Abstract
Loss of β cell identity, the presence of polyhormonal cells, and reprogramming are emerging as important features of β cell dysfunction in patients with type 1 and type 2 diabetes. In this study, we have demonstrated that the transcription factor NKX2.2 is essential for the active maintenance of adult β cell identity as well as function. Deletion of Nkx2.2 in β cells caused rapid onset of a diabetic phenotype in mice that was attributed to loss of insulin and downregulation of many β cell functional genes. Concomitantly, NKX2.2-deficient murine β cells acquired non–β cell endocrine features, resulting in populations of completely reprogrammed cells and bihormonal cells that displayed hybrid endocrine cell morphological characteristics. Molecular analysis in mouse and human islets revealed that NKX2.2 is a conserved master regulatory protein that controls the acquisition and maintenance of a functional, monohormonal β cell identity by directly activating critical β cell genes and actively repressing genes that specify the alternative islet endocrine cell lineages. This study demonstrates the highly volatile nature of the β cell, indicating that acquiring and sustaining β cell identity and function requires not only active maintaining of the expression of genes involved in β cell function, but also continual repression of closely related endocrine gene programs.
Introduction
Type 1 and type 2 diabetes mellitus (T1D and T2D) are chronic conditions in which glycemic control becomes severely dysregulated. Although there are many causes of diabetes, one of the main contributors to the progression of disease is the loss of β cell function and β cell mass, which ultimately leads to an inability to meet metabolic demand (1). Recently, Talchai et al. (2) proposed that loss of β cell identity rather than β cell death accounted for a significant portion of the β cell loss reported during diabetes progression. This landmark study raised the possibility that permanent regulatory mechanisms must be sustained to maintain the fully differentiated functional state of the β cell. Therefore, understanding the mechanisms required to actively maintain β cell identity during adverse metabolic conditions that could lead to diabetes will be essential for the development of interventions and/or therapies to treat the disease.
Studies from many laboratories have identified the extrinsic signaling pathways and intrinsic transcriptional networks needed to generate functional insulin-producing β cells in vitro and in vivo (reviewed in ref. 3). More recently, there has been increasing evidence that several of the developmental regulatory factors that are essential for endocrine lineage specification, including NKX6.1, NEUROD1, and RFX6, are also required for the maintenance of β cell function in the adult (4–9). This suggests that many of the transcriptional regulatory networks that are necessary for the initial specification of β cells may continue to be expressed in the adult β cell to actively maintain β cell identity and function.
We have previously demonstrated that NKX2.2 is absolutely required for the formation of all β cells during development; however, it is not known whether NKX2.2 is also important for β cell function in the adult. NKX2.2 is a highly conserved homeobox transcription factor that regulates cell fate decisions in several tissues, including the pancreas, intestine, and CNS (10–17). During embryogenesis, NKX2.2 is expressed throughout the developing pancreatic epithelium and gradually becomes restricted to the neurogenin 3–expressing (NGN3-expressing) endocrine progenitor population, and subsequently to the α, β, PP, and ε cell lineages (18). Within the adult islet, NKX2.2 expression is maintained in the α, β, and PP cells. Global deletion of Nkx2.2 in mice demonstrated that NKX2.2 is essential for endocrine lineage specification; deletion of Nkx2.2 resulted in decreased formation of α and PP cells, as well as a complete abrogation of β cell specification, leading to severe hyperglycemia and neonatal lethality (10, 11). In humans, loss-of-function mutations in NKX2.2 also resulted in the development of permanent neonatal diabetes, suggesting that NKX2.2 is also important for β cell formation during human fetal development (19).
Although these studies demonstrated a critical role for NKX2.2 in β cell development in mice and humans, the complete absence of β cells precluded assessment of its function in adult islets. To determine whether NKX2.2 is necessary for β cell maturation and/or function, we generated mouse models that allowed constitutive and inducible deletion of the Nkx2.2 gene. Disruption of Nkx2.2 in maturing β cells resulted in the rapid development of diabetes, with a significant decrease in insulin expression and content. Strikingly, the loss of genes associated with β cell identity and function was accompanied by increased expression of genes from alternative islet cell fates. Furthermore, β cells appeared to transdifferentiate to acquire other non–β cell endocrine identities. Deletion of Nkx2.2 in fully differentiated adult β cells also resulted in the very rapid onset of diabetes, and the islets of these mice were also characterized by a loss of β cell identity and the acquisition of δ cell characteristics, confirming the importance of NKX2.2 in both establishing and maintaining β cell identity. Notably, loss of NKX2.2 in human islets also led to a significant dysregulation in the expression of genes known to be essential in glucose homeostasis and β cell identity. Overall, direct regulation of these functions by NKX2.2 confirmed the conservation that exists between mice and humans. These results demonstrate that NKX2.2 is a master regulator that plays a critical role in mouse and human β cells to maintain β cell identity and function in the adult by directly activating β cell genes and preventing reprogramming by actively repressing non–β cell endocrine genes.
Results
Deletion of Nkx2.2 in β cells causes glucose intolerance and impaired insulin secretion.
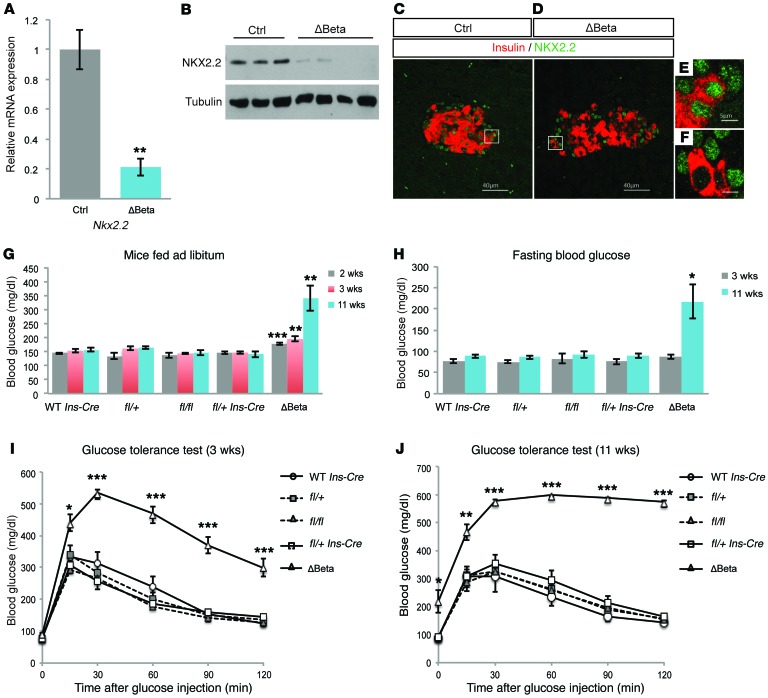
To determine the function of NKX2.2 specifically in β cells, we generated RIP- Nkx2.2fl/fl (hereafter referred to as Nkx2.2ΔBeta) mice (20, 21). For the metabolic studies, we included cohorts of mice carrying each single allele as controls to ensure these individual mutations were not contributing to the observed metabolic defects (Figure 1). Nkx2.2ΔBeta mice displayed efficient deletion of Nkx2.2; reduction of Nkx2.2 transcript can be detected as early as postnatal day 0 (P0), and greater than 80% of the Nkx2.2 RNA transcript was deleted in the insulin-producing cells of 4-week-old Nkx2.2ΔBeta mice versus littermate controls (Figure 1A). Western blot analysis confirmed there was a corresponding reduction of NKX2.2 protein (Figure 1B). Since NKX2.2 is also expressed in several islet endocrine cell populations, we used coimmunofluorescence staining for NKX2.2 and insulin in adult islets to demonstrate the efficient recombination of NKX2.2 specifically in the β cell lineage (Figure 1, C–F).
Figure 1. Deletion of Nkx2.2 in β cells results in diabetes.
(A) Quantitative reverse transcriptase PCR (qRT-PCR) analysis of Nkx2.2 expression in islets isolated from Nkx2.2ΔBeta (ΔBeta) and control (Ctrl) mice at 4 weeks of age (n = 4). **P ≤ 0.01; 2-tailed Student’s t test. (B) Western Blot analysis of isolated islets from Nkx2.2ΔBeta and control mice confirms a reduction of NKX2.2 protein in islets isolated from 4 individual adult Nkx2.2ΔBeta mice. (C and D) Immunofluorescence staining of insulin (red) and NKX2.2 (green) demonstrates the loss of NKX2.2 expression in β cells from Nkx2.2ΔBeta compared with control mice at 4 weeks of age. The white boxes indicate regions of the islet that are shown in higher magnification in E and F. (G) Ad libitum blood glucose levels in 2-week-old male Nkx2.2ΔBeta mice compared with controls (n = 3–16), in 3-week-old mice (n = 5–22), and in 11-week-old mice (n = 6–18). **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test. Each control genotype was examined separately to ensure that the individual Cre and floxed alleles did not cause metabolic phenotypes. (H) Higher fasting blood glucose levels are evident in 11-week-old Nkx2.2ΔBeta mice compared with controls (3-week-old mice: n = 6–23; 11-week-old mice: n = 8–21). *P ≤ 0.05; 2-tailed Student’s t test. (I) Glucose intolerance is observed in Nkx2.2ΔBeta male mice compared with controls at 3 weeks of age (n = 6–23). *P ≤ 0.05, ***P ≤ 0.001; 2-tailed Student’s t test. (J) Glucose intolerance becomes more severe at 11 weeks of age in Nkx2.2ΔBeta male mice compared with control mice (n = 8–21). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test.
In contrast to Nkx2.2-null mice, which develop severe hyperglycemia and die shortly after birth (10), both female and male Nkx2.2ΔBeta mice survive into adulthood. The adult mice appear overtly indistinguishable from their littermate controls and exhibit no apparent difference in BW (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI88017DS1). However, by 2 weeks of age, the male Nkx2.2ΔBeta mice displayed significantly higher blood glucose levels than controls. The hyperglycemic phenotype became exacerbated with age, reaching mean glucose levels of 339.3 mg/dl at 11 weeks (Figure 1G). Fasting blood glucose levels in the Nkx2.2ΔBeta mice were also significantly elevated (≥217.3 mg/dl) at 11 weeks of age (Figure 1H).
Consistent with the elevated ad libitum and fasting blood glucose levels, 3-week-old Nkx2.2ΔBeta mice were unable to effectively clear a glucose bolus (Figure 1I). The metabolic impairment worsened with age, suggesting there was a progressive loss of β cell function in Nkx2.2ΔBeta mice (Figure 1J). Similar phenotypes were observed in female Nkx2.2ΔBeta mice; however, hyperglycemia and glucose intolerance were manifested at slightly older ages and with less intensity (Supplemental Figure 1, C and D). To eliminate the possibility that the observed glucose intolerance was due to an inability of the peripheral tissues to take up glucose, we performed insulin tolerance tests at 3 weeks of age. As expected for a β cell–specific genetic modification, no difference in insulin resistance was detected between Nkx2.2ΔBeta mice and controls (Supplemental Figure 1E).
Nkx2.2ΔBeta mice display reduced numbers of insulin-producing cells.
