Abstract
MicroRNA-187 (miR-187) has been reported to be involved in the occurrence and development of several types of cancers; however, a role for miR-187 in osteosarcoma (OS) has not yet been reported. Here, miR-187 was found to be significantly downregulated in OS cell lines and tissue samples, and decreased miR-187 expression was shown to be correlated closely with the TNM stage and lymph node metastasis. miR-187 overexpression suppressed OS cell proliferation, colony formation, migration, invasion, and epithelial-mesenchymal transition (EMT). Mechanically, zinc finger E-box binding homeobox 2 (ZEB2) was shown to serve as a direct target of miR-187 in OS cells and the overexpression of ZEB2 rescued the miR-187-induced suppression of proliferation, colony formation, migration, and invasion in OS cells. In clinical OS specimens, ZEB2 expression levels were elevated and were inversely correlated with miR-187 expression. These results suggest that miR-187 functions as a tumor suppressor in OS, partially by targeting ZEB2, and that miR-187 can serve as a promising candidate for OS.
Keywords: Osteosarcoma, miR-187, ZEB2, proliferation
Introduction
Osteosarcoma (OS) is the most common primary malignant bone tumor affecting rapidly growing bones of children and young adults [1]. Despite many strategies being used to treat this disease, the clinical outcomes and prognosis of OS patients has remained poor over the past few decades [2,3]. Hence, there is a critical need to elucidate the potential mechanism that mediates the initiation and progression of OS so that novel prognostic biomarkers and targeted therapies for treatment of the disease can be developed.
MicroRNAs (miRNAs) are an abundant class of endogenous, small (18-25-nucleotide long), noncoding, single-stranded RNA molecules that regulate multiple target mRNAs through binding to their 3’-untranslated regions (3’UTR) in a sequence-specificmanner [4,5]. It has been shown that miRNA alterations and dysfunctionare involved in tumorigenesis and tumor progression via the regulation of cancer cell proliferation, differentiation, apoptosis, migration, and invasion [6-8]. Indeed, aberrant miRNA expression in OS has been shown to play crucial roles in OS development and progression by target molecule regulation [9,10], which emphasizes the importance of miRNAs in diagnosis, therapy, and prognosis of OS.
miR-187, located on chromosome18q12.2, has been found to be downregulated and function as a tumor suppressor in several types of cancer, such as non-small cell lung cancer [11], colorectal cancer [12], B-cell lymphoma [13], prostate cancer [14], and clear cell renal cell carcinoma [15]. Until now, the expression status, functional role, and underlying molecular mechanism of miR-187 in OS remain unknown. In the present study, the expression of miR-187 in human OS cells and tissues was assessed, after which, the effects of the miRNA on cell proliferation, migration, and invasion were investigated. The molecular mechanism underlying the function of miR-187 in OS was assessed in this study, the results of which may provide a better understanding of OS development and progression.
Materials and methods
Patients and tissue samples
Primary OS tissues and their corresponding adjacent normal tissues (ANT) were obtained from 40 patients who underwent OS tissues resection at the Department of Orthopaedic Surgery, China-Japan Union Hospital of Jilin University (Changchun, China) between July 2014 and July 2015. Adjacent normal tissues were collected at a distance of 5 cm from the OS tissues. All tissue biopsies were immediately frozen in liquid nitrogen following surgery, and stored at -80°C until RNA extraction. None of the patients had received chemotherapy or other therapy prior to surgery. Clinical-pathologic features of OS patients were recorded and are shown in Table 1. Prior to any data and sample collection, written informed consent was obtained from all patients. This study was approved by the ethics committee of Jilin University (Changchun, China).
Table 1.
Correlation between clinicopathological features and miR-187 expression in OS tissues
| Variables | No. of cases | miR-187 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n %) | High (n %) | |||
| Age (years) | P > 0.05 | |||
| < 20 | 28 | 16 (57.1) | 12 (42.9) | |
| ≥ 20 | 12 | 6 (50.0) | 6 (50.0) | |
| Gender | P > 0.05 | |||
| Male | 21 | 12 (57.1) | 9 (42.9) | |
| Female | 19 | 10 (52.6) | 9 (47.4) | |
| TNM stage | P < 0.01 | |||
| I-II | 32 | 14 (43.8) | 18 (56.2) | |
| III-IV | 8 | 8 (100.0) | 0 (0.0) | |
| Tumor size | P > 0.05 | |||
| < 5 | 30 | 16 (53.3) | 14 (46.7) | |
| ≥ 5 | 10 | 6 (60.0) | 4 (40.0) | |
| Lymph node metastasis | P < 0.01 | |||
| No | 31 | 15 (48.4) | 16 (51.2) | |
| Yes | 9 | 7 (77.8) | 2 (22.2) | |
Cell lines and transfection
Four human OS cell lines (HOS, Saos-2, U2OS, and MG-63) and a normal human osteoblast cell line (hFOB 1.19) were brought from the Shanghai Institute for Biological Sciences (Shanghai, China) and were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Gaithersburg, MD), supplemented with 10% fetal bovine serum (FBS, HyClone, South Logan, UT), 100 U/mL penicillin, and 100 mg/mL streptomycin at 37°C in a humidified incubator containing 5% CO2.
