Abstract
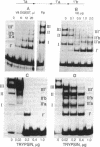
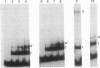
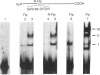
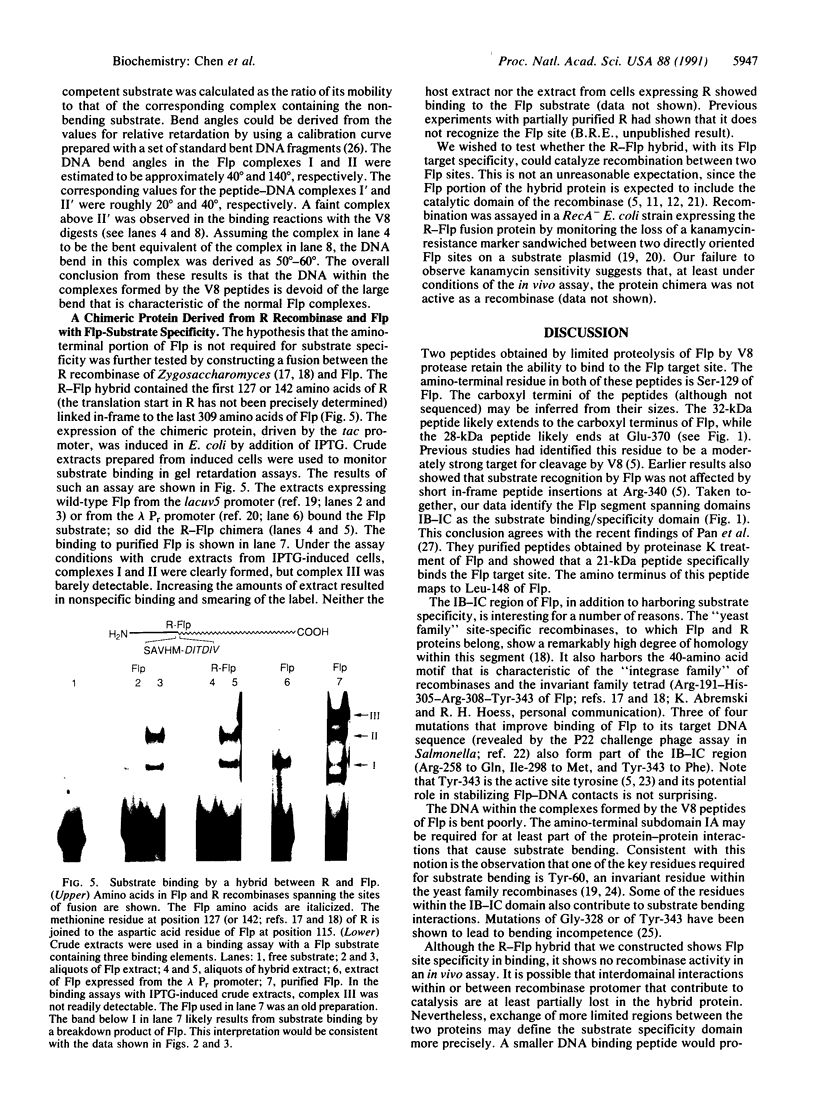
Binding of a partial proteolytic digest by V8 enzyme of the yeast site-specific recombinase Flp to its target site gives rise to DNA-protein complexes that are smaller than those produced by the full-sized protein. The smallest of these complexes (occupancy of one peptide monomer per site) contains either one of two polypeptides (32 and 28 kDa) of the V8 digestion mixture. The amino termini of both polypeptides map to Ser-129 of Flp, corresponding to V8 cleavage at Glu-128. The relative mobilities of the complexes formed by the V8 peptides indicate that they lack the sharp substrate bend that is characteristic of Flp-derived complexes. A hybrid protein consisting of the amino-terminal one-third of the R recombinase (from Zygosaccharomyces rouxii) and the carboxyl-terminal two-thirds of Flp recognizes the Flp target site.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amin A. A., Sadowski P. D. Synthesis of an enzymatically active FLP recombinase in vitro: search for a DNA-binding domain. Mol Cell Biol. 1989 May;9(5):1987–1995. doi: 10.1128/mcb.9.5.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki H., Jearnpipatkul A., Tatsumi H., Sakurai T., Ushio K., Muta T., Oshima Y. Molecular and functional organization of yeast plasmid pSR1. J Mol Biol. 1985 Mar 20;182(2):191–203. doi: 10.1016/0022-2836(85)90338-9. [DOI] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. W., Evans B. R., Zheng L., Jayaram M. Tyr60 variants of Flp recombinase generate conformationally altered protein-DNA complexes. Differential activity in full-site and half-site recombinations. J Mol Biol. 1991 Mar 5;218(1):107–118. doi: 10.1016/0022-2836(91)90877-9. [DOI] [PubMed] [Google Scholar]
- Evans B. R., Chen J. W., Parsons R. L., Bauer T. K., Teplow D. B., Jayaram M. Identification of the active site tyrosine of Flp recombinase. Possible relevance of its location to the mechanism of recombination. J Biol Chem. 1990 Oct 25;265(30):18504–18510. [PubMed] [Google Scholar]
- Futcher A. B. Copy number amplification of the 2 micron circle plasmid of Saccharomyces cerevisiae. J Theor Biol. 1986 Mar 21;119(2):197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- Govind N. S., Jayaram M. Rapid localization and characterization of random mutations within the 2 micron circle site-specific recombinase: a general strategy for analysis of protein function. Gene. 1987;51(1):31–41. doi: 10.1016/0378-1119(87)90471-9. [DOI] [PubMed] [Google Scholar]
