Abstract
Protein ligand charge can impact physiological delivery with charge reduction often benefiting performance. Yet neutralizing mutations can be detrimental to protein function. Herein, three approaches are evaluated to introduce charged-to-neutral mutations of three cations and three anions within an affibody engineered to bind epidermal growth factor receptor. These approaches – combinatorial library sorting or consensus design, based on natural homologs or library-sorted mutants – are used to identify mutations with favorable affinity, stability, and recombinant yield. Consensus design, based on 942 affibody homologs, yielded a mutant of modest function (Kd = 11 ±4 nM, Tm = 62 °C, and yield = 4.0 ±0.8 mg/L as compared to 5.3 ±1.7 nM, 71 °C, and 3.5 ±0.3 mg/L for the parental affibody). Extension of consensus design to ten additional mutants exhibited varied performance including a substantially improved mutant (Kd = 6.9 ±1.4 nM, Tm = 71 °C, and 12.7 ±0.9 mg/L yield). Sorting a homolog-based combinatorial library of 7×105 mutants generated a distribution of mutants with lower stability and yield, but did identify one strongly binding variant (Kd = 1.2 ±0.3 nM, Tm = 69 °C, and 6.0 ±0.4 mg/L yield). Synthetic consensus design, based on the amino acid distribution in functional library mutants, yielded higher affinities (p=0.05) with comparable stabilities and yields. The best of four analyzed clones had Kd = 1.7 ±0.5 nM, Tm = 68 °C, and 7.0 ±0.5 mg/L yield. While all three approaches were effective in creating targeted affibodies with six charged-to-neutral mutations, synthetic consensus design proved to be the most robust. Synthetic consensus design provides a valuable tool for ligand engineering, particularly in the context of charge manipulation.
Keywords: affibody, charge, consensus design, epidermal growth factor receptor
Introduction
Ligand charge impacts in vivo performance, particularly physiological distribution. Cationic and highly anionic polypeptides clear the blood more rapidly than their neutral counterparts (Pimm et al., 1995). Additionally, positively charged proteins have increased vascular permeability and extravasate more rapidly in solid tumors (Dellian et al., 2000). Escalation in the number of positively or negatively charged residues increases kidney signal at early time points for multiple ligands (García Garayoa et al., 2008; Tran et al., 2007; Tran et al., 2008). In particular, reduction in the number of charged residues has been effective to improve biodistribution for several scaffolds including the affibody (Tran et al., 2007; Tran et al., 2008), knottin (Kimura et al., 2012), and fibronectin type III domain (Hackel et al., 2012). Thus, charge modulation – in particular, reduction – warrants further investigation for improved ligand delivery.
Yet charge modification can significantly impact protein solubility, stability, affinity, specificity, and production. Introducing charged residues on the surface of a protein is generally well-tolerated and in some cases beneficial to stability and solubility (Pace, 1990). However, removal of charged residues can be more problematic (Hackel et al., 2012; Pace, 1990). Numerous systems have exhibited reduced solubility upon charged-to-neutral residue replacements: two independent mutations (E48N or D130N) in type S1 dihydrofolate reductase (Dale et al., 1994), a combination of five neutralizing mutations in the KcsA potassium channel (Slovic et al., 2004), K185F mutation in HIV integrase (Jenkins et al., 1995), K435L mutation in viral reverse transcriptase (Das and Georgiadis, 2001), and E100W mutation in leptin (Zhang et al., 1997). Some previous efforts to modify charge for pharmacokinetic impact studied large molecules, such as antibody Fc (Boswell et al., 2010; Igawa et al., 2010) in which a few amino acid mutations are less likely to affect protein properties. Conversely, a similar number of charge mutations within a small ligand represents a greater fraction of the total protein and may therefore be more impactful.
The current study evaluates three different protein engineering approaches to modulate charge of the affibody domain. Affibodies are 58-residue, three-helix bundles based on the B domain of staphylococcal protein A (Nilsson et al., 1987). Directed evolution of 13 residues on the surface of two helices has yielded molecular targeting agents for imaging and therapy, as well as scientific reagents (Nygren, 2008). Several affibodies have been engineered to bind selectively to epidermal growth factor receptor (EGFR) including ZEGFR:1907, with 3 nM affinity (Friedman et al., 2008). EGFR is overexpressed in numerous cancers (Hynes and MacDonald, 2009) and improved affinity ligands could have utility in molecular imaging and targeted therapy. The presented analyses were performed in the context of a hybrid affibody with the 13 paratope mutations of ZEGFR:1907 and framework mutations engineered, in the context of ZHER2:342, (Feldwisch et al., 2010) for improved synthesis and stability while maintaining binding affinity but without direct consideration of charge.
The use of natural homolog sequences to predict mutational tolerance has been useful (Cochran et al., 2006; Steipe et al., 1994; Steipe, 2004), albeit inconsistent (Durani and Magliery, 2013; Sullivan et al., 2011), for general mutations. We aimed to study the ability of homolog sequences to predict tolerance of charged-to-neutral mutations. Yet, consensus design is limited by the ability of nature to sample sequence space as well as the requisite retention of functional activity, which is different than our current activity of interest, i.e., EGFR binding. Thus, considering the potential limitations of consensus design, we allowed homolog sequences to guide our selection of mutations while extending beyond natural consensus design. High throughput sorting of a combinatorial library – using the genotype-phenotype linkage of yeast display (Boder and Wittrup, 1997) and flow cytometry – expands the number of mutants that can be efficiently evaluated albeit with potential limitations on the characterization of some phenotypes. The two approaches can be combined to overcome the limitations of natural consensus design. High throughput sorting can efficiently identify a population of functional sequences to serve as a synthetic basis for consensus design, which has been effectively implemented in evolving the chorismate mutase enzyme (Jäckel et al., 2010).
Herein, these three approaches – consensus design, combinatorial library screening, and synthetic consensus design – were used to introduce numerous variations of six simultaneous charged-to-neutral mutations within an EGFR-binding affibody. Mutant analysis at the clonal and population levels provides significant insight into the impact of charged-to-neutral mutations within this affibody domain. While all three approaches yielded a range of affinities, stabilities, and recombinant yields including several highly functional mutants, synthetic consensus design was the most effective. A mutant with 3.2 ±1.9-fold enhanced affinity (1.7 ±0.5 nM Kd), comparable stability (68 °C Tm), and improved yield (7.0 ±0.5 mg/L), despite neutralization of six charged residues, was engineered.
