ABSTRACT
Histone deacetylase 9 (HDAC9) is expressed in B cells, and its overexpression has been observed in B-lymphoproliferative disorders, including B-cell non-Hodgkin lymphoma (B-NHL). We examined HDAC9 protein expression and copy number alterations in primary B-NHL samples, identifying high HDAC9 expression among various lymphoma entities and HDAC9 copy number gains in 50% of diffuse large B-cell lymphoma (DLBCL). To study the role of HDAC9 in lymphomagenesis, we generated a genetically engineered mouse (GEM) model that constitutively expressed an HDAC9 transgene throughout B-cell development under the control of the immunoglobulin heavy chain (IgH) enhancer (Eμ). Here, we report that the Eμ-HDAC9 GEM model develops splenic marginal zone lymphoma and lymphoproliferative disease (LPD) with progression towards aggressive DLBCL, with gene expression profiling supporting a germinal center cell origin, as is also seen in human B-NHL tumors. Analysis of Eμ-HDAC9 tumors suggested that HDAC9 might contribute to lymphomagenesis by altering pathways involved in growth and survival, as well as modulating BCL6 activity and p53 tumor suppressor function. Epigenetic modifications play an important role in the germinal center response, and deregulation of the B-cell epigenome as a consequence of mutations and other genomic aberrations are being increasingly recognized as important steps in the pathogenesis of a variety of B-cell lymphomas. A thorough mechanistic understanding of these alterations will inform the use of targeted therapies for these malignancies. These findings strongly suggest a role for HDAC9 in B-NHL and establish a novel GEM model for the study of lymphomagenesis and, potentially, preclinical testing of therapeutic approaches based on histone deacetylase inhibitors.
KEY WORDS: HDAC9, Lymphoma, Transgenic mouse
Summary: This study demonstrates that aberrant expression of HDAC9 in B cells promotes development of lymphoproliferative disease and lymphoma through altering expression of genes involved in the cell cycle and survival, and modulating the activity of key B-lineage factors such as BCL6 and p53.
INTRODUCTION
Non-Hodgkin lymphoma (NHL) are a heterogeneous group of cancers of B, T or natural-killer cells, and constitute 4-5% of all cancers (Ferlay et al., 2013; Siegel et al., 2015), with diffuse large B-cell lymphoma (DLBCL) being the most common subtype, accounting for 31% of all adult NHLs (Martelli et al., 2013). Overall, approximately five cases of NHL per 100,000 individuals are identified annually (rising to 12 per 100,000 in North America), with incidence increasing, especially in developed countries. Similarly, NHL is the 11th most common cause of cancer death worldwide, resulting in around 200,000 deaths in 2012. Despite improvements in 5-year relative survival rates to 70% over the last four decades (largely due to the use of antibodies and antibody–drug conjugates directed against cell-surface antigens), patients with relapsed or refractory disease continue to have poor outcomes (Ansell, 2015; Grover and Park, 2015). New approaches are therefore required in NHLs, and therapeutic targeting of epigenetic modifiers, including histone deacetylases (HDACs), holds great promise (Hassler et al., 2013).