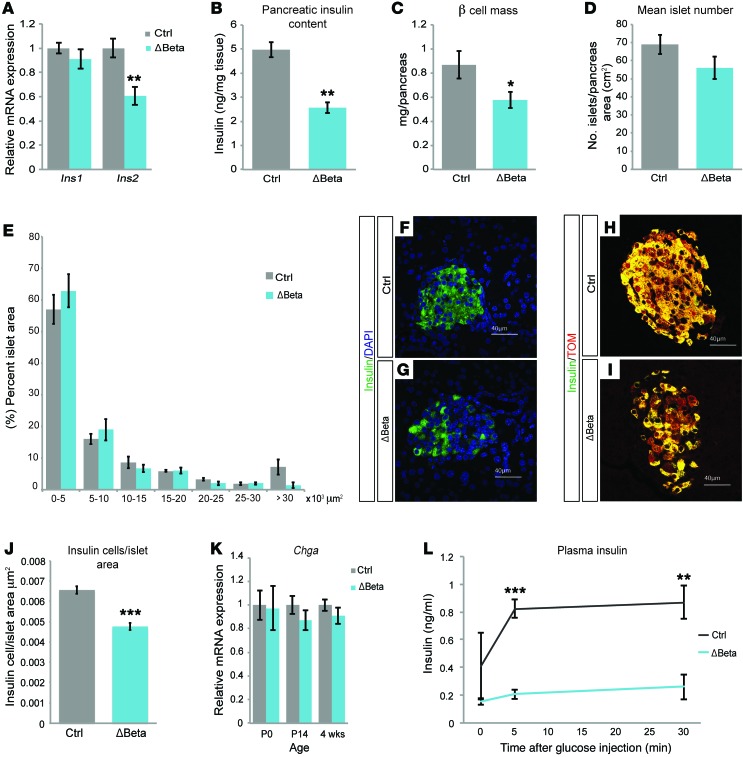
Consistent with previous studies demonstrating that NKX2.2 directly regulates insulin expression (22), β cell deletion of Nkx2.2 resulted in a significant reduction of insulin 2 gene expression and total insulin content in 4-week-old Nkx2.2ΔBeta mice (Figure 2, A and B). Insulin 1 gene expression was unaffected, suggesting that NKX2.2 is dispensable for its expression and the reduction in insulin content is primarily due to the loss of insulin 2. There was also a 33.6% decrease in β cell mass in the Nkx2.2ΔBeta mice compared with their littermate controls (Figure 2C); however, this did not appear to be due to decreases in the number of islets or islet size (Figure 2, D and E). We did, however, observe normal-sized islets containing a disproportionate number of cells lacking insulin (Figure 2, F and G). To determine whether the loss of insulin content without a corresponding change in islet size was due to the retention of a β cell population that no longer produced insulin, we introduced the R26R-Tomato allele into the Nkx2.2ΔBeta mice to genetically label the β cell lineage regardless of its insulin-producing status. Although the Tomato reporter was coexpressed with insulin in the majority of control β cells, the Nkx2.2ΔBeta mice contained a significant number of Tomato-expressing cells that had significantly less insulin expression (Figure 2, H and I). Furthermore, enumeration of the number of insulin-expressing cells per islet area confirmed the loss of insulin without a corresponding reduction in islet cell numbers (Figure 2J), suggesting that a loss of insulin content per cell and not cell loss causes the observed decrease in β cell mass. Consistent with a lack of islet cell loss, we also did not detect any reduction in expression of the pan-endocrine marker chromogranin A (Chga) at any ages tested (Figure 2K).
Figure 2. Lineage tracing in Nkx2.2ΔBeta mice indicates loss of insulin expression in Nkx2.2ΔBeta β cells.
(A) qPCR analysis of insulin 1 (Ins1) and insulin 2 (Ins2) mRNA expression in islets from Nkx2.2ΔBeta 4-week-old mice (ΔBeta) compared with controls (n = 4). **P ≤ 0.01; 2-tailed Student’s t test. (B) Nkx2.2ΔBeta 4-week-old mice have an approximately 50% reduction in pancreas insulin content compared with controls (n = 3). **P ≤ 0.01; 2-tailed Student’s t test. (C) Loss of NKX2.2 results in decreased β cell mass at 4 weeks of age (n = 3). *P ≤ 0.05; 2-tailed Student’s t test. (D) Nkx2.2ΔBeta 4-week-old mice have no statistical difference in islet numbers compared with controls (n = 3); 2-tailed Student’s t test. (E) Islet size of Nkx2.2ΔBeta mice compared with controls was not statistically different at 4 weeks of age (n = 3); 2-tailed Student’s t test. There was a trend down in the number of largest islets, but the difference did not reach significance. (F and G) Immunofluorescence staining of insulin (green) and DAPI (blue) shows a decrease of insulin expression in islet cells from Nkx2.2ΔBeta 4-week-old mice compared with controls. (H and I) Immunofluorescence staining of insulin (green) and Tomato (red) shows a decrease or absence of insulin expression in Tomato-expressing β cell lineages. (J) Nkx2.2ΔBeta 4-week-old mice have fewer insulin-positive cells per islet area compared with controls (n = 3). ***P ≤ 0.001; 2-tailed Student’s t test. (K) qPCR analysis of chromogranin A (Chga) mRNA expression shows no statistical significance in pancreas from Nkx2.2ΔBeta mice compared with controls at P0 and 2 weeks of age (n = 4), as well as in islets from Nkx2.2ΔBeta 4-week-old mice compared with controls (n = 4–7). (L) Insulin levels are significantly lower in Nkx2.2ΔBeta mice after a glucose stimulus compared with controls (n = 3–4). **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test.
Although Nkx2.2ΔBeta mice display a significant reduction in the number of insulin-producing cells, this degree of β cell loss does not usually result in hyperglycemia (23, 24). To determine whether islet function was affected, we performed glucose-stimulated insulin secretion assays on young and old Nkx2.2ΔBeta mice. Surprisingly, although these islets contain significantly fewer insulin-producing cells and less overall insulin content, they were able to secrete normal amounts of insulin during a 1-hour glucose challenge in vitro (Supplemental Figure 1F); however, the ability of Nkx2.2ΔBeta islets to secrete basal insulin declined with age (Supplemental Figure 1G). Furthermore, although the Nkx2.2ΔBeta islets were able to compensate in response to a single dose of glucose in vitro, they were unable to mount an insulin response to an in vivo glucose challenge (Figure 2L), indicating that Nkx2.2ΔBeta mutant β cells have additional defects that contribute to the severe metabolic dysfunction in the mutant animals (see Discussion).
Nkx2.2 is essential for the acquisition of monohormonal β cell identity.
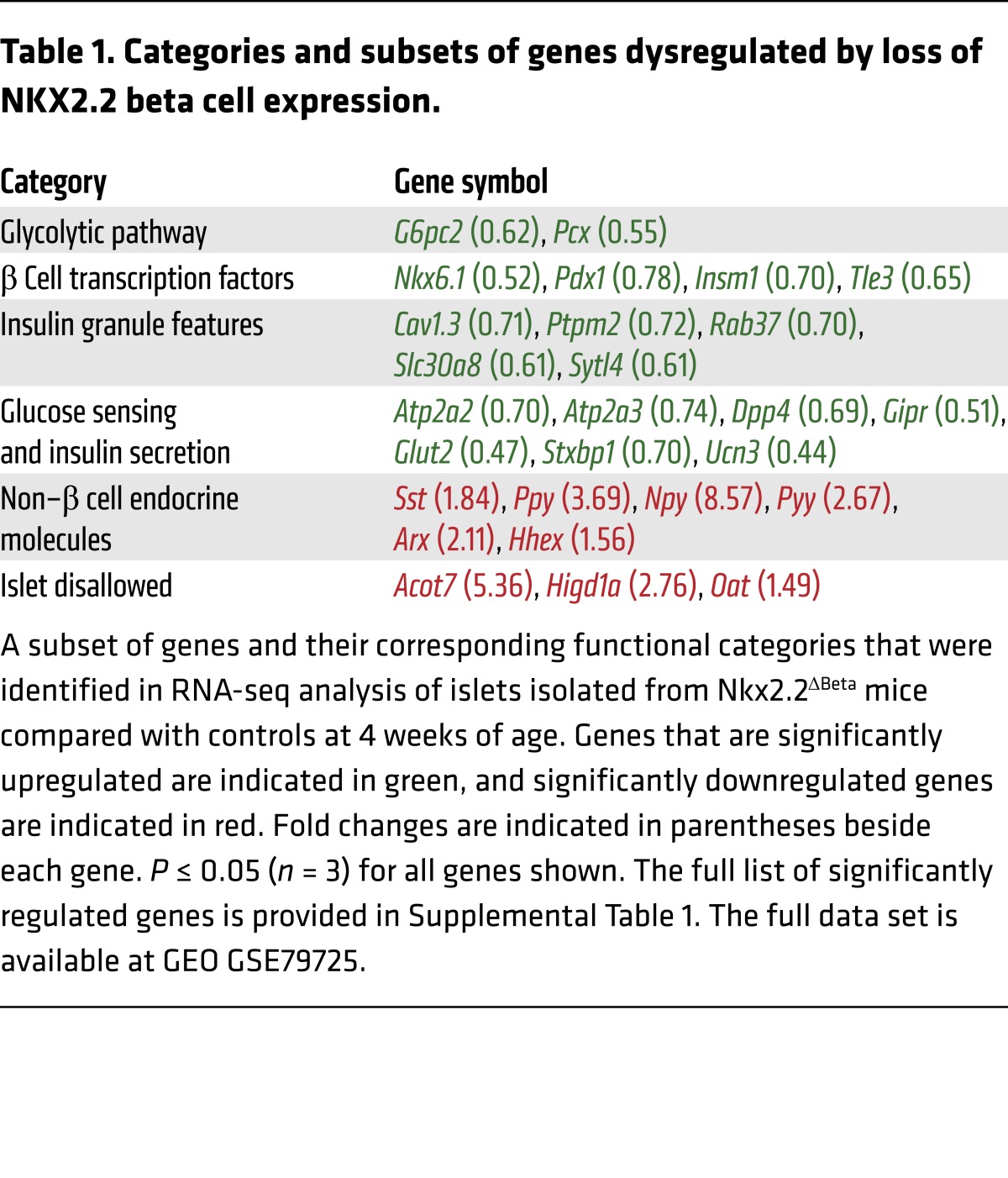
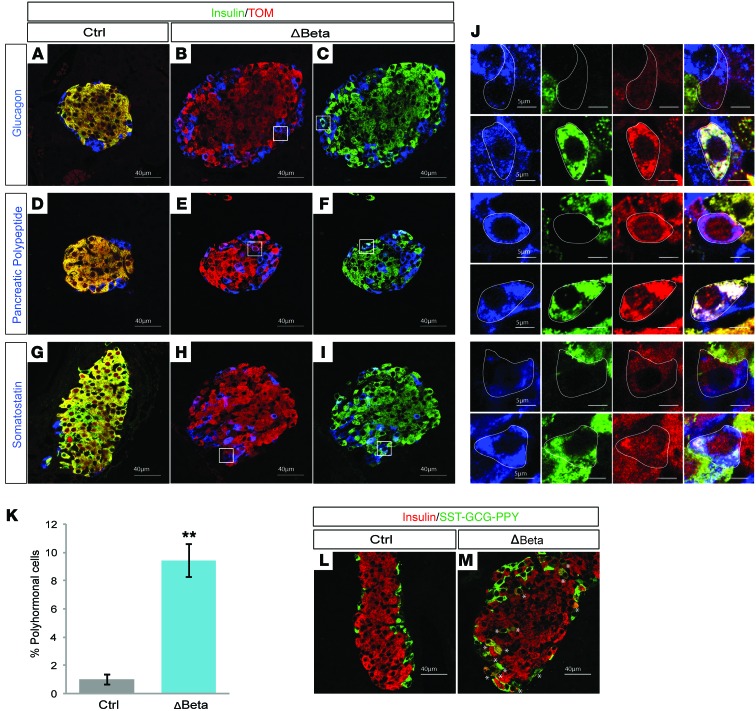
To identify the primary molecular changes that were causing β cell dysfunction after deletion of Nkx2.2, we assessed the transcriptome of isolated islets extracted from Nkx2.2ΔBeta mice and their littermate controls at 4 weeks of age. This analysis was performed shortly after the onset of hyperglycemia to minimize secondary effects caused by a chronically impaired metabolic environment. Consistent with the observed β cell dysfunction, we found several essential β cell genes, including those encoding β cell transcription factors, glucose transporter 2 (Glut2), Slc30a8, insulin granule proteins, and additional components of the insulin secretory pathway, to be significantly downregulated (Table 1). We validated a subset of these gene changes in islets at the protein level, including the reduction of 2 of the most functionally important β cell factors, GLUT2 and NKX6.1 (Supplemental Figure 2, A–F). Although NKX6.1 could still be detected in a number of insulin-expressing cells, it appeared to be expressed at lower levels. Notably, in addition to the loss of many essential β cell genes in the Nkx2.2ΔBeta mice, there was a significant upregulation of the non–β cell pancreatic endocrine hormones, including somatostatin (SST), pancreatic polypeptide, and glucagon (Table 1). Since we had not observed an overall increase in islet size that could account for increased numbers of the non-β endocrine cells that express these hormones, we immunostained islets with insulin and SST, pancreatic polypeptide, or glucagon to determine their cell localization. In addition, we lineage-traced the mutant β cells using the R26R-Tomato reporter to assess whether the β cell population in the Nkx2.2ΔBeta mice had acquired expression of the non–β cell endocrine hormones and/or novel cellular identities. This analysis revealed that not only were the Nkx2.2ΔBeta islets losing β cell gene expression, but a significant number of β cells were coexpressing insulin and either glucagon, pancreatic polypeptide, or SST. Of particular note, NKX6.1 expression became extinguished from these bihormonal cells (Supplemental Figure 2F). In addition, there were populations of lineage-labeled β cells that appeared to be completely reprogrammed; they no longer expressed insulin, but now expressed an alternative endocrine hormone (Figure 3, A–J). These data revealed that loss of Nkx2.2 is sufficient to cause partial and/or complete transdifferentiation of β cells into the other islet cell types.