Materials and Methods
Pfam Homolog Consensus
Affibody EA68 (Figure 1, origin described in Results) was used as the seed sequence in a search of the Pfam database (Finn et al., 2014). Proteins with less than 42 residues were discarded resulting in 942 homologs. Sequences were aligned to EA68 using a block substitution (BLOSUM) scoring matrix (Henikoff and Henikoff, 1992). Amino acid frequencies were calculated at each site with a modification to account for repeat sequences without allowing their redundancy to dominate sequence statistics: identical sequences were counted as the square root of the number of occurrences of the sequence.
Figure 1.
(A) Schematic of affibody domain variant (PDB: 1h0t (Wahlberg et al., 2003)). (B) Sequences of different affibody variants used as the basis for this study. ZHER2:342 is an evolved HER2 binder with the original framework sequence. ABY-025 (Feldwisch et al., 2010) is a mutant with an optimized framework. ZEGFR:1907 is a mutant with 13 sites evolved to bind EGFR. EA68 is a combined mutant with the EGFR binding mutations and the optimized framework, with five exceptions: three A to S mutants – at sites 42, 46, and 54 – were retained as A. These mutations were selected for protein synthesis but slightly destabilize the protein; the S33K mutation was avoided because of the introduction of charged K and S33A was chosen, which provides 10 °C with no impact on HER2 affinity and reduced Ig binding; the N43E mutation was avoided because of the introduction of charged E. Negatively and positively charged residues are colored in blue and red, respectively, in the sequence. The corresponding sites in the structure are also colored, but note that the structure does not correspond to this amino acid sequence, but rather a different variant.
Clone Production and Yield Quantification
Genes were constructed by overlap extension PCR (Pogulis et al., 1996) of eight oligonucleotides (Integrated DNA Technologies) per clone, digested by NheI-HF and BamHI-HF restriction enzymes (New England Biolabs), and ligated into a pET-22b vector containing a C-terminal hepta-histidine (Novagen, EMD Millipore) with T4 DNA ligase (New England Biolabs). Plasmids were transformed via heat-shock into high efficiency T7 E. coli (New England Biolabs), and proper transformants were selected on lysogeny broth (10 g/L tryptone, 5.0 g/L yeast extract, 10.0 g/L sodium chloride in water) plates (15 g/L agar) containing kanamycin (50 mg/L). Two mL of saturated culture was added to 100 mL of lysogeny broth medium in a 250 mL shake flask, incubated at 37 °C, 250 rpm until an optical density at 600 nm between 0.5 and 1 units was achieved and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside for 2 hours at 25 – 28 °C. Cells were pelleted, resuspended in lysis buffer (50 mM sodium phosphate (pH 8.0), 0.5 M sodium chloride, 5% glycerol, 5 mM 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate, and 25 mM imidazole), and underwent four freeze-thaw cycles. The soluble fraction was isolated from cell lysate via centrifugation at 12,000g for 10 min. Affibodies were purified by immobilized metal affinity chromatography on 0.2 mL resin volume Ni-NTA HisPur spin columns (Thermo Fisher Scientific) and concentration assessed using absorbance at 280 nm on a Synergy H1 microplate reader (BioTek). Clones were further purified by reversed-phase high-performance liquid chromatography with a C18 column using a 15-minute gradient of 25% to 80% elution buffer (90% acetonitrile, 9.9% water, 0.1% trifluoroacetic acid) and the remaining solution composed of running buffer (99.9% water, 0.1% trifluoroacetic acid). Isolated affibodies were lyophilized. Yields are presented as the average ± standard error of eight or nine production runs.
Library Construction and Sorting
Genes for four charge-reduced affibody sub-libraries were constructed via overlap extension PCR of 12–15 oligonucleotides per sub-library, with degenerate codons present at six of the following positions: 2, 4, 7, 8, 15, 37, 49, 53, or 58 (Table S1). PCR reaction products and linearized pCT surface display vector were homologously recombined upon transformation via electroporation into EBY100 yeast (Chao et al., 2006). Transformants were allowed to multiply in SD-CAA (0.07 M sodium citrate (pH 5.3), 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, and 20 g/L glucose in water) at 30 °C, 250 rpm until log phase growth, then pelleted and resuspended in SG-CAA (0.1M sodium phosphate (pH 6.0), 6.7 g/L yeast nitrogen base, 5 g/L casamino acids, 19 g/L galactose, and 1 g/L glucose in water) and shaken at 30 °C, 250 rpm for 8–24 hr to induce cell surface affibody expression. Libraries were then pooled for fluorescence-activated cell sorting experiments. Two populations of 6.7 × 106 (10× theoretical diversity) cells were pelleted, washed with 4 °C PBS containing 1% bovine serum albumin (PBSA), and heated to 70 °C for 30 minutes. Cells were kept at 4 °C from this point forward. The yeast were then washed with cold PBSA, pelleted, and resuspended in PBSA with 20 mg/L mouse anti-c-MYC antibody (clone 9E10, Covance) with or without 20 nM biotinylated EGFR ectodomain (Prospec). After incubating on ice for 10 minutes, the populations were washed, pelleted, and resuspended in PBSA, streptavidin-Alexa Fluor 647 (Life Technologies), and goat anti-mouse-FITC antibody (Sigma-Aldrich). Yeast were sorted on a FACSAria II (BD Biosciences) flow cytometer with the top 5% of cells, in terms of EGFR binding : surface expression ratio, collected. Plasmids from collected yeast were recovered with a Zymoprep kit (Zymo Research) following manufacturer’s protocol and affibody-encoding genes amplified by PCR. Resultant PCR products were homologously recombined with pCT vector upon electroporation into EBY100 yeast. Two additional sorts were performed following this procedure, with the exception that in those cases cells were heat-treated at 80 °C rather than 70 °C. Encoding regions from recovered yeast from the final sort were PCR amplified, ligated into pET-22b vector, transformed into high efficiency 5-alpha E. coli (New England Biolabs) and grown on plates (lysogeny broth with 15 g/L agar and 50 mg/L kanamycin). Genes from fifty-four plate colonies were sequenced and found to all encode for affibodies. Sixteen sequences had combinations of charge mutations not designed into the sub-libraries – in some cases more neutralizations and in other cases less – and were discarded. The remaining thirty-eight sequences were used to create synthetic consensus frequencies.
