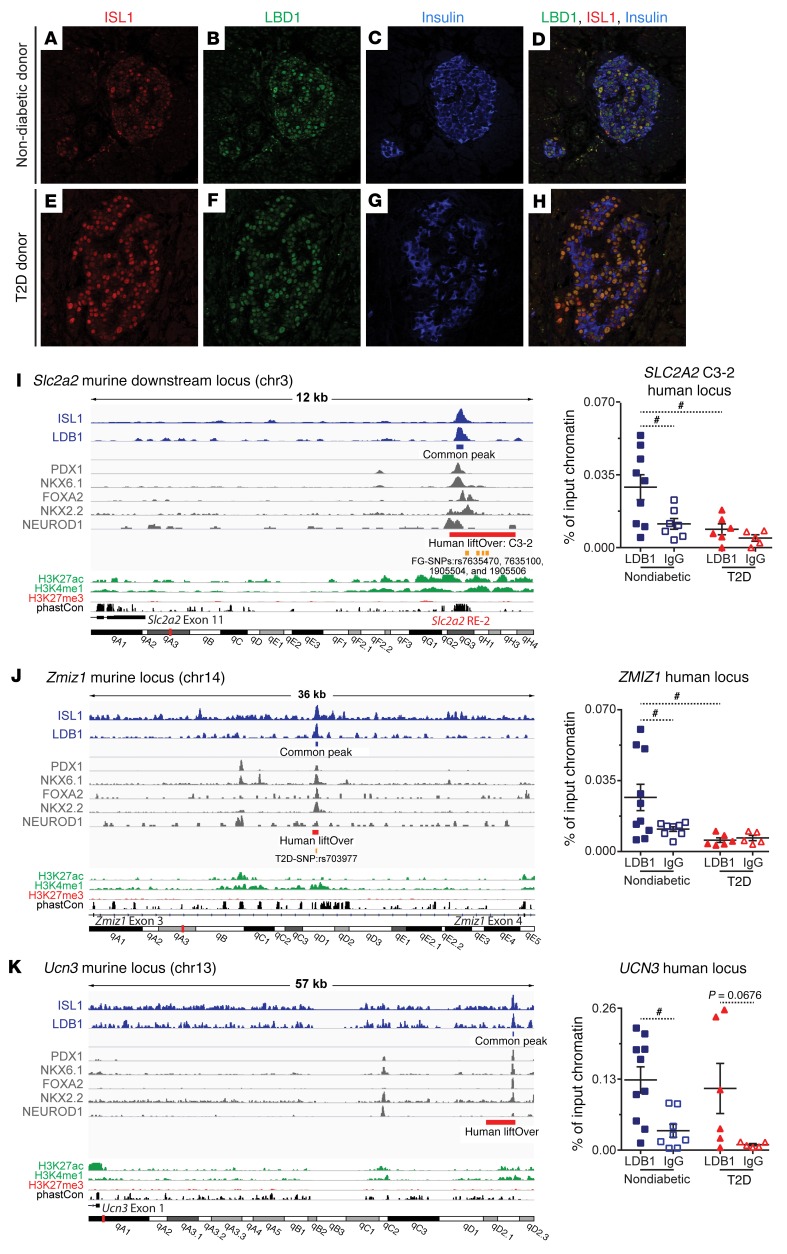
The histone h3 modifications enriched in proximity to the LDB-ISL1 common peaks are indicative of active enhancers. Specifically, the LDB-ISL1 common peaks are located in genomic regions devoid of inactive chromatin modifications (H3K27me3) and enriched for the histone modifications associated with active regulatory elements (H3K27ac and H3K4me1) but not active promoters (H3K4me3) (Figure 5C) (56, 58). In light of recent evidence reporting that human islet–specific, active enhancers are more likely to be coenriched for PDX1, NKX6.1, NKX2.2, and/or FOXA2 (59), our combination of de novo motif, cistromic colocalization, and IP data are significant. Taken together, these observations indicate that LDB1-ISL1 complexes are an integral component of active enhancers in murine islets and suggest a similar role in human islets. Indeed, LDB1 and ISL1 are coexpressed in the β cells of adult nondiabetic and type 2 diabetic donors (Figure 6, A–H, and Supplemental Table 2). Furthermore, our analysis of a published human islet RNA-seq data set incorporating 77 donors revealed a weak negative correlation (R2 = 0.087; P = 0.009) between ISL1 expression with increasing HbA1c levels (Supplemental Figure 8A); whereas, there was no correlation between LDB1 expression and HbA1c levels (Supplemental Figure 8B) (60).
Figure 6. LDB1-mediated complexes occupy active enhancers in murine and human islets.