Table 1. Categories and subsets of genes dysregulated by loss of NKX2.2 beta cell expression.
Figure 3. NKX2.2 is essential for the establishment of β cell identity.
(A–J) Immunofluorescence staining of pancreata from Nkx2.2ΔBeta (ΔBeta) and control mice at 4 weeks of age. White boxes indicate regions that are magnified in the panels in J. (A, D, and G) Representative images of control islets colabeled with insulin (green), Tomato (TOM; red), and either glucagon (A), pancreatic polypeptide (D), or somatostatin (G) (blue). (B, E, and H) Representative images of Nkx2.2ΔBeta islets colabeled with Tomato (TOM; red) and either glucagon (B), pancreatic polypeptide (E), or somatostatin (H) (blue). (C, F, and I) Representative images of Nkx2.2ΔBeta islets colabeled with insulin (green) and either glucagon (C), pancreatic polypeptide (F), or somatostatin (I) (blue). (J) Images of representative single lineage-labeled insulin-positive β cells expressing either glucagon, pancreatic polypeptide, or somatostatin and insulin-negative β cells expressing either glucagon, pancreatic polypeptide, or somatostatin. The channels are separated out from left to right: glucagon, pancreatic polypeptide, and somatostatin (blue), insulin (green), and Tomato (red). The final column shows the merged channels. (K) Quantification analysis shows that an average of 10% of β cells in Nkx2.2ΔBeta compared with control mice at 4 weeks of age coexpress insulin and either glucagon, somatostatin (Sst), or pancreatic polypeptide (n = 3). **P ≤ 0.01; 2-tailed Student’s t test. (L and M) Representative images showing immunofluorescence analysis of pancreata from Nkx2.2ΔBeta and control mice stained with guinea pig anti-insulin antibody (red) and a combination of rabbit anti–pancreatic polypeptide (PPY), rabbit anti-somatostatin (SST), and rabbit anti-glucagon (GCG) antibodies (green). Asterisks indicate the location of polyhormonal cells, which are present at both the mantle and the core of the islet.
It is important to note that, unlike in the Nkx2.2-null mice, RNA sequencing (RNA-seq) analysis did not reveal an increase in ghrelin expression. Immunofluorescent staining confirmed there was no change in numbers of ghrelin cells at birth or in 2-week-old mice (Supplemental Figure 2, G–J). In older mice, ghrelin-expressing cells were not evident in either WT or mutant mice. These results confirm our previous studies of the Nkx2.2-null mice that indicated that the observed increase in ghrelin-producing cells at the expense of β cells was due to a cell fate lineage decision during development and there was no evidence of β cells converting into ghrelin cells (10, 14).
Quantification of the bihormonal and polyhormonal populations at 4 weeks of age revealed that approximately 10% of the β cells had acquired multihormonal features (Figure 3, K–M). Detailed immunofluorescence analysis of the Nkx2.2ΔBeta mutant islets also revealed extraordinary heterogeneity in the Nkx2.2ΔBeta islets. At any given age, we observed constant relative ratios of the different populations of Tomato lineage-labeled β cells that were either expressing only insulin, expressing no insulin, expressing insulin with other hormones, or expressing other endocrine hormones without insulin (Figure 3J). Notably, we did not observe evidence of β cell dedifferentiation; Ngn3, Sox9, or other progenitor markers were not upregulated in the β cell lineage (Supplemental Table 1).
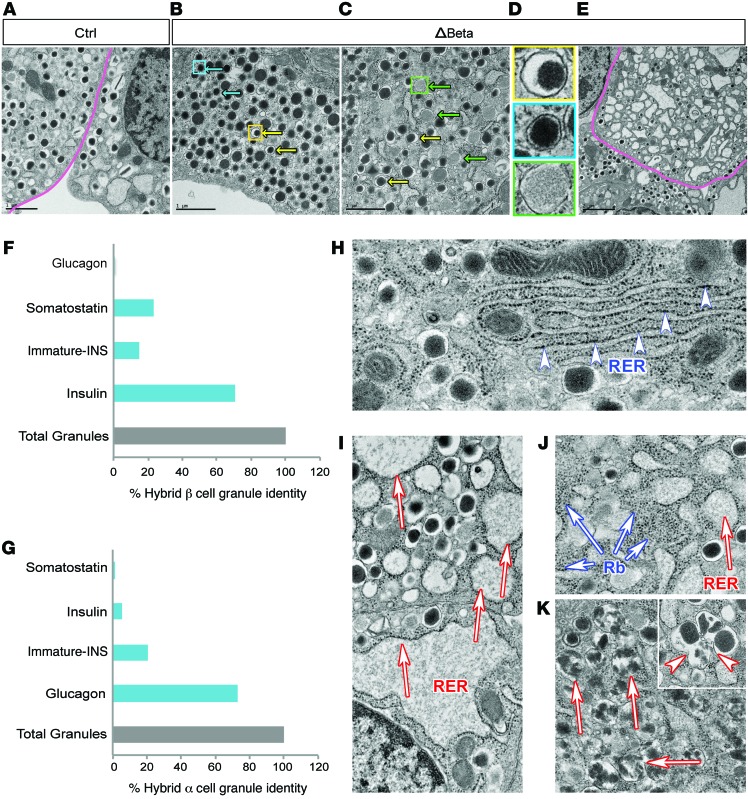
To characterize the different β cell populations at the single-cell level, electron microscopy was performed on control and Nkx2.2ΔBeta mutant islets. As predicted by the light microscopy analysis, control islets contained endocrine cells displaying insulin granules with characteristic electron-dense crystal cores (Figure 4A). In contrast, the mutant islets were populated by approximately 11.3% of β cells containing both insulin granules and either glucagon- or SST-like granules (Figure 4, B–D). Quantification of granule identity within these hybrid β cells confirmed that at least 20% of the granules appeared to be SST-like, and 0.5% appeared to be glucagon-like (Figure 4F). Additionally, we quantified the granule content of hybrid α cells, and found that 5.2% of these cells contained insulin-appearing granules and 1.5% contained SST-appearing granules (Figure 4G). Remarkably, these results suggest that many of the Nkx2.2ΔBeta β cells have acquired a fully hybrid identity that is characterized not only by the expression of more than 1 endocrine hormone, but also by the likely expression of their corresponding granule packaging machinery.
Figure 4. Nkx2.2ΔBeta islet cells exhibit ultrastructural alterations compatible with a disrupted secretory granule identity and morphology, accompanied by ER stress.
(A–E) Transmitted electron microscopy (TEM) of pancreatic islets reveals a combination of insulin and either glucagon- or somatostatin-like granules in the same cell in Nkx2.2ΔBeta (ΔBeta) mice. Magenta lines outline individual β cells. (B and C) Granule identity is indicated by colored arrows: glucagon (blue), insulin (yellow), and somatostatin (green). (D) Framed insets indicate regions magnified in B (yellow-framed inset shows an insulin granule; blue-framed inset shows a glucagon granule) and C (green-framed inset shows a somatostatin granule). (E) Representative image of a β cell from Nkx2.2ΔBeta islets with severely altered secretory granules. (F) Quantification of individual granule identity in hybrid β cells from Nkx2.2ΔBeta mice (n = 93 hybrid β cells with a total of 10,533 granules counted). (G) Quantification of individual granule identity in hybrid α cells from Nkx2.2ΔBeta mice (n = 31 hybrid α cells with 3,297 total granules counted). (H–K) TEM images of single cells from control and Nkx2.2ΔBeta mice. (H) Representative image of normal-looking rough endoplasmic reticulum (RER) in control islets (arrowheads). (I and J) Islet cells from Nkx2.2ΔBeta mice show enlarged RER cisternae (red-stroked arrows) and increased free ribosomes not associated with RER (blue-stroked arrows). (K) β Cells in Nkx2.2ΔBeta mice contained large populations of insulin granules that exhibited an altered crystallization pattern (red-stroked arrows), and numerous coalescent granules (inset, arrowheads).