HDACs catalyze deacetylation of acetylated lysine residues on histones, and are also being found to act on a growing number of non-histone proteins (Haberland et al., 2009; Yang and Seto, 2008). As such, the functional interaction networks of HDACs encompass many biological and cellular processes beyond chromatin modification and gene regulation. In humans, there are 11 canonical HDACs grouped into three major classes: class I comprises HDACs 1, 2, 3 and 8; class II comprises HDACs 4, 5, 6, 7, 9 and 10; and class IV is represented by HDAC11. HDAC9, alongside HDACs 4, 5 and 7, forms the class IIa subfamily, and these proteins are key transcriptional co-regulators in development and differentiation (Martin et al., 2009). Mutation or aberrant expression of HDAC9 has been implicated in diverse conditions, including ischemic stroke, schizophrenia and obesity (Bellenguez et al., 2012; Chatterjee et al., 2014; Lang et al., 2012), and also as a maker of poor outcome in cancer (Milde et al., 2010; Moreno et al., 2010). HDAC9, which is subject to complex regulation via differential promoter usage and alternative splicing, is preferentially expressed in the lymphoid lineage within the hematopoietic system (Petrie et al., 2003). HDAC9 is highly expressed in B-lymphoproliferative disorders, including in B-cell non-Hodgkin lymphoma (B-NHL) cell lines and patient samples, suggesting that its deregulation might lead to abnormal B-cell proliferation (Petrie et al., 2003; Sun et al., 2011). These findings are supported by the recurrent amplification of the HDAC9 locus (chr. 7p21.1) in B-NHL (Bea et al., 2005; Bentz et al., 1999, 1996; Monni et al., 1996; Rubio-Moscardo et al., 2005; Tagawa et al., 2005). Additionally, a number of HDAC inhibitors have been shown to induce cell death in B-NHL cells (Haery et al., 2015; Lemoine and Younes, 2010). Although several in vivo mouse models examining the biological functions of the class I and II HDACs are available (Witt et al., 2009), a role for HDAC9 or other family members in B-NHL has not been examined in vivo. The study of Hdac9–/– knockout mice has, however, highlighted HDAC9 as an important factor in inhibiting the generation and function of regulatory T (Treg) cells (Tao et al., 2007; Yan et al., 2011).
Underlining a potential role in B-NHL, HDAC9 interacts with BCL6 (Basso et al., 2010; Miles et al., 2005; Petrie et al., 2003), a transcriptional repressor that is crucial for germinal center (GC) formation (Basso and Dalla-Favera, 2012). Transgenic mice that constitutively express BCL6 in B cells develop a lymphoproliferative syndrome that culminates with the development of B-NHL (Cattoretti et al., 2005). BCL6 directly recruits class-II HDACs through its zinc-finger domain (Lemercier et al., 2002), and its transcriptional targets in GC B cells include TP53, thus modulating DNA-damage-induced apoptotic responses (Phan and Dalla-Favera, 2004). Evidence for a major role for defective acetylation in the pathogenesis of B-NHL is supported by the frequent occurrence of structural alterations inactivating CREBBP and EP300, genes encoding two highly related histone acetyltransferases (HATs) and non-HATs (Pasqualucci and Dalla-Favera, 2015; Pasqualucci et al., 2011). These mutations lead to aberrant activation and deactivation, respectively, of BCL6 and p53 (Pasqualucci et al., 2011), and we hypothesized that aberrant HDAC9 expression could also interfere with p53 and/or BCL6. We therefore sought to characterize HDAC9 expression in human B-cell lymphomas and establish whether aberrant expression can drive B-cell lymphoma in a genetically engineered mouse (GEM) model. Here, we report the development of a GEM in which an HDAC9 transgene was constitutively expressed in B cells under the control of the immunoglobulin heavy chain (Eμ) enhancer (Eµ-HDAC9). Eµ-HDAC9 mice developed B-lymphoproliferative disorders with progression towards B-NHL. This is consistent with the hypothesis that deregulated protein acetylation plays a pathological role in B-NHL, and provides a model for preclinical evaluation of HDAC inhibitors (HDACIs).