- Jayaram M., Crain K. L., Parsons R. L., Harshey R. M. Holliday junctions in FLP recombination: resolution by step-arrest mutants of FLP protein. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7902–7906. doi: 10.1073/pnas.85.21.7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram M. Two-micrometer circle site-specific recombination: the minimal substrate and the possible role of flanking sequences. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5875–5879. doi: 10.1073/pnas.82.17.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lebreton B., Prasad P. V., Jayaram M., Youderian P. Mutations that improve the binding of yeast FLP recombinase to its substrate. Genetics. 1988 Mar;118(3):393–400. doi: 10.1093/genetics/118.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Meyer-Leon L., Huang L. C., Umlauf S. W., Cox M. M., Inman R. B. Holliday intermediates and reaction by-products in FLP protein-promoted site-specific recombination. Mol Cell Biol. 1988 Sep;8(9):3784–3796. doi: 10.1128/mcb.8.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R. L., Evans B. R., Zheng L., Jayaram M. Functional analysis of Arg-308 mutants of Flp recombinase. Possible role of Arg-308 in coupling substrate binding to catalysis. J Biol Chem. 1990 Mar 15;265(8):4527–4533. [PubMed] [Google Scholar]
- Parsons R. L., Prasad P. V., Harshey R. M., Jayaram M. Step-arrest mutants of FLP recombinase: implications for the catalytic mechanism of DNA recombination. Mol Cell Biol. 1988 Aug;8(8):3303–3310. doi: 10.1128/mcb.8.8.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad P. V., Young L. J., Jayaram M. Mutations in the 2-microns circle site-specific recombinase that abolish recombination without affecting substrate recognition. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2189–2193. doi: 10.1073/pnas.84.8.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A. E., Murray A. W., Szostak J. W. Roles of the 2 microns gene products in stable maintenance of the 2 microns plasmid of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Oct;7(10):3566–3573. doi: 10.1128/mcb.7.10.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. DNA binding by proteins. Science. 1988 Sep 2;241(4870):1182–1187. doi: 10.1126/science.2842864. [DOI] [PubMed] [Google Scholar]
- Schwartz C. J., Sadowski P. D. FLP protein of 2 mu circle plasmid of yeast induces multiple bends in the FLP recognition target site. J Mol Biol. 1990 Nov 20;216(2):289–298. doi: 10.1016/s0022-2836(05)80320-1. [DOI] [PubMed] [Google Scholar]
- Schwartz C. J., Sadowski P. D. FLP recombinase of the 2 microns circle plasmid of Saccharomyces cerevisiae bends its DNA target. Isolation of FLP mutants defective in DNA bending. J Mol Biol. 1989 Feb 20;205(4):647–658. doi: 10.1016/0022-2836(89)90310-0. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utatsu I., Sakamoto S., Imura T., Toh-e A. Yeast plasmids resembling 2 micron DNA: regional similarities and diversities at the molecular level. J Bacteriol. 1987 Dec;169(12):5537–5545. doi: 10.1128/jb.169.12.5537-5545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert F. C., Broach J. R. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986 Aug 15;46(4):541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]