Abstract
Human NK cell deficiencies are rare yet result in severe and often fatal disease, particularly as a result of viral susceptibility. NK cells develop from hematopoietic stem cells, and few monogenic errors that specifically interrupt NK cell development have been reported. Here we have described biallelic mutations in IRF8, which encodes an interferon regulatory factor, as a cause of familial NK cell deficiency that results in fatal and severe viral disease. Compound heterozygous or homozygous mutations in IRF8 in 3 unrelated families resulted in a paucity of mature CD56dim NK cells and an increase in the frequency of the immature CD56bright NK cells, and this impairment in terminal maturation was also observed in Irf8–/–, but not Irf8+/–, mice. We then determined that impaired maturation was NK cell intrinsic, and gene expression analysis of human NK cell developmental subsets showed that multiple genes were dysregulated by IRF8 mutation. The phenotype was accompanied by deficient NK cell function and was stable over time. Together, these data indicate that human NK cells require IRF8 for development and functional maturation and that dysregulation of this function results in severe human disease, thereby emphasizing a critical role for NK cells in human antiviral defense.
Introduction
NK cell deficiency (NKD) is an inborn or primary immunodeficiency causing susceptibility to severe and often fatal viral infection and malignancy (1–5). NKD can be classified as classical, arising from profound or complete absence of the NK cell population or major subsets of peripheral NK cells, or functional, arising from impaired NK cell function with normal peripheral NK cell counts (5, 6). There are presently 2 genetically defined causes of classical (MCM4, GATA2) and 1 of functional NKD (FCGR3A) (2–4, 7–9).
Human NK cell development results in generation of distinct phenotypic and functional subsets found in peripheral blood: namely the CD56bright and CD56dim subsets. While identified primarily by the density of CD56 on the cell surface, these subsets include the coordinated expression of cell surface and intracellular markers that reflect each subset’s unique function. CD56bright NK cells are considered immunomodulatory and are described phenotypically as CD16−/loCD57−perforinlo, CD117+, CD62Lhi, and CD94/NKG2A+. While they constitute the minority of NK cells in peripheral blood, they represent the dominant population in secondary lymphoid tissue (10, 11). CD56dim NK cells, which constitute more than 90% of peripheral blood NKs, are CD56dimCD16hiperforinhiCD57+/− killer-cell immunoglobulin-like receptor+ (KIR+), and are the primary mediators of NK cell–mediated cytotoxicity. The relationship between the subsets is not completely understood, but both in vivo and ex vivo evidence in humans suggests that the CD56bright subset is a developmental precursor of CD56dim, and it is hypothesized that secondary lymphoid tissue serves as a site of NK cell terminal differentiation that is required for acquisition of cytotoxic function (12–15).
Specific loss of NK cells or NK cell subsets as occurs in classical NKD is rare but has been reported. Homozygous MCM4 mutation causes adrenal insufficiency, decreased numbers of circulating NK cells, and impaired terminal maturation, likely as a result of chromosomal aberrations and impaired proliferation of the CD56bright subset (2, 3). These patients show distinctive loss of the CD56dim subset and correlative relative increase in the CD56bright subset (2). In contrast, GATA2 mutations result in specific loss of CD56bright NK cells and decreased frequency of NK cells overall in circulating blood. While GATA2 patients have CD56dim NK cells that vary in number, they are largely nonfunctional, and affected patients develop severe herpesvirus and papillomavirus infections (4). NKD has also been reported in 1 isolated case of a child with a mutation in RTEL1, encoding regulator of telomere elongation helicase, although NK cell subsets were not examined in this case (16, 17).
Interferon regulatory factor 8 (IRF8; also known as interferon consensus binding protein, ICSBP) is a member of the IRF family of transcription factors that help shape the inflammatory response, particularly to viral infections. These transcription factors are also becoming increasingly recognized for their roles in human hematopoiesis through rare, naturally occurring germline mutations.
Mutations of IRF8 in mice and humans cause cellular deficiency ranging from severe defects of monocyte and DC production with marked myeloproliferation to more subtle alterations of specific DC subsets with normal leukocyte counts (18–20). Gene dosage and mutation site–specific effects have been observed, suggesting that the level of IRF8 activity is critical to normal immune cell development and function. In addition to developmental aberrations, mature cell function is affected as exemplified by defects in macrophage function (21). In humans, heterozygous IRF8 T80A mutations have been identified as a cause of primary immunodeficiency leading to specific loss of the CD11c+CD1c+ circulating DC subset (19). K108E mutations of IRF8 have an even more profound impact, leading to the loss of DCs as well as monocytes (19).
Before recent advances in genomic sequencing technology, NKD was reported in a number of pediatric cases. Perhaps the most renowned was an isolated case of NKD and viral susceptibility ultimately proven as resulting from a GATA2 mutation (4, 22). The original NKD case, however, was reported in 1982 as a familial, non–X-linked susceptibility to EBV infection that led to the death of 2 affected siblings and ongoing illness in a third (1). Here we report that biallelic mutations in IRF8 cause intrinsic NKD with decreased NK cell number, decreased frequency of the CD56dim subset, and accompanying susceptibility to viral and mycobacterial infection. In doing so we define and characterize IRF8 mutations in this original, unsolved family with NKD and identify NKD in 2 unrelated patients with biallelic IRF8 mutations. These data identify IRF8 as a critical regulator of NK cell terminal maturation and define a novel cause of a rare human inborn NK cell developmental defect.
Results
Patients and clinical history.
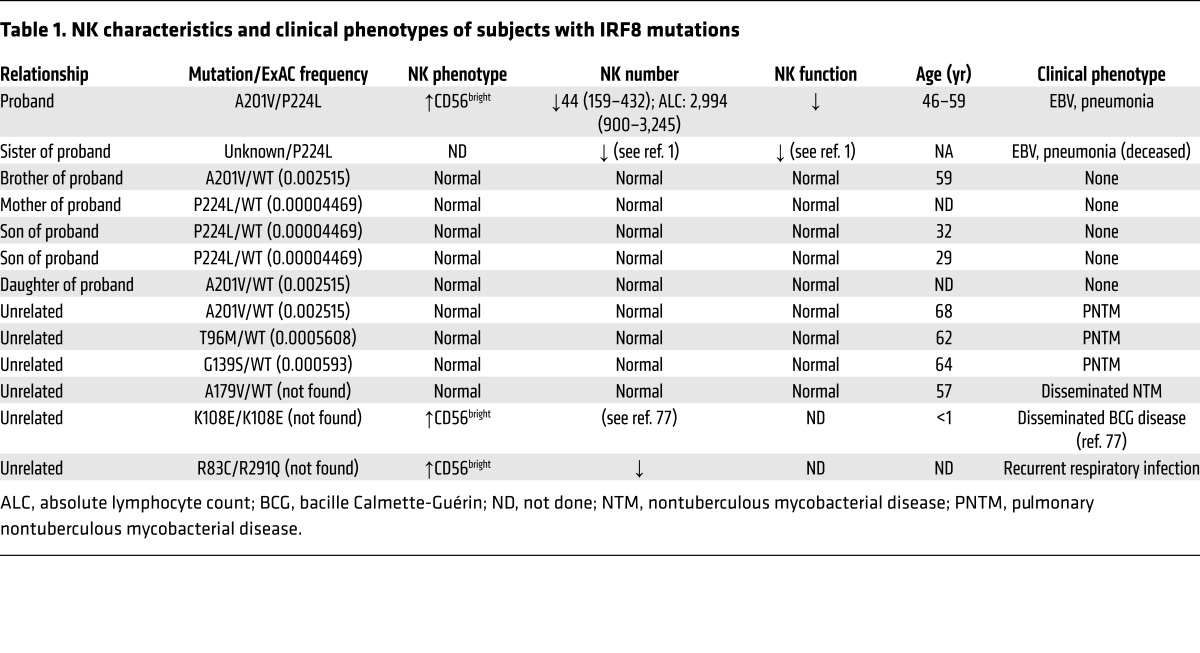
The index case proband (II.2) presented with a history of sinopulmonary infections during childhood and severe infectious mononucleosis at age 22 years requiring prolonged hospitalization (1). Born to non-consanguineous parents, his brother (II.6) died at age 16 years of multiple organ failure following severe EBV mononucleosis, and his sister (II.5) died at age 38 years due to progressive pulmonary disease. She also had prolonged hospitalization at age 17 years for severe EBV mononucleosis. Both siblings had lymphadenopathy and bronchiectasis beginning in childhood. Notably, the 3 siblings’ EBV-related illnesses occurred in separate calendar years. A fourth sibling (brother, II.3) had EBV mononucleosis at age 21 years at the same time as the proband, but had a typical course of illness and did not require hospitalization. The unusual nature of disease and clinical presentation in the proband prompted immunological evaluation and was described as a novel, non–X-linked immune deficiency leading to susceptibility to EBV disease associated with a deficiency of NK cell cytotoxicity (1). Since the original description, the patient has continued to have fairly regular, recurrent episodes of sinusitis and has mucosal changes consistent with recurrent and chronic disease as well as presumably respiratory viral infections. He has no evidence of bronchiectasis or lymphadenopathy. T cell and B cell counts have been consistently normal, and immunoglobulins are present in normal quantities and quality. His surviving brother (II.3), mother (I.1), children (III.1–3), and nephew (III.4) have unremarkable medical histories. Other unrelated patients are summarized in Table 1.
Table 1. NK characteristics and clinical phenotypes of subjects with IRF8 mutations.
Genetics.