RESULTS
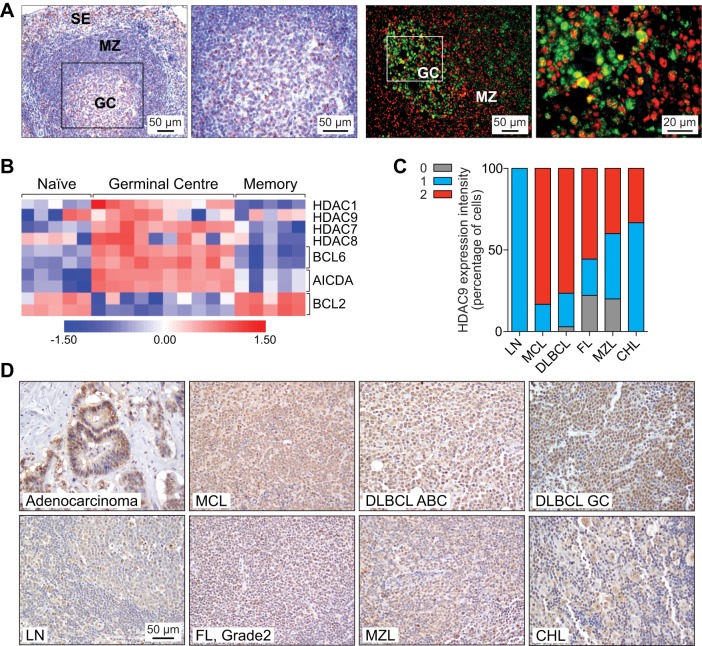
Within the immune system, a role for HDAC9 in the control of Treg cell function has previously been described (Beier et al., 2012; de Zoeten et al., 2010; Parra, 2015; Tao et al., 2007), and we found that, in normal human mature B cells, HDAC9 mRNA expression is significantly upregulated in the GC (Petrie et al., 2003) (Fig. 1A). HDAC9 protein is detected in a subset of GC cells, where it is co-expressed with BCL6 (Fig. 1A), as well as in a subset of lymphoid cells in the mantle zone and paracortex (Klein et al., 2003) (Fig. 1B). High HDAC9 gene expression in B-lymphoproliferative disorders, including B-NHL cell lines and patient samples, has pointed to a potential role in these diseases (Petrie et al., 2003; Sun et al., 2011). In line with these findings, we detected high HDAC9 protein levels among various lymphoma entities, including DLBCL (n=34), marginal zone lymphoma (MZL) (n=5), follicular lymphoma (FL) (n=9), classical Hodgkin lymphoma (CHL) (n=3) and mantle cell lymphoma (MCL) (n=6). Highest levels of HDAC9 expression were observed in the most aggressive lymphomas, such as DLBCL (both GC and non-GC subtypes) and MCL (77% and 83%, respectively, P=1.0, Fisher's exact test). In contrast, low-grade B-cell lymphomas, as well as CHL, showed low HDAC9 expression in tumor cells when nuclear intensity was compared with that of adenocarcinoma cells as a positive control (P=0.004, Fisher's exact test) (Fig. 1C). In addition to high HDAC9 expression, frequent amplification of the HDAC9 locus (chr. 7p21.1) has been observed in B-NHL (Bea et al., 2005) and, consistent with these results, we found copy number gains of HDAC9, including high-level amplifications, in 46.3% (25/54) of DLBCL patients (Fig. S1). A total of 46% (13/28) of samples with HDAC9 copy number gains presented trisomy 7 (Fig. S1A), whereas 43% (12/28) of cases reported with smaller regions of amplification within the chromosome that contained the HDAC9 gene (Fig. S1B). Here, one case displayed a specific amplification of HDAC9 (18,409,840-18,605,177 bp) (Fig. S1C, Table S1).
Fig. 1.