(A–H) Co-IF for Insulin, LDB1, and ISL1 using human pancreatic sections. n = 1 nondiabetic donor; n = 1 type 2 diabetic donor (T2D). All images were captured at ×20 zoom. (I) Murine Slc2a2 downstream locus and ChIP-qPCR at the human SLC2A2 C3-2 locus. (J) Murine Zmiz1 locus and ChIP-qPCR at the human ZMIZ1 locus. (K) Murine Ucn3 locus and ChIP-qPCR at the human UCN3 locus. (I–K) IGV windows of murine loci. LDB1 and ISL1 ChIP-seqs labeled in blue. LDB1-ISL1 common peaks are indicated by blue line segments. PDX1, NKX6.1, FOXA2, NKX2.2, and NEUROD1 ChIP-seqs are labeled in gray. Human islet regulome active enhancers: human liftOvers (indicated by red line segments); human SNP liftOvers (indicated by orange line segments). FG, fasting glycemia associated and T2D, type 2 diabetes associated. H3K27ac and H3K4me1 ChIP-seqs are labeled in green; H3K27me3 ChIP-seq is labeled in red; PhastCon (evolutionary conservation) is labeled in black. Published cis-elements are indicated in red text. Nondiabetic donor ChIPs are indicated by blue and white squares: LDB1 IP = blue squares (n = 10); normal goat IgG IP = white squares (n = 8). Type 2 diabetic donor ChIPs are indicated by red and white triangles: LDB1 IP = red triangles (n = 6); normal goat IgG IP = white triangles (n = 5). Pooled data represent the mean ± SEM. #P < 0.05 by 1-way ANOVA with Holm-Sidak correction for the following post-hoc comparisons: nondiabetic LDB1 versus nondiabetic normal goat IgG; nondiabetic LDB1 versus type 2 diabetic LDB1; type 2 diabetic LDB1 versus type 2 diabetic normal goat IgG; and nondiabetic normal goat IgG versus type 2 diabetic normal goat IgG. Absent statistical annotation indicates comparisons were not significant. See also Supplemental Figures 8–10 and Supplemental Tables 1–4. chr, chromosome.
To determine whether LDB1 is enriched at active enhancer elements in human islets, we performed ChIP–quantitative PCR (ChIP-qPCR) using whole-islet chromatin from nondiabetic and type 2 diabetic donors. We consulted the Human Islet Regulome Browser (59) and selected genomic loci designated C3-accessible chromatin, i.e., active enhancers, that also batch coordinate converted (UCSC liftOver, https://genome.ucsc.edu/cgi-bin/hgLiftOver) to loci co-occupied by LDB1, ISL1, PDX1, and NKX6.1 in murine islets. The 10 loci we selected batch coordinate converted in proximity to misregulated genes from Table 1 or harbored converted regions with fasting glycemia– or type 2 diabetes–associated SNPs (Supplemental Table 1 and Figure 6, I–K, and Supplemental Figure 9, A–C, E, and F) (59). For example, an active enhancer that is conserved from humans to mice in proximity to SLC2A2 met both criteria (Figure 6I). Of the 10 loci tested, we observed statistically significant enrichment for LDB1 over the IgG control at the SLC2A2 C3-2, ZMIZ1, and UCN3 loci as well as trending enrichment at the MAFA and GLIS3 C3-2 loci (Figure 6, I–K, and Supplemental Figure 9, A and B). Comparably little LDB1 enrichment was observed at the DDX18 negative control locus as well as at the PDX1, SLC2A2 C3-1, GLIS3 C3-1, and both MNX1 loci (Supplemental Figure 9, B–F, Supplemental Figure 10B, and data not shown). Notably, the active enhancers in proximity to SLC2A2 C3-2 and MAFA correspond to the characterized murine cis-regulatory elements that are direct ISL1 targets: Slc2a2-Re2 and Mafa-Region3 (Figures 6I and Supplemental Figure 9A) (30, 53, 61). It is also worth noting that LDB1 enrichment at the UCN3 locus in nondiabetic adults displayed a strong negative correlation (R2 = 0.569) with donor age (Supplemental Figure 10C). Conversely, LDB1 enrichment at the PDX1 locus in nondiabetic adults displayed an equally strong but positive correlation (R2 = 0.597) with donor age (Supplemental Figure 10D). No such correlations between LDB1 enrichment and age in non-diabetics were discovered at the DDX18 , MAFA , SLC2A2 C3-2, ZMIZ1 , and GLIS3 C3-2 loci (Supplemental Figure 10, B, E–H). These findings indicate that LDB1 enrichment at certain active enhancers fluctuates with age.
Remarkably, in the type 2 diabetic donor islets, LDB1 enrichment was depleted at the SLC2A2 C3-2 and ZMIZ1 loci, and there was a trend toward reduced occupancy at the MAFA locus (Figure 6, I and J and Supplemental Figure 9A). The SLC2A2 C3-2 and ZMIZ1 loci contained SNPs associated with fasting glycemia levels and type 2 diabetes, respectively (Figure 6, I and J, and Supplemental Table 1). In the type 2 diabetic islets, LDB1 enrichment at the SLC2A2 C3-2 and ZMIZ1 loci was not correlated with donor age (Supplemental Figure 10, F and G). Considering that the overall age distribution of the nondiabetic and type 2 diabetic donors was not significantly different (Supplemental Figure 10A and Supplemental Tables 3 and 4), our findings suggest that LDB1 occupancy at the SLC2A2 C3-2 and ZMIZ1 loci reflects type 2 diabetes status independent of donor age. Taken together, these human data indicate that LDB1 complexes are indeed enriched at certain active enhancer elements in islets and can become dysregulated in type 2 diabetes.
Discussion
The importance of LDB1 in developmental populations, including pancreatic endocrine progenitors, is well established (13, 14, 16, 25, 27, 29, 62). Here, we provide insights into the role of LDB1 in a terminally differentiated cell type. Lineage tracing revealed that depletion of LDB1 in the mature β cell precipitated the collapse of its terminally differentiated state, highlighted by the concomitant reduction in prominent markers of β cell identity and the activated expression of the endocrine progenitor transcription factor NEUROG3. Reversion of the mature β cells to a progenitor-like state resulted in impaired glucose homeostasis due to significantly reduced insulin expression and secretion. Importantly, our cistromic and transcriptomic analyses revealed that a coordinated functional relationship between LDB1 and ISL1, heretofore only described in progenitor cell populations, is critical for maintaining the terminally differentiated status of mature β cells. Finally, we demonstrated that LDB1-mediated complexes are incorporated into the trans-regulatory components that occupy active enhancers in both murine and human islets and that these complexes are disrupted at several loci in the setting of type 2 diabetes.