The mutant islets also contained a large number of cells that were devoid of granules and displayed features of altered endoplasmic reticulum with numerous enlarged cisternae (Figure 4, E and I–K, red-stroked arrows), whose membranes exhibited reduced ribosome content and increased frequency of free ribosomes in the cytosol (Figure 4J, blue-stroked arrows) compared to control cells (Figure 4H). Many β cells from Nkx2.2ΔBeta mice also contained insulin granules that exhibited an altered crystallization pattern (Figure 4K, red stroked arrows), and numerous coalescent granules (Figure 4K, inset, arrowheads), suggesting early stages of increased secretory vesicle autophagy. Collectively, these ultrastructural alterations indicate that in the absence of NKX2.2, many of the β cells became severely dysfunctional. It was not possible to determine whether these cells derived from the polyhormonal populations or arose independently. Surprisingly, despite the high degree of observed β cell dysmorphogenesis and apparent dysfunction, there was little evidence of β cell death by caspase-3 staining at 4 weeks (GDG, unpublished observations), which was consistent with the maintenance of normal islet size in the Nkx2.2ΔBeta mice (Figure 2E) and the preservation of WT levels of Chga expression (Figure 2K).
Nkx2.2 directly activates β cell genes and represses non–β cell features.
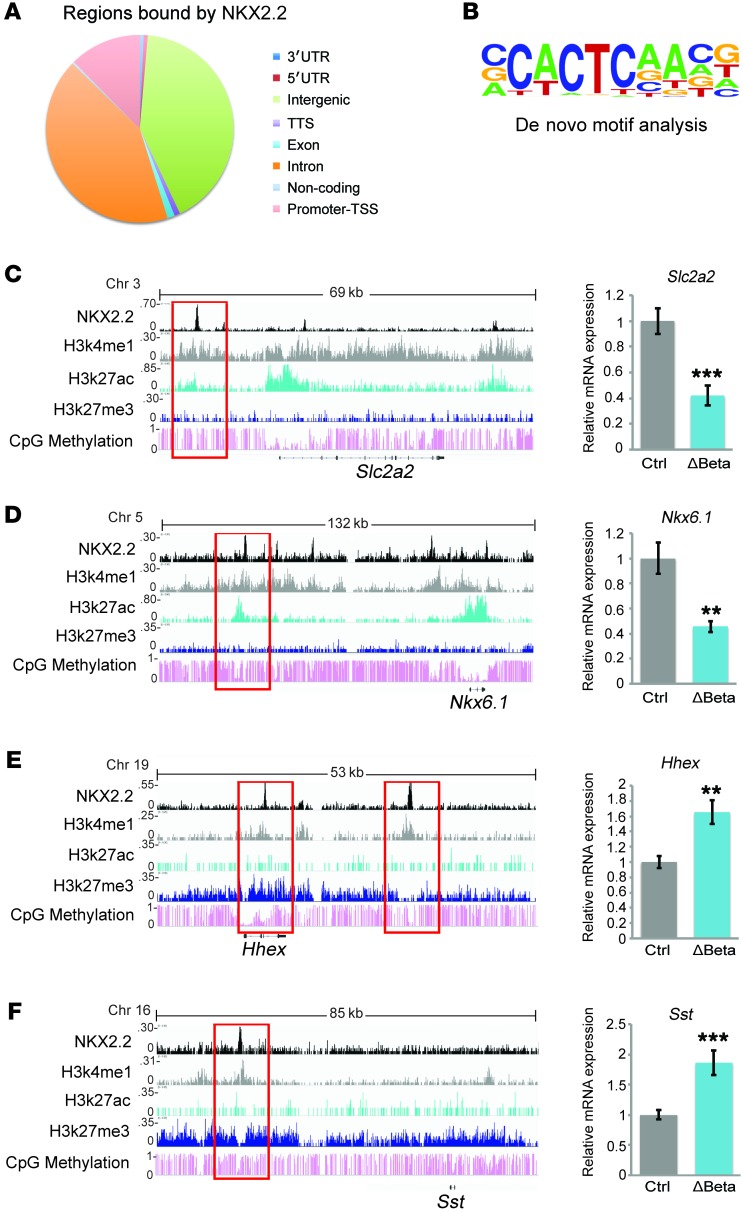
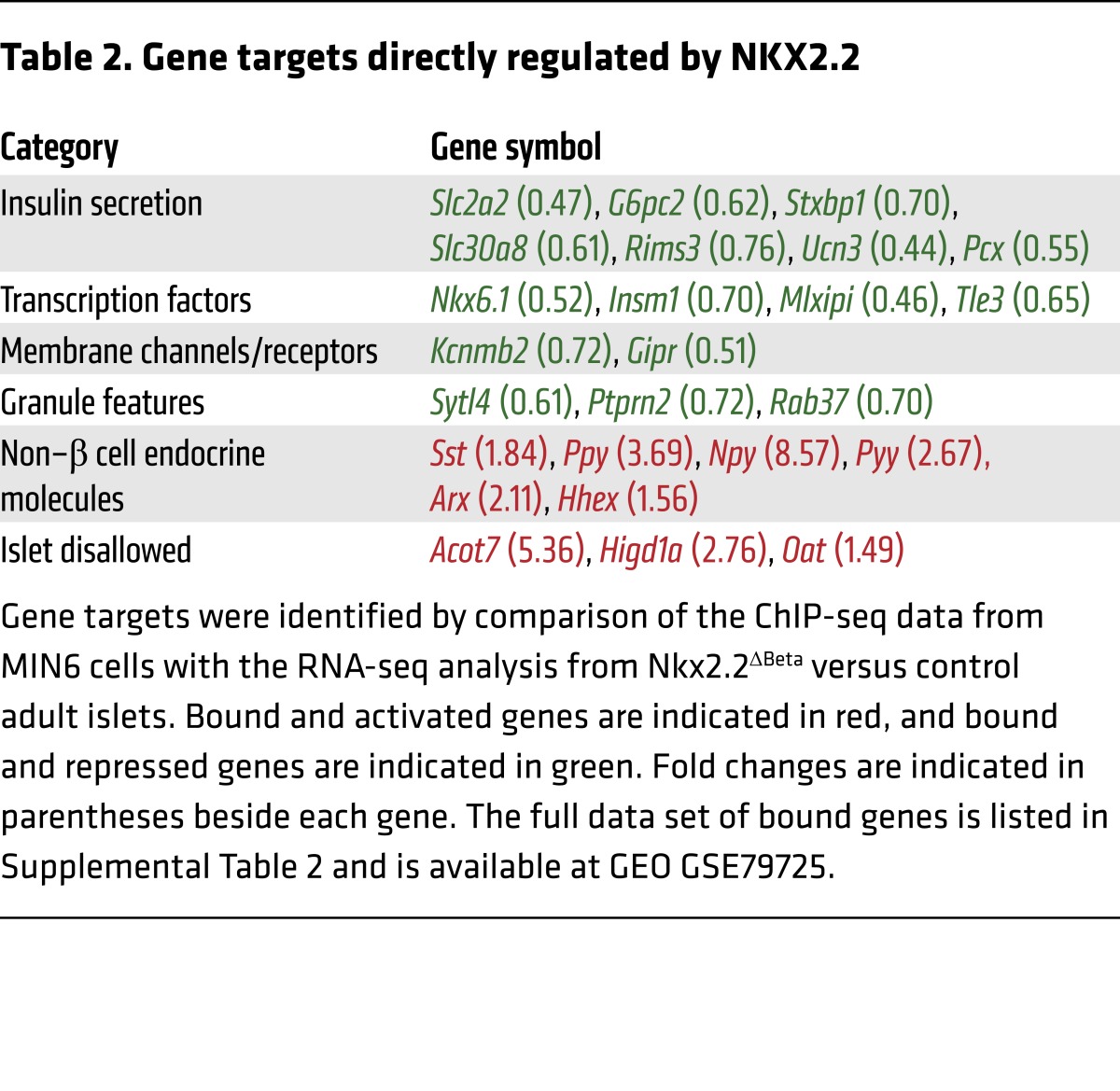
The transdifferentiation phenomenon observed in the Nkx2.2ΔBeta mice could be directly caused by the deletion of Nkx2.2 or could be secondary to the loss of β cell identity. To distinguish these possibilities, we performed ChIP sequencing (ChIP-seq) analysis in MIN6 cells (25) to determine how NKX2.2 directly impacts the regulatory networks required to maintain β cell identity. Similarly to previous studies in human β cells (26), NKX2.2 predominantly bound distal intergenic regions of the genome, rather than at promoter elements (Figure 5A). De novo motif analysis showed CACTC to be the core motif sequence that is preferably bound by NKX2.2 in 46.42% of its targets (P < 1 × 10–217) (Figure 5B). Consistent with the Nkx2.2ΔBeta phenotype, NKX2.2 binds and activates β cell genes that are essential for glucose uptake and insulin secretion (Slc2a2, G6pc2, Ucn3, and Slc30a8), insulin granule homeostasis (Sytl4, Ptprn2, and Rab37), and key β cell transcription factors (Nkx6.1, Insm1, and Tle3). Interestingly, NKX2.2 was also found to directly bind and repress non–β cell endocrine genes, including endocrine hormones (Sst, Ppy, Pyy, and Npy) and islet-disallowed genes (Oat, Acot7, and Higd1a), as well as the α and δ cell determination factors Arx and Hhex (Table 2 and Figure 5, C–F). In agreement with previous studies that have suggested that NKX2.2 functions as an activator and repressor (27, 28), this analysis revealed that within the β cell, NKX2.2 simultaneously activates β cell gene programs and represses non–β cell programs. These data are consistent with the polyhormonal phenotype observed in the Nkx2.2ΔBeta mice and clarify that NKX2.2 maintains β cell identity and function by directly activating β cell genes and repressing non–β cell endocrine genes.
Figure 5. NKX2.2 directly activates important β cell genes and actively represses non-β islet endocrine genes.
(A) Distribution of regions bound by NKX2.2 within the genome obtained from ChIP-seq analysis performed in a MIN6 cell line (n = 3). (B) The consensus NKX2.2 binding motif identified through de novo motif analysis. (C–F) ChIP-seq analysis identified binding of NKX2.2 to active or repressed enhancers. qRT-PCR analysis confirms differential expression of direct NKX2.2 gene targets in islets from Nkx2.2ΔBeta (ΔBeta) compared with control mice (n = 5–7). **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test.
Table 2. Gene targets directly regulated by NKX2.2.
The ability of NKX2.2 to function as both an activator and a repressor in a single cell may be dependent on the underlying chromatin features present in the vicinity of NKX2.2 binding. To characterize the regions that appeared to be activated or repressed by NKX2.2, we compared the association of NKX2.2 binding peaks with chromatin marks present in β cells isolated from young mice (29). Interestingly, consistent with the relatively equal number of activated (49.2%) and repressed (50.8%) targets (Supplemental Figure 3A), NKX2.2 appeared to equally associate with active enhancers (H3K4me+ and H3K27ac+) and poised enhancers (H3K4me+, H3K27ac–, H3K27me3–). Only a small number of NKX2.2-bound regions were associated with repressive marks (H3K4me+ and H3K27me3+) (Supplemental Figure 3B). This would suggest, not unexpectedly, that NKX2.2 binds to active enhancers to regulate the expression of β cell–specific genes. However, it also suggests that NKX2.2 function is equally necessary to maintain repression of the lineage-related non–β cell endocrine genes that are associated with poised enhancers that have been implicated in facilitating islet cell plasticity (30).
Nkx2.2 maintains β cell function and identity in mature β cells.