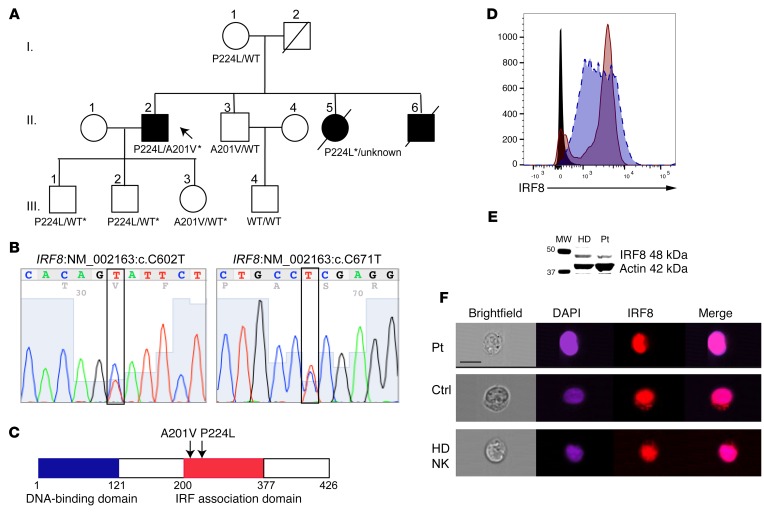
The 1982 description of the proband’s family is without follow-up until now and did not include any genetic or genomic analysis. Thus, whole exome sequencing of genomic DNA was performed using blood samples collected from the proband; his son, mother, and surviving brother; and the son of the brother (Figure 1A). Excellent average per-base coverage was observed for all samples (proband, 105X; mother, 124X; brother, 116X; son, 114X; fraternal nephew, 90X). The exome data suggested disease-related segregation of mutations in IRF8 (NM_002163:c.602C>T, 39 variant reads of 112 total in the proband; NM_002163:c.671C>T, 44 variant of 72 total proband reads) predicted by conceptual translation to result in amino acid changes p.A201V and p.P224L, respectively. Of those mutations predicted to be damaging by SIFT, PolyPhen, LRT, and MutationTaster algorithms, few aside from IRF8 were expressed highly in immune cells, particularly NK cells (see Supplemental Table 1 for list of variants identified by whole exome sequencing; supplemental material available online with this article; doi:10.1172/JCI86276DS1). Sanger sequencing of IRF8 in the proband’s sons and daughter confirmed the presence of both alleles in his children and the absence of either variant in his wife (Figure 1B and Supplemental Figure 1). The P224L variant was present in his mother, and the A201V variant was present in a surviving brother. Therefore, IRF8 mutations were confirmed with familial segregation in accordance with Mendelian expectations. In addition, the P224L mutation was confirmed from paraffin-embedded tissue from the patient’s deceased sister (Supplemental Figure 1); degradation of the DNA extracted from the same sample did not permit amplification and sequencing of the A201V mutation. The P224L mutation is reported 5 times in the Exome Aggregation Consortium (ExAC) database (allelic frequency of 4.4 × 10–5) with no homozygous cases. The A201V mutation is present at an allelic frequency of 0.002515 with no homozygotes. Likewise, within the Baylor-Hopkins Center for Mendelian Genomics database of over 6,000 exomes, no homozygous or compound heterozygous cases of IRF8 variants are found (23). Both mutations map to exon 7, in the region of IRF8 known as the IRF association domain (Figure 1C).
Figure 1. Linkage analysis of IRF8 compound heterozygous mutations in familial NKD and genetic confirmation of mutation.
(A) Pedigree of affected family. Roman numerals indicate generations. Arrow points to proband. Shown are amino acid variants in IRF8 as detected by whole exome sequencing. Asterisk indicates variant confirmed by Sanger sequencing. (B) Confirmation of proband’s IRF8 variants by Sanger sequencing. (C) Schematic of human IRF8 showing affected residues found within the IRF association domain. (D) Protein expression of IRF8 in EBV-transformed patient B cell line (maroon, solid line) relative to normal control (blue, dashed line). Isotype control is shown in black. Shown is 1 representative experiment from 3 independent repeats. (E) Western blot of IRF8 (48 kDa) in healthy donor (HD) or patient (Pt) lymphoblastoid cell lines with actin (42 kDa) as a loading control. Shown is 1 representative experiment from 3 independent repeats. (F) Nuclear localization of IRF8 in patient (Pt) or control (Ctrl) B cell lines or healthy donor NK cells. Cells were fixed and stained for IRF8 (eFluor 710, red) followed by counterstaining with DAPI (blue). Scale bar: 10 μm. Shown is 1 representative experiment from 3 independent repeats.
Evaluation of the patient’s protein expression by flow cytometry in lymphoblastoid cell lines showed normal levels of expression (Figure 1D), and Western blot analysis of IRF8 from patient EBV-transformed B cell lines confirmed protein expression at the expected molecular weight of full-length protein, as did healthy donor (Figure 1E; see complete unedited blots in the supplemental material). The only previously reported homozygous mutation in IRF8, K108E, was shown to impair nuclear translocation of IRF8 in transfected cells (21). To determine whether the novel compound heterozygous alleles found in our proband prevent the nuclear recruitment of IRF8, we evaluated the patient B cell line by imaging flow cytometry. In the absence of stimulation, IRF8 was detected in both the cytoplasm and the nucleus of patient and control B cell lines. This was also observed for healthy donor NK cells, with colocalization between DAPI and IRF8 as calculated by similarity score (24) of 3.4 for patient cells, 2.2 for control B cells, and 2.2 for primary human NK cells (median values) (Figure 1F). Stimulation with IL-15 did not increase localization of IRF8 in primary human NK cells (similarity score 2.2, not shown). Together these data confirm novel compound heterozygous mutations in IRF8 that do not abrogate protein expression or nuclear localization.
Biallelic IRF8 mutation leads to impaired NK cell cytotoxic function.
To specifically test the effect of biallelic mutations on NK cell cytotoxicity, we performed standard 51Cr release assays. As previously described, the proband has consistently low NK cell functional activity against the prototypical K562 target (ref. 1 and Figure 2A). The proband’s cytotoxic function was evaluated multiple times during a 10-year time interval and consistently showed profoundly decreased activity relative to normal controls with specific lysis (at maximal effector-to-target ratio) that was 0% and 31% of normal control levels (Figure 2B). The patient’s children, carriers of the P224L or A201V mutations, had slightly decreased but normal cytotoxic function, as did his nephew and mother (Figure 2, A and C, and not shown). Unrelated carriers of the A201V mutation from a separate family of a different nationality similarly had normal NK cell cytotoxic function (Figure 2C), despite having late onset of pulmonary nontuberculous mycobacterial disease (Table 1). Other heterozygous patients (G139S/WT, A179/WT, T96M/WT) also had normal NK cell function despite the presence of pulmonary nontuberculous mycobacterial infection (Figure 2C and Table 1). To determine whether decreased NK cell lytic function in the proband was due to low NK cell frequency within the lymphocyte population, cytotoxicity assays using purified NK cells were performed, and showed significantly decreased lysis of targets (Figure 2D), suggesting that an intrinsic defect in NK cell cytotoxic function underlies the stably decreased impairment observed. Therefore, the compound heterozygous A201V/P224L mutation results in low/absent NK cell cytotoxic function, an effect that is repeatedly observed over extended time and likely accounts for the proband’s profound susceptibility to viral disease.
Figure 2. NK cell functional deficiency specifically occurs in IRF8 compound-heterozygous patient and is stable over time.
NK cell cytotoxicity was evaluated by standard 51Cr release assay against K562 targets. (A) Shown is a representative assay including our proband (A201V/P224L), his healthy brother (A201V/WT), and his healthy son (P224L/WT). The gray filled area represents the range of 10 healthy donors. ***P < 0.0001, Student’s unpaired t test against mean of all healthy donors. Patient data are representative of 6 independent repeats. (B) The proband’s NKD is stable over time as assessed by 51Cr release assay against K562 targets. Shown is relative specific lysis by patient NK cells at the highest effector-to-target ratio (dashed line, circles) compared with corresponding healthy donor control (solid line, squares) at dates from 2004 to 2012 (x axis). (C) 51Cr release assay of unrelated A201V/WT (daughter of proband or unrelated as indicated), P224L/WT (son of proband), G139S/WT, A179V/WT, and T96M/WT patients (circles). The gray filled area represents the range of 10 normal donors. Each condition was performed in triplicate and is shown as the mean. (D) 51Cr release assay of freshly isolated purified patient (circles) and healthy donor (squares) NK cells against K562 target cells. Shown is 1 representative of 2 independent experiments with each condition performed in triplicate.
Biallelic IRF8 mutation results in decreased NK cell number and impaired NK cell terminal differentiation.
We next functionally examined NK cells from probands with 2 different combinations of compound heterozygous and a homozygous (i.e., biallelic) variant allele: A201V/P224L, R83C/R291Q, and homozygous K108E. Human NK cells in peripheral blood fall largely within 2 dominant subsets with unique phenotypic and functional signatures. High density of CD56 and the presence of CD94/NKG2A, CD117, and CD62L on the cell surface classify the CD56bright subset phenotypically. These cells are considered the most potent cytokine producers upon stimulation, primarily of TNF-α and IFN-γ. The CD56dim subset is characterized by markers of terminal differentiation such as CD57, activating and inhibitory KIRs, and CD16, and is thought to be derived from the CD56bright subset (13). In addition, CD56dim NK cells express large quantities of perforin and granzymes; they are specialized for contact-mediated target cell lysis.