Circular Dichroism
Purified, lyophilized affibody clones were resuspended in PBS to a concentration of 0.5 – 1 g/L. Ellipticity was measured between 200 and 260 nm wavelengths at room temperature on a Jasco J-815 spectrophotometer in a quartz cuvette with 1 mm path length. Thermal denaturation was performed by measuring ellipticity at 220 nm from 20 to 98 °C (2 °C/min). The midpoint of thermal denaturation (Tm) was calculated using a two-state unfolding model. Heated samples were cooled to 20 °C and subjected to a post-heating wavelength scan.
Affinity Measurement
A431 epidermoid carcinoma cells, which express approximately 2.8 × 106 EGFR per cell (Spangler et al., 2010), were provided by Dr. Daniel Vallera (University of Minnesota). Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 1% penicillin streptomycin at 37 °C in humidified air with 5% CO2. At 70–80% confluence, cells were detached with trypsin-ethylenediaminetetraacetic acid, pelleted, and resuspended in cold PBSA to prevent EGFR internalization prior to use. Fifty thousand cells were washed with PBSA and incubated with varying concentrations of ligand. Cells were pelleted, washed again with PBSA, and incubated with fluorescein-conjugated anti-His6 antibody (Abcam) in PBSA. Cells were washed and analyzed using flow cytometry on an Accuri C6 (BD Biosciences). Median fluorescence values at each ligand concentration, corresponding to binding fraction, were used to determine affinity values with least squares regression assuming a 1:1 binding interaction. Equilibrium dissociation constants, Kd, are presented as the average ± standard error of three to five titrations.
Size Exclusion Chromatography and Target Specificity Assay
Purified and lyophilized affibody clones, aprotinin, and cytochrome C were resuspended in PBS to a concentration of 1 mg/mL. 200 uL of each sample was run through a Superdex75 10/300 GL size exclusion column on an ӒKTAprime Plus chromatography system (GE Healthcare Life Sciences) at a flow rate of 0.5 mL/min for 1 hour. Retention volumes were determined by monitoring light absorbance at 280 nm.
MCF-7 breast adenocarcinoma cells which express fewer than 5 × 103 EGFR per cell, graciously provided by Dr. Deepali Sachdev (University of Minnesota), were used as pseudo-negative controls for target specificity assays. MCF-7 cells were cultured, detached, and processed identically to A431. After detachment and PBSA washing, fifty thousand A431 or MCF-7 cells were incubated with 500 nM affibody ligand. Upon binding equilibrium, cells were pelleted, washed with PBSA and incubated with fluorescein-conjugated anti-His6 antibody is PBSA. Cells were pelleted, washed in PBSA again then analyzed via flow cytometry on an Accuri C6. Median fluorescence with background signal subtracted are presented as the mean ± standard error of three replicates.
Results
Hybridizing EGFR Targeting Mutations with an Improved Framework
Affibody ZEGFR:1907 engineered by Friedman et al. (Friedman et al., 2008) to target EGFR was the partial basis for our starting ligand. Seven framework mutations, V1A, D2E, N3A, F5Y, N6A, N23T, and S33A, identified by Feldwisch et al. (Feldwisch et al., 2010) in the context of a HER2 targeting affibody ZHER2:342, were implemented to improve stability and reduce immunoglobulin binding (Figure 1). There are five exceptions from the optimized framework as published: three A to S mutants – at sites 42, 46, and 54 – were retained as alanine. These mutations were selected to improve protein synthesis at the expense of a slight reduction in stability; the S33K and N43E mutations were avoided because of the introduction of charged groups; S33A was chosen to provide stabilization (Tm increases 10 °C) with no impact on HER2 affinity and reduced Ig binding. This hybrid molecule, termed EA68 for EGFR-targeting affibody with six positively charged residues and eight negatively charged residues, was the basis for our protein engineering efforts. EA68 exhibits binding affinity to EGFR-overexpressing A431 cells with Kd = 5.3 ±1.7 nM (Figure 2A), which is comparable to ZEGFR:1907 (2.8 nM). The molecule exhibits the expected α-helical secondary structure (Figure 2B). The midpoint of thermal denaturation is 71 °C (Figure 2C), which is comparable to the HER2 binding affibody with a similarly optimized framework (Tm = 69 °C) but different sequence in the binding paratope. Unfolding is only partially reversible as α-helical secondary structure, but varied signal, is observed upon cooling to 25 °C after thermal denaturation (Figure 2B). The recombinant yield from E. coli is 3.5 ±0.3 mg/L in unoptimized shake flask productions. Thus, EA68 serves as a useful baseline to initiate charge engineering.
Figure 2. Characterization of EA68 affibody.
(A) A431 cells overexpressing EGFR were incubated on ice with the indicated concentration of purified EA68 followed by fluorescein-conjugated anti-His6 antibody, and analyzed by flow cytometry. The equilibrium dissociation constant (Kd) is calculated by minimizing sum of squared errors between experiment and a theoretical 1:1 binding model. (B,C) Purified EA68 was analyzed by circular dichroism spectroscopy by (B) scanning wavelengths from 200 to 260 nm at 20 °C. Triplicate scans were collected before and after thermal denaturation and cooling and (C) scanning temperatures from 20 °C to 98 °C (2 °C/min.) at 220 nm. The midpoint of thermal denaturation was calculated from a two-state model.
Charge Removal by Consensus Design
A consensus design approach was implemented to identify tolerable mutations to remove charged residues. Natural homologs to the engineered affibody were identified from the Pfam (Finn et al., 2014) database using EA68 as the basis sequence. Amino acid frequencies were calculated at each site based on the 942 homologs in the family. Identical sequences were partially discounted to account for their multiple natural appearances without allowing their redundancy to dominate sequence statistics. The charged framework sites exhibit variable conservation with a frequency range of 5–92%, median of 64%, and standard deviation of 30% (Table I). Frequencies of neutral residues range from 2% to 72%. The median number of different amino acids observed at each charged site was 6.5. The median Shannon entropy (Shannon, 1948) was 1.2 for all sites and 1.4 at charged framework sites. These values indicate a modest level of diversity from which to design mutants. Three positive and three negative residues were mutated to neutral within the framework of EA68. Consensus design identified the set of six charged-to-neutral mutations whose summed natural frequencies were highest: E2Q (24%), K4N (38%), E8A (21%), E15Q (30%), K49Q (21%), and K58N (62%). A clone incorporating all six mutations, named EA35, exhibited a comparable affinity (11 ±4 nM) and recombinant yield (4.0 ±0.8 mg/L) to EA68 but a 9.6 °C reduction in thermal stability (Figure 3, Figure S1, and Figure S2).