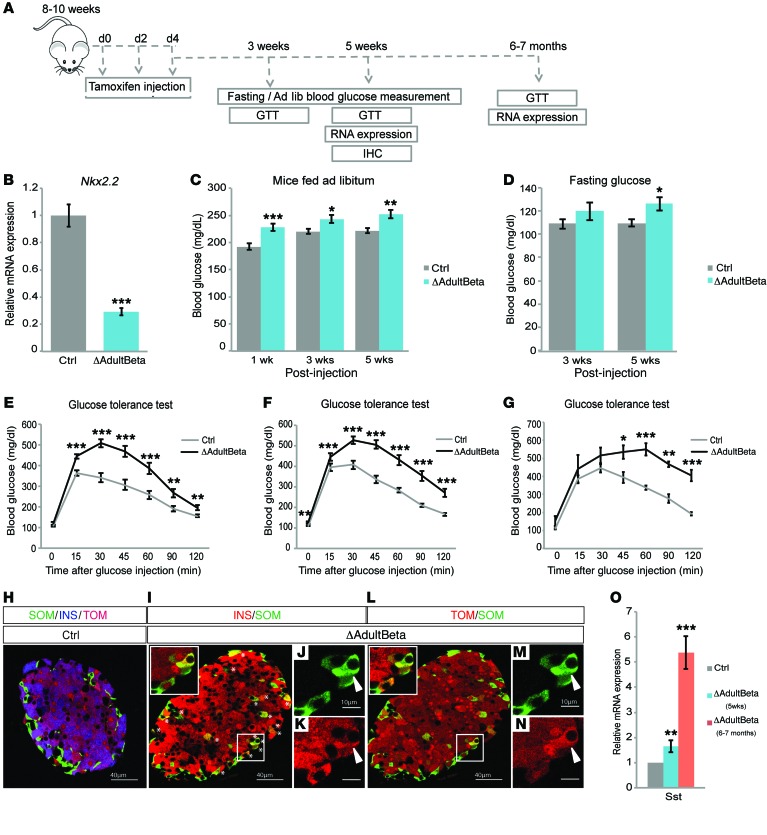
Although analysis of the Nkx2.2ΔBeta mice was performed in adult mice, it is possible that constitutive deletion of Nkx2.2 in the β cells during their normal maturation process compromises the initial acquisition and “locking in” of a stable β cell identity. To assess the importance of NKX2.2 in maintaining β cell function and identity not only during the maturation process of the β cell but also after they have fully differentiated, we deleted Nkx2.2 in adult β cells. Adult Nkx2.2fl/fl MIP-CreERT (31) (hereafter referred to as Nkx2.2ΔAdultBeta) mice were given i.p. injections of tamoxifen every other day for 5 days (Figure 6A). We were able to consistently achieve approximately 70% deletion of Nkx2.2 mRNA from the treated mice (Figure 6B).
Figure 6. NKX2.2 is essential for the maintenance of β cell identity and function in adult β cells.
(A) Schematic of experimental design. (B) qRT-PCR analysis demonstrates significant deletion of Nkx2.2 in isolated islets from Nkx2.2ΔAdultBeta mice compared with controls 5 weeks after the last injection (n = 5–7). ***P ≤ 0.001; 2-tailed Student’s t test. (C) Deletion of Nkx2.2 increases ad libitum blood glucose in Nkx2.2ΔAdultBeta compared with controls starting at 1 week after the last injection (n = 16–20). *P ≤ 0.05, **P = ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test. (D) Fasting blood glucose levels become significantly increased 5 weeks after the last injection in Nkx2.2ΔAdultBeta compared with controls (n = 12–15). *P ≤ 0.05; 2-tailed Student’s t test. (E) Nkx2.2ΔAdultBeta mice are glucose intolerant at 3 weeks after the last injection compared with controls (n = 12–15). **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test. (F) Glucose intolerance becomes more severe 5 weeks after the last injection in Nkx2.2ΔAdultBeta compared with controls (n = 12–15). **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test. (G) Glucose intolerance continues to worsen with age; glucose tolerance test at 6–7 months after the last injection in Nkx2.2ΔAdultBeta compared with controls (n = 3–4). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test. (H–N) Immunofluorescence staining of pancreata from Nkx2.2ΔAdultBeta and control mice 5 weeks after the last injection. Insets are magnifications of selected areas indicated by white boxes showing coexpression of insulin (INS) and somatostatin (SST) (J and K) and coexpression of SST and Tomato (TOM) (M and N). The images in I–N show the same islet that was costained with insulin, somatostatin, and Tomato. The channels were separated out and false-colored to more clearly indicate coexpressed cells. qRT-PCR analysis confirms the increase in somatostatin over time in Nkx2.2ΔAdultBeta compared with controls (n = 4–6). Asterisks indicate the location of insulin/somatostatin-copositive cells. (O) qRT-PCR analysis confirms the increase in somatostatin over time in Nkx2.2ΔAdultBeta compared to controls (n = 4-6). **P ≤ 0.01, ***P ≤ 0.001; 2-tailed Student’s t test.
Interestingly, depletion of Nkx2.2 in the adult β cell resulted in the rapid onset of diabetes. Ad libitum blood glucose levels of Nkx2.2ΔAdultBeta mice were significantly increased within 1 week of the last injection, and continued to worsen with age (Figure 6C). Correspondingly, a significant increase in the fasting blood glucose levels was observed, although, similarly to that in the Nkx2.2ΔBeta mice (Figure 1H), this increase occurred at a slightly later stage (Figure 6D). Furthermore, the Nkx2.2ΔAdultBeta mice were unable to effectively clear glucose following a glucose challenge (Figure 6E). This phenotype also became more severe with age (Figure 6, F and G).
Despite the defective glucose response in the Nkx2.2ΔAdultBeta mice, there was no obvious change in islet size or number of islets (Figure 6, H–N). Similar to that in the Nkx2.2ΔBeta mice, there was also decreased expression of several important β cell genes, including Nkx6.1 and Glut2, and increased expression of islet-disallowed genes such as Acot7 (Supplemental Figure 4). To investigate whether the observed islet dysfunction in the Nkx2.2ΔAdultBeta mice was also due to the development of polyhormonal cells, we introduced the R26R-Tomato reporter to lineage-trace the β cell lineage and immunostained the mutant islets with insulin and either glucagon, SST, or pancreatic polypeptide. Surprisingly, we were only able to identify cells coexpressing insulin and SST (Figure 6, I–K); there were no cells coexpressing insulin and the other endocrine hormones. Consistently, we also observed increased Sst gene expression with age (Figure 6O). These data suggest that NKX2.2 is essential in the acquisition and maintenance of β cell identity, during development and in the adult. However, they also imply that the level of β cell plasticity becomes more restricted with age, and only retains the ability to acquire δ cell characteristics.
Nkx2.2 function is conserved in human islets.
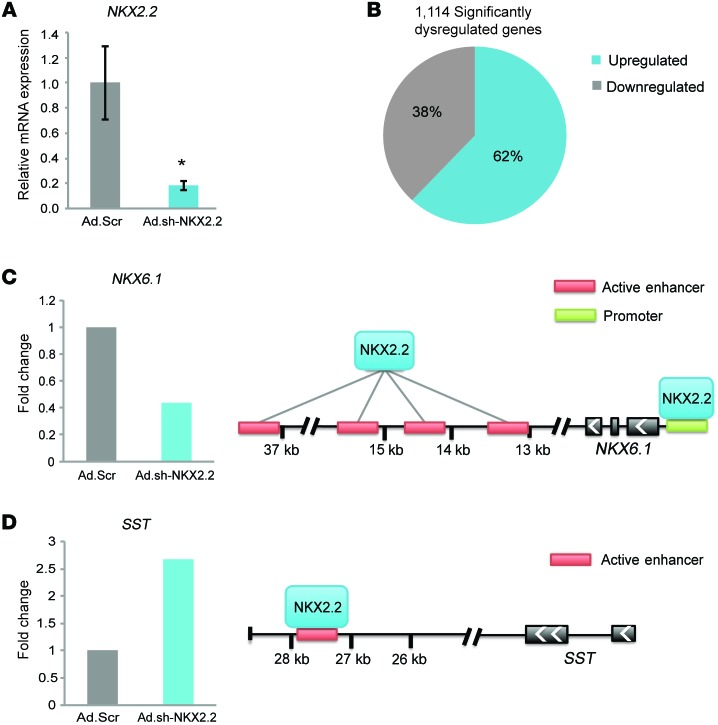
To investigate whether NKX2.2 is also important for the function and identity of human β cells, islets from 3 individuals were infected with adenovirus encoding an shRNA directed against human NKX2.2 or a scrambled shRNA control. We were able to achieve an 80% reduction in NKX2.2 expression, which led to significant (P < 0.05) gene expression changes of which approximately 38% were downregulated and approximately 62% were upregulated (Figure 7, A and B). Since NKX2.2 is removed from all islet cells, the higher ratio of repressed versus activated genes in the human islets compared with the Nkx2.2ΔBeta mice may indicate that NKX2.2 functions predominantly as a repressor in the other islet cell types, including the α cells, which express high levels of NKX2.2. Future studies to delete Nkx2.2 specifically in the postnatal α cells will help clarify the molecular activity of NKX2.2 in the α cell population.
Figure 7. NKX2.2 function is conserved in human islets.
(A) qRT-PCR analysis reveals significant deletion of NKX2.2 in human islets transduced with Ad.sh-NKX2.2 compared with scramble shRNA control vector (Ad.Scr) (n = 3). *P ≤ 0.05; 2-tailed Student’s t test. (B) One thousand one hundred fourteen genes were differentially regulated in human islets when NKX2.2 was reduced. The graph indicates the percentage distribution of genes differentially expressed (P ≤ 0.05) from adenoviral-mediated deletion of NKX2.2 in human islets compared with scramble control. (C and D) Binding sites shown to be activated or repressed in human islets through ChIP-seq analysis show direct targets of NKX2.2 that become dysregulated after its deletion. Fold change values obtained from RNA-seq data (n = 3). T2D-associated genes (43).