In the broader context of LDB1 functional paradigms, our cistromic analysis indicates that LDB1 is almost invariably coenriched with ISL1, suggesting shared similarities to the LDB1, LIM-HD paradigms described in differentiating neuronal cell populations (16, 29). In these neuronal cell populations, the relative stoichiometries of LIM-HD species dictate the resulting LDB1-mediated complex (12, 63). In ISL1-enriched neuronal populations, ISL1 functions in obligatory heterodimers with either LIM-HD or paired-like homeobox factors (64). The resulting LDB1-bound heterodimers recognize unique bipartite motifs, ensuring the initiation of distinct transcriptional programs (64). Our unbiased motif analysis of the LDB1, ISL1 common peaks failed to identify bipartite elements, suggesting that in mature β cells, the LDB1-ISL1 complexes do not incorporate other LIM-HD components (Figure 7). This is not unexpected, given the relatively low expression of other LIM-HD species in β cells.
Figure 7. Working model of LDB1-mediated complexes in terminally differentiated β cells.
Schematic of the LDB1-ISL1 complex at a generalized active enhancer in a terminally differentiated β cell. The dashed-line box indicates a speculative role for LDB1-ISL1 complexes mediating the higher-order chromatin structures necessary for appropriate gene regulation. Flanking the central model are schematics representing the status of LDB1-ISL1 complexes in the respective loss-of-function models. In the Ldb1fl/fl MIP-CreERTM schematic, the described intramolecular binding between the tandem LIM domains and the ISL1 LIM-interacting domain (LID) is indicated. SAD, self-association domain; HD, homeo-domain; LIM1,2, tandem LIM domains; TFs, transcription factors; H3, histone h3.
Correspondingly, LDB1-mediated complexes are sensitized to the relative stoichiometric levels of their components (12). Lowly expressed and redundant LDB1-binding partners can serve as buffers to fluctuations from steady state (65). This scenario is exemplified by LMO4 in neuronal progenitors (63). While the Ldb1fl/fl MIP-CreERTm mice essentially model a scenario prohibiting LDB1-mediated complexes, the Isl1fl/fl MIP-CreERTm mouse model disrupts LDB1-ISL1 complex formation but does not prohibit the formation of LDB1-mediated complexes with other LMO and LM-HD factors (Figure 7). It is notable that the magnitude of the transcriptomic effects of LDB1 and ISL1 depletion is often distinct between the 2 loss-of-function models (Figure 3D and Table 1). In ISL1-depleted β cells, LDB1 interactions with residual binding partners may, in part, explain the discrepancies observed between the 2 loss-of-function transcriptomes (Figure 7).
A recent study characterizing the human islet regulome revealed that chromatin regions coenriched for FOXA2, PDX1, NKX6.1, NKX2.2, and MAFB were more likely to be islet-specific, active enhancers (59). Here, we have connected LDB1 and ISL1 to this critical set of pancreatic endocrine transcription factors, a finding that also supports recently published studies describing an ISL1-SET7/9-PDX1-NEUROD1 complex (66, 67). LDB1, unlike these five trans-acting factors, has no DNA-binding capacity and, importantly, mediates promoter-enhancer looping (12, 26). The role for LDB1 in mediating promoter-enhancer looping has been thoroughly documented in hematopoiesis/erythropoiesis (62) and most recently in corticotrope and cardiac progenitor differentiation (25, 27). LDB1-mediated looping has even been manipulated to reactivate developmentally silenced β-globin genes (68). Interestingly, the human islet regulome study also revealed that islet-specific, active enhancers were more likely to be involved in higher-order chromatin architecture (59). Given the results of our multi–data set cistromic analysis and the human islet regulome study (59), we speculate that LDB1, in complex with ISL1, plays a critical role in establishing a higher-order chromatin structure in islets (Figure 7). In this scenario, the collapse of the terminally differentiated state in LDB1-depleted β cells likely reflects widespread disruption of chromatin architecture.
The collapse of the terminally differentiated state in LDB1-depleted β cells showed key hallmarks described in models of murine β cell dedifferentiation: loss of insulin, concomitant with activation of NEUROG3 (49, 69). In contrast to some of these models of murine β cell dedifferentiation, LDB1-depleted β cells did not reprogram into other endocrine lineages, an observation that may relate to the critical role of LDB1-mediated complexes in the developing endocrine pancreas. Ablation of Ldb1 in PAX6+ endocrine precursors giving rise to non–ε cell lineages halts the maturation of the α, β, and δ cell lineages (13). Considering that LDB1 and ISL1 remain ubiquitously expressed in the mature endocrine pancreas, our findings broadly suggest that LDB1-mediated complexes underpin the terminally differentiated status of the α and δ cell lineages as well. This may explain why Ldb1-depleted β cells do not reprogram in spite of marked reductions in PDX1 and MNX1 — factors necessary for the repression of β cell reprograming to α and δ cell fates, respectively (10, 70). Thus, prohibiting the formation of LDB1-mediated complexes in mature β cells leads to a loss of β cell identity but simultaneously precludes activating aspects of the other terminally differentiated pancreatic endocrine cell types.