Table I.
Natural amino acid frequencies at each charged site (excluding sites 14 and 17 engineered for binding). Amino acids for clone EA68 and the mutant that removes three acidic and three basic residues while maximizing natural neutral frequency (Max Neutral, also known as EA35) are reported. The amino acids of mutants are noted with matches to EA35indicated as •.
Figure 3. Thermal stability (black diamonds), affinity (gray circles), and recombinant yield (white squares) for EA68, EA35, and EA35 variants.
The midpoints of thermal denaturation (Tm) were measured by circular dichroism spectroscopy of purified protein (n=1–2 with a mean difference of 7%). The equilibrium dissociation constants were measured by titration of affibody mutants on A431 cells using flow cytometry to detect binding (n=3–5). Recombinant yield is the amount of purified affibody recovered per L of culture (n=8–9). Points and error bars represent means and standard errors.
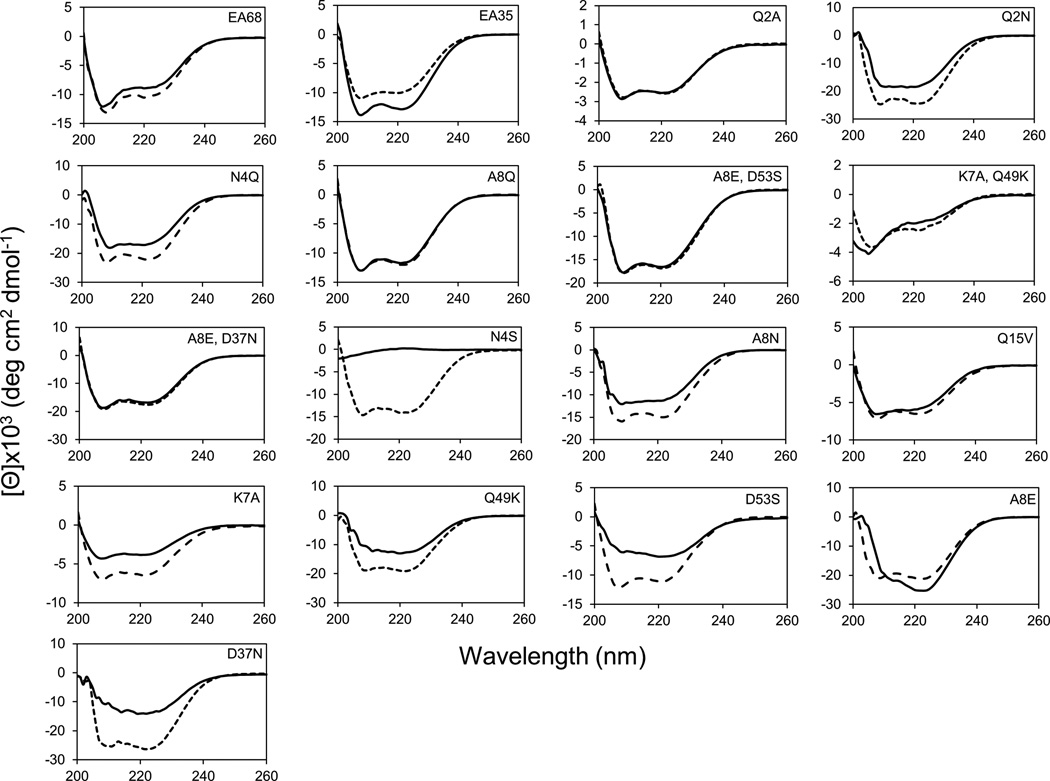
An additional ten mutants, similarly selected based on summed natural amino acid frequencies, were produced and evaluated (Figure 3). Overall, all but one mutant (EA35 A8Q) had affinities within 3-fold of EA68 (range: 6.9 ±1.4 nM to 20 ±7 nM versus 5.3 ±1.7 nM). All recombinant yields were comparable or better than EA68 (range: 4.0 ±0.8 mg/L to 16.6 ±0.8 mg/L versus 3.5 ±0.3 mg/L). However, 9 of 11 thermal stabilities decreased by at least 5 °C (range: 59 °C to 71 °C versus 71 °C). The reversibility of unfolding varies between mutants in regards to secondary structure as assessed by circular dichroism spectroscopy (Figure 4). The optimal combination of affinity and stability is EA35 A8E/D37N: 6.9 ±1.4 nM affinity and 71 °C Tm, which had the fifth highest sum of natural frequencies.
Figure 4. Circular dichroism spectroscopy of mutants.
Purified affibody domains were analyzed by circular dichroism spectroscopy by scanning wavelengths from 200 to 260 nm. Triplicate scans were collected before (- - -) and after (–––) thermal denaturation and cooling.
Several sets of mutants differ by the choice of neutral amino acid at a single site, which enables site-wise analysis of amino acid choices at sites 2, 4, 8, and 15 (Figure 3 and Figure S3). At site 2, A, N, and Q yield indistinguishable affinities while Q2A provides modestly elevated stability. At site 4, N, Q, and S yield similar affinities and stabilities; N4Q and N4S provide elevated yields. At site 8, A8N provides the optimal affinity whereas A8Q provides the highest stability; A8Q and A8N provide elevated yields. Q and V, at site 15, provide similar affinities, stabilities, and yields. Moreover, three mutants have a charge reversion relative to EA35 as well as a neutralizing mutation (i.e. A8E/D53S, K7A/Q49K, and A8E/D37N) which required the production and analysis of five additional mutants to allow for individual site comparisons (Figure 3 and Figure S3). EA35 K7A/Q49K maintains affinity and stability with high yield. Analysis of the individual mutations to EA35 revealed that both EA35 K7A and EA35 Q49K had improved affinity, moderate stability, and high yield. EA35 A8E/D53S has modest affinity but improved stability and yield. Single mutant EA35 A8E had high affinity and stability and reasonable recombinant yield while EA35 D53S bound with 6.4 ±2.9 nM affinity but was less stable. While EA35 A8E/D37N was the most stable EA35 mutant (Tm = 71 °C), EA35 A8E was only slightly more stable than EA35 (65 °C) and EA35 D37N was less stable (58 °C). The synergy of these mutations is reasonable given their proximity (11.2 Å, on average, in affibody variants with solved structures.). Yet binding and yield lack these synergies as EA35 A8E provides the strongest binding (2.8 ±1.0 nM) while EA35 D37N has the highest yield (32.3 ±2.4 mg/L).