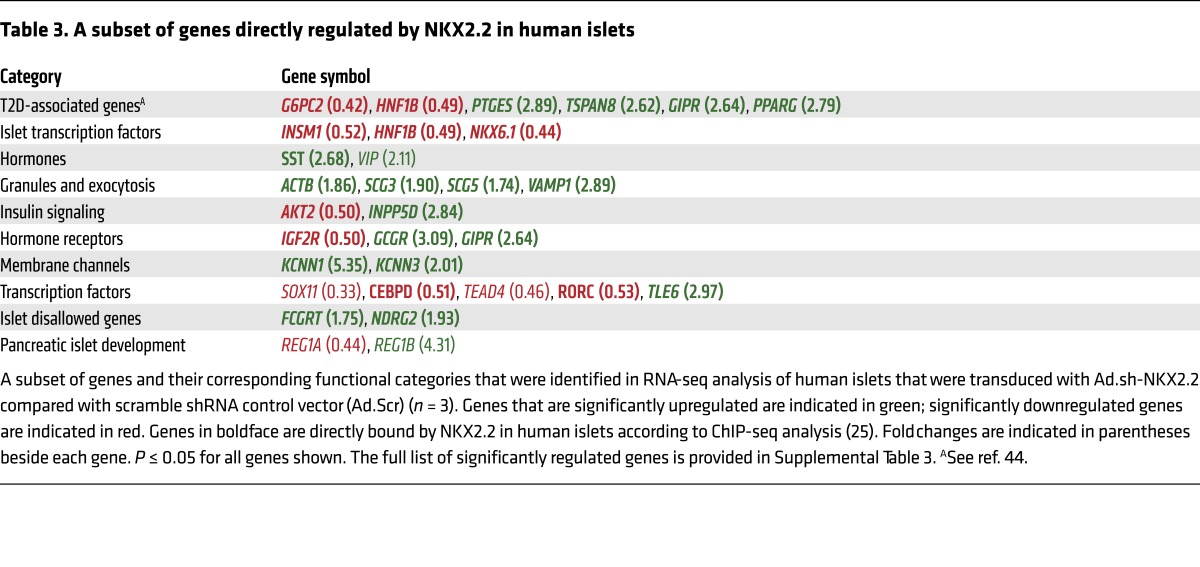
Comparison of the gene expression changes with previously published ChIP-seq analysis of NKX2.2 binding sites in human islets (26) demonstrated that, similarly to mouse NKX2.2, human NKX2.2 functions as both an activator and a repressor. Furthermore, it confirmed its importance in the regulation of factors essential for glucose homeostasis and insulin secretion, as well as its regulation of genes associated with T2D (Table 3). Notably, several important NKX2.2 gene targets were conserved, including NKX6.1, which was bound and downregulated (Figure 7C), and SST, which was bound and upregulated after loss of NKX2.2 (Figure 7D). Collectively, these data demonstrate a conserved role for NKX2.2 in maintaining β cell function and identity by activating important β cell genes and actively repressing non–β cell features.
Table 3. A subset of genes directly regulated by NKX2.2 in human islets.
Discussion
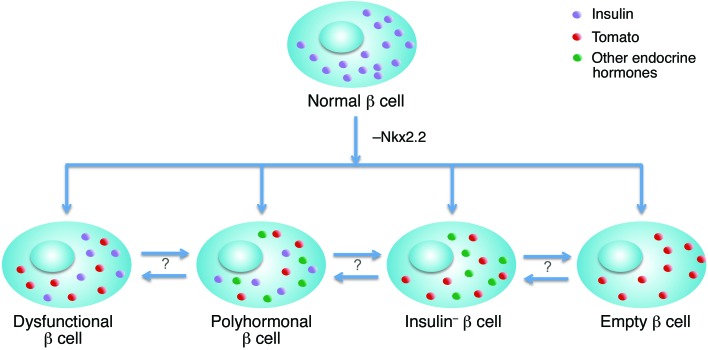
There is increasing evidence that loss of β cell identity and the acquisition of bihormonal and polyhormonal cells are occurring in the islets of patients with type 1 and type 2 diabetes (32–34). However, the molecular basis of these reprogramming events has not been elucidated, and it is not known whether these events participate in the initiation of disease or represent a condition that is secondary to disease progression. In the present study, we uncover the essential role of NKX2.2 in the maturation and maintenance of adult mouse and human β cell function and identity. β Cell deletion of Nkx2.2 destabilized the β cell phenotype, leading to a reduction in insulin expression and content, and the loss of some β cell functional features (Figure 2, A–C), which unexpectedly was not accompanied by cell death. Instead, we observed the presence of at least 4 different β cell subpopulations within the mutant islets (Figure 3, A–J): (a) single-hormone-positive insulin expressing cells; (b) β cells that coexpress insulin and other islet hormones, suggesting the occurrence of a partial transdifferentiation event; (c) cells that have lost insulin expression and only express other islet endocrine hormones, perhaps indicative of a complete cellular transdifferentiation; and (d) cells that do not express any hormones. The existence of these different β cell populations suggests that β cells could gradually traverse stepwise through partial reprogramming, then to complete transdifferentiation, and finally to a complete loss of β cell identity (Figure 8). However, we did not observe changes in the relative ratios of these 4 different subpopulations with age or pregnancy, perhaps suggesting that the islet contains heterogeneous β cell populations that differentially respond to the loss of NKX2.2. We cannot, however, rule out the possibility that there is some interconversion between the different phenotypic states (Figure 8).
Figure 8. Model of NKX2.2 function in the adult β cell.
NKX2.2 appears to be a master regulatory protein that is essential for the acquisition and maintenance of β cell identity. Loss of NKX2.2 in β cell results in the formation of different β cell subpopulations that were identified by the expression of the β cell–specific Tomato reporter. Dysfunctional β cells: This category includes lineage-labeled (red dots) insulin-expressing cells (purple dots) unable to mount a proper insulin response and represents the largest population of mutant β cells. Polyhormonal β cells: Lineage-labeled (red dots), insulin-positive (purple dots) β cells that are misexpressing the other islet endocrine hormones (green dots). It is possible that these cells represent incomplete transdifferentiation. Insulin-negative β cells: Lineage-labeled (red dots) β cells that have lost insulin expression, but have acquired the expression of other non-β endocrine hormones (green dots). Empty β cells: A rarer population of lineage-labeled cells (red dots) without expression of any endocrine hormones. It is unknown whether these populations originate independently or are able to interconvert. Arrows represent the possible transitioning between the different populations.
In contrast to previous studies that suggest that β cells undergo dedifferentiation prior to reprogramming (2), we did not observe a significant increase in the expression of progenitor markers to suggest that deletion of Nkx2.2 causes β cell dedifferentiation. Instead, our data suggest that loss of Nkx2.2 results in direct reprogramming through the primary regulation of specific gene sets. Interestingly, we also observed more restricted reprogramming potential in adult versus young β cells. Deletion of Nkx2.2 in β cells shortly after the activation of insulin resulted in all combinations of polyhormonal cell populations; however, deletion of Nkx2.2 in adult β cells only led to the formation of insulin- and SST-coexpressing cells (Figure 6, I–K), implying that the plasticity of the β cell is diminished over time. The role of NKX2.2 function in maintaining β cell identity by activating β cell gene programs and repressing non–β cell programs appears to be conserved in human islets. Similarly to loss of Nkx2.2 in adult mouse β cells, inactivation of NKX2.2 in human islets predominantly led to the upregulation of SST (Figure 7D). Interestingly, the predominance of insulin- and SST-coexpressing cell populations has been described in several recent studies reporting the presence of polyhormonal cells in islets derived from diabetic individuals (33, 35).
One intriguing feature of the Nkx2.2ΔBeta mice was their impaired ability to mount a response to glucose in vivo, even though there was an enhanced response in vitro. This phenotype is remarkably similar to that seen with a β cell deletion of Slc30a8, the gene that encodes the zinc transporter ZNT8 (36). Mice lacking ZNT8 specifically in their β cells had lower peripheral insulin concentrations, but increased insulin secretion in vitro due to a key role for islet-derived ZNT8 in regulating insulin degradation by the liver. Since Slc30a8 is significantly downregulated in Nkx2.2ΔBeta islets (P < 1.65 × 10–8), this may contribute to the severe metabolic impairment observed in vivo. In addition, we noted a modest increase in serum levels of SST (Supplemental Figure 2K) and significantly increased expression of somatostatin receptor 1 (Sstr1) (2.42-fold; P < 0.0009) in the β cells of Nkx2.2ΔBeta mice. Since SST is a potent inhibitor of insulin secretion (37), this could also explain why there is a more severe impairment of insulin secretion in vivo. Ultimately, these results indicate that NKX2.2 is essential for regulating many aspects of β cell function that have a profound effect on maintaining glucose homeostasis in the body.
We have determined that NKX2.2 plays an essential role in regulating β cell function and identity. Although previous studies have demonstrated the ability of NKX2.2 to function as an activator and repressor in certain cellular and genomic contexts, to our knowledge this is the first evidence that NKX2.2 simultaneously functions in the β cell to directly activate the β cell program and repress non-β endocrine genes. The unexpected finding that 47% of NKX2.2 binding peaks were present at poised rather than repressed enhancers also provides an explanation for why β cells are only able to revert to closely related endocrine cell fates, rather than a completely unrelated cell identity. Furthermore, it suggests a mechanism whereby NKX2.2 is involved in the priming of enhancers during development and may be essential later for the direct activation of the genes involved in β cell function as well as the active repression of gene sets that would otherwise activate non-β islet-related lineages. The direct regulation of both β cell and non–β cell gene programs by NKX2.2 also suggests that the presence of bihormonal and/or polyhormonal cells is not secondary to the loss of β cell identity. Importantly, these studies also demonstrate the highly unstable nature of the β cell; there is an ongoing need to actively maintain the activation of genes involved in β cell function, but to also maintain repression of closely related gene programs. Therefore, it is possible that in conditions of metabolic stress, disruption of a single regulatory pathway will cause a simultaneous loss of β cell function and identity.
A potential caveat regarding the study is that MIN6 cells were used to identify direct targets of NKX2.2. Although it would have been optimal to perform ChIP-seq analysis on purified adult β cells, we were not able to extract sufficient chromatin from the primary tissue. To overcome any potential artifacts that may be associated with using an immortalized cell line, we designated genes to be valid direct targets only if they were both bound by NKX2.2 in MIN6 cells and shown to be regulated by NKX2.2 in vivo in the Nkx2.2ΔBeta mice. Furthermore, many of the direct targets we identified were shown to be bound by NKX2.2 in human islets (26). The correlation of these NKX2.2 binding sites with enhancers and repressors identified in young β cells (29) further demonstrates an association of NKX2.2 with existing regulatory elements. Additional studies will be performed in the future to determine whether NKX2.2 plays a direct role in influencing the chromatin structure and maintaining chromatin marks to preserve the gene regulation in the β cell.
The development of polyhormonal cells during T1D and T2D has been postulated to be indicative of a more primitive endocrine cell phenotype that is known to exist during human islet cell development (38). Interestingly, during the early stages of human pancreas formation, the endocrine cells are initially polyhormonal and only gradually resolve into monohormonal cell identities (39). Intriguingly, unlike in mouse pancreatic development, where NKX2.2 is expressed early and there are only minor populations of polyhormonal cells, in humans NKX2.2 is not detected until late in pancreas formation, during the stage when monohormonal cells are acquired (40). This suggests that there may be an intrinsic function of NKX2.2 for acquiring a functional monohormonal identity.
The role of NKX2.2 in maintaining β cell identity in T2D remains relatively unexplored. We have determined that NKX2.2 expression is downregulated more than 5-fold (P adjusted < 1.19 × 10–7) in db/db mouse islets, and a recent study demonstrated that NKX2.2 expression was reduced in the diabetic Goto-Kakizaki rat model (41). Reduced NKX2.2 expression has also been reported in primate models of diabetes induced by a high-fat/sugar diet (42). In humans, because of the variability that exists between T2D patient samples, the expression of NKX2.2 remains controversial. In some patient cohorts, there was little or no change in NKX2.2 expression levels (43, 44). However, in other studies, diabetic individuals had an downward trend of NKX2.2 expression (45, 46). Given the polygenic nature of T2D, it remains possible that larger sample sizes will identify NKX2.2 dysregulation in subclasses of T2D patients. In support of this, Pasquali et al. (26) identified a large number of T2D SNPs present in islet enhancers that were occupied by NKX2.2.