While β cell dedifferentiation is increasingly implicated in the progression of type 2 diabetes (71, 72), our human islet investigation identified 2 putative active enhancer elements that are depleted of LDB1 enrichment in islets from type 2 diabetic donors. Remarkably, these 2 putative active enhancers were in proximity to SLC2A2 and ZMIZ1 and contained SNPs associated with fasting glycemia levels and type 2 diabetes, respectively. Similarly, we identified 2 putative active enhancers in respective proximity to UCN3 and PDX1, for which the level of LDB1 enrichment correlated with age. While our human analysis was limited to a small sample size of islet donors and only targeted a set of 7 genes, our results suggest that the formation and/or positioning of LDB1-mediated complexes occupying functional enhancers within human islets are sensitive to type 2 diabetes and aging.
We believe that our study addresses a knowledge gap of critical significance to islet biologists — the biology that underpins the terminally differentiated status of the β cell. In both mice and humans, it is increasingly appreciated that an important period of functional maturation occurs between β cell specification and terminal differentiation (73). Here, we have demonstrated that the LDB1-mediated complexes required for pancreatic endocrine maturation are necessary to maintain the mature, functional status of β cells in adult animals. The recognition of β cell dedifferentiation in type 2 diabetes and the improving but still limited ability of directed differentiation protocols to generate functionally mature β cells (71, 74–76) support the translational relevance of our findings. Considering that LDB1-mediated complexes recruit chromatin-remodeling factors and establish chromatin architecture (21–24, 26), LDB1 represents an attractive therapeutic target positioned at the intersection of the transcriptional networks and epigenetics governing the identities of the pancreatic endocrine lineages.
Methods
Animal models.
The Ldb1fl, Isl1fl, MIP-CreERTm, and Rosa26LSLeYFP strains have been described previously (32–34, 36, 77). Mice were maintained on a mixed SV/129, C57Bl/6 background. At P28, male mice, while briefly anesthetized using inhaled isoflurane, received an oral gavage of Tm (Sigma-Aldrich; T5648) at a dose of 150 μg/g BW, with the Tm prepared at 50-mg/ml in 90% sunflower seed oil and 10% EtOH (v/v). Oral gavages were supplied at 24-hour intervals over a 5 day period. Unless otherwise indicated, mice were analyzed at P56. For male P28 Isl1fl/fl Rosa26LSLeYFP/LSLeYFP MIP-CreERTm, Isl1fl/fl MIP-CreERTm, and Isl1fl/fl mice, Tm administration was reduced to 100 μg/g BW for 5 oral gavages at 24-hour intervals. Unless otherwise indicated, mice were analyzed at P49.
IHC and IF.
Pancreata were dissected, fixed in 10% formalin solution (pH 7.0) (Sigma-Aldrich; HT501128) for 17 hours at 4°C and embedded in paraffin. Sections were blocked with CAS-Block (Invitrogen, Thermo Fisher Scientific; 008120), and primary antisera were applied overnight at 4°C. Fluorophore-conjugated and biotinylated secondary antisera were applied for 60 minutes and 40 minutes at 37°C, respectively, and nuclei were counterstained with DAPI (Vector Laboratories; H-1200). IHC signal was detected using a standard Vectastain Elite ABC Kit (Vector Laboratories; PK-6100) with Peroxidase Substrate Kit DAB (Vector Laboratories; SK-4100) and a Vectastain ABC-AP Kit (Vector Laboratories; AK-5000) with a BCIP/NBT AP Substrate Kit (Vector Laboratories; SK-5400), and nuclei were counterstained with hematoxylin or nuclear fast red. When Tyramide Signal Amplification (TSA) (PerkinElmer) was used, HRP-conjugated secondary antisera were applied for 40 minutes at 37°C and a TSA Plus Cyanine 3 System (PerkinElmer; NEL744001KT) was used at 1:250 for 10 minutes. For NEUROG3 IF, dissected pancreata were fixed for 2 hours at 4°C, cryoprotected overnight in 30% sucrose v/v 1× PBS, and embedded in OCT compound (Tissue-Tek; 4583). Heat-induced epitope retrieval using sodium citrate buffer (pH 6.0) and a pressure cooker was performed for all IHC and IF procedures. See the Supplemental Table 5 for details on the antisera and dilutions.
Signal was visualized with a Leica DM6000 B or Nikon Eclipse E600 microscope, and images were captured using Leica LAS AF software and a Leica DFC300 FX digital camera or using MetaMorph software and a QImaging Q/Click digital camera, respectively. To quantify IHC staining, slides were digitally scanned by an Aperio Scanscope CS2 and analyzed using Aperio’s ImageScope. To quantify IF staining, the MetaMorph software suite was used for scanning and analysis. Insulin+ and ChgA+ cell mass was calculated as the antigen+ area over the total pancreatic tissue area, multiplied by the pancreatic wet weight that was determined immediately after dissection. Islet area was the average of ChgA+ islet size over an entire tissue section. Islet density is the reciprocal of the number of islets per pancreatic tissue area. The number of glucagon+ and somatostatin+ cells was normalized to the total ChgA+ area of each section.
Glucose and insulin assays.
Glucose tolerance, insulin tolerance, glucose-stimulated plasma insulin levels, total pancreatic insulin, and glucagon content were assessed as described previously (30). Insulin concentration was measured by an Ultrasensitive Mouse Insulin ELISA (Crystal Chem; 90080). Random-fed plasma glucose levels were assessed between 10:00 am and 11:00 am.
Static islet incubations.