HDAC9 is highly expressed in human B-cell lymphomas. (A) HDAC9 expression in germinal center (GC) lymphatic nodules of normal human tonsils. Left panels, immunohistochemical staining for HDAC9 (red). Cells were nuclear counterstained with hematoxylin (blue). Right panels, immunofluorescent analysis of HDAC9 (red) and BCL6 (green) co-expression. SE, subepithelial cells; MZ, marginal zone. (B) Expression of HDAC family members in purified mature B-cell subpopulations (naive, GC, memory). Expression patterns of BCL6, AICDA and BCL2 are shown as controls. Individual columns correspond to independent samples. The color scale reflects the range in expression values after log2 transformation (0, mean expression level; red, high expression; blue, low expression). Expression data from Klein et al. (2003). (C) Average signal intensity of HDAC9 staining in the indicated samples was scored as negative (0; gray), low (1; blue) or high (2; red) relative to rectal adenocarcinoma cells expressing HDAC9. Samples containing cells expressing on average HDAC9 with equal or higher intensity were scored as 2 and samples with lower expression were scored as 1. Cells lacking expression of HDAC9 were scored as 0. (D) Representative images of HDAC9 expression. Expression of HDAC9 in rectal adenocarcinoma is shown as a positive control. DLBCL, diffuse large B-cell lymphoma; GC, germinal cell; ABC, activated B-cell; CHL, classical Hodgkin lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; MZL, marginal zone lymphoma; LN, reactive lymph node.
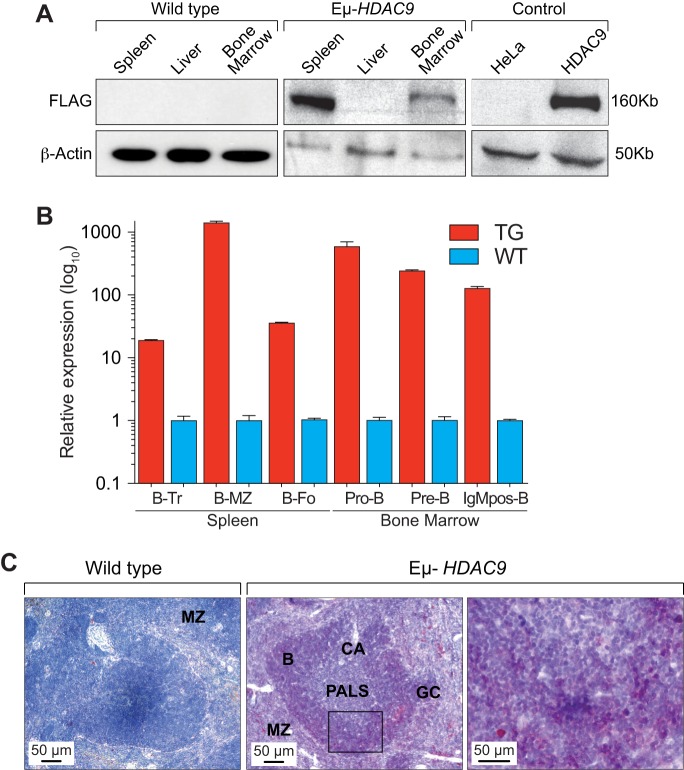
Although several in vivo mouse models examining the biological functions of the class I and II HDACs are available (Parra, 2015), a role for HDAC9 or other class-IIa family members in B-NHL has never been examined in vivo. We therefore expressed a human HDAC9 transgene (HDAC9TG) in the B-cell compartment from an early stage of B-cell development under the control of the immunoglobulin heavy chain (IgH) enhancer (Eμ) (Fig. S2A). We generated three independent transgenic lines (designated as 1468, 1469 and 1839) (Fig. S2B) and monitored a total of 124 mice (78 Eµ-HDAC9 and 46 wild type) for tumor formation and overall survival. We found expression of the HDAC9TG in the bone marrow and spleen but not in the liver (Fig. 2A). We detected expression of HDAC9TG throughout all B-cell stages in the bone marrow (pro-B, pre-B and naive-B) and spleen (transitional, marginal zone and follicular), with greatest expression of HDAC9TG found in the splenic marginal zone (Fig. 2B,C). When analyzed between 6 and 12 months of age, a fraction (3/17, 18%) of Eµ-HDAC9 mice exhibited splenomegaly (Fig. S3A,B), compared to 0/10 wild-type littermates. Histopathology and fluorescence-activated cell sorting (FACS) analysis revealed evidence of abnormal B-cell expansion in the spleen, compatible with the development of lymphoproliferative disorder (LPD) (n=1), and splenic MZL (SMZL) (n=2); no abnormalities were observed in control mice (0/10) (P<0.0001) (Fig. S3A-C). Analysis of immunoglobulin (Ig) gene rearrangements in these mice confirmed monoclonal expansions of B-cell populations in 2/3 young-adult Eµ-HDAC9 mice at 8 months of age (Table 1). These results closely mirror those for a GEM model constitutively expressing BCL6 in B cells under the control of the immunoglobulin heavy chain (IgH) Iµ promoter (Cattoretti et al., 2005). A remarkably similar fraction of these mice (4/24, 17%) also displayed LPD at 6 months of age, representing early stages of lymphomagenesis before the evolution and onset of a B-cell neoplasm later in life (Cattoretti et al., 2005).