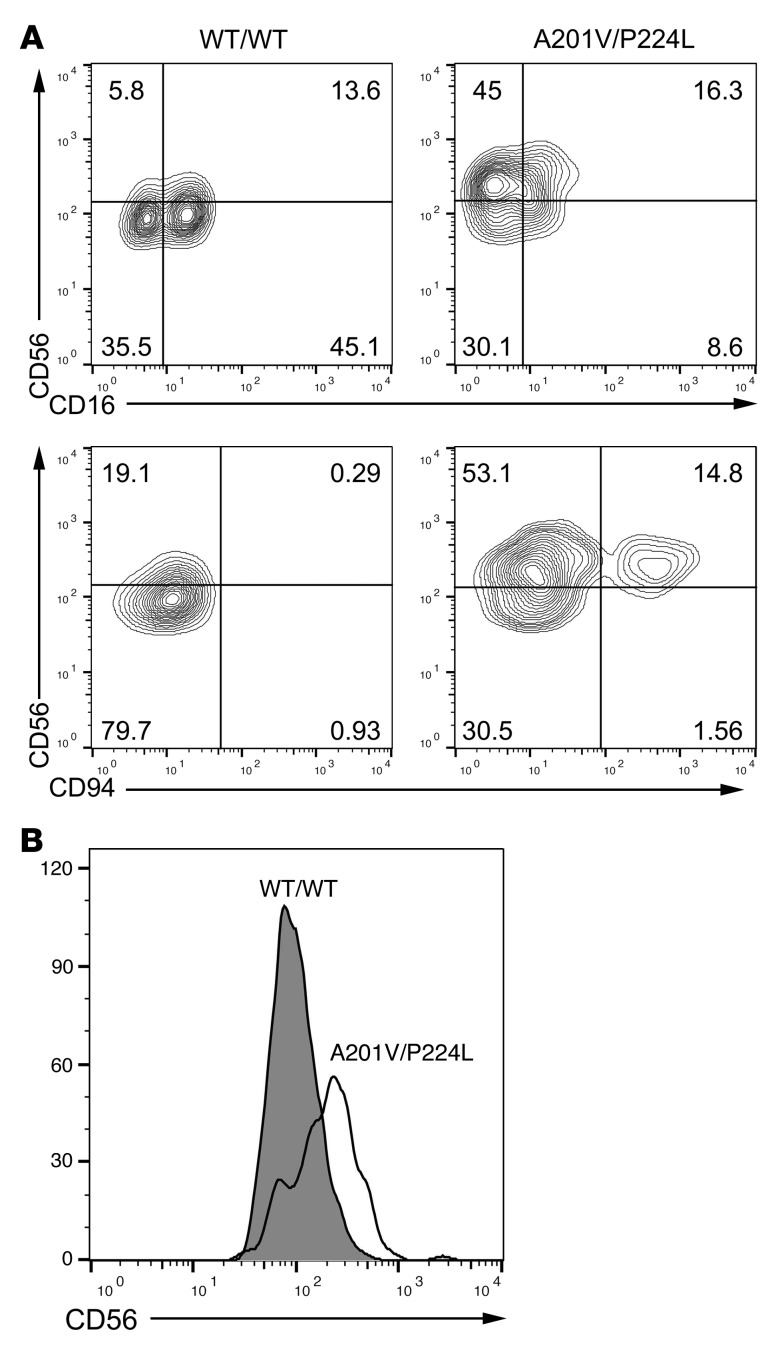
Consistently over the time of evaluation (>10 years) the patient had decreased frequency and absolute number of CD56+CD3− NK cells within the lymphocyte population (1.7%–6.0% vs. 4.6%–13.7% in healthy controls) (Figure 3, A–C, and Table 1). Evaluation of the phenotypic subsets within peripheral blood showed a relative paucity of the CD56dim subset in the patient, accompanied by an overrepresentation of the CD56bright subset. From our initial evaluation in 2004 and at all subsequent times evaluated, patient 1’s CD56bright subset was between 30% and 40% (27.9%–40.3%) of the NK cell population, in contrast to controls, which fell within the previously described range (4.1%–8.4%) (Figure 3, A–C, and ref. 25). Evaluation of heterozygous A201V/WT subjects (both unrelated and related) showed normal frequencies of both NK cells within the peripheral blood mononuclear cell (PBMC) population (13.9%) and CD56bright NK cells (3.7%). Further, evaluation of other heterozygous individuals (T96M/WT, G139S/WT, A179V/WT) showed frequencies of NK cells and CD56bright NK cells consistent with normal controls (Figure 3, B–D). In contrast, the R83C/R291Q and the homozygous K108E patient had low overall numbers of NK cells (3.9% and 5.4%, respectively) and a significantly expanded CD56bright population consistent with our proband (65.8% and 72.7%; Figure 3, B–D). Therefore, compound heterozygous or homozygous mutations in IRF8 selectively result in aberrant generation of the terminally mature subsets of NK cells and a decreased frequency of NK cells within the lymphocyte population.
Figure 3. Stable impairment in NK cell terminal differentiation in patients with biallelic IRF8 mutations.
NK cell phenotype was evaluated by multiparametric FACS phenotyping. (A) Representative FACS plot of healthy donor control (WT/WT) and our proband (A201V/P224L). Lymphocytes were gated on FSC/SSC. Data shown are representative of 6 independent repeats. (B) Stable NKD (left) and increased CD56bright population (right) in our proband shown over time (dashed line) with healthy donor shown as reference (solid line) at multiple dates from 2004 to 2015 (x axis). Single time point analysis of A201V/WT (unrelated, open diamond), A179V/WT (open triangle), an unrelated R83C/R291Q compound-heterozygous patient (solid inverted triangle), and a K108E/K108E patient (solid diamond) is also shown. (C) Percentage of CD56+CD3− NK cells within the lymphocyte gate is shown for 4 normal controls, our proband at 4 independent time points, 3 unrelated heterozygous IRF8 mutation patients, an unrelated compound-heterozygous patient, and a K108E/K108E patient. (D) Percentage of CD56bright NK cells within the NK cell compartment is shown for the cohort described in C. (E) FACS phenotyping of heterozygous, compound-heterozygous, and homozygous IRF8 patients. For intracellular staining, mononuclear cells were stimulated (S) with PMA and ionomycin in the presence of brefeldin A or incubated without stimulation (U) before fixation, permeabilization, and detection of intracellular effector molecules. (F) Expression of IFN-γ (after PMA/ionomycin stimulation) and perforin and CD16 in unstimulated NK cells from patients and healthy donors gated on CD56dim and CD56bright subsets. Shown is mean ± SD of 3 independent experiments for perforin and CD16 in healthy donor and A201V/P224L patient; a single experiment is shown for the K108E/K108E patient and IFN-γ detection from all subjects. *P < 0.05 by unpaired Student’s t test. (G) Cytokine secretion from healthy donor (HD) or A201V/P224L patient (Pt) detected by cytokine bead array from PBMCs stimulated for 4 hours by nonautologous EBV-transformed B cells in lytic cycle.
Because of the patient’s consistently low NK cell cytotoxic function and abnormal NK cell subset distribution, we performed multiparametric flow cytometric immune phenotyping of NK cell subsets, including intracellular cytokines IFN-γ and TNF-α as well as perforin and granzymes. We evaluated the patient, his clinically unaffected single-heterozygous family members, an unrelated heterozygous A201V/WT patient with pulmonary nontuberculous mycobacterial infection, a heterozygous T96M/WT patient, an unrelated compound-heterozygous patient (R83C/R291Q), and a homozygous K108E patient (Table 1).
Multiparametric flow cytometric analysis of the proband’s lymphocytes showed that NK cell markers were consistent with the expanded CD56bright subset but otherwise showed no aberrant expression of markers of activation, inhibition, or granzyme B (Figure 3E). Despite decreased representation of the CD56dim subset, markers of NK cell terminal differentiation and memory (CD57, NKG2C) were expressed, as were activating receptors (NKG2D) and inhibitory receptors (NKG2A). Interestingly, however, our proband had consistently low levels of perforin at multiple time points tested, as did the homozygous K108E patient (Figure 3E). Upon stimulation, the proband, as well as those patients with compound heterozygous variants in IRF8, expressed normal or higher than normal levels of IFN-γ and TNF-α measured both as percentage of the population expressing cytokine and on a per-cell basis (Figure 3E and not shown). The production of IFN-γ was comparable between patient and control CD56bright NK cells, which are generally the most potent producers of IFN; however, the patient CD56dim NK cells had increased IFN-γ production relative to control. This accounted for the increase in IFN-γ production in his NK cells when considered as a total population (Figure 3F). The patient with K108E/K108E mutations had impaired IFN-γ production upon stimulation from both NK cell subsets, potentially underscoring the different effects of the different mutations on immune function even in the context of similar NK cell phenotype. The observed decrease in perforin expression in our patient with A201V/P224L mutations and the patient with homozygous K108E mutations was confined to the CD56dim subset, the primary cytolytic effectors. CD16 (FcγRIIIA) was expressed at normal levels on CD56bright and CD56dim NK cells from the patients with A201V/P224L and R83C/R291Q mutations (Figure 3, E and F). Low expression of CD16 on NK cells from the K108E patient was restricted to the CD56dim NK cell subset (Figure 3F).
Given the important role that cytokine production plays in the control of the viral immune response and previously described DC deficiency in patients with IRF8 mutations, we sought to determine whether cytokine production in the context of costimulation by accessory cells was impaired in our patient. To do so, we incubated PBMCs from healthy donor or patient with a nonautologous EBV-transformed B cell line that had been induced to lytic cycle. Secreted cytokines (IL-12p40, IFN-γ, TNF-α, and IL-10) were measured by flow cytometric bead array. As suggested by the intracellular flow cytometry data, the coculture-induced production of IFN-γ and TNF-α was not impaired in the patient, and, similarly to that observed in cytokine-stimulated NK cells, the production of IFN-γ in these cocultures was increased (Figure 3G). Interestingly, elevated secretion of both IL-12 and IL-10 was also observed from patient cells.