Islets were isolated as described previously (30) and cultured for 2 days in islet culture media (RPMI 1640, 10% FBS v/v, 1× penicillin-streptomycin, 1× glutamine, and 10 mM glucose) at 37°C and 5% CO2. Islets were washed with glucose-free Krebs-Ringer bicarbonate buffer (KRBB) with 0.25% BSA and allowed to equilibrate for 30 minutes in glucose-free KRBB. Islets were then incubated in batch at 2, 5, 8, and 16 mM glucose ± 50 nM Ex-4. Each individual condition was performed in triplicate and consisted of 5 handpicked, similarly sized islets. Following a 30-minute incubation period, the culture media was collected, and insulin levels were measured as described above. For the Ldb1fl/fl and Ldb1fl/fl MIP-CreERTm mice that provided islets for static incubations, the initial gavage was at 5.5 weeks of age, and the total pulse-chase period was 9 and 7 weeks for the Ldb1fl/fl and Ldb1fl/fl MIP-CreERTm mice, respectively.
Cell sorting and RNA-seq library construction, deep sequencing, and analysis.
Islets from induced lineage–tracing animals were isolated and dispersed into single cells using 0.05% Trypsin-EDTA (Gibco, Thermo Fisher Scientific; 25300-054). A Beckman Coulter MoFlo Astrios was used to enrich for single, eYFP+ cells directly into TRIzol (Invitrogen, Thermo Fisher Scientific; 15596018). Each sort reflected islets isolated from an individual mouse. When extracting RNA, sorts with low cell recovery were pooled in their entirety to achieve sufficient RNA for downstream applications, with a minimum threshold of 25,000 cells. Total RNA was extracted using RNAeasy Micro Columns (QIAGEN; 74004), then DNAse treated with the TURBO DNA-Free Kit (Ambion, Thermo Fisher Scientific; AM1907) and purified by an RNAeasy Micro Column. Libraries of total RNA were generated using ScriptSeq Complete Gold – Low Input (Illumina; SCL6G). RNA-seq library sequencing and preprocessing were performed by the Functional Genomics Core at the University of Pennsylvania. Reads were aligned to UCSC mouse genome mm9, and gene expression levels were quantified using RUM pipeline (78). Differential gene expression analysis was done by comparing the WT transcriptome with the individual loss-of-function transcriptomes using edgeR (79). Genes with an FDR of 0.01 or less were selected as differentially expressed.
IP and Western blot analysis.
Nuclear lysates were obtained from Min6 insulinoma cells as described previously (80). The Min6 cells were a gift of J. Miyazaki (81). Lysates were incubated with 10 μg IP antibody diluted in PBS overnight at 4°C. Dynabeads Protein G (Novex; 10003D) were added to the lysate and allowed to immunoprecipitate for 2 hours at 4°C. Immunoprecipitated eluates were resolved by 10% SDS-PAGE and transferred to nitrocellulose. Membranes were blocked with milk in PBS plus Tween-20 (PBST) (5% w/v) and blotted. Signal was detected with Luminata Crescendo Western HRP Substrate (EMD Millipore; WBLUR0100) using the Bio-Rad ChemiDoc Touch Imaging System. See Supplemental Table 6 for details on the antisera used.
ChIP-qPCR and ChIP-seq library preparation, sequencing, and analysis.
Human and 8- to 10-week-old male CD1 mouse islet sonicates were prepared, and ChIP was performed as previously described (30). For the ChIPs, CLIM-2 antibody (N-18)-X (Santa Cruz Biotechnology Inc.; sc11198 X); anti–islet 1 antibody (EP4182) (Abcam; ab109517); and normal goat IgG (Santa Cruz Biotechnology Inc.; 2028) were used. See Supplemental materials for information on islet active enhancers (Supplemental Table 1), human islet donors (Supplemental Tables 2–4), and qPCR primers (Supplemental Table 7).
ChIP-seq libraries were prepared with a NEBNext Ultra DNA Library Prep Kit for Illumina (New England BioLabs; E7370S) according to their low-input ChIP-seq protocol. All the libraries were sequenced by the Functional Genomics Core at the University of Pennsylvania and aligned to UCSC mouse genome mm9 using Bowtie (82). To obtain adequate read depth, biological replicates were pooled together by selecting unique reads for ISL1 and LDB1, respectively. Peak calling was performed using the “findPeak” command in Homer (83). For a comparative analysis of the ISL1 and LDB1 cistromes, peaks were pooled to prepare a master peak set, in which overlapping peaks were merged for a center distance of less than 100 bp. ISL1 and LDB1 occupancy was measured in master peaks as normalized tag counts (reads per million). Peaks with an ISL1 signal 2-fold over LDB1 signal were defined as ISL1-alone binding sites. A de novo motif search was performed within a 200-bp window using “findMotifsGenome.pl.” Motif density was examined for the top enriched motifs within 400 bp around peak centers using the “annotatePeaks.pl” command in Homer and a weighed-moving-average with a window size of 20 taken for smooth visualization of motif density.
Accession numbers for previously published and high-throughput data sets.
To determine the correlation between HbA1c levels in human islets, we reanalyzed a published data set in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE50244) (60). PDX1, NKX6.1, FOXA2, NEUROD1, H3K27me3, H3K27ac, H3K4me1, and H3K4me3 islet cistrome data were downloaded from the GEO database (GEO GSE40975, GSE68618, and GSE30298); the European Molecular Biology Laboratory – European Bioinformatics Institute (EMBL-EBI) ArrayExpress database (E-MTAB-1143); and the NCBI’s Sequence Read Archive (SRA008281) in fastq format and reprocessed using the same pipeline with ISL1 and LDB1 data for consistency (54–58). Integrative Genomics Viewer (IGV) software was used to visualize cistromic data (84). The NCBI’s GEO accession number for the RNA-seq and ChIP-seq data sets reported herein is GSE84759.