The properties of the collection of consensus mutants were analyzed to assess the utility of natural sequence frequency to guide mutational design. No significant correlation was observed between performance (affinity, stability, or yield) and natural sequence frequency as assessed by the change in natural frequency between pairwise mutants (Figure S4,A–C) or the cumulative frequencies of neutral mutant amino acids (Figure S4,D–F).
Charge Removal in a Combinatorial Library
Based on the variability of consensus design for EA68 and other molecules (Durani and Magliery, 2013; Sullivan et al., 2011), a parallel approach was used that expanded the evaluated sequence space. Combinatorial libraries were created in which three positively charged sites and three negatively charged sites were mutated to neutral amino acids (Table S2). The six mutated sites were chosen again based on natural homolog sequence frequencies, but now using cumulative neutral residue frequency as the metric. Thus, the first library design diversified E2, E8, and E15 (56%, 37%, and 42% neutral naturally) while conserving D36, D37, E47, and D53 (2%, 22%, 7%, 18% neutral naturally); analogously, K4, K7, and K58 (59%, 33%, and 72% neutral) were diversified while K49 and K50 (25% and 10%) were conserved. Three variations were also considered in which an alternative site was diversified and, concomitantly to retain the same overall charge, a previously diversified site was conserved as charged: (Library 1) K49 was diversified and K7 conserved; (Library 2) D37 was diversified and E8 conserved; or (Library 3) D53 diversified and E8 conserved. The permitted neutral residues were chosen as degenerate codons that incorporated the naturally frequent amino acids while also permitting additional chemical functionalities yet excluding charged residues. Five to nine amino acids were permitted at each site to yield theoretical diversities of ≤2×105 mutants. The four libraries were constructed within a yeast surface display system with 2.1×107 to 3.4×107 transformants per library, yielding >99% sampling of theoretical sequence space (Patrick et al., 2003).
The combinatorial libraries were pooled and sorted for stability and binding affinity by thermally treating the yeast-displayed proteins at 70–80 °C and, after cooling, testing binding to 20 nM EGFR ectodomain. Note that thermal denaturation of EA35 and its variants is not fully reversible in the yeast displayed context, which enables this thermal stress to be effectively used (Figure 5A). Mutants exhibiting the best retention of thermal stability and binding were collected and sequenced. Site-wise amino acid frequencies (Figure 5B,C) exhibit a range of behaviors. Amino acids dominant in stable binders are frequent in natural homologs although the inverse is not always true. For example, site 58 is 86% N when naturally neutral, but only 5% N in synthetically sorted mutants. No synthetically dominant residue is rare naturally, although several residues absent naturally are observed at reasonable percentages in sorted binders (maximum: 27% and median: 4% for amino acids not observed in natural homologs).
Figure 5. Combinatorial library analysis.
(A) Yeast displaying affibodies (left: parental EA68, right: enriched stable binding mutants) were treated at 20 °C or 80 °C for 30 minutes, cooled, and incubated with 20 nM biotinylated EGFR ectodomain. Binding was detected by streptavidin-AlexaFluor 647, and affibody display was quantified with anti-c-myc antibody with anti-mouse-AlexaFluor 488 using flow cytometry for quantification. (B) Amino acid frequencies at each diversified site in the combinatorial libraries and enriched stable binders. (C) The observed sequence frequencies in the stable binders relative to the amino acid frequencies in natural affibody homologs. Squares represent K7 and K49 for which the synthetic sequence frequencies sum to, at most, 100% because of the library design requirement to maintain three cations. Circles indicate E8, D37, and D53 for which the synthetic sequence frequencies sum to, at most, 200%.
Four mutants from the sorted population were produced recombinantly and evaluated for yield, affinity, stability, and secondary structure (Figure 6). Three of the four clones have thermal stabilities lower than any of the consensus designed mutants (Figure 7). The average Tm of mutants selected from the library is 57 ±4 °C versus 64 ±1 °C for consensus designed mutants (p=0.23 for two-tailed Student t-test). Production yields were slightly lower for library mutants (6.7 ±2.0 mg/L) than consensus mutants (9.8 ±1.2 mg/L; p=0.24). Affinities were similar between groups (12 ±6 nM for library mutants and 11 ±1 nM for consensus mutants). Notably, though, one of the mutants (EA35 Q2A/K7T/A8E/Q49K/D53Y/N58Y) exhibited very good stability (69 °C) and affinity (1.2 ±0.3 nM) with an acceptable yield (6.0 ±0.4 mg/L).
Figure 6. Analysis of clones from combinatorial library analysis and synthetic consensus design.
The midpoints of thermal denaturation (Tm) were measured by circular dichroism spectroscopy of purified protein. The equilibrium dissociation constants were measured by titration of affibody mutants on A431 cells using flow cytometry to detect binding (n=3). Recombinant yield is the amount of purified affibody recovered per L of culture (n=9). Points and error bars represent means and standard error.
Figure 7. Thermal stability (left), affinity (center), and recombinant yield (right) comparison across design strategies.
Consensus Design from a Synthetic Population
The amino acid frequencies observed in the stable, binding population were used as the basis for synthetic consensus design. Such an approach – using a synthetic source of diversity for consensus design – was successful for enzyme engineering (Jäckel et al., 2010). As in the consensus design from natural homolog sequences above, mutants were designed to neutralize three negatively charged residues and three positively charged residues with a maximal sum of observed frequencies. The top four synthetic consensus designs were produced and evaluated (Figure 6). Thermal stabilities were comparable to consensus design (65 ±1 °C versus 64 ±1 °C); yields were also similar (11 ±3 mg/L versus 9.8 ±1.2 mg/L). Affinities were significantly improved (p=0.05) with two improved mutants (EA35 N4Y/K7A/A8Q/Q49K/N58Y with Kd = 1.8 ±0.4 nM and EA35 N4P/K7P/A8Q/Q49K/N58Y with Kd = 1.7 ±0.5 nM) and two mutants with affinities similar to natural consensus designs. Notably, EA35 N4P/K7P/A8Q/Q49K/N58Y also has strong stability (68 °C) and acceptable yield (7.0 ±0.5 mg/L).
Cellular Binding Specificity, Protein Oligomerization and Solubility
To address the possibility of non-specific binding, A431 (~3 million EGFR per cell) and MCF-7 (<5,000 EGFR per cell) cell lines were incubated with EA68 and six representative charge-reduced clones at 500 nM and analyzed using flow cytometry (Figure 8). For all clones tested, median fluorescence of MCF-7 samples were not statistically different from those containing 0 nM ligand (p = 0.15). Additionally, at 500 nM ligand, fluorescent signal was 2–3 orders of magnitude higher for A431 cells than MCF-7 in all cases.