Fig. 2.
Characterization of Eµ-HDAC9 transgenic mice. (A) Western blot analysis for detection of HDAC9TG expression with monoclonal anti-FLAG M2 antibody. HeLa cells, which lack expression of FLAG, and HeLa cells transfected with FLAG-tagged HDAC9 are shown as negative and positive controls, respectively. (B) HDAC9TG (TG) transcript expression in B-cell subsets. mRNA transcripts for HDAC9 were analyzed by quantitative reverse transcription PCR (RT-qPCR) from HDAC9TG (TG, red) spleen (left) or bone marrow (right) versus controls from wild-type littermates (WT, blue). RNA from three individuals (spleen) or five individuals (bone marrow) was pooled and values are expressed as the fold change in transcript abundance (±s.d.) compared with values for WT mice. Transitional B-cell (B-Tr), P<0.0001; marginal zone B-cell (B-MZ), P<0.0001; follicular B-cell (B-Fo), P<0.0001; Pro B-cell (Pro-B), P=0.0010; Pre B-cell (Pre-B), P<0.0001; IgM-positive/mature B-cell (IgMpos-B), P<0.0001. (C) Immunohistochemical analysis of HDAC9TG expression in spleen from two Eµ-HDAC9 mice and a wild-type littermate control. HDAC9 expression was detected with anti-FLAG M2 antibody and stained red with AEC (3-amino-9-ethylcarbazole). Cells were nuclear counterstained with hematoxylin. In Eµ-HDAC9 spleen, expression of HDAC9TG is highest in the marginal zone and white pulp (B-cell zone). MZ, marginal zone; CA, central arteriole; PALS, periarteriolar lymphoid sheath (T-cell zone); GC, germinal center; B, primary follicles B-cell rich (B-cell zone).