Statistics.
Pooled data represent the mean ± SEM. GraphPad Prism 7 (GraphPad Software)was used to perform statistical analyses. Outliers were identified by the ROUT method at Q = 10% and removed. Statistical significance was determined by 2-tailed Student’s t test (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001); repeated-measures or non–repeated-measures 1-way ANOVA with Holm-Sidak correction (#P < 0.05, ##P < 0.01, ###P < 0.001, and ####P < 0.0001); repeated-measures, 2-way ANOVA with Bonferroni’s or Holm-Sidak correction (†P < 0.05, ††P < 0.01, †††P < 0.001, and ††††P < 0.0001); or Pearson’s product-moment correlation coefficient in combination with linear regression analysis (§P < 0.05 and §§P < 0.01; see supplemental materials). A P value of less than 0.05 was considered statistically significant.
Study approval.
The IACUCs of Children’s Hospital of Philadelphia, the University of Pennsylvania, and Vanderbilt University Medical Center approved the experimental methods involving animals.
Author contributions
Conceptualization: BNE, CLM, and DAS; methodology: BNE, HWL, CJ, C. Li, KJW, CLM, and DAS; software: HWL; formal analysis: BNE and HWL; investigation: BNE, CJ, DNG, LQW, JHL, BLT, ERW, CSH, and C. Li; resources: GD, BLT, VK, JY, C. Liu, CSH, KHK, AN, C. Liu, MS, RS, GD, and LS; data curation: HWL; writing of the original draft: BNE and DAS; writing , review, and editing: BNE, HWL, CSH, CL, MS, CLM, and DAS; visualization: BNE, HWL, and CJ; supervision: CLM and DAS; project administration: BNE, CLM, and DAS; funding acquisition: CLM and DAS.
Supplementary Material
Acknowledgments
We thank Sylvia Evans (University of California San Diego Skaggs School of Pharmacy and Pharmaceutical Sciences, San Diego, CA, USA), Heiner Westphal (Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, USA), and Louis Philipson (The University of Chicago Medicine Kovler Diabetes Center, Chicago, IL, USA) for mouse strains; Paul Love (Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, USA) and Chris Wright (Vanderbilt University School of Medicine, Nashville, TN, USA) for antisera; and Gerd Blobel (Children’s Hospital of Pennsylvania, Philadelphia, PA, USA) for helpful discussions. Funding for this work was provided by the NIH (R01-DK068157, to DAS; DK078606 and DK019525, to CLM; T32-GM07229 and T32-HD007516-15, to BNE; and R01-DK068471, to MS) and the Juvenile Diabetes Research Foundation (2-2007-730, to CLM). These studies were also supported by cores of the Penn Diabetes Research Center (NIH P30-DK19525), including the Functional Genomics and Radioimmunoassay and Biomarkers Cores, the Center for Islet Transplantation, the Molecular Pathology and Imaging Core (NIH P01-DK049210), and the Children’s Hospital of Philadelphia Pathology and Flow Cytometry Core. Some human islets were obtained through the Integrated Islet Distribution Program.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(1):215–229. doi:10.1172/JCI88016.
See the related Commentary at β Cells led astray by transcription factors and the company they keep.
Contributor Information
Christine Juliana, Email: cjul@mail.med.upenn.edu.
Giselle Dominguez, Email: gd2321@columbia.edu.
Jin-Hua Liu, Email: jin-hua.liu@Vanderbilt.Edu.
Vasumathi Kameswaran, Email: vasukam@gmail.com.
Juxiang Yang, Email: juxiang@mail.med.upenn.edu.
Chengyang Liu, Email: chliu@mail.med.upenn.edu.
Ali Naji, Email: ali.naji@uphs.upenn.edu.
Changhong Li, Email: li@email.chop.edu.
Maike Sander, Email: masander@ucsd.edu.
Roland Stein, Email: roland.stein@vanderbilt.edu.
Lori Sussel, Email: lgs2@columbia.edu.