miR-187 mimic and corresponding negative control (miR-NC) miRNAs were purchased from RiboBio (Guangzhou, China) and the ZEB2 expression vector (pCDNA3.1-ZEB2) was provided by Dr. Ju Peng (Jilin University). Transfection of miRNA or ZEB2 was performed using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cell lines and tissues specimens using TRIzol reagent (Invitrogen, Carlsbad, CA) and the resulting RNA quantity and integrity were determined using Nanodrop and Agilent 2100 Bioanalyzer systems. For miR-187 expression level analysis, reverse transcription and real time quantitative PCR were performed using the TaqMan MicroRNA Reverse Transcription Kit, the TaqMan miRNA assay (Applied Biosystems, Foster City, CA), and primers specific to miR-187 and U6 (used as an internal reference) (Applied Biosystems). An ABI 7900 Sequence Detection System (Life Technologies, NY) was used for these experiments. For the quantification of ZEB2 mRNA expression, reverse transcription and real time quantitative PCR were performed using the PrimeScript RT reagent Kit (Takara, Dalian, China), Real-time PCR Mixture Reagent (Takara), and the ABI 7900 Fast system. The primers for ZEB2 and GAPDH (internal reference) used in this study were described previously [16]. The relative expression levels of ZEB2 mRNA and miR-187 were determined using the 2-∆∆Ct method.
Cell proliferation and colony formation assays
An MTT assay was performed to assess cell proliferation. Briefly, MG63 or U2OS cells were seeded on a 96-well plate at a concentration of 1 × 104 cells/well. The following day, miR-187 mimic, miR-NC, or ZEB2 plasmid (pCDNA3.1-ZEB2) was transfected into the cells using Lipofectamine 2000 according to the manufacturer’s protocol. After 72 h of transfection, medium containing sterile MTT dye (10 μL; 5 mg/mL) was added to each well, after which the cells were incubated for a further 4 h. Finally, dimethyl sulfoxide (150 μL; DMSO, Sigma-Aldrich) was added into each well andthe absorbance at 570 nm was measured in a microtiter plate reader (Molecular Devices, Menlo Park, CA).
For colony formation assays, transfected cells were harvested, seeded onto 6-well plates at a density of 1000 cells/well and then incubated at 37°C and 5% CO2 in a humidified incubator for 14 days. The resulting colonies were fixed in 4% paraformaldehyde for 5 min before being stained with 1.0% crystal violet for 1 min and then for 30 s. The colonies were photographed and counted under a light microscope (Olympus, Tokyo, Japan).
Cell migration and invasion assays
A wound-healing assay was performed to assess cell migration. Briefly, transfected cells were seeded onto six-well plates at a density of 1 × 104 cells/well and grown to 80-100% confluence. A wound was then created across the confluent cell monolayer using a sterile plastic micropipette tip. Cells were rinsed with phosphate-buffered saline (PBS) and cultured for another 24 h with serum-free medium. Images of wound closure were captured at different time points (0 and 24 h) under a light microscope to assess the rate of gap closure.
For invasion assays, 2 × 104 transfected cells suspended in serum-free medium were added to the top chamber of a transwell plate precoated with Matrigel (BD Biosciences), and DMEM supplemented with 10% FBS was added to the lower chamber. The cells were cultured for 48 h at 37°C in a humidified incubator containing 5% CO2. The cells remaining on the upper surface of the membrane were removed using sterile cotton, while the cells that invaded the lower chamber were fixed in 90% alcohol and stained with 0.1% crystal violet. Photographs were taken and the number of invaded cells was counted in five randomly selected fields under a light microscope (200 ×, Olympus, Tokyo, Japan).