Table 1.
Tumor type and IgH rearrangements in Eμ-HDAC9 mice
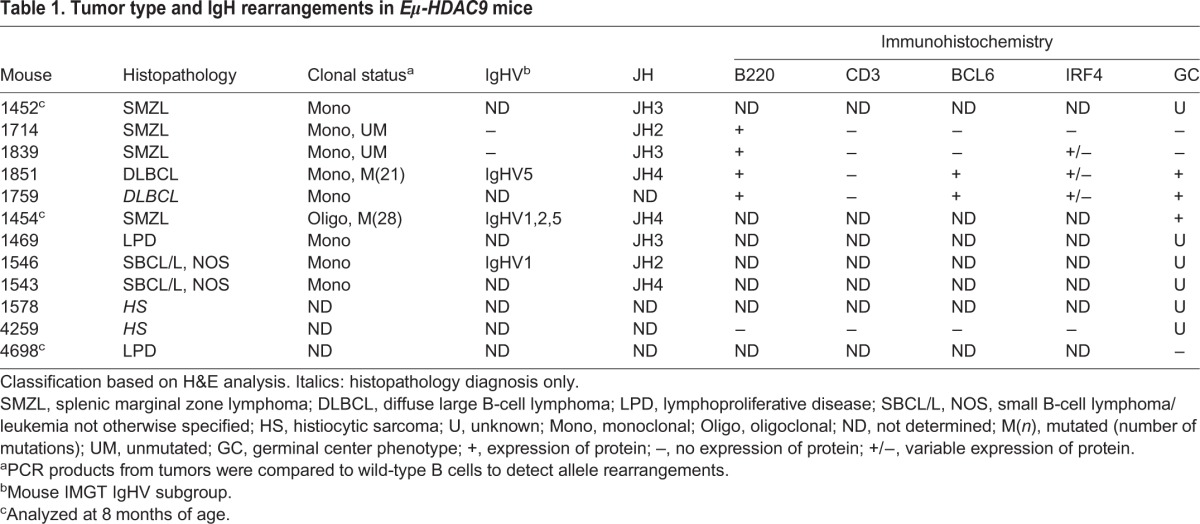
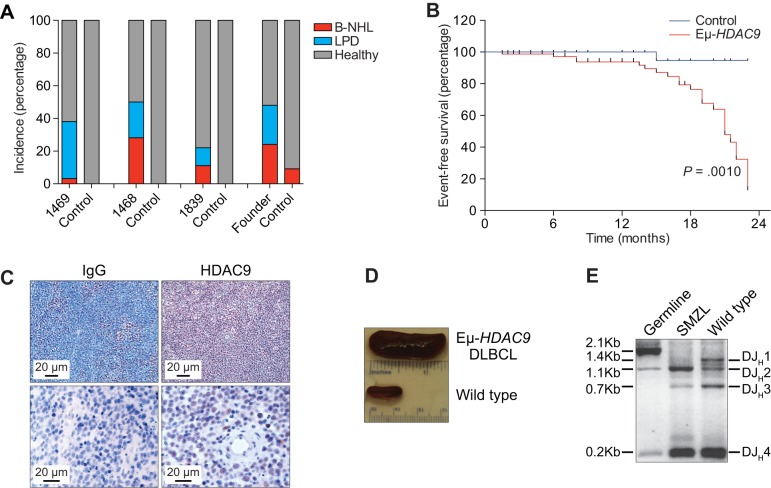
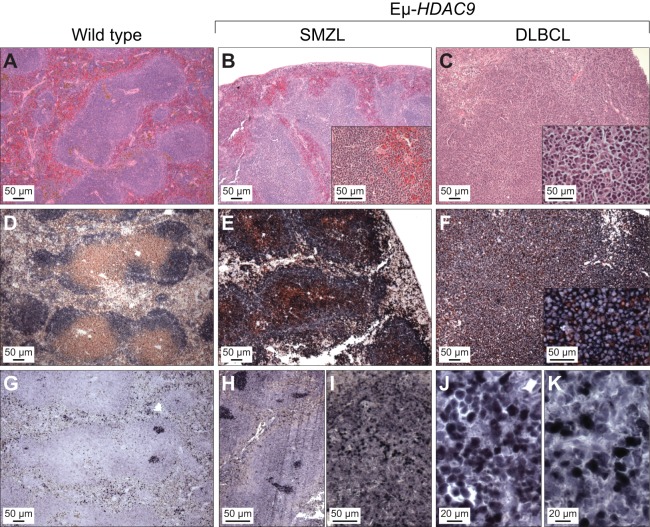
With age (i.e. past 14 months), Eµ-HDAC9 mice developed a significantly higher frequency of lymphoproliferations compared to control animals, such that, by 21 months of age, 48% of Eµ-HDAC9 mice survived compared with 95% of wild-type control mice (P=0.0010, Fig. 3A). Phenotypic analysis in a subset of animals revealed that approximately 40% (line 1469), 50% (line 1468) and 20% (line 1839) of Eµ-HDAC9 mice developed B-cell lymphomas, compared to a minor fraction (5%) of wild-type littermates that exhibited evidence of B cell malignancies (P<0.0001) (Fig. 3B). Flow cytometric analysis of tumors from spleens of adult Eµ-HDAC9 mice displayed a mature B-cell immunophenotypic profile (B220dull, IgDlow/neg, IgMhigh, CD23neg and CD21neg) (Fig. S4). Eµ-HDAC9 B-cell tumors were primarily of splenic origin (Fig. 3C,D), with or without nodal involvement, expressed B220, indicating B-cell derivation, and were histologically defined as LPD, SMZL and DLBCL (Table 1). An additional 20% of Eµ-HDAC9 mice either developed splenic B-cell lymphoma/leukemia (SBCL/L) or another hematological malignancy, such as histiocytic sarcoma (Table 1).