References
- 1.van Belle TL, Coppieters KT, von Herrath MG. Type 1 diabetes: etiology, immunology, and therapeutic strategies. Physiol Rev. 2011;91(1):79–118. doi: 10.1152/physrev.00003.2010. [DOI] [PubMed] [Google Scholar]
- 2.Rutter GA, Pullen TJ, Hodson DJ, Martinez-Sanchez A. Pancreatic β cell identity, glucose sensing and the control of insulin secretion. Biochem J. 2015;466(2):203–218. doi: 10.1042/BJ20141384. [DOI] [PubMed] [Google Scholar]
- 3.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 4.Bramswig NC, Kaestner KH. Epigenetics and diabetes treatment: an unrealized promise? Trends Endocrinol Metab. 2012;23(6):286–291. doi: 10.1016/j.tem.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver-Krasinski JM, Stoffers DA. On the origin of the beta cell. Genes Dev. 2008;22(15):1998–2021. doi: 10.1101/gad.1670808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gannon M, et al. pdx-1 function is specifically required in embryonic beta cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev Biol. 2008;314(2):406–417. doi: 10.1016/j.ydbio.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101(9):2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang C, et al. MafA is a key regulator of glucose-stimulated insulin secretion. Mol Cell Biol. 2005;25(12):4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raum JC, et al. Tshz1 regulates pancreatic β-cell maturation. Diabetes. 2015;64(8):2905–2914. doi: 10.2337/db14-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao T, et al. Pdx1 maintains β cell identity and function by repressing an α cell program. Cell Metab. 2014;19(2):259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bramswig NC, et al. Epigenomic plasticity enables human pancreatic α to β cell reprogramming. J Clin Invest. 2013;123(3):1275–1284. doi: 10.1172/JCI66514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4(12):1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter CS, et al. Islet α-, β-, and δ-cell development is controlled by the Ldb1 coregulator, acting primarily with the islet-1 transcription factor. Diabetes. 2013;62(3):875–886. doi: 10.2337/db12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay M, et al. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130(3):495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- 15.Jurata LW, Kenny DA, Gill GN. Nuclear LIM interactor, a rhombotin and LIM homeodomain interacting protein, is expressed early in neuronal development. Proc Natl Acad Sci U S A. 1996;93(21):11693–11698. doi: 10.1073/pnas.93.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110(2):237–249. doi: 10.1016/S0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- 17.Gill GN. Decoding the LIM development code. Trans Am Clin Climatol Assoc. 2003;114:179–189. [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, et al. Nuclear adaptor Ldb1 regulates a transcriptional program essential for the maintenance of hematopoietic stem cells. Nat Immunol. 2011;12(2):129–136. doi: 10.1038/ni.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L, Lee JY, Gross J, Song SH, Dean A, Love PE. A requirement for Lim domain binding protein 1 in erythropoiesis. J Exp Med. 2010;207(12):2543–2550. doi: 10.1084/jem.20100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Omari K, et al. Structural basis for LMO2-driven recruitment of the SCL:E47bHLH heterodimer to hematopoietic-specific transcriptional targets. Cell Rep. 2013;4(1):135–147. doi: 10.1016/j.celrep.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goardon N, et al. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25(2):357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier N, et al. Novel binding partners of Ldb1 are required for haematopoietic development. Development. 2006;133(24):4913–4923. doi: 10.1242/dev.02656. [DOI] [PubMed] [Google Scholar]
- 23.Krivega I, Dale RK, Dean A. Role of LDB1 in the transition from chromatin looping to transcription activation. Genes Dev. 2014;28(12):1278–1290. doi: 10.1101/gad.239749.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galloway JR, Bethea M, Liu Y, Underwood R, Mobley JA, Hunter CS. SSBP3 Interacts with Islet-1 and Ldb1 to impact pancreatic β-cell target genes. Mol Endocrinol. 2015;29(12):1774–1786. doi: 10.1210/me.2015-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caputo L, et al. The Isl1/Ldb1 complex orchestrates genome-wide chromatin organization to instruct differentiation of multipotent cardiac progenitors. Cell Stem Cell. 2015;17(3):287–299. doi: 10.1016/j.stem.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol Cell. 2007;28(5):810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, et al. Enhancer-bound LDB1 regulates a corticotrope promoter-pausing repression program. Proc Natl Acad Sci U S A. 2015;112(5):1380–1385. doi: 10.1073/pnas.1424228112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. Ldb1-nucleated transcription complexes function as primary mediators of global erythroid gene activation. Blood. 2013;121(22):4575–4585. doi: 10.1182/blood-2013-01-479451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho HH, et al. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 2014;10(4):e1004280. doi: 10.1371/journal.pgen.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ediger BN, et al. Islet-1 Is essential for pancreatic β-cell function. Diabetes. 2014;63(12):4206–4217. doi: 10.2337/db14-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thor S, Ericson J, Brännström T, Edlund T. The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron. 1991;7(6):881–889. doi: 10.1016/0896-6273(91)90334-V. [DOI] [PubMed] [Google Scholar]
- 32.Tamarina NA, Roe MW, Philipson L. Characterization of mice expressing Ins1 gene promoter driven CreERT recombinase for conditional gene deletion in pancreatic β-cells. Islets. 2014;6(1):e27685. doi: 10.4161/isl.27685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, et al. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc Natl Acad Sci U S A. 2007;104(32):13182–13186. doi: 10.1073/pnas.0705464104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wicksteed B, et al. Conditional gene targeting in mouse pancreatic ß-Cells: analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010;59(12):3090–3098. doi: 10.2337/db10-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greig NH, et al. Once daily injection of exendin-4 to diabetic mice achieves long-term beneficial effects on blood glucose concentrations. Diabetologia. 1999;42(1):45–50. doi: 10.1007/s001250051111. [DOI] [PubMed] [Google Scholar]
- 36.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11(11):1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, et al. LIM-homeodomain transcription factor Isl-1 mediates kisspeptin’s effect on insulin secretion in mice. Mol Endocrinol. 2014;28(8):1276–1290. doi: 10.1210/me.2013-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henseleit KD, Nelson SB, Kuhlbrodt K, Hennings JC, Ericson J, Sander M. NKX6 transcription factor activity is required for alpha- and beta-cell development in the pancreas. Development. 2005;132(13):3139–3149. doi: 10.1242/dev.01875. [DOI] [PubMed] [Google Scholar]