Luciferase assays
The 3’-untranslated regions (UTRs) of human ZEB2 were amplified from human cDNA using PCR and cloned into the XhoI and NotI sites downstream of the luciferase reporter gene in the pGL3-control vector (Ambion, Austin, TX). The resulting plasmid was named WT-ZEB2-3’UTR. For mutagenesis of the miR-187-binding site, the QuikChange site-directed Mutagenesis Kit (Agilent Technologies, Palo Alto, CA) was used according to the manufacturer’s instructions, and the mutagenesis product was named Mut-ZEB2-3’UTR. For luciferase assays, cells were co-transfected with 100 ng wide-type or mutant ZEB2 3’UTR reporter plasmids and 5 ng pRL-SV40 Renilla luciferase construct (for normalization), and 100 nM miR-187 mimic or miR-NC using Lipofectamine 2000 according to the manufacturer’s protocol. Firefly and Renilla luciferase activities were measured after 48 h of transfection by using a Dual-Luciferase Reporter Assay System (Promega, MA). Renilla luciferase activity was normalized to that of firefly luciferase.
Western blot analysis
Total protein was extracted from cultured cells using RIPA buffer containing a protease inhibitor (Beyotime, Jiangsu, China) and was quantified using a bicinchoninic acid (BCA) kit (Pierce, Rockford, IL). Proteins (50 μg per sample) were separated by 10% SDS/PAGE before being transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA) at 80 V for 2 h at 4°C. The membranes were blocked in 5% nonfat dry milk in TBS (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) at room temperature for 1 h and were then incubated with antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) against ZEB2 (1:1000), E-cadherin (1:1000), N-cadherin (1:1500), vimentin (1:500), or GAPDH (1:3000) in TBS overnight at 4°C. After three washes in TBST (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20), the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (1:5000, Santa Cruz) for 1 h at room temperature. Immunoreactive bands were detected by chemiluminescence using the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, USA) and X-ray film. GAPDH served as a normalization control.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5 (GraphPad Software, Inc., San Diego, CA) and SPSS 17 software (SPSS, Chicago, IL). All data are reported as mean ± SD (standard deviation) and all experiments were repeated at least three times. Differences were assessed using a two-tailed student’s t-test or one-way ANOVA and the correlation between miR-187 and ZEB2 expression was analyzed with Pearson’s correlation. In all cases, differences with P < 0.05 were considered statistically significant.
Results
miR-187 expression is downregulated in OS tissues and cell lines
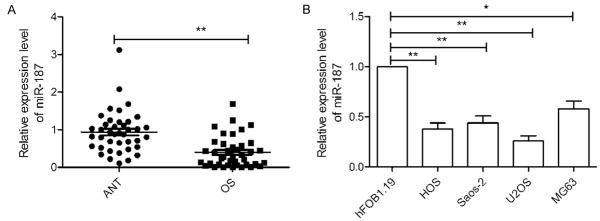
Initially, the expression of miR-187 was evaluated in 40 OS tissues and adjacent normal tissues (ANT). qRT-PCR analysis demonstrated that miR-187 expression was significantly downregulated in OS tissues compared to that in adjacent normal tissues (P < 0.05, Figure 1A). To determine the clinicopathological significance of miR-187 aberrations, the median value (0.384) of miR-187 expression was used as the cut-off point for dividing OS tissues into two groups: low expression (< median, n = 22) and high expression (> median, n = 18). Further analysis demonstrated that miR-187 expression in OS tissues was associated with TNM stage and lymph node metastasis (P < 0.05; Table 1). No significant differences were observed between miR-187 expression and patient’s gender, age, or tumor size (P > 0.05; Table 1). miR-187 expression was examined in four OS cell lines using quantitative real-time PCR (qRT-PCR). As shown in Figure 1B, four OS cell lines displayed lower levels of miR-187 than the normal human osteoblastic cell line, hFOB 1.19 (P < 0.05; Figure 1B). Among these OS cell lines, U2OS (lowest endogenous miR-187 expression) and MG-63 (highest endogenous miR-187 expression) cells were used for subsequent experiments.
Figure 1.
miR-187 expression is downregulated in OS tissues and cell lines. A. Relative miR-187 level in 40 pairs of OS- and adjacent normal bone tissues (ANT) was assessed by quantitative RT-PCR (qRT-PCR). B. Relative miR-187 levels were determined via qRT-PCR in four osteosarcoma cell lines (HOS, Saos-2, U2OS, and MG-63) and a human normal osteoblastic cell line (hFOB1.19). *P < 0.05, **P < 0.01.
miR-187 suppresses OS cell proliferation and colony formation
To explore the potential role of miR-187 in OS pathogenesis, U2OS and MG63 cells were transfected with miR-187 mimic or miR-NC, after which miR-187 expression was assayed using qRT-PCR. The results showed that the expression level of miR-187 was significantly upregulated in the miR-187 mimic group compared to the miR-NC group (Figure 2A), which suggests that miR-187 expression was successfully enhanced in the U2OS and MG63 cells. The results of the MTT assay demonstrated that the upregulation of miR-187 significantly suppresses the proliferation of U2OS and MG63 cells compared with miR-NC-transfected cells (Figure 2B). Colony formation analysis revealed that miR-187 overexpression significantly suppresses colony formation by U2OS and MG63 cells (Figure 2C). These results suggest that miR-187 inhibits OS growth in vitro.