Fig. 3.
Frequency of lymphoproliferative disease (LPD) and B-cell lymphoma (B-NHL) in Eµ-HDAC9 mice. (A) Incidence of B-NHL (red) and LPD (blue) in three Eµ-HDAC9 lines [1469 (n=28), 1468 (n=18) and 1839 (n=8)] and founder mice (n=17) aged between 14 and 23 months as compared to wild-type (control) littermates [1469 (n=18), 1468 (n=8), 1839 (n=9) and founder mice (n=11)]. Statistical analysis was performed using; Chi-square (χ2) test on combined value for Eμ-HDAC9 lines and founders versus wild-type controls (P<0.0001). (B) Kaplan–Meier plot of event-free (B-NHL) survival of transgenic versus wild-type littermate controls. Eµ-HDAC9 (n=78) and control (n=46) mice, P=0.0010, log-rank (Mantel–Cox) test. (C) Immunohistochemical (IHC) analysis of HDAC9 expression (red, anti-HDAC9 antibody) in Eµ-HDAC9 diffuse large B-cell lymphoma (DLBCL) lymph nodes. Staining for IgG (blue) was used as a control. (D) Representative Eµ-HDAC9 spleen infiltrated by DLBCL (top) compared to wild-type littermate control spleen (bottom). (E) Analysis of rearranged IgH genes in Eµ-HDAC9 splenic marginal zone lymphoma (SMZL). Shown is a monoclonal D-JH rearrangement confirmed by sequencing (D-JH2, 1.1 kb). Also shown is polyclonal rearrangement of a wild-type littermate control. Tail DNA was used for germline configuration.
Molecular analysis of the rearranged IgH genes from Eµ-HDAC9 B-NHL samples confirmed their monoclonal origin (Fig. 3E) with evidence of somatic hypermutation (SHM) of IgV genes in 4/7 (57%) of tumors (Fig. S5). Thus, we concluded that Eµ-HDAC9 lymphomas and LPDs were derived from B-cell precursors, with evidence of transit through the GC or having experienced the GC reaction. In mice, the lymphomas and LPDs exhibited both GC and post-/non-GC immunophenotypes (Table 1). Immunohistochemical analysis revealed heterogeneous expression of BCL6, including cases that were below the detection level by immunohistochemistry (IHC), as previously observed in Iμ-BCL6-derived lymphomas (Cattoretti et al., 2005). Eµ-HDAC9 tumors were also found to express variable levels of IRF4 (MUM1), a marker for GC B cells and plasma cells, and typically found in non-GC-type DLBCL (Fig. 4 and Table 1) (Falini et al., 2000).
Fig. 4.
Histopathological analysis of mature B-cell lymphomas in Eµ-HDAC9 mice. Representative images showing disorganization of lymphoid tissue and expression of B220, CD3 and BCL6. Mouse spleen sections from wild type, splenic marginal zone lymphoma (SMZL) and diffuse large B-cell lymphoma (DLBCL) cases (as indicated) were stained with hematoxylin-eosin (A-C), doubled stained with B220 (blue)/CD3 (brown) (D-F), or BCL6 (blue) (G-J). IRF4 expression is shown in panels I and K.