- 40.Schaffer AE, et al. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic beta cell identity. PLoS Genet. 2013;9(1):e1003274. doi: 10.1371/journal.pgen.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soyer J, et al. Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development. 2010;137(2):203–212. doi: 10.1242/dev.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellitzer G, et al. IA1 is NGN3-dependent and essential for differentiation of the endocrine pancreas. EMBO J. 2006;25(6):1344–1352. doi: 10.1038/sj.emboj.7601011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Arber S, Jessell TM, Edlund H. Selective agenesis of the dorsal pancreas in mice lacking homeobox gene Hlxb9. Nat Genet. 1999;23(1):67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- 44.Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Pancreas dorsal lobe agenesis and abnormal islets of Langerhans in Hlxb9-deficient mice. Nat Genet. 1999;23(1):71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 45.Wang S, et al. Sustained Neurog3 expression in hormone-expressing islet cells is required for endocrine maturation and function. Proc Natl Acad Sci U S A. 2009;106(24):9715–9720. doi: 10.1073/pnas.0904247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, et al. Neurog3 gene dosage regulates allocation of endocrine and exocrine cell fates in the developing mouse pancreas. Dev Biol. 2010;339(1):26–37. doi: 10.1016/j.ydbio.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao X, et al. No evidence for β cell neogenesis in murine adult pancreas. J Clin Invest. 2013;123(5):2207–2217. doi: 10.1172/JCI66323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Courtney M, et al. The inactivation of Arx in pancreatic α-cells triggers their neogenesis and conversion into functional β-like cells. PLoS Genet. 2013;9(10):e1003934. doi: 10.1371/journal.pgen.1003934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49(5):825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain D, Baldi S, Zabel A, Straub T, Becker PB. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res. 2015;43(14):6959–6968. doi: 10.1093/nar/gkv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du A, et al. Islet-1 is required for the maturation, proliferation, and survival of the endocrine pancreas. Diabetes. 2009;58(9):2059–2069. doi: 10.2337/db08-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khoo C, et al. Research resource: the pdx1 cistrome of pancreatic islets. Mol Endocrinol. 2012;26(3):521–533. doi: 10.1210/me.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic beta cells. Cell Rep. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffman BG, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20(8):1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tennant BR, et al. Identification and analysis of murine pancreatic islet enhancers. Diabetologia. 2013;56(3):542–552. doi: 10.1007/s00125-012-2797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Avrahami D, et al. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab. 2015;22(4):619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fadista J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A. 2014;111(38):13924–13929. doi: 10.1073/pnas.1402665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raum JC, et al. FoxA2, Nkx2.2, and PDX-1 regulate islet beta-cell-specific mafA expression through conserved sequences located between base pairs -8118 and -7750 upstream from the transcription start site. Mol Cell Biol. 2006;26(15):5735–5743. doi: 10.1128/MCB.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Love PE, Warzecha C, Li L. Ldb1 complexes: the new master regulators of erythroid gene transcription. Trends Genet. 2014;30(1):1–9. doi: 10.1016/j.tig.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song MR, Sun Y, Bryson A, Gill GN, Evans SM, Pfaff SL. Islet-to-LMO stoichiometries control the function of transcription complexes that specify motor neuron and V2a interneuron identity. Development. 2009;136(17):2923–2932. doi: 10.1242/dev.037986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazzoni EO, et al. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat Neurosci. 2013;16(9):1219–1227. doi: 10.1038/nn.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gadd MS, et al. Structural basis for partial redundancy in a class of transcription factors, the LIM homeodomain proteins, in neural cell type specification. J Biol Chem. 2011;286(50):42971–42980. doi: 10.1074/jbc.M111.248559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z, et al. ISL-1 promotes pancreatic islet cell proliferation by forming an ISL-1/Set7/9/PDX-1 complex. Cell Cycle. 2015;14(24):3820–3829. doi: 10.1080/15384101.2015.1069926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W, et al. PDX1 and ISL1 differentially coordinate with epigenetic modifications to regulate insulin gene expression in varied glucose concentrations. Mol Cell Endocrinol. 2016;428:38–48. doi: 10.1016/j.mce.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 68.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158(4):849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z, York NW, Nichols CG, Remedi MS. Pancreatic β cell dedifferentiation in diabetes and redifferentiation following insulin therapy. Cell Metab. 2014;19(5):872–882. doi: 10.1016/j.cmet.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan FC, Brissova M, Powers AC, Pfaff S, Wright CV. Inactivating the permanent neonatal diabetes gene Mnx1 switches insulin-producing β-cells to a δ-like fate and reveals a facultative proliferative capacity in aged β-cells. Development. 2015;142(21):3637–3648. doi: 10.1242/dev.126011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cinti F, et al. Evidence of β-cell dedifferentiation in human type 2 diabetes. J Clin Endocrinol Metab. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo S, et al. Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest. 2013;123(8):3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacovetti C, Matkovich SJ, Rodriguez-Trejo A, Guay C, Regazzi R. Postnatal β-cell maturation is associated with islet-specific microRNA changes induced by nutrient shifts at weaning. Nat Commun. 2015;6:8084. doi: 10.1038/ncomms9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pagliuca FW, et al. Generation of functional human pancreatic β cells in vitro. Cell. 2014;159(2):428–439. doi: 10.1016/j.cell.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rezania A, et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat Biotechnol. 2014;32(11):1121–1133. doi: 10.1038/nbt.3033. [DOI] [PubMed] [Google Scholar]
- 76.Hrvatin S, et al. Differentiated human stem cells resemble fetal, not adult, β cells. Proc Natl Acad Sci U S A. 2014;111(8):3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11(11):1283–1293. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grant GR, et al. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27(18):2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17(15):6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miyazaki J, et al. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 82.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.