In summary, it is becoming increasingly clear that β cell death may not be the primary cause of the loss of β cell mass during diabetes progression. Instead, there is emerging evidence that fully differentiated mature β cells in mice and humans retain much more plasticity than previously appreciated. In the present study, we identified NKX2.2 as master regulator of such plasticity, actively repressing non–β cell lineages and directly activating β cell functional programs. Understanding the molecular mechanisms allowing for the retained plasticity will be important for deriving fully differentiated β cells with a “locked-in” identity in vitro. Furthermore, these findings indicate that blocking or reversing β cell reprogramming could represent viable therapies for treating diabetes.
Methods
Mouse strains
Mice carrying a conditional allele of Nkx2.2fl/fl (21) were bred to RIP-Cre mice (20) and to tamoxifen-inducible MIP-CreERT mice (31). To induce the MIP-CreERT allele, tamoxifen (Sigma-Aldrich) was dissolved in corn oil, and a dose of 100 mg/kg BW was given via i.p. injection every other day for 3 days. All mice used in physiological tests were male, unless otherwise stated in the text. It is important to note that deletion of the genomic fragment encoding the Nkx2.2 gene did not delete the associated long noncoding RNA (lncRNA) and deletion of Nkx2.2 did not alter expression of the associated lncRNA.
Tissue preparation and immunohistochemistry
Mouse pancreata were fixed either for 2 hours or overnight in 10% neutral-buffered formalin (VWR) at 4°C. Tissue was then transferred to 70% ethanol and subsequently embedded in paraffin and sectioned at 5 μm thickness. Antigen retrieval was performed using 10 mM sodium citrate buffer, pH = 6, at boiling temperature in a water bath for 15 minutes. After a 20-minute cool-down period at room temperature, samples were incubated with primary antibodies overnight and further stained with secondary antibodies the following day. Details of primary and secondary antibodies used are listed in Supplemental Tables 4 and 5, respectively.
Transmitted electron microscopy
Ultrastructural analysis of WT and Nkx2.2 mutant pancreatic islets was performed by transmitted electron microscopy, as previously described with minor modifications (47, 48). Briefly, islets were fixed immediately after isolation at 4°C in 0.1 M sodium cacodylate buffer, pH = 7.4, 2% paraformaldehyde, 2.5% glutaraldehyde (Electron Microscopy Sciences), and 3 μM CaCl2. Samples were then postfixed with osmium tetraoxide (1% wt/vol in H2O) and counterstained with uranyl acetate (2% wt/vol in H2O). After gradual dehydration in ethanol, samples were embedded in Durcupan resin (Sigma-Aldrich) and polymerized overnight at 60°C and –20 mmHg, as previously described (49). Ultrathin sections (70 nm) were then cut using a 35° angle Diatome diamond knife, and mounted on 300 mesh gold grids (Electron Microscopy Sciences). After counterstaining with uranyl acetate (1% wt/vol in H2O) and Sato lead (1% wt/vol in H2O), sections were imaged at 80 keV using an electron microscope (JEOL JEM-1400), equipped with a Gatan Ultrascan 1000XP with 2,000 × 2,000 resolution CCD camera, using the image acquisition software Gatan Digital Micrograph (Gatan Inc.).
ChIP and sequencing
MIN6 cells (25) were grown to confluence in a 15-cm culture dish and were formaldehyde–cross-linked for 10 minutes. Cross-linked chromatin was fragmented by sonication using the Diagenode BioRuptor for 3 cycles of 5 minutes each (30 seconds on/off). Two micrograms of rabbit anti-NKX2.2 antiserum was added to 80 μg of sheared chromatin. The antibody/chromatin complex was left to rotate end to end overnight at 4°C, and subsequently pulled with protein G Dynabeads (Thermo Fisher Scientific). Chromatin was washed, eluted, and reverse cross-linked, followed by protease treatment.
ChIP-seq libraries were prepared according to the instructions of the KAPA Hyper Prep kit for Illumina. Sequencing was performed using the Illumina HiSeq 2500 system according to the manufacturer’s instructions. Peak annotation was performed using the MACS peak caller (version 1.4.2 20120305). The q value cutoff we used was 0.01. Binding peaks were annotated with genes with a transcription start site present within 100 kb of an identified NKX2.2 binding peak. De novo motif analysis was performed with Hypergeometric Optimization of Motif EnRichment (HOMER version 4.6). The dataset is available at NCBI’s Gene Expression Omnibus (GEO) database under the accession number GSE79725.
Western blot analysis
Pancreatic islets were isolated out from approximately 21-week-old mice. Protein was extracted using radioimmunoprecipitation assay (RIPA) buffer during a 5-minute sonication period (30 seconds on/off). Thirteen micrograms of protein was used for analysis as previously described (50). Primary and secondary antibodies used are listed in Supplemental Tables 4 and 5, respectively.
Glucose tolerance test, plasma insulin, and insulin content
Glucose tolerance tests.
Mice were fasted overnight, followed by an i.p. injection with glucose (2 mg/g BW). Tail vein blood samples were collected at several time points after the injection. Glucose concentration was determined using the Accu-Chek Compact Plus Blood Glucose Meter (Roche).
Acute plasma insulin and somatostatin assays.
Mice were fasted overnight, followed by i.p. injections with glucose (2 mg/g BW). For insulin measurements, blood plasma samples were collected at different time points after the injection through heart puncture, and individual cohorts were used per time point. Somatostatin (SST) was measured by collection of blood plasma samples 30 minutes after the injection through heart puncture. Plasma insulin was measured by ultrasensitive insulin ELISA kit (Crystal Chem), and SST was measured by SST ELISA kit (Phoenix Pharmaceuticals).
Pancreatic insulin content.
Pancreata were dissected and disrupted using a homogenizer. Insulin was extracted by acid ethanol. The insulin concentration was analyzed by insulin ELISA (Mercodia) and normalized to pancreas weight (g).
Islet isolation and glucose stimulated insulin secretion assays
Islet isolation.
Mouse pancreatic islets were isolated by perfusion of the pancreata with Collagenase P (Roche) through the common hepatic bile duct at a concentration of 1 mg/ml of M199 medium (Invitrogen). Pancreata were removed and dissociated at 37°C for 16 minutes. After several washes using M199 medium supplemented with 10% FBS (Gemini Bio Products), islets were separated onto a gradient using a combination of serum-free M199 medium and Histopaque (Sigma-Aldrich). Islets were then hand-picked to avoid exocrine contamination and processed for different applications.
Human islets from 3 donors were obtained through the NIH-supported Integrated Islet Distribution Program via Mount Sinai Human Islet Core facility. The islets were harvested from deceased donors without any identifying information at NIH-approved centers with informed consent and IRB approval at the islet isolation centers. Donors ranged in age from 32 to 64 years old (mean 45.7); 2 were female and 1 was male. Mean BMI was 28.03 (range 23–32.8).
Glucose stimulated insulin secretion assays.
Twenty islets per well were cultured in Krebs buffer with a glucose concentration of 2.8 mM for 1 hour in a 12-well plate; supernatant was collected to measure basal insulin secretion. Islets were then transferred to Krebs buffer with 16.7 mM glucose, and the supernatant was collected after 1 hour. Islets were disrupted using a homogenizer in 50 μl of lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 1% Octylphenoxy poly(ethyleneoxy)ethanol [IGEPAL]), and insulin content was extracted by acid ethanol. Insulin in supernatant and islet lysates was measured by ultrasensitive insulin ELISA (Crystal Chem). Secreted insulin was calculated as percentage of total insulin content per hour.
Quantitative real-time reverse transcriptase PCR and RNA-seq
Total mRNA was prepared from P0 pancreata or isolated islets at 4 weeks of age using the RNeasy Micro Kit (QIAGEN) and reverse transcribed with SuperScript (Roche). Real-time reverse transcriptase PCR was carried out with either SYBR Green (Bio-Rad) or TaqMan probes (Applied Biosystems) and measured on a Bio-Rad CFX96 Real Time System. Expression levels were normalized to cyclophilin B for mouse samples and to 36B4 for human samples. Primers and probes used for mouse and human samples are listed in Supplemental Tables 6 and 7, respectively.
For RNA-seq analysis, RNA from mouse and human islets was obtained using the RNeasy Micro Kit (QIAGEN). RNA quantity and quality were assessed using the Agilent 2100 Bioanalyzer, and 400 ng of total RNA was sent to the Columbia University Genome Center for library preparation, sequencing (Illumina 2500), and bioinformatics analysis. Expression changes were considered significant if they had a P value of at least 0.05 and a log2 fold change of at least 0.5. The GEO accession number for this data set is GSE79725.
Adenovirus
Ad.shRNA directed against human NKX2.2, targeting GCCGACGAGTCACCGGACAA, was obtained from Welgen Inc. Ad.Scrambled was provided by Andrew Stewart (Icahn School of Medicine at Mount Sinai). Adenovirus was packaged and produced in HEK-293A cells. Titers were determined by plaque assay (PFU). Dispersed human islets were transduced with either experimental or control (Ad.Scrambled) at 100 and 300 MOI in serum-free media for 2 hours. Transduction was terminated by addition of complete medium containing 10% FCS and cultured for 96 hours.
Cell lines
The adenovirus packaging cell line, HEK-293A (Life Technologies), was cultured in DMEM supplemented with 10% FCS, 1% penicillin-streptomycin, and 1× MEM containing nonessential amino acids. The MIN6 cell line was cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin.
Morphometric analysis
For all morphometric analysis, quantification was done on 6 evenly spaced sections through the whole pancreas. To determine β cell mass, percentage islet area, and islet number, tiles of each section were imaged at ×10 using a Leica fluorescent microscope (DM5500). To calculate β cell mass, each section was stained with insulin (β cell area) and amylase (pancreas area). Islet area was divided by the pancreas area and then multiplied by the weight of the pancreas (g). To analyze percentage islet area, each section from the different levels was stained with insulin and DAPI. Islet size was measured manually based on morphology and guided by insulin and DAPI staining. Islets were arranged by size, and the total islet number was used to calculate the percentage represented in each size group. To determine the islet cell numbers, all islets were quantified in each section and then divided by the total pancreas area (cm2). To determine the number of insulin cells per islet area as well as the percentage of polyhormonal cells, images of 5 islets per section in 6 different levels were imaged at ×40 on a Zeiss Confocal LSM 710 microscope. The number of insulin-positive cells was counted as a function of total islet area. Percentage of polyhormonal cells was calculated by division of the number of cells copositive for insulin and either SST, glucagon, or pancreatic polypeptide by the number of insulin-positive cells and multiplied by 100. At least 3,000 cells were counted to calculate insulin cells per islet area, and at least 3,500 for the percentage of polyhormonal cells. All areas and cell quantification were processed with ImageJ 1.46R software (NIH).
Image analysis
All immunofluorescent images were processed using Adobe Photoshop CS5.1 and ImageJ 1.46R. Only brightness and contrast were adjusted in accordance with the Journal of Cell Metabolism figure manipulation guidelines.
Statistics
Statistical analysis was calculated using a 2-tailed Student’s unpaired t test. Results are expressed as the mean ± SEM, and a P value of 0.05 or less was considered significant.
Animal approvals
Mice were housed and treated in accordance with the animal care protocol (AAAG3206) approved by the IACUC of Columbia University.