Figure 8. Cellular EGFR binding specificity.

MCF7 (EGFR-) and A431 (EGFR+) cell lines were incubated on ice with 500 nM affibody ligand, labeled with fluorescein-conjugated anti-His6 antibody, and median fluorescence determined by flow cytometry (n=3). Bars and error bars represent means and standard error.
EA68 and seven EA35 affibodies were analyzed with size exclusion chromatography to assess their degree of oligomerization (Figure 9). Aprotinin (6.5 kDa) and cytochrome c (12.3 kDa), used as standards, eluted in 15.9 and 13.9 mL retention volumes respectively. EA68 eluted closest to aprotinin at 16.2 mL with all EA35 clones eluting between 17.0 and 20.1 mL of PBS with an average ± standard deviation of 18.0 ± 1.1 mL. Modest peak broadening was also observed. For all proteins tested, no aggregation was visible up to and including concentrations of 1 mg/mL in PBS.
Figure 9. Size exclusion chromatography of mutant subset.

Lyophilized proteins were resuspended in PBS to 1 mg/mL of which 0.2 mg was passed through a Superdex 75 30/100 size exclusion column at 0.5 mL/min and elution monitored by absorbance at 280 nm. Aprotinin and cytochrome c were used as molecular weight standards.
Discussion
Consensus design was successful in achieving comparable affinity (Kd = 11 ±4 nM for EA35 vs. 5 ±2 nM for EA68) and recombinant yield (4.0 ±0.8 mg/L vs. 3.5 ±0.3 mg/L) when neutralizing six charged residues from the parental affibody, but exhibited significantly reduced thermal stability (Tm = 62 °C vs. 71 °C) (Figure 3). The results reveal that it is valuable to explore various mutants near the consensus design as four of the other ten homolog-guided mutants had higher stabilities, three had nominally higher affinities (albeit with differences of modest statistical power), and eight of ten mutants had higher yields. In particular, EA35 A8E/D37N provides 10 °C higher thermal stability, 1.6 ±0.9-fold stronger affinity, and 3.2 ±0.9-fold increased recombinant yield relative to the consensus mutant EA35. In fact, EA35 A8E/D37N is comparable to the original EA68 despite the neutralization of six charged residues. Thus, while consensus design is a validated approach, it does not always yield the optimal solution. Amino acid frequency in natural homologs can be useful for identifying tolerated amino acids, but their exact choice should not be dictated by the precise natural frequency as evidenced by the lack of correlation between performance (affinity, stability, or yield) and natural frequency for the frequently observed mutations (Figure S4).
The analysis of mutants based on homolog frequencies can be broadened using combinatorial library selections provided an efficient assay exists. For example, thermal stability can be enriched by heating yeast-displayed proteins (Pavoor et al., 2012). While cell death occurs in response to the thermal stress, structural integrity is sufficiently maintained to allow for precise discrimination via fluorescence-activated cell sorting; plasmids can then be recovered and amplified by PCR. In addition, affinity selections are straightforward. Libraries were synthesized that include 7×105 mutants using degenerate codons including amino acids frequent in homologs as well as rare or unrepresented residues. Combinatorial library sorting enabled identification of a mutant with higher affinity than the top 11 consensus mutants (9.2 ±5.7-fold stronger than EA35) and thermal stability superior to all but one variant (7 °C higher than EA35). Yet the other three selected mutants were comparably or less effective than the consensus mutants. The ability of the thermal and binding selection pressure to enrich the highly functional mutant from a collection of poorly functional mutants (Figure 5A) highlights the power of the approach. Yet, the inability of the selection to deplete some of the modestly effective clones relative to the highly effective clones highlights the limitations of the technique. The selection of comparable affinities to the consensus approach is expected given the selection design: affibodies were incubated with 20 nM EGFR ectodomain to focus selections on stability with retention of good affinity.
Despite the lack of correlation of performance and amino acid frequency in the consensus family of mutants (Figure S4), in the combinatorial library selections, highly enriched mutations were only observed for naturally frequent mutations (Figure 5) thereby indicating no dominant false negatives from consensus design. There were some modest false negatives such as E8Q (16% in natural homologs to 43% in selected mutants), K58Y (0% naturally to 27% in selected mutants), and eight other mutations absent naturally but present at least 10% in selected mutants. As in the direct consensus design of the top 11 clones (Figure 3), there are significant false positives: K4N (93% naturally to 24% in selected mutants) and K58N (86% naturally to 5% in selected mutants).
Beyond identification of a mutant with improved affinity, the combinatorial library selection – and subsequent sequence analysis of functional mutants – provides an additional amino acid distribution to guide consensus design. This ‘synthetic consensus design’ identified a mutant – from only four tested – with 6 °C higher thermal stability, 6.4 ±4.3-fold stronger affinity, and 1.8 ±0.5-fold higher recombinant yield than EA35. In fact, this mutant had higher yield, 3.2 ±1.9-fold (p=0.05) higher affinity, and comparable stability to parental EA68 despite the neutralization of six charged residues. A second mutant also had high affinity (1.8 ±0.4 nM), and the other two synthetic mutants had comparable affinities to the natural consensus series resulting in a set of mutants with significantly improved (p=0.05) affinities relative to natural consensus design (Figure 7). It is noteworthy, that the top clones from the combinatorial library selection and the synthetic consensus design had two and three mutations (D53Y/K58Y or K4P/K7P/K58Y), respectively, that are rare in natural homologs (0–2%). Thus, broadened searches of sequence space were needed to identify such mutants.
This EGFR-binding affibody proved to be amenable to all three modes of engineering charge. The optimal mode for engineering other molecules could be dependent on the molecule of interest as well as the desired properties of interest, but the results herein provide valuable guidance. The synthetic consensus approach proved to be the most effective (highest affinities and comparable stabilities to natural consensus, which were superior to combinatorial library) albeit with modest additional effort relative to natural consensus design. Nevertheless, it is a straightforward approach. Moreover, the synthetic consensus approach is highly valuable in instances in which natural homolog data is not abundant, such as synthetic proteins, or highly conserved at the sites of interest (Jäckel et al., 2010). In cases where natural homolog data is abundant and a rapid, efficient engineering approach is especially valued, natural consensus design can be effective for engineering charge neutralization. Based on the results with EA68, however, it would be recommended to test several mutants of high natural frequency. This is evidenced by the improvement of several consensus mutants relative to EA35, in particular EA35 A8E/D37N.