Figure 2.
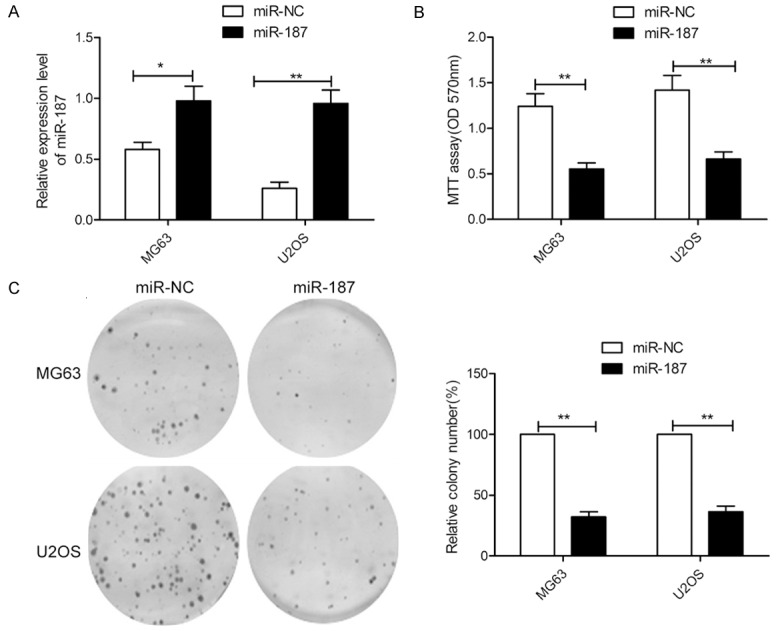
miR-187 inhibits proliferation and colony formation in OS cells. A. Relative miR-187 levels were assessed in MG63 and U2OS cells after transfection with miR-187 or miR-NC. B, C. Cell proliferation and colony formation were measured in MG63 and U2OS cells after transfection with miR-187 or miR-NC. *P < 0.05, **P < 0.01.
miR-187 suppresses OS cell migration and invasion
To further assess the effects of miR-187 on migration and invasion, wound healing and transwell invasion assays were performed. These experiments revealed that ectopic expression of miR-187 significantly inhibits the migration and invasion of both OS cell lines (Figure 3A and 3B), which indicates that miR-187 inhibits metastasis of OS cells in vitro.
Figure 3.
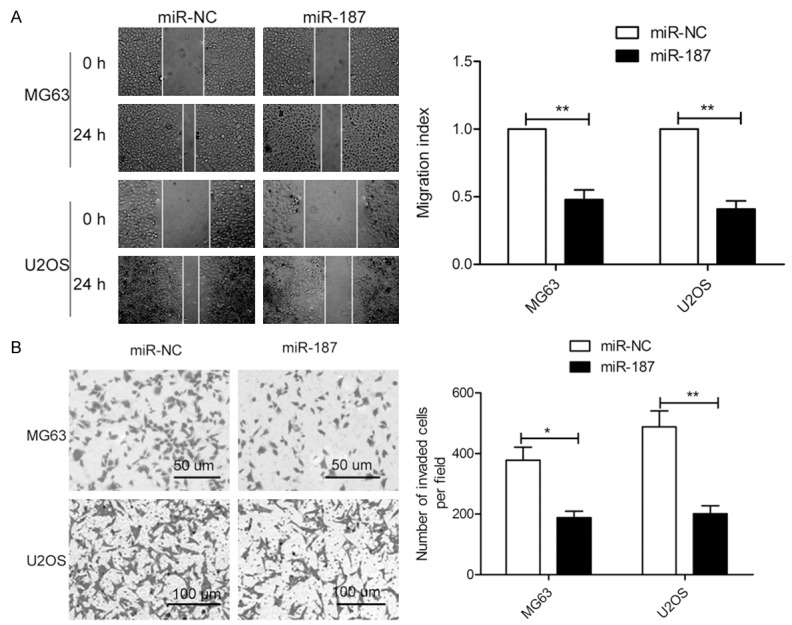
miR-187 inhibits migration and invasion in OS cells. A. Cell migration was assessed using a wound healing assay for MG63 and U2OS cells after transfection with miR-187 or miR-NC. B. Cell invasion was investigated by transwell invasion assays for MG63 and U2OS cells after transfection with miR-187 or miR-NC. *P < 0.05, **P < 0.01.