As IRF8 serves a previously identified role in DC development and IRF8 deficiency has been shown to lead to a variable loss or absence of DC subsets, we performed phenotypic analysis of monocyte and DC populations. A subtle deficiency of plasmacytoid dendritic cell (pDCs) and CD141+ DCs was observed in the index patient relative to heterozygous family members and a wild-type control (Supplemental Figure 2). These differences were within the scope of those previously ascribed to human IRF8 mutations and were not as severe as those observed in the patient with homozygous K108E mutation (19).
Our findings, however, suggested a novel NK cell phenotype of arrested terminal maturation in the presence of biallelic IRF8 mutations. To further determine the role of IRF8 in NK cell maturation, we performed phenotypic analysis of NK cells from Irf8–/–, Irf8+/–, and Irf8+/+ mice by flow cytometry. While murine NK cells do not express CD56, and not all transcription factor functions are evolutionarily conserved between human and murine NK cells, the developmental stages of murine NK cells can be stratified according to expression of CD27 and CD11b (Mac-1) (26). As previously reported (27–29), Irf8–/– mice had significant splenomegaly and lymphocytosis, leading to increased absolute NK cell number in both spleen (14 × 106 ± 5.7 × 106) and blood (1.2 × 106 ± 0.7 × 106) relative to that of Irf8+/+ controls (spleen 2.5 × 106 ± 1.2 × 106, blood 0.89 × 106 ± 0.6 × 106). Despite this, as seen in Figure 4, NK cells from blood and spleen of Irf8–/–, but not Irf8+/–, mice had an increased frequency of immature Mac-1loCD27hi NK cells (22.5% ± 2.7%) within the NK cell population when compared with wild-type controls (9.7% ± 1.8%). This was also reflected by absolute numbers of these subsets (blood 0.45 × 106 ± 0.4 × 106, spleen 3.6 × 106 ± 1.9 × 106) when compared with wild-type controls (blood 0.36 × 106 ± 0.5 × 106, spleen 0.31 × 106 ± 0.3 × 106). Conversely, Irf8–/– mice had decreased frequency of mature Mac-1hiCD27lo NK cells (48.3% ± 3.9%) compared with wild-type controls (64.3% ± 3.9%). This was despite increased absolute cell number (blood 1 × 106 ± 0.6 × 106, spleen 5.7 × 106 ± 2.1 × 106) compared with controls (blood 0.7 × 106 ± 0.5 × 106, spleen 1.4 × 106 ± 0.6 × 106). Together, these demonstrate impairment in terminal maturation in mice with homozygous, but not heterozygous, Irf8 loss-of-function mutations, suggesting an important parallel to our patients with biallelic IRF8 mutations and increased frequency of immature NK cells.
Figure 4. Irf8–/– mice have a block in terminal NK cell maturation.
Spleen and blood from Irf8–/–, Irf8+/– littermates, and Irf8+/+ controls were analyzed by flow cytometry for markers of NK cell differentiation. (A) Representative FACS of spleen (top) and blood (bottom) from the same mouse. Cells were gated on single cells, then NK1.1+CD3−. (B) Pooled spleen and blood from 4–6 mice per condition are shown. Data shown are mean ± SD. *P < 0.05 by Student’s unpaired t test.
IRF8 is required for human NK cell development in a cell-intrinsic manner.
Given the broad expression of IRF8 within immune cell populations, we sought to determine whether the observed effect on NK cell function was due to NK cell–intrinsic mechanisms or representative of the need for other immune or nonimmune cell types in NK cell development. Thus, we evaluated the ability of patient CD34+ hematopoietic stem cells (HSCs) to generate NK cells in isolation. HSCs were sorted from our proband’s peripheral blood and incubated on OP9-DL1 stromal cells for 28 days in the presence of IL-7, FLT3L, c-Kit, and IL-15, as this in vitro experimental system has been previously demonstrated to promote NK cell development (14, 30, 31). As shown in Figure 5A, patient NK cells derived from CD34+ HSCs were largely CD56bright (76% vs. 19.7% from healthy donor control). The mean fluorescence intensity of CD56 was significantly higher on patient cells versus the normal control (224 vs. 112 in healthy donor) (Figure 5B), consistent with immaturity and incomplete maturation. In addition, while healthy donor–derived cells acquired CD16 (58%), the patient-derived cells had decreased CD16 expression yet retained CD94, a phenotype that is reflective of an immature state. These results show that the role for IRF8 is cell intrinsic and that IRF8 is functioning directly in the generation of appropriate and mature NK cell subsets during development.
Figure 5. Impairment in terminal NK cell differentiation is NK cell intrinsic.
CD34+ hematopoietic stem cells were sorted from the proband and a healthy control, and NK cells were generated by coculture with OP9-DL1 stromal cells for 35 days. Cells were harvested and analyzed in parallel with freshly isolated PBMCs from the proband or the same healthy donor. (A) In vitro–generated NK cells from healthy donor (left, WT/WT) or our proband (right, A201V/P224L) were analyzed by FACS for CD56 expression and CD16 (top) and CD94 (bottom) expression. (B) Histogram showing mean fluorescence intensity of CD56 expression on in vitro–derived NK cells from healthy donor (gray histogram) or patient 1 (empty histogram).
IRF8 is expressed in human NK cell subsets and is upregulated upon IL-15 stimulation.
Expression of IRF8 in human NK cell developmental subsets has not been previously determined. We performed real-time PCR and intracellular flow cytometry to evaluate the expression of IRF8 in human NK cell developmental subsets. While human peripheral blood contains only the CD56bright and CD56dim NK cell subsets, we used human tonsil as a source of developing NK cells and evaluated these cells based on known markers of differentiation (stages 1–5) (32). As shown in Figure 6A, healthy human donors expressed IRF8 in all developmental subsets as measured by intracellular flow cytometry. Interestingly, expression was highest in stage 1 (CD34+) NK cell precursors; however, all developmental subsets isolated from tonsil, as well as those from peripheral blood, expressed significant quantities of IRF8. Further, stimulation of primary human NK cells with IL-15 for as few as 6 hours resulted in an upregulation of IRF8 as measured by quantitative PCR (qPCR) (3-fold; Figure 6B) and flow cytometry (1.3-fold; Figure 6, C and D). Therefore, IRF8 is expressed in human NK cells and is responsive to IL-15 stimulation. This observation is consistent with a critical role for IRF8 during NK cell development.
Figure 6. IRF8 is expressed in all NK cell developmental subsets and is upregulated upon IL-15 stimulation.
(A) NK cell developmental subsets were isolated from tonsil as described in Methods, and IRF8 was detected by intracellular flow cytometry. Shown is 1 representative experiment from 3 independent experiments from 3 normal donors. FMO, fluorescence minus one; PB, peripheral blood. (B) NK cells were incubated for 6 hours with 10 ng/ml of IL-15 or vehicle control, and then RNA was extracted and qPCR performed as described in Methods. ***P < 0.001 by Student’s t test. Shown are the means of 3 independent experiments performed on 3 donors, each done in quadruplicate. (C) NK cells were stimulated as in B and subjected to flow cytometric analysis as in A. Dashed line, IL-15 stimulated; solid line dark histogram, unstimulated; solid line light histogram unstained. (D) Shown is the mean ± SD of 3 independent experiments. *P < 0.05 by unpaired Student’s t test. MFI, mean fluorescence intensity.
IRF8 is required for NK cell expression of transcription factors and NK cell effector molecules.
Previously reported homozygous K108E mutations in IRF8 lead to impaired synthesis of IL-12 due to abrogated IRF1-dependent transcription of the IL12B gene (19). To determine whether the mutations in our proband affected functional binding of IRF8 with cofactors IRF1 and PU.1, we performed luciferase assays with each allelic variant (WT, A201V, and P224L). As seen in Figure 7A, neither of our patient’s variants affected transcriptional activity dependent on PU.1 or IRF1. This is consistent with the observation that DC development was only partially affected in the patient and contrasts with biallelic K108E mutation, in which transactivation with IRF1 was severely impaired and monocytes and DCs were absent (19). Collectively, this underscores the variability of mutational impact on the DC phenotype. To determine the effect that the IRF8 mutations have on human NK cell function, we performed NanoString gene expression analysis (Human Immunology v2 gene set) on isolated NK cells from our proband, his family members, and 3 healthy donor controls. We identified 403 genes with a greater than 1.5-fold up- or downregulation in the patient relative to all 3 healthy donors (Supplemental Table 2). These included 18 of particular relevance to NK cell development, cytotoxicity, or homeostasis (Figure 7B), including transcription factors NFIL3, IKZF2, TBX21, EOMES, and STAT5A/B. Differentially expressed effector molecules and cell surface receptors included PRF1, GZMA, GZMB, CCR7, CD27, CD2, HAVCR2, and KLRG1. Ingenuity Pathway Analysis of all genes differentially up- or downregulated more than 5-fold revealed transcription factors, growth factors, kinases, and transmembrane receptors (Figure 7C). To provide additional evidence that the genes differentially expressed in NK cells might be direct transcriptional targets of the mutant IRF8 proteins, we compared genes that consistently had an absolute fold change of ≥2 with published ChIP sequencing data (33). This analysis revealed a significant overlap of differentially expressed genes with loci associated with 1 or more IRF8 peaks (Supplemental Table 3). There was no preferential association of up- or downregulated transcripts with the presence of IRF8 binding. To further biologically validate differential expression of potential direct targets, we focused on NFIL3 and PRDM1, where IRF8 binding was observed in the study by Langlais et al. (33) (Supplemental Table 3), and IRF4, which also has evidence of IRF8 binding by ChIP sequencing (34). We performed qPCR on healthy donor– or patient-derived B cell lines and found decreased expression of each in the patient relative to 3 healthy donors (Figure 7D). Re-expression of WT IRF8 protein in the patient B cell line rescued the expression of IRF4 and PRDM1 and partially rescued expression of NFIL3. Therefore, biallelic IRF8 deficiency results in decreased expression of human NK cell transcriptional regulators and effector molecules as well as genes not previously associated with NK cell regulation. Complementation of the patient B cell line with WT IRF8 restores expression of these, demonstrating a direct effect of compound heterozygous A201V/P224L mutations on the expected IRF8-mediated gene expression profile.
Figure 7. Deregulation of multiple genes in A201V/P224L IRF8 patient NK cells.
(A) Promoter assays were performed in HeLa cells with WT or mutant alleles as indicated. Mutant and WT IRF8 showed indistinguishable regulation of PU.1- and IRF1-dependent genes. Results are displayed as fold increase relative to cells cotransfected with the empty vector. These data are derived from triplicate samples and are displayed as the mean ± SD. Data were analyzed using Student’s unpaired t test (**P < 0.01, ***P < 0.001). (B) Selected NK cell genes up- or downregulated in IRF8 A201V/P224L patient and heterozygous family members identified by NanoString gene expression analysis. (C) Selected immune genes classified by Ingenuity Pathway Analysis that are up- or downregulated more than 3-fold in the A201V/P224L patient relative to all 3 healthy donors (total NK cells). (D) Expression of selected genes validated by qPCR of B cell lines from 2 healthy donors (HD), patient (Pt), or patient cells stably transfected with WT IRF8 (Pt+IRF8). Mean ± SD of 1 representative experiment of 4 (performed in quadruplicate) is shown normalized to the mean of HD. *P < 0.05, ***P < 0.0001 by ordinary 1-way ANOVA with Tukey’s multiple comparisons.
Discussion
Genetic causes of aberrations in NK cell development are exceedingly rare but confer familial susceptibility to disease, particularly viral infection. Here we identify a novel human NK cell developmental defect and cause of NKD due to biallelic mutation in the IRF8 gene. Interestingly, while homozygous and heterozygous IRF8 mutations lead to a spectrum of DC deficiency ranging from mild to severe, biallelic variants in our patient and 2 additional unrelated patients result in NKD as a result of a block in terminal maturation. In our patient this phenotype occurs in the context of normal activation of PU.1 and IRF1, underscoring the varying impact of IRF8 mutations.
Our proband was originally diagnosed in 1980 and reported in 1982 with the first case of non–X-linked susceptibility to EBV as a result of NKD (1), and as such is within the first reported cohort of patients with NKD. Having the unique opportunity to study the 1 surviving affected patient from this family longitudinally has underscored the stability of the NK cell phenotype, namely decreased NK cell number with a paucity of CD56dim NK cells and interrupted maturation.
The IRF family plays a well-documented role in inflammation, and these transcription factors are also becoming increasingly recognized for their importance in hematopoietic development (35, 36). In particular, a critical requirement for IRF8 in DC development is described in mice with both hypomorphic and knockout mutations of IRF8, as well as previously described patients with heterozygous and homozygous mutations (19). Patients with homozygous K108E mutation are close in phenotype to the knockout mouse, with severe deficits of monocytes and DCs and a myeloproliferative disorder (29, 37–39). This homozygous mutation in the DNA-binding domain of IRF8 abrogates nuclear translocation but not protein expression, leading to impaired function and defective IL-12, IFN-γ, and TNF-α production (21). In contrast, the spontaneous heterozygous T80A mutation leads to specific loss of the CD1c+ compartment of CD11c+ DCs but retention of CD141+ DCs, monocytes, and CD123+ plasmacytoid DCs (19). Relative to disease caused by autosomal recessive K108E mutation, the T80A heterozygous mutation leads to a less severe clinical course. IL-12 production, but not type I IFN, is affected. In both cases, both B and T cell numbers are largely normal; however, CD25hiFoxP3+ Treg numbers are decreased in patients with K108E mutation (39).
The mechanism for the differential impact of IRF8 mutations on DC and NK cell homeostasis remains unclear. Gene dosage and mutation site–specific effects are commonly observed in transcriptions factors such as IRF8 that are regulated by superenhancer structures (40). As opposed to A201V/P224L, the K108E mutation is unable to activate IRF1- and PU.1-dependent transcription and, in the biallelic state, is associated with monocyte and DC deficiency and a very marked distortion of NK cell development in the ratio of CD56bright to CD56dim cells. The patient described here with A201V/P224L mutation has an NK phenotype with only subtle DC deficiency, in keeping with the ability of the 2 variants to activate transcription with PU.1 and IRF1. As these processes are required for DC development and function, these data suggest that additional IRF8-dependent mechanisms are required for normal NK cell homeostasis.
While all of our patient’s surviving single-heterozygous family members have unremarkable medical history, an unrelated patient with heterozygous A201V/WT mutation of IRF8 and late-onset pulmonary nontuberculous mycobacterial infection was identified, although a role for this IRF8 change in her disease state is unclear. The late onset of disease in an unrelated patient with A201V mutation suggests the relatively mild impact of this variant and that careful monitoring of heretofore clinically unaffected heterozygous family members may be prudent.
The apparent requirement for IRF8 in an NK cell–intrinsic manner, as we show using in vitro differentiation experiments, points to a direct role in the generation of appropriately distributed CD56bright and CD56dim NK cells and the subsequent acquisition of effector function. As with GATA2 mutations, it is intriguing that perturbation of development that allows for the presence of CD56dim NK cells in peripheral blood nevertheless significantly affects the function of these cells. The profound disease in our patient and the consistent absence of NK cell cytotoxic function as a result of this developmental block speak to the strict regulation of this process and support a model in which CD56bright cells are the precursors of CD56dim. The impaired terminal maturation of NK cells from Irf8–/–, but not Irf8+/–, mice suggests a conserved requirement for IRF8 in the final stages of NK cell development and supports a linear relationship between the subsets. As with GATA2 mutations, CD56+CD3+ T cells are present in our patient, often at higher frequencies than in normal controls.
IRF8 is known to play a role in both cell fate decision and functional control of viral infection. IRFs are well-described downstream mediators of TLR signaling, and IRF8 mediates TLR9 signaling through TRAF6 and NF-κB (41, 42). Production of IFN-γ and TNF-α was normal in our patient in NK cells stimulated by PMA/ionomycin or EBV-transformed B cells, suggesting that disruption of cytokine production was not the primary mechanism of viral susceptibility in our patient. Elevated levels of IFN-γ (both intracellular and secreted in response to stimuli) were measured from our patient at multiple times of assessment. While the clinical significance of this is unclear, it may be related to PRDM1 downregulation (detected by gene expression array and qPCR of patient B cell lines), as PRDM1 has been previously shown to directly bind and negatively regulate the IFN-γ locus in human NK cells (43). In contrast, as previously reported in PBMCs (19), K108E/K108E NK cells did not produce IFN-γ in response to stimulation, suggesting that the differential effect of the mutations on IRF1 activation may include differing effects on IFN-γ production in NK cells. In primary NK cells or NK-92 cell lines, cytokine stimulation increases levels of nuclear IRF8 and increased binding to DNA (43, 44). In CD8+ T cells, common γ chain signaling in combination with CD3 cross-linking results in increased IRF8 expression in concert with JAK/STAT signaling and leads to differentiation of CD8+ effectors from naive precursors. Deletion of IRF8 results in decreased, but not absent, T lymphocyte cytotoxicity, and it is hypothesized that presentation of γc cytokine, namely IL-15, in combination with antigen by neighboring DCs could drive CD8+ differentiation (45). IL-15 is unique among cytokines in its absolute requirement for NK cell development and homeostasis in both mice and humans (46, 47). Therefore, the positioning of IRF8 in this critical signaling pathway speaks to its importance in generating mature, functional human NK cells, and the significant downregulation of JAK3 and STAT5A and -B in our patient relative to 3 healthy donors supports this hypothesis.
While we show that the NK cell developmental defect is intrinsic, an additional model for the requirement of IRF8 in NK cell development and function would be through the trans-presentation of IL-15 by DCs, which would potentially make the loss of DCs and functional defect of NK cells in IRF8 patients particularly damaging. Our data support a link between IL-15, IRF8, and NK cell development, as we show that IL-15 increases IRF8 protein expression in as few as 6 hours in ex vivo NK cells. This observation suggests a need for the IL-15–induced proliferation of CD56bright NK cells in the generation of the CD56dim subset, as proposed in the report of MCM4 deficiency, the only other reported immune deficiency to result in this distinctive NK cell phenotype (2). This concept was originally proposed by Lutz et al. after they showed that CD56bright NK cells undergo significantly greater proliferation than the CD56dim subset (48). In addition, FLT3L-mediated development of DCs is dependent on IRF8 signaling, and FLT3L-deficient mice have decreased NK cell number and impaired NK cell development (49). Therefore, our data could suggest a similar requirement for IRF8 in FLT3L-mediated signaling in NK cells.