In all, several mutants were engineered with a reduced number of charged residues relative to the parental EA68 while maintaining stability, recombinant yield, and affinity (which in some cases was actually improved). Receptor specificity was retained, demonstrated by binding comparisons between low (MCF-7) and high (A431) EGFR-expressing cell lines for a representative subset of clones. All mutants tested exhibited preferential binding to A431 cells two to three orders of magnitudes higher than MCF-7 as represented by median fluorescence. Size exclusion chromatography indicated that clones were monomeric at experimentally relevant concentrations. Retention volumes were higher than predicted by molecular weight alone and peaks were broadened somewhat, but these results may be due to a generally ‘stickiness’ between affibody and column resin. Taken together such results validate these molecules as useful variants to examine the impact of charge on physiological distribution.
On a different note, it is worth highlighting that the initial experiment in this study examined the ability to merge an evolved framework and an evolved paratope in the affibody domain. A subset of the optimized framework mutations – originally identified in the context of a HER2 binding affibody (Feldwisch et al., 2010) – were efficiently integrated with a paratope evolved at 13 sites to bind EGFR without detriment to affinity or stability. This, along with a similar result observed with a HER3-binding paratope (Malm et al., 2013), suggests a modular capability of these particular framework mutations, although paratope- and target-dependent impacts could be expected in some cases.
Conclusions
We were able to apply a selection of the improved affibody framework mutations to an evolved EGFR-binding affibody domain and further mutate the molecule for charged-to-neutral mutations of six residues. The molecule was relatively robust to charge removal yet showed differential retention of stability, affinity, and yield dependent upon the site and residue of mutation. A consensus design approach of 11 charge-reduced clones yielded decreased affinity (median 2.0-fold Kd increase) and stability (median ΔTm = 9 °C) with yields exceeding that of parental affibody (median 2.8-fold higher). A broader search of sequence space via a homolog-biased combinatorial library yielded comparable affinities (12 ±6 nM vs. 11 ±1 nM), but lower stabilities (57 ±4 °C vs. 64 ±1 °C) and recombinant yields (6.7 ±2.0 mg/L vs. 9.8 ±1.2 mg/L). Consensus design from the sequences evolved in the synthetic combinatorial library yielded similar results to the natural consensus design with modestly improved affinity (to 1.7 ±0.5 nM). In all, the three methods of charge reduction produced a range of phenotypes including several promising lead molecules with strong affinity, stability, and recombinant yield. Moreover, the comparison of charge modulation techniques provides valuable insight for future charge engineering.
Supplementary Material
Table II.
Amino acid diversities allowed in the four sub-libraries for selection of EA68 mutants with removal of three acidic and three basic residues.
| Sub- library |
E2 | K4 | K7 | E8 | E15 | D36 | D37 | E47 | K49 | K50 | D53 | K58 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ASTIFVQ | FLSYPHITN | ASTFLPIV | ASTIFVQ | VALSQ | D | D | E | K | K | D | FLSYPHITN |
| 2 | ASTIFVQ | FLSYPHITN | K | ASTIFVQ | VALSQ | D | D | E | VALSQ | K | D | FLSYPHITN |
| 3 | ASTIFVQ | FLSYPHITN | ASTFLPIV | E | VALSQ | D | FLSYPHITN | E | K | K | D | FLSYPHITN |
| 4 | ASTIFVQ | FLSYPHITN | ASTFLPIV | E | VALSQ | D | D | E | K | K | FLSYPHITN | FLSYPHITN |
Acknowledgments
Experimental assistance by Nicole Olson is appreciated. This work was supported by the University of Minnesota and the National Institutes of Health (R21 EB019518).
References
- Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Boswell CA, Tesar DB, Mukhyala K, Theil FP, Fielder PJ, Khawli LA. Bioconjug. Chem. Department of Pharmacokinetic and Pharmacodynamic Sciences, Genentech Research and Early Development, South San Francisco, California 94080, USA: American Chemical Society; 2010. Effects of charge on antibody tissue distribution and pharmacokinetics. [DOI] [PubMed] [Google Scholar]
- Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Cochran JR, Kim YS, Lippow SM, Rao B, Wittrup KD. Improved mutants from directed evolution are biased to orthologous substitutions. Protein Eng. Des. Sel. 2006;19:245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]
- Dale GE, Broger C, Langen H, D’Arcy A, Stüber D. Improving protein solubility through rationally designed amino acid replacements: solubilization of the trimethoprim-resistant type S1 dihydrofolate reductase. Protein Eng. 1994;7:933–939. doi: 10.1093/protein/7.7.933. [DOI] [PubMed] [Google Scholar]
- Das D, Georgiadis MM. A directed approach to improving the solubility of Moloney murine leukemia virus reverse transcriptase. Protein Sci. 2001;10:1936–1941. doi: 10.1110/ps.16301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellian M, Yuan F, Trubetskoy VS, Torchilin VP, Jain RK. Vascular permeability in a human tumour xenograft: molecular charge dependence. Br. J. Cancer. 2000;82:1513–1518. doi: 10.1054/bjoc.1999.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durani V, Magliery TJ. Protein engineering and stabilization from sequence statistics: Variation and covariation analysis. Methods Enzymol. 2013;523:237–256. doi: 10.1016/B978-0-12-394292-0.00011-4. [DOI] [PubMed] [Google Scholar]
- Feldwisch J, Tolmachev V, Lendel C, Herne N, Sjöberg A, Larsson B, Rosik D, Lindqvist E, Fant G, Höidén-Guthenberg I, Galli J, Jonasson P, Abrahmsén L. Design of an optimized scaffold for affibody molecules. J. Mol. Biol. 2010;398:232–247. doi: 10.1016/j.jmb.2010.03.002. http://www.ncbi.nlm.nih.gov/pubmed/20226194. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M. Pfam: The protein families database. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M, Orlova A, Johansson E, Eriksson TLJ, Höidén-Guthenberg I, Tolmachev V, Nilsson FY, Ståhl S. Directed evolution to low nanomolar affinity of a tumor-targeting epidermal growth factor receptor-binding affibody molecule. J. Mol. Biol. 2008;376:1388–1402. doi: 10.1016/j.jmb.2007.12.060. [DOI] [PubMed] [Google Scholar]