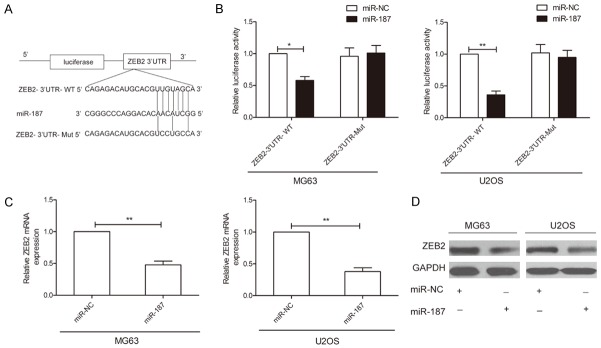
ZEB2 is a direct target of miR-187 in OS
Using the TargetScan and miRanda algorithms, ZEB2 was selected as a candidate target of miR-187 since the 3’UTR of ZEB2 mRNA contains a complementary site for the seed region of miR-187 (Figure 4A). To further confirm ZEB2 as a direct target of miR-187, a dual-luciferase reporter assay was performed and miR-187 overexpression was shown to significantly decrease the luciferase activity of wild-type ZEB2-3’UTR in MG63 and U2OS cells, while it failed to repress the mutated ZEB2 3’UTR (Figure 4B). To determine whether miR-187 regulates endogenous ZEB2 expression in OS cell lines, the mRNA and protein expression levels of ZEB2 were assessed by qRT-PCR and western blot, respectively. Overexpression of miR-187 was found to result in a significant decrease in ZEB2 mRNA and protein levels in MG63 and U2OS cells (Figure 4C and 4D). From these results, we conclude that ZEB2 is a direct downstream target of miR-187 in OS cells.
Figure 4.
ZEB2 is a direct target for miR-187 in OS. A. The predicted binding sites for miR-187 in the 3’UTR of ZEB2 and the mutations in the binding sites are shown. B. MG63 and U2OS cells were cotransfected with miR-187 mimic or miR-NC and wild-type or mutant-type ZEB2 3’UTR reporter plasmid. Luciferase activity was measured 48 h after transfection. WT: Wild-type, Mut: Mutant-type. C, D. ZEB2 mRNA and protein expression levels were measured in MG63 and U2OS cells after transfection with miR-187 mimic or miR-NC by qRT-PCR and western blot analysis, respectively. GAPDH was used to an internal control. *P < 0.05, **P < 0.01.
miR-187 regulates epithelial-mesenchymal transition in OS cells
It has been demonstrated that ZEB2 caninduce epithelial-mesenchymal transition (EMT), which is known to contribute to cancer cells metastasis [16-18]. Since it was shown in this study that ZEB2 is a target of miR-187, it is possible that miR-187 may play a role in regulating EMT. Accordingly, EMT markers were assessed by western blot analysis in OS cells transfected with miR-187 mimic or miR-NC. As shown in Figure 5, overexpression of miR-187 led to increase E-cadherin expression and decrease N-cadherin and vimentin expression. These findings suggest that miR-187 overexpression may inhibit EMT.
Figure 5.
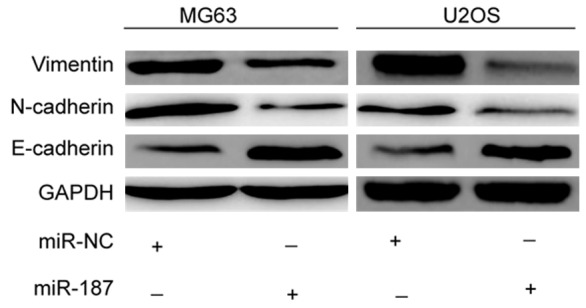
miR-187 inhibits epithelial-to-mesenchymal transition (EMT) in OS. Western blot analysis of E-cadherin, N-cadherin, and vimentin expression in MG63 and U2OS cells after transfection with miR-187 or miR-NC. GAPDH was used as a loading control. *P < 0.05, **P < 0.01.
ZEB2 overexpression reverse inhibitory effects of miR-187 in OS
To further verify the functional connection between miR-187 and ZEB2, MG63 and U2OS cells were cotransfected with ZEB2 expression plasmids and miR-187 mimic or miR-NC.In the case of both MG63 and U2OS cells, the expression of ZEB2 was found to be significantly increased after cotransfection with miR-187 and pcDNA-ZEB2 compared with cells transfected with only miR-187 (Figure 6A and 6B). Overexpression of ZEB2 was furthermore shown to reverse the inhibitory effects of miR-187 on cell proliferation, colony formation, migration, and invasion in OS cells (Figure 6C-F).These results suggest that miR-187 inhibits cell proliferation, colony formation, migration, and invasion in OS cells via downregulation of ZEB2.