Author contributions
LS conceived the mouse study. GDG and TLM collected and analyzed the mouse data. GDG conceived and executed the human islet study. ASB assisted with the human islet study. VC performed the electron microscopic studies and assisted with their interpretation. SMK and AT assisted with the computational analysis of ChIP-seq and RNA-seq data. KHK provided the raw data for the mouse chromatin marks for comparative analysis. LS and GDG assembled and analyzed the data and wrote the manuscript. All authors discussed the results and edited the manuscript.
Supplementary Material
Acknowledgments
We thank P. Herrera (University of Geneva) for the RIP-Cre mouse line and L. Philipson (University of Chicago) for the MIP-CreERT2 mouse line. We thank members of the Sussel laboratory for helpful discussions and Anthony Romer for assisting with the db/db mouse RNA-seq experiment. We thank the Columbia University Genome Center for performing the RNA sequencing and the New York University (NYU) Genome Technology Center (GTC) for expert library preparation and ChIP sequencing. The GTC shared resource is partially supported by the Cancer Center Support Grant (P30 CA016087) at the Laura and Isaac Perlmutter Cancer Center. We also thank the NYU Applied Bioinformatics Center for providing bioinformatics support and helping with the analysis and interpretation of the data. This work has used computing resources at the High Performance Computing Facility of the Center for Health Informatics and Bioinformatics at the NYU Langone Medical Center. We are grateful to Bobbie Schneider for assistance with transmitted electron microscopy sample preparation at the Electron Microscopy Resource of the Fred Hutchinson Cancer Research Center, Seattle, Washington, USA. Studies in human islets were performed in the Human Islet and Adenovirus Core of the Einstein–Mount Sinai Diabetes Research Center (2 P30 DK020541-38). Additional core facility support was provided by the Columbia Diabetes Endocrinology Research Center (DERC) (NIH P30 DK063608) and the Herbert Irving Comprehensive Cancer Center. Funding for the project was provided by NIH R01 DK082590 (to LS), R01 DK088383 (to KHK), a Consejo Nacional de Ciencia y Tecnología grant from the Mexican government (to GDG), the Naomi Berrie Fellowship in Diabetes Research (to TLM), and JDRF postdoctoral fellowship 3-2101-791 (to TLM). Work in the laboratory of VC was supported by Life Sciences Discovery Fund grant 4553677 and JDRF grant 17-2011-620.
Footnotes
TLM’s present address is: Center for Diabetes and Metabolic Diseases, Indiana University School of Medicine and Indiana Biosciences Research Institute, Indianapolis, Indiana, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(1):244–259. doi:10.1172/JCI88017.
See the related Commentary at β Cells led astray by transcription factors and the company they keep.
Contributor Information
Aaron S. Bender, Email: aaron.bender@mssm.edu.
Vincenzo Cirulli, Email: vcirulli@uw.edu.
Lori Sussel, Email: lgs2@columbia.edu.
References
- 1.Prentki M, Nolan CJ. Islet β cell failure in type 2 diabetes. J Clin Invest. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talchai C, Xuan S, Lin HV, Sussel L, Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240(3):530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 4.Schaffer AE, et al. Nkx6.1 controls a gene regulatory network required for establishing and maintaining pancreatic β cell identity. PLoS Genet. 2013;9(1):e1003274. doi: 10.1371/journal.pgen.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor BL, Liu FF, Sander M. Nkx6.1 is essential for maintaining the functional state of pancreatic β cells. Cell Rep. 2013;4(6):1262–1275. doi: 10.1016/j.celrep.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naya FJ, et al. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11(18):2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu C, et al. Pancreatic β cells require NeuroD to achieve and maintain functional maturity. Cell Metab. 2010;11(4):298–310. doi: 10.1016/j.cmet.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SB, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463(7282):775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piccand J, et al. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 2014;9(6):2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sussel L, et al. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic β cells. Development. 1998;125(12):2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 11.Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L. Ghrelin cells replace insulin-producing β cells in two mouse models of pancreas development. Proc Natl Acad Sci U S A. 2004;101(9):2924–2929. doi: 10.1073/pnas.0308604100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desai S, et al. Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev Biol. 2008;313(1):58–66. doi: 10.1016/j.ydbio.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson KR, White P, Kaestner KH, Sussel L. Identification of known and novel pancreas genes expressed downstream of Nkx2.2 during development. BMC Dev Biol. 2009;9:65. doi: 10.1186/1471-213X-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papizan JB, et al. Nkx2.2 repressor complex regulates islet β-cell specification and prevents β-to-α-cell reprogramming. Genes Dev. 2011;25(21):2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mastracci TL, et al. Nkx2.2 and Arx genetically interact to regulate pancreatic endocrine cell development and endocrine hormone expression. Dev Biol. 2011;359(1):1–11. doi: 10.1016/j.ydbio.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark JK, O’keefe A, Mastracci TL, Sussel L, Matise MP, Kucenas S. Mammalian Nkx2.2+ perineurial glia are essential for motor nerve development. Dev Dyn. 2014;243(9):1116–1129. doi: 10.1002/dvdy.24158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Q, et al. Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development. 2014;141(3):548–555. doi: 10.1242/dev.095323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arnes L, Leclerc K, Friel JM, Hipkens SB, Magnuson MA, Sussel L. Generation of Nkx2.2:lacZ mice using recombination-mediated cassette exchange technology. Genesis. 2012;50(8):612–624. doi: 10.1002/dvg.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flanagan SE, et al. Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab. 2014;19(1):146–154. doi: 10.1016/j.cmet.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127(11):2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 21.Mastracci TL, Lin CS, Sussel L. Generation of mice encoding a conditional allele of Nkx2.2. Transgenic Res. 2013;22(5):965–972. doi: 10.1007/s11248-013-9700-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cissell MA, Zhao L, Sussel L, Henderson E, Stein R. Transcription factor occupancy of the insulin gene in vivo. Evidence for direct regulation by Nkx2.2. J Biol Chem. 2003;278(2):751–756. doi: 10.1074/jbc.M205905200. [DOI] [PubMed] [Google Scholar]
- 23.Liu YQ, Nevin PW, Leahy JL. β-Cell adaptation in 60% pancreatectomy rats that preserves normoinsulinemia and normoglycemia. Am J Physiol Endocrinol Metab. 2000;279(1):E68–E73. doi: 10.1152/ajpendo.2000.279.1.E68. [DOI] [PubMed] [Google Scholar]
- 24.Peshavaria M, et al. Regulation of pancreatic β-cell regeneration in the normoglycemic 60% partial-pancreatectomy mouse. Diabetes. 2006;55(12):3289–3298. doi: 10.2337/db06-0017. [DOI] [PubMed] [Google Scholar]
- 25.Ishihara H, et al. Pancreatic β cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia. 1993;36(11):1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
- 26.Pasquali L, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle MJ, Loomis ZL, Sussel L. Nkx2.2-repressor activity is sufficient to specify α-cells and a small number of β-cells in the pancreatic islet. Development. 2007;134(3):515–523. doi: 10.1242/dev.02763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doyle MJ, Sussel L. Nkx2.2 regulates β-cell function in the mature islet. Diabetes. 2007;56(8):1999–2007. doi: 10.2337/db06-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avrahami D, et al. Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab. 2015;22(4):619–632. doi: 10.1016/j.cmet.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang A, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16(4):386–399. doi: 10.1016/j.stem.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wicksteed B, et al. Conditional gene targeting in mouse pancreatic β-cells: analysis of ectopic Cre transgene expression in the brain. Diabetes. 2010;59(12):3090–3098. doi: 10.2337/db10-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White MG, et al. Expression of mesenchymal and α-cell phenotypic markers in islet β-cells in recently diagnosed diabetes. Diabetes Care. 2013;36(11):3818–3820. doi: 10.2337/dc13-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoneda S, et al. Predominance of β-cell neogenesis rather than replication in humans with an impaired glucose tolerance and newly diagnosed diabetes. J Clin Endocrinol Metab. 2013;98(5):2053–2061. doi: 10.1210/jc.2012-3832. [DOI] [PubMed] [Google Scholar]
- 34.Piran R, Lee SH, Li CR, Charbono A, Bradley LM, Levine F. Pharmacological induction of pancreatic islet cell transdifferentiation: relevance to type I diabetes. Cell Death Dis. 2014;5:e1357. doi: 10.1038/cddis.2014.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cinti F, et al. Evidence of β-cell dedifferentiation in human type 2 diabetes. J Clin Endocrinol Metab. 2016;101(3):1044–1054. doi: 10.1210/jc.2015-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamaki M, et al. The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J Clin Invest. 2013;123(10):4513–4524. doi: 10.1172/JCI68807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strowski MZ, Parmar RM, Blake AD, Schaeffer JM. Somatostatin inhibits insulin and glucagon secretion via two receptors subtypes: an in vitro study of pancreatic islets from somatostatin receptor 2 knockout mice. Endocrinology. 2000;141(1):111–117. doi: 10.1210/endo.141.1.7263. [DOI] [PubMed] [Google Scholar]
- 38.Riopel M, Li J, Fellows GF, Goodyer CG, Wang R. Ultrastructural and immunohistochemical analysis of the 8-20 week human fetal pancreas. Islets. 2014;6(4):e982949. doi: 10.4161/19382014.2014.982949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bocian-Sobkowska J, Zabel M, Wozniak W, Surdyk-Zasada J. Polyhormonal aspect of the endocrine cells of the human fetal pancreas. Histochem Cell Biol. 1999;112(2):147–153. doi: 10.1007/s004180050401. [DOI] [PubMed] [Google Scholar]
- 40.Jennings RE, et al. Development of the human pancreas from foregut to endocrine commitment. Diabetes. 2013;62(10):3514–3522. doi: 10.2337/db12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mollet IG, Malm HA, Wendt A, Orho-Melander M, Eliasson L. Integrator of stress responses calmodulin binding transcription activator 1 (Camta1) regulates miR-212/miR-132 expression and insulin secretion. J Biol Chem. 2016;291(35):18440–18452. doi: 10.1074/jbc.M116.716860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiori JL, et al. Resveratrol prevents β-cell dedifferentiation in nonhuman primates given a high-fat/high-sugar diet. Diabetes. 2013;62(10):3500–3513. doi: 10.2337/db13-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo S, et al. Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest. 2013;123(8):3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taneera J, et al. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 2012;16(1):122–134. doi: 10.1016/j.cmet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Dominguez V, et al. Class II phosphoinositide 3-kinase regulates exocytosis of insulin granules in pancreatic β cells. J Biol Chem. 2011;286(6):4216–4225. doi: 10.1074/jbc.M110.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marselli L, et al. Gene expression profiles of β-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaferia GR, et al. β1 Integrin is a crucial regulator of pancreatic β-cell expansion. Development. 2013;140(16):3360–3372. doi: 10.1242/dev.098533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller R, et al. Switching-on survival and repair response programs in islet transplants by bone marrow-derived vasculogenic cells. Diabetes. 2008;57(9):2402–2412. doi: 10.2337/db08-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staubli V, Loustalot P. Electron microscopy of transplantable melanotic and amelanotic hamster melanomas. Cancer Res. 1962;22:84–88. [PubMed] [Google Scholar]
- 50.Borok MJ, Papaioannou VE, Sussel L. Unique functions of Gata4 in mouse liver induction and heart development. Dev Biol. 2016;410(2):213–222. doi: 10.1016/j.ydbio.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.