- García Garayoa E, Schweinsberg C, Maes V, Brans L, Bläuenstein P, Tourwe Da, Schibli R, Schubiger PA. Influence of the molecular charge on the biodistribution of bombesin analogues labeled with the [99mTc(CO)3]-core. Bioconjug. Chem. 2008;19:2409–2416. doi: 10.1021/bc800262m. [DOI] [PubMed] [Google Scholar]
- Hackel BJ, Sathirachinda A, Gambhir SS. Designed hydrophilic and charge mutations of the fibronectin domain: towards tailored protein biodistribution. Protein Eng. Des. Sel. 2012;25:639–647. doi: 10.1093/protein/gzs036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U. S. A. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes NE, MacDonald G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009;21:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, Nanami M, Sekimori Y, Nabuchi Y, Aso Y, Hattori K. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng. Des. Sel. 2010;23:385–392. doi: 10.1093/protein/gzq009. [DOI] [PubMed] [Google Scholar]
- Jäckel C, Bloom JD, Kast P, Arnold FH, Hilvert D. Consensus protein design without phylogenetic bias. J. Mol. Biol. 2010;399:541–546. doi: 10.1016/j.jmb.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TM, Hickman AB, Dyda F, Ghirlando R, Davies DR, Craigie R. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6057–6061. doi: 10.1073/pnas.92.13.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura RH, Teed R, Hackel BJ, Pysz Ma, Chuang CZ, Sathirachinda A, Willmann JK, Gambhir SS. Pharmacokinetically stabilized cystine knot peptides that bind alpha-v-beta-6 integrin with single-digit nanomolar affinities for detection of pancreatic cancer. Clin. Cancer Res. 2012;18:839–849. doi: 10.1158/1078-0432.CCR-11-1116. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3271184&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malm M, Kronqvist N, Lindberg H, Gudmundsdotter L, Bass T, Frejd FY, Höidén-Guthenberg I, Varasteh Z, Orlova A, Tolmachev V, Ståhl S, Löfblom J. Inhibiting HER3-Mediated Tumor Cell Growth with Affibody Molecules Engineered to Low Picomolar Affinity by Position-Directed Error-Prone PCR-Like Diversification. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein a. Protein Eng. Des. Sel. 1987;1:107–113. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- Nygren P-A. Alternative binding proteins: affibody binding proteins developed from a small three-helix bundle scaffold. FEBS J. 2008;275:2668–2676. doi: 10.1111/j.1742-4658.2008.06438.x. [DOI] [PubMed] [Google Scholar]
- Pace CN. Measuring and increasing protein stability. TRENDS Biotechnol. 1990;8:93–98. doi: 10.1016/0167-7799(90)90146-o. [DOI] [PubMed] [Google Scholar]
- Patrick WM, Firth AE, Blackburn JM. User-friendly algorithms for estimating completeness and diversity in randomized protein-encoding libraries. Protein Eng. 2003;16:451–457. doi: 10.1093/protein/gzg057. [DOI] [PubMed] [Google Scholar]
- Pavoor TV, Wheasler JA, Kamat V, Shusta EV. An enhanced approach for engineering thermally stable proteins using yeast display. Protein Eng. Des. Sel. 2012;25:625–630. doi: 10.1093/protein/gzs041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimm M, Gribben S, Bogdán K, Hudecz F. The effect of charge on the biodistribution in mice of branched polypeptides with a poly (l-lysine) backbone labelled with 125 I, 111 In or. J. Control. Release. 1995;37:161–172. [Google Scholar]
- Pogulis RJ, Vallejo a N, Pease LR. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol. Biol. 1996;57:167–176. doi: 10.1385/0-89603-332-5:167. [DOI] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell Syst. Technol. J. 1948;27:379, 423, 623–656. [Google Scholar]
- Slovic AM, Kono H, Lear JD, Saven JG, DeGrado WF. Computational design of water-soluble analogues of the potassium channel KcsA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1828–1833. doi: 10.1073/pnas.0306417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler JB, Neil JR, Abramovitch S, Yarden Y, White FM, Lauffenburger Da, Wittrup KD. Combination antibody treatment down-regulates epidermal growth factor receptor by inhibiting endosomal recycling. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13252–13257. doi: 10.1073/pnas.0913476107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steipe B, Schiller B, Plückthun A, Steinbacher S. Sequence statistics reliably predict stabilizing mutations in a protein domain. J. Mol. Biol. 1994;240:188–192. doi: 10.1006/jmbi.1994.1434. [DOI] [PubMed] [Google Scholar]
- Steipe B. Methods Enzymol. Ontario, Canada: University of Toronto, Program in Proteomics and Bioinformatics, Department of Biochemistry; 2004. Consensus-based engineering of protein stability: From intrabodies to thermostable enzymes. [DOI] [PubMed] [Google Scholar]
- Sullivan BJ, Durani V, Magliery TJ. Triosephosphate isomerase by consensus design: Dramatic differences in physical properties and activity of related variants. J. Mol. Biol. 2011;413:195–208. doi: 10.1016/j.jmb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Tran TA, Ekblad T, Orlova A, Sandström M, Feldwisch J, Wennborg A, Abrahmsén L, Tolmachev V, Eriksson Karlström A. Effects of lysine-containing mercaptoacetyl-based chelators on the biodistribution of 99mTc-labeled anti-HER2 Affibody molecules. Bioconjug. Chem. 2008;19:2568–2576. doi: 10.1021/bc800244b. [DOI] [PubMed] [Google Scholar]
- Tran T, Engfeldt T, Orlova A. 99mTc-maEEE-ZHER2: 342, an Affibody molecule-based tracer for the detection of HER2 expression in malignant tumors. Bioconjugate Chem. 2007;2:1956–1964. doi: 10.1021/bc7002617. [DOI] [PubMed] [Google Scholar]
- Wahlberg E, Lendel C, Helgstrand M, Allard P, Dincbas-Renqvist V, Hedqvist A, Berglund H, Nygren P-A, Härd T. An affibody in complex with a target protein: structure and coupled folding. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3185–3190. doi: 10.1073/pnas.0436086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Basinski MB, Beals JM, Briggs SL, Churgay LM, Clawson DK, DiMarchi RD, Furman TC, Hale JE, Hsiung HM, Schoner BE, Smith DP, Zhang XY, Wery JP, Schevitz RW. Crystal structure of the obese protein leptin-E100. Nature. 1997;387:206–209. doi: 10.1038/387206a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.