Figure 6.
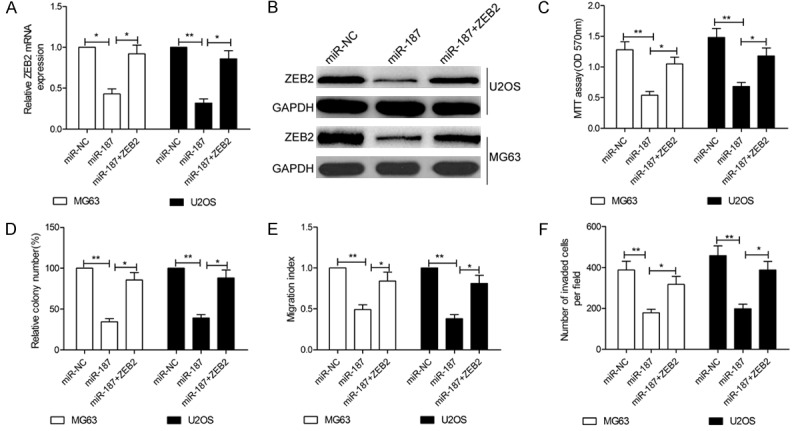
ZEB2 overexpression reverses the inhibitory effects of miR-187 in OS. MG63 and U2OS cells were transfected with miR-187 mimic or miR-NC together with a ZEB2 overexpression plasmid (pcDNA3.1-ZEB2). A, B. ZEB2 expression in the abovementioned cells was assessed at the mRNA and protein levels. GAPDH was used as a loading control. C-F. Cell proliferation, colony formation, migration, and invasion were measured in the indicated cells. *P < 0.05, **P < 0.01.
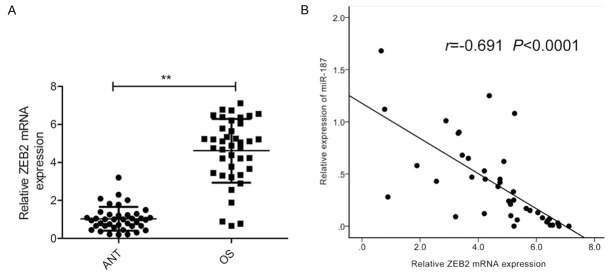
ZEB2 is upregulated in OS tissues and is inversely correlated with miR-187 expression
Finally, ZEB2 mRNA expression levels were assessed in OS specimens and adjacent normal bone tissues. qRT-PCR analysis revealed significantly higher ZEB2 mRNA expression levels in OS tissues than in normal bone tissues (Figure 7A). Pearson’s correlation analysis furthermore confirmed an inverse correlation between miR-187 expression and ZEB2 mRNA expression in OS tissues (r = -0.691, P < 0.0001; Figure 7B).
Figure 7.
ZEB2 is upregulated in OS tissues and is inversely correlated with miR-187 expression. A. ZEB2 mRNA levels in 40 OS specimens and adjacent normal bone tissues (ANT) were assessed by qRT-PCR. B. Pearson’s correlation analysis between miR-187 expression and ZEB2 mRNA level. *P < 0.05, **P < 0.01.
Discussion
Dysregulation of miRNAs has been reported to play a role in OS progression and development [9,10,19] and an improved understanding of biological functions of different miRNAs may therefore contribute to the development of more effective biomarkers and therapeutic targets for OS patients. In this study, we present preliminary evidence that miR-187 expression is downregulated in OS tissues and cell lines and is associated with TNM stage and lymph node metastasis. We furthermore report findings to suggest that miR-187 plays a negative regulatory role in OS growth and metastasis via ZEB2 repression.
Recently, miR-187 was found to be downregulated and to function as a tumor suppressor in several types of cancers [11-15]. Mao and colleagues, for example, reported that miR-187 is significantly downregulated in lung tumors and that miR-187 overexpression inhibits cell proliferation, invasion, and migration as well as inducing cell apoptosis by repressing CYP1B1 expression [11]. Qin et al found that miR-187 acted as a tumor suppressor in colorectal cancer by inhibiting cell proliferation, apoptosis, and invasive capacity via CD276 [12]. Zhao et al demonstrated that miR-187 inhibits clear cell renal cell carcinoma proliferation in vitro, inhibits in vivo tumor growth, and decreases motility by repressing the B7 homolog 3 (B7-H3) [15]. On the contrary, miR-187 has been shown to be upregulated and function as an oncogenein thyroid cancer [20], breast cancer [21], oral carcinoma [22], and acute lymphoblastic leukemia [23]. These reports suggest that the functional role of miR-187 in different cancers may be paradoxical; however, the role of miR-187 in OS has not yet been reported.