While a direct role for IRF8 in NK cell development has not been described, IRF2-deficient mice have reduced NK cell number and, as a result, impaired NK cell cytotoxic function (50, 51). Further, IRF2-deficient bone marrow is enriched for immature NK cells (selective loss of the CD11bhiDX5hi subset) with higher rates of apoptosis and decreased sensitivity to IL-15, a defect that is cell intrinsic (51). Whether the loss of CD56dim NK cells in IRF8 deficiency is a result of accelerated apoptosis of this subset, as is the case for IRF2 mice, or a result of impairment in the transition from CD56bright to CD56dim, remains to be seen. While IRF1-deficient mice also have decreased NK cell number and function, IRF1 is required for IL-15 production by bone marrow stromal cells, and the NKD in these mice is due to the loss of IL-15 in the niche (52).
IRF8 plays a role in lineage commitment, particularly of DCs and B cells, and can substitute for the highly related factor IRF4 in some circumstances (18, 40, 53, 54). Both IRF4 and IRF8 have the capacity to act as either transcriptional activators or repressors but require other lineage-specific transcription factors as binding partners to achieve the appropriate outcome. For example, IRF4/IRF8 can interact with PU.1 or SPI-B at cis-regulatory regions termed Ets-IRF composite elements (EICEs) in B cells, DCs, and macrophages. While PU.1 is not required for NK cell development, PU.1-deficient NK cells in mice showed reduced proliferation both at baseline and in response to IL-12 and IL-2 (55). However, PU.1 is difficult to detect in mature NK cells and is hypothesized to be required exclusively for lineage commitment (56). While EICEs are fairly well characterized, genetic approaches in T cells and DCs have recently revealed novel motifs recognized by IRF4/IRF8, such as AP-1–IRF composite elements bound by BATF-IRF complexes (57, 58). Given the normal regulation of PU.1 and IRF1 elements by our patient’s IRF8 variants, work is currently underway to identify novel binding partners of IRF8 in NK cells.
Recent high-resolution analysis of the ontology of both mouse and human ILC and NK cell lineages shows IRF8 highly expressed specifically in NK cells and, in the case of human cells, intraepithelial ILC1s (59, 60). Our gene expression analysis of purified NK cells suggests that IRF8 plays a direct role in the regulation of NK cell genes required for maturation and function of human NK cells. These include NFIL3 (encoding E4BP4), PRDM1 (encoding BLIMP-1), TBX21 (encoding T-bet), and IKZF2 (encoding HELIOS), all previously shown to be required for mature NK cell generation and/or homeostasis (43, 61–66). E4bp4–/– mice fail to generate even immature NK cell precursors, indicating a very early blockade in development, and E4BP4 is considered a master regulator of NK cell fate (61, 64, 65). The previously demonstrated gene dosage effect of E4BP4 suggests that repression but not absence of NFIL3, as seen in our patient, may lead to decreased mature NK cell numbers while not completely abrogating development (61), and placing NFIL3 downstream of IRF8 would be consistent with previous reports showing normal IRF8 expression in Nfil3–/– mice (67). Similarly, Tbx21–/– mice fail to generate mature NK cells and T-bet expression increases with NK cell maturation, with T-bet highly expressed in the CD56dim NK cell subset (62, 68). Both PRDM1 and HELIOS tune NK cell responsiveness; PRDM1 is a transcriptional repressor that binds directly to TNF and IFNG loci to regulate cytokine secretion in human NK cells (43) and is bound directly by IRF8 in B cells and macrophages (34), whereas HELIOS regulation is a mechanism of modulation of NK cell activity mediated through NKp46 ligation (69). Finally, the NK cell defect observed in our patient may be attributable to the significant downregulation of STAT5a and STAT5b, in concert with JAK3 and perforin. This signaling axis has been shown to be critical for NK cell survival and function, and STAT5b regulates NK cell cytolytic activity by binding directly to the perforin promoter (70, 71). Collectively, our data suggest that IRF8 may be playing a role in both NK cell lineage commitment and effector responses, which is in keeping with the functional and developmental defects observed in NK cells from patients with biallelic IRF8 mutations.
In summary, we have defined a novel NKD and primary immunodeficiency that occurs specifically as a result of biallelic mutations in IRF8 encoding the transcription factor IRF8. These alterations result in impaired NK cell function, decreased NK cell number, and loss of the CD56dim subset of human NK cells, reflecting impaired NK cell terminal maturation also observed in Irf8–/–, but not Irf8+/–, mice. Immunodeficiency occurring in association with IRF8 mutation is likely to involve defects of NK cells in addition to previously described loss of monocytes, DCs, and macrophage function. These findings define a novel, critical role for IRF8 function in NK cell development, extending the previously described range of phenotypes associated with IRF8 mutation in humans and identifying IRF8 mutation as the fourth genetic explanation for classical NKD leading to aberrant NK cell development and association with susceptibility to viral disease.
Methods
Human NK cell preparation and target cell lines.
PBMCs were isolated by density centrifugation over Ficoll-Paque lymphocyte isolation medium according to the manufacturer’s protocol (Amersham). For enrichment of NK cells for qPCR analysis, whole blood was incubated with RosetteSep (STEMCELL Technologies) before density centrifugation. For isolation of cells from tonsils, tissue was obtained from routine tonsillectomies and mechanically separated by passing through a 40-μm strainer prior to isolation of secondary lymphoid cells by density centrifugation. Human K562 target cells were used for 51Cr release assays and maintained in supplemented RPMI media as previously described (72). K562 cells were acquired from American Type Culture Collection and routinely confirmed to be mycoplasma negative.
Murine NK cell phenotyping.
Irf8–/– mice were generated as described previously (29) and were a gift from Keiko Ozato (NIH, Bethesda, Maryland, USA). Male and female mice between 9 and 12 weeks of age were euthanized upon arrival in accordance with Baylor College of Medicine’s animal care guidelines. Spleens were removed and single-cell suspensions prepared by mechanical separation and density centrifugation as previously described (73). Blood was obtained by heart puncture immediately following euthanasia. All cells were maintained in PBS/2% FCS at 4°C throughout the staining procedure. Cells were stained for viability with LIVE/DEAD cell staining kit according to the manufacturer’s instructions (Thermo Fisher). Rat anti-mouse Fc Block (Becton Dickinson) was added for 10 minutes on ice, and then fluorophore-conjugated specific antibodies against extracellular markers were added for 30 minutes (antibodies listed in Supplemental Table 4). Cells were washed once in 2% FCS/PBS, then fixed in a final concentration of 1% paraformaldehyde. Cells were acquired and data analyzed as described for human flow cytometry and immune phenotyping.
Generation of lymphoblastoid cell lines.
Lymphoblastoid cell lines were generated from patient PBMCs by addition of EBV supernatant as previously described (74). Briefly, PBMCs were transduced with a B95-8 EBV supernatant and cultured in RPMI medium with 10% FBS, 2 mM glutamine, and 1 μg/ml cyclosporin A. Cell lines were confirmed mycoplasma negative before their use in experiments.
NK cell functional assays.
Standard 51Cr release assays were performed in round-bottomed plates against K562 target cells as previously described (75).
Flow cytometry and immune phenotyping.
Fluorophore-conjugated antibodies used for multiparametric flow cytometry analysis of patient PBMCs are listed in Supplemental Table 4. Where indicated, stimulation for the detection of intracellular cytokines was by PMA/ionomycin for 4 hours in the presence of brefeldin A followed by fixation and permeabilization with BD Cytofix/Cytoperm. For intracellular staining of IRF8 by flow cytometry, cells were permeabilized using the FoxP3 transcription buffer staining kit (eBioscience) followed by incubation with IRF8 eFluor 710 (clone V3GYWCH, eBioscience). Flow cytometry was acquired on a BD Fortessa configured for 14 colors. All analysis was done in FlowJo X (Tree Star). Positive populations were identified as those with fluorescence greater than that of isotype or fluorescence-minus-one controls. Cytokine bead arrays were performed with custom BD Biosciences CBA Flex Sets using IL-12, IFN-γ, TNF-α, and IL-10 probes. PBMCs (2 × 106) were incubated for 4 hours with 105 EBV-transformed B cells previously induced to lytic cycle with 200 ng/ml phorbol 12-myristate 13-acetate. Data were analyzed with FlowJo X (Tree Star), and statistical analysis was performed and graphed using Prism 6.0 software (GraphPad Software).
Imaging flow cytometry.
Cells were fixed and permeabilized using FoxP3 transcription buffer (eBioscience) followed by incubation with IRF8 eFluor 710 (clone V3GYWCH, eBioscience) and DAPI. Samples were acquired on an ImageStream MkII (EMD Millipore) with a ×40 objective. Spectral compensation was applied using single-stained controls. Images were processed and exported using IDEAS software (EMD Millipore). Similarity score was calculated in IDEAS as a measure of colocalization.
Western blotting.
Lymphoblastoid cell lines (3 × 105) from patient or healthy donor were lysed, separated by 4%–10% SDS-PAGE gel, then transferred to nitrocellulose membrane, and blocked with skim milk. IRF8 was detected with anti-ICSBP (clone C-19, Santa Cruz Biotechnology) and actin with anti-actin (catalog A2066, Sigma-Aldrich) followed by goat anti-rabbit IRDye 700 secondary antibody. Proteins were detected on a LiCor Odyssey.
Quantitative reverse transcriptase PCR.
RNA was extracted from enriched NK cells or secondary lymphoid cells (SLC) using DirectZol (Genesee Scientific) or RNeasy (Qiagen), and cDNA was made using VILO (Invitrogen). IRF8, PRDM1, NFIL3, and IRF4 were amplified on a Roche Light Cycler by Taqman gene expression array. GAPDH was used to normalize expression for each sample. Each condition was performed in quadruplicate. Analysis was performed with Roche Light Cycler software and exported to Excel before graphing with Prism 6.0 (GraphPad software).
Genetic analyses.
Genomic DNA was extracted from EDTA-preserved nonheparinized blood samples using the Gentra PureGene Blood Kit (Qiagen). Whole exome sequencing was conducted as previously described (23). Briefly, exonic portions of genomic DNA fragments were captured in solution using custom VCrome probes (NimbleGen). Resulting DNA was sequenced using Illumina HiSeq 2500 equipment. Data processing and variant annotation were performed per standard pipeline analyses. Variants were evaluated using both recessive and dominant models. Bioinformatic filters were placed using a minor allelic frequency less than 0.005 in recessive models within the Baylor-Hopkins Center for Mendelian Genomics (18), ESP5400, 1000 Genomes, and ExAC databases. Potential recessive variants were also excluded if present in the ExAC database with a homozygote or hemizygote count of 10 or greater. For compound heterozygous analyses, variants were excluded if both variants were present in the maternal exome or in the exome of the unaffected brother. For autosomal dominant model analyses, variants were excluded with minor allelic frequencies greater than 0.001 in the same databases and if present in the ExAC database with an allele count greater than 5. Variants were confirmed by Sanger sequencing; primers were constructed using Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi), and amplified DNA constructs were sequenced by the Baylor DNA Sequencing Core Facility. Sequences were visualized using 4Peaks software (Nucleobytes). For the deceased sister, formalin-fixed paraffin-embedded gallbladder tissue slices were retrieved from the Pathology Department at the University of Pennsylvania (Philadelphia, Pennsylvania, USA). DNA was extracted by the Baylor Human Genome Sequencing Center for Sanger sequencing.
In vitro NK cell differentiation.
CD34+ precursors were isolated from peripheral blood of the patient or healthy donor and highly purified by fluorescence-based cell sorting. Following purification, cells were incubated on mitotically inactivated OP9-DL1 stromal cells (a gift from J.C. Zúñiga-Pflücker, University of Toronto, Toronto, Ontario, Canada) in media containing 5 ng/ml IL-3, 20 ng/ml IL-7, 10 ng/ml FLT3L, and 10 ng/ml IL-15 for 4 weeks. After culture, cells were phenotyped by FACS in parallel with control cells from a healthy donor.
RNA isolation and gene expression analysis.
NK cells were isolated by FACS sorting to greater than 95% purity following enrichment by RosetteSep negative selection (STEMCELL Technologies). Fifty to a hundred nanograms of RNA was analyzed with the Human Immunology v2 probe set using a NanoString nCounter. Background correction was done using the mean of 8 negative control probes. Normalization was performed by calculation of the geometric mean of positive controls and content normalization parameters (15 housekeeping genes). Data processing was performed using nSolver software, and data were exported to Ingenuity Pathway Analysis for further analyses. Genes up- and downregulated greater than 1.5-fold in samples relative to the mean of all 3 healthy donor controls were considered significant.
Luciferase assays.
Constructs encoding WT human IRF8 or allelic variants A201V and P224L were generated in the pIRES2-EGFP expression vector (Clontech). These were cotransfected with vectors encoding IRF1 or PU.1 and reporter luciferase vectors harboring interferon-stimulated response element (ISRE) or EICE motifs and quantified using the Promega dual luciferase assay system. Details of the PSMB8 and TAPASIN promoter luciferase vectors have been described previously (76).
Statistics.
Statistical analysis was performed using Prism 6.0 (GraphPad Software). Unpaired 2-tailed Student’s t test was used to compare data sets. Ordinary 1-way ANOVA with Tukey’s multiple comparisons test was performed for multiple comparisons. For all data shown, error bars denote SD. A P value less than 0.05 was considered significant.
Study approval.
All studies were performed in accordance with the Declaration of Helsinki with the written and informed consent of all participants under the guidance of Children’s Hospital of Philadelphia, NIH, University of Newcastle, and Baylor College of Medicine Institutional Review Boards. Animal studies were performed in accordance with the Baylor College of Medicine and NIH Animal Care Committees.
Author contributions
The study was conceptualized by EMM, JRL, and JSO. Methodology was developed by EMM, VB, GMD, MC, SMH, M.A. Caligiuri, JRL, RAG, SP, and JSO. Investigation was performed by EMM, MDK, IKC, DFL, SNJ, DMM, ASP, ZCA, TNC, JTG, SP, LSA, GDH, JBS, MHS, LBW, DTL, ST, M.A. Care, SM, AGF, and EPS. EMM and JSO wrote the original draft of the manuscript. EMM, JRL, VB, MC, GMD, MDK, and JSO reviewed and edited the manuscript. Funding acquisition was by JSO and JRL. Clinical care was provided by JSO, SMH, SS, MDK, AJC. Resources were provided by KO.
Supplementary Material
Acknowledgments
The authors thank all the patients and their families, particularly the proband’s, for their engagement in this study. The authors also thank Linda Monaco-Shawver for technical assistance. This project was supported by the Genomic and RNA Profiling Core at Baylor College of Medicine and the expert assistance of the core director, Lisa D. White, and Mylinh Bernardi. This work was supported by NIH–National Institute of Allergy and Infectious Diseases grants R01AI067946 and R01 AI120989 to JSO, the Jeffrey Modell Foundation, and NIH (National Human Genome Research Institute/National Heart, Lung, and Blood Institute) grant U54HG006542 to the Baylor-Hopkins Center for Mendelian Genomics.
Footnotes
DFL’s present address is: Allergic Disease Associates, Philadelphia, Pennsylvania, USA.
EPS’s present address is: Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
GDH’s present address is: Washington University School of Medicine, St. Louis, Missouri, USA.
Conflict of interest: J.R.Lupski has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc. is a member of the Scientific Advisory Board of Baylor Genetics, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting (5294533; 5306616; 5523217; 5599920; 5667968; 5780223; 6132954; 6713300; 7141420; 7189511; 7192579; 7273698; 7537899; 8129353; 9365899; 0424473; 0610396; 0989805). The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis (CMA) and clinical exome sequencing offered in Baylor Genetics (BMGL: http://www.bmgl.com/BMGL/Default.aspx).
Reference information:J Clin Invest. 2017;127(1):306–320. doi:10.1172/JCI86276.
Contributor Information
Venetia Bigley, Email: Venetia.Bigley@newcastle.ac.uk.
Matthew A. Care, Email: matthew.care@gmail.com.
Sheetal Maisuria, Email: S.Maisuria@leeds.ac.uk.
Sumihito Togi, Email: sumihito.togi@nih.gov.
Zeynep Coban Akdemir, Email: Zeynep.CobanAkdemir@bcm.edu.
Sinisa Savic, Email: sinisa.savic@nhs.net.
Matthew Collin, Email: matthew.collin@newcastle.ac.uk.
Richard A. Gibbs, Email: agibbs@bcm.edu.
Michael A. Caligiuri, Email: michael.caligiuri@osumc.edu.
Keiko Ozato, Email: ozatok@dir6.nichd.nih.gov.
Silke Paust, Email: silke.paust@bcm.edu.
James R. Lupski, Email: jlupski@bcm.edu.
References
- 1.Fleisher G, Starr S, Koven N, Kamiya H, Douglas SD, Henle W. A non-x-linked syndrome with susceptibility to severe Epstein-Barr virus infections. J Pediatr. 1982;100(5):727–730. doi: 10.1016/S0022-3476(82)80572-6. [DOI] [PubMed] [Google Scholar]
- 2.Gineau L, et al. Partial MCM4 deficiency in patients with growth retardation, adrenal insufficiency, and natural killer cell deficiency. J Clin Invest. 2012;122(3):821–832. doi: 10.1172/JCI61014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hughes CR, et al. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122(3):814–820. doi: 10.1172/JCI60224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mace EM, et al. Mutations in GATA2 cause human NK cell deficiency with specific loss of the CD56(bright) subset. Blood. 2013;121(14):2669–2677. doi: 10.1182/blood-2012-09-453969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orange JS. Natural killer cell deficiency. J Allergy Clin Immunol. 2013;132(3):515–525. doi: 10.1016/j.jaci.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orange JS. Unraveling human natural killer cell deficiency. J Clin Invest. 2012;122(3):798–801. doi: 10.1172/JCI62620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries E, et al. Identification of an unusual Fcγ receptor IIIa (CD16) on natural killer cells in a patient with recurrent infections. Blood. 1996;88(8):3022–3027. [PubMed] [Google Scholar]
- 8.Grier JT, et al. Human immunodeficiency-causing mutation defines CD16 in spontaneous NK cell cytotoxicity. J Clin Invest. 2012;122(10):3769–3780. doi: 10.1172/JCI64837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jawahar S, Moody C, Chan M, Finberg R, Geha R, Chatila T. Natural Killer (NK) cell deficiency associated with an epitope-deficient Fc receptor type IIIA (CD16-II) Clin Exp Immunol. 1996;103(3):408–413. doi: 10.1111/j.1365-2249.1996.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22(11):633–640. doi: 10.1016/S1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 11.Fehniger TA, et al. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101(8):3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 12.Chan A, et al. CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J Immunol. 2007;179(1):89–94. doi: 10.4049/jimmunol.179.1.89. [DOI] [PubMed] [Google Scholar]
- 13.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 14.Freud AG, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203(4):1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Romagnani C, et al. CD56brightCD16– killer Ig-like receptor– NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178(8):4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 16.Etzioni A, Eidenschenk C, Katz R, Beck R, Casanova JL, Pollack S. Fatal varicella associated with selective natural killer cell deficiency. J Pediatr. 2005;146(3):423–425. doi: 10.1016/j.jpeds.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 17.Hanna S, Béziat V, Jouanguy E, Casanova JL, Etzioni A. A homozygous mutation of RTEL1 in a child presenting with an apparently isolated natural killer cell deficiency. J Allergy Clin Immunol. 2015;136(4):1113–1114. doi: 10.1016/j.jaci.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 18.Aliberti J, et al. Essential role for ICSBP in the in vivo development of murine CD8α+ dendritic cells. Blood. 2003;101(1):305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 19.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tailor P, Tamura T, Morse HC, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111(4):1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salem S, et al. Functional characterization of the human dendritic cell immunodeficiency associated with the IRF8(K108E) mutation. Blood. 2014;124(12):1894–1904. doi: 10.1182/blood-2014-04-570879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biron CA, Byron KS, Sullivan JL. Severe herpesvirus infections in an adolescent without natural killer cells. N Engl J Med. 1989;320(26):1731–1735. doi: 10.1056/NEJM198906293202605. [DOI] [PubMed] [Google Scholar]
- 23.Lupski JR, et al. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5(6):57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.George TC, et al. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311(1–2):117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Angelo LS, et al. Practical NK cell phenotyping and variability in healthy adults. Immunol Res. 2015;62(3):341–356. doi: 10.1007/s12026-015-8664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176(3):1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 27.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22(1):145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 28.Masumi A, Tamaoki S, Wang IM, Ozato K, Komuro K. IRF-8/ICSBP and IRF-1 cooperatively stimulate mouse IL-12 promoter activity in macrophages. FEBS Lett. 2002;531(2):348–353. doi: 10.1016/S0014-5793(02)03556-1. [DOI] [PubMed] [Google Scholar]
- 29.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87(2):307–317. doi: 10.1016/S0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 30.De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 2005;106(10):3498–3506. doi: 10.1182/blood-2005-02-0496. [DOI] [PubMed] [Google Scholar]
- 31.Jaleco AC, et al. Differential effects of Notch ligands Delta-1 and Jagged-1 in human lymphoid differentiation. J Exp Med. 2001;194(7):991–1002. doi: 10.1084/jem.194.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freud AG, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22(3):295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Langlais D, Barreiro LB, Gros P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med. 2016;213(4):585–603. doi: 10.1084/jem.20151764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu H, et al. Regulation of bifurcating B cell trajectories by mutual antagonism between transcription factors IRF4 and IRF8. Nat Immunol. 2015;16(12):1274–1281. doi: 10.1038/ni.3287. [DOI] [PubMed] [Google Scholar]
- 35.Collin M, Bigley V, Haniffa M, Hambleton S. Human dendritic cell deficiency: the missing ID? Nat Rev Immunol. 2011;11(9):575–583. doi: 10.1038/nri3046. [DOI] [PubMed] [Google Scholar]
- 36.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 37.Giese NA, et al. Interferon (IFN) consensus sequence-binding protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J Exp Med. 1997;186(9):1535–1546. doi: 10.1084/jem.186.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turcotte K, Gauthier S, Tuite A, Mullick A, Malo D, Gros P. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia-like syndrome in BXH-2 mice. J Exp Med. 2005;201(6):881–890. doi: 10.1084/jem.20042170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bigley V, et al. The human syndrome of dendritic cell, monocyte, B and NK lymphoid deficiency. J Exp Med. 2011;208(2):227–234. doi: 10.1084/jem.20101459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grajales-Reyes GE, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16(7):708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujimura H, et al. Toll-like receptor 9 signaling activates NF-κB through IFN regulatory factor-8/IFN consensus sequence binding protein in dendritic cells. J Immunol. 2004;172(11):6820–6827. doi: 10.4049/jimmunol.172.11.6820. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, et al. IRF-8/interferon (IFN) consensus sequence-binding protein is involved in Toll-like receptor (TLR) signaling and contributes to the cross-talk between TLR and IFN-γ signaling pathways. J Biol Chem. 2006;281(15):10073–10080. doi: 10.1074/jbc.M507788200. [DOI] [PubMed] [Google Scholar]
- 43.Smith MA, et al. PRDM1/Blimp-1 controls effector cytokine production in human NK cells. J Immunol. 2010;185(10):6058–6067. doi: 10.4049/jimmunol.1001682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lehtonen A, Lund R, Lahesmaa R, Julkunen I, Sareneva T, Matikainen S. IFN-α and IL-12 activate IFN regulatory factor 1 (IRF-1), IRF-4, and IRF-8 gene expression in human NK and T cells. Cytokine. 2003;24(3):81–90. doi: 10.1016/j.cyto.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Miyagawa F, Zhang H, Terunuma A, Ozato K, Tagaya Y, Katz SI. Interferon regulatory factor 8 integrates T-cell receptor and cytokine-signaling pathways and drives effector differentiation of CD8 T cells. Proc Natl Acad Sci U S A. 2012;109(30):12123–12128. doi: 10.1073/pnas.1201453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy MK, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191(5):771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma A, Koka R, Burkett P. Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol. 2006;24:657–679. doi: 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 48.Lutz CT, et al. Human NK cells proliferate and die in vivo more rapidly than T cells in healthy young and elderly adults. J Immunol. 2011;186(8):4590–4598. doi: 10.4049/jimmunol.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95(11):3489–3497. [PubMed] [Google Scholar]
- 50.Lohoff M, et al. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J Exp Med. 2000;192(3):325–336. doi: 10.1084/jem.192.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taki S, Nakajima S, Ichikawa E, Saito T, Hida S. IFN regulatory factor-2 deficiency revealed a novel checkpoint critical for the generation of peripheral NK cells. J Immunol. 2005;174(10):6005–6012. doi: 10.4049/jimmunol.174.10.6005. [DOI] [PubMed] [Google Scholar]
- 52.Ogasawara K, et al. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature. 1998;391(6668):700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- 53.Lu R, Medina KL, Lancki DW, Singh H. IRF-4,8 orchestrate the pre-B-to-B transition in lymphocyte development. Genes Dev. 2003;17(14):1703–1708. doi: 10.1101/gad.1104803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura T, et al. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J Immunol. 2005;174(5):2573–2581. doi: 10.4049/jimmunol.174.5.2573. [DOI] [PubMed] [Google Scholar]
- 55.Colucci F, Samson SI, DeKoter RP, Lantz O, Singh H, Di Santo JP. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97(9):2625–2632. doi: 10.1182/blood.V97.9.2625. [DOI] [PubMed] [Google Scholar]
- 56.Hesslein DG, Lanier LL. Transcriptional control of natural killer cell development and function. Adv Immunol. 2011;109:45–85. doi: 10.1016/B978-0-12-387664-5.00002-9. [DOI] [PubMed] [Google Scholar]
- 57.Glasmacher E, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338(6109):975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tussiwand R, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490(7421):502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koues OI, et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell. 2016;165(5):1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shih HY, et al. Developmental acquisition of regulomes underlies innate lymphoid cell functionality. Cell. 2016;165(5):1120–1133. doi: 10.1016/j.cell.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gascoyne DM, et al. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10(10):1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 62.Gordon SM, et al. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36(1):55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenne CN, et al. T-bet-dependent S1P5 expression in NK cells promotes egress from lymph nodes and bone marrow. J Exp Med. 2009;206(11):2469–2481. doi: 10.1084/jem.20090525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kamizono S, et al. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206(13):2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Male V, Nisoli I, Gascoyne DM, Brady HJ. E4BP4: an unexpected player in the immune response. Trends Immunol. 2012;33(2):98–102. doi: 10.1016/j.it.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 66.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Vα14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 67.Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 2011;117(23):6193–6197. doi: 10.1182/blood-2010-07-295873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Knox JJ, Cosma GL, Betts MR, McLane LM. Characterization of T-bet and eomes in peripheral human immune cells. Front Immunol. 2014;5:217. doi: 10.3389/fimmu.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Narni-Mancinelli E, et al. Tuning of natural killer cell reactivity by NKp46 and Helios calibrates T cell responses. Science. 2012;335(6066):344–348. doi: 10.1126/science.1215621. [DOI] [PubMed] [Google Scholar]
- 70.Imada K, et al. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188(11):2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu CR, Ortaldo JR, Curiel RE, Young HA, Anderson SK, Gosselin P. Role of a STAT binding site in the regulation of the human perforin promoter. J Immunol. 1999;162(5):2785–2790. [PubMed] [Google Scholar]
- 72.Banerjee PP, Pandey R, Zheng R, Suhoski MM, Monaco-Shawver L, Orange JS. Cdc42-interacting protein-4 functionally links actin and microtubule networks at the cytolytic NK cell immunological synapse. J Exp Med. 2007;204(10):2305–2320. doi: 10.1084/jem.20061893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paust S, et al. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol. 2010;11(12):1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tosato G, Cohen JI. Generation of Epstein-Barr Virus (EBV)-immortalized B cell lines. Curr Protoc Immunol. 2007;Chapter 7:Unit 7.22. doi: 10.1002/0471142735.im0722s76. [DOI] [PubMed] [Google Scholar]
- 75.Sanborn KB, et al. Myosin IIA associates with NK cell lytic granules to enable their interaction with F-actin and function at the immunological synapse. J Immunol. 2009;182(11):6969–6984. doi: 10.4049/jimmunol.0804337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doody GM, Stephenson S, McManamy C, Tooze RM. PRDM1/BLIMP-1 modulates IFN-γ-dependent control of the MHC class I antigen-processing and peptide-loading pathway. J Immunol. 2007;179(11):7614–7623. doi: 10.4049/jimmunol.179.11.7614. [DOI] [PubMed] [Google Scholar]
- 77.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.