In this study, the expression of miR-187 in OS tissues and cell lines was initially found to be significantly downregulated in OS tissues and cell lines compared with corresponding normal bone tissues and the normal cell line. The biological functions of miR-187 in OS were then investigated, and it was found that miR-187 suppresses cell proliferation, colony formation, migration, invasion, and EMT in OS. These results suggest that miR-187 may function as a tumor suppressor in OS.
It is well known that miRNA function relies primarily on miRNA target gene(s) [6,7,19]. To explore the potential mechanism underlying the role of miR-187 in OS progression, bioinformatic analysis was conducted and identified ZEB2 as a putative miR-187 target gene, where ZEB2 (zinc finger E-box-binding homeobox 2 protein) is a member of the delta-EF1 (TCF8)/Zfh1 family of 2-handed zinc finger/homeodomain proteins [24]. Recent studies have demonstrated that ZEB2 may be an important oncogenic transcription factor contributing to tumor progression in multiple cancers including non-small lung cancer[18], gastric cancer[17], ovarian cancer[25], and liver cancer[16]. In the case of OS, ZEB2 has been shown to be involved in OS progression by regulating miR-141[26] and miR-200[27]. In the present study, we further confirmedZEB2 as a direct target of miR-187 in OS using a luciferase reporter assay, qRT-PCR, and western blot analysis. Moreover, ZEB2 overexpression was found to reverse the tumor-suppressive effects of miR-187 on OS cells. Notably, ZEB2 expression is upregulated in OS tissues and is negatively correlated with miR-187 expression level. Collectively, these findings may suggest that the anti-tumor effect of miR-187 in OS occurs in part by targeting ZEB2.
It is well known that epithelial- mesenchymal transition (EMT) is involved in cancer cell invasion and migration during the development and progression of cancer and metastasis [28]. In the EMT process, cells acquire morphological and molecular alterations that lead to downregulation of adhesion molecules such as E-cadherin and upregulation of mesenchymal markers such as N-cadherin, thereby facilitating tumor metastasis and progression [29]. Accordingly,EMT has been shown to play an important role in invasion and migration of OS cells [30]. It has been reported that ZEB2 binds to the E-BOX sequence in the E-cadherin promoter, leading to downregulation of E-cadherin expression, thereby inducing EMT [31]. In this study, ZEB2 was shown to be a functional target of miR-187 in OS cells, and thus we investigated whether miR-187 may affect EMT in OS cells. Our findings revealed that overexpression of miR-187 leads to increased expression of E-cadherin and decreased expression of N-cadherin and vimentin, which suggests that miR-187 may inhibit EMT in OS.
Overall, we demonstrated in this study that miR-187 expression is downregulated in OS tissues and cell lines and that decreased miR-187 expression in OS is closely associated with TNM stage and lymph node metastasis. Moreover, we also showed that upregulation of miR-187 suppresses OS cell proliferation, colony formation, migration, invasion, and EMT via the downregulation of ZEB2. Our experimental data therefore suggest that miR-187 may be a promising therapeutic target for OS.
Disclosure of conflict of interest
None.
References
- 1.Rytting M, Pearson P, Raymond AK, Ayala A, Murray J, Yasko AW, Johnson M, Jaffe N. Osteosarcoma in preadolescent patients. Clin Orthop Relat Res. 2000:39–50. doi: 10.1097/00003086-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest. 2001;19:292–315. doi: 10.1081/cnv-100102557. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Malan-Muller S, Hemmings SM, Seedat S. Big effects of small RNAs: a review of microRNAs in anxiety. Mol Neurobiol. 2013;47:726–739. doi: 10.1007/s12035-012-8374-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 7.Farazi TA, Spitzer JI, Morozov P, Tuschl T. MiRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garzon R, Marcucci G. Potential of microRNAs for cancer diagnostics, prognostication and therapy. Curr Opin Oncol. 2012;24:655–659. doi: 10.1097/CCO.0b013e328358522c. [DOI] [PubMed] [Google Scholar]
- 9.Nugent M. MicroRNA and bone cancer. Adv Exp Med Biol. 2015;889:201–230. doi: 10.1007/978-3-319-23730-5_11. [DOI] [PubMed] [Google Scholar]
- 10.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and potential targets in osteosarcoma: review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao M, Wu Z, Chen J. MicroRNA-187-5p suppresses cancer cell progression in non-small cell lung cancer (NSCLC) through down-regulation of CYP1B1. Biochem Biophys Res Commun. 2016;478:649–655. doi: 10.1016/j.bbrc.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Wang ZS, Zhong M, Bian YH, Mu YF, Qin SL, Yu MH, Qin J. MicroRNA-187 inhibits tumor growth and invasion by directly targeting CD276 in colorectal cancer. Oncotarget. 2016;7:44266–4427. doi: 10.18632/oncotarget.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang F, Jin Y, Wei Y. MicroRNA-187 induces diffuse large B-cell lymphoma cell apoptosis via targeting BCL6. Oncol Lett. 2016;11:2845–2850. doi: 10.3892/ol.2016.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casanova-Salas I, Masia E, Arminan A, Calatrava A, Mancarella C, Rubio-Briones J, Scotlandi K, Vicent MJ, Lopez-Guerrero JA. MiR-187 targets the androgen-regulated gene ALDH1A3 in prostate cancer. PLoS One. 2015;10:e0125576. doi: 10.1371/journal.pone.0125576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Lei T, Xu C, Li H, Ma W, Yang Y, Fan S, Liu Y. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem Biophys Res Commun. 2013;438:439–444. doi: 10.1016/j.bbrc.2013.07.095. [DOI] [PubMed] [Google Scholar]
- 16.Pang X, Huang K, Zhang Q, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272–3279. doi: 10.3892/or.2015.4321. [DOI] [PubMed] [Google Scholar]
- 17.Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5’-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, Yang Z, Zhang P, Liu Y, Shao G. MiR-154 inhibits migration and invasion of human non-small cell lung cancer by targeting ZEB2. Oncol Lett. 2016;12:301–306. doi: 10.3892/ol.2016.4577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE. MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab. 2008;93:1600–1608. doi: 10.1210/jc.2007-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulrane L, Madden SF, Brennan DJ, Gremel G, McGee SF, McNally S, Martin F, Crown JP, Jirstrom K, Higgins DG, Gallagher WM, O’Connor DP. MiR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin Cancer Res. 2012;18:6702–6713. doi: 10.1158/1078-0432.CCR-12-1420. [DOI] [PubMed] [Google Scholar]
- 22.Lin SC, Kao SY, Chang JC, Liu YC, Yu EH, Tseng SH, Liu CJ, Chang KW. Up-regulation of miR-187 modulates the advances of oral carcinoma by targeting BARX2 tumor suppressor. Oncotarget. 2016 doi: 10.18632/oncotarget.11349. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lou Y, Liu L, Zhan L, Wang X, Fan H. MiR-187-5p regulates cell growth and apoptosis in acute lymphoblastic leukemia via DKK2. Oncol Res. 2016;24:89–97. doi: 10.3727/096504016X14597766487753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang SB, He XJ, Xia YJ, Hu WJ, Luo JG, Zhang J, Tao HQ. MicroRNA-145-5p inhibits gastric cancer invasiveness through targeting N-cadherin and ZEB2 to suppress epithelial-mesenchymal transition. Onco Targets Ther. 2016;9:2305–2315. doi: 10.2147/OTT.S101853. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Wang Q, Jiang H, Deng X, Fang W, Guo S. [Expressions of ZEB2 and C-myc in epithelial ovarian cancer and their clinical significance] . Nan Fang Yi Ke Da Xue Xue Bao. 2015;35:1765–1769. [PubMed] [Google Scholar]
- 26.Xu H, Mei Q, Xiong C, Zhao J. Tumor-suppressing effects of miR-141 in human osteosarcoma. Cell Biochem Biophys. 2014;69:319–325. doi: 10.1007/s12013-013-9801-7. [DOI] [PubMed] [Google Scholar]
- 27.Mei Q, Li F, Quan H, Liu Y, Xu H. Busulfan inhibits growth of human osteosarcoma through miR-200 family microRNAs in vitro and in vivo. Cancer Sci. 2014;105:755–762. doi: 10.1111/cas.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sekhon K, Bucay N, Majid S, Dahiya R, Saini S. MicroRNAs and epithelial-mesenchymal transition in prostate cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11708. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in cancer. Molecules. 2016;21 doi: 10.3390/molecules21070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang G, Yuan J, Li K. EMT transcription factors: implication in osteosarcoma. Med Oncol. 2013;30:697. doi: 10.1007/s12032-013-0697-2. [DOI] [PubMed] [Google Scholar]
- 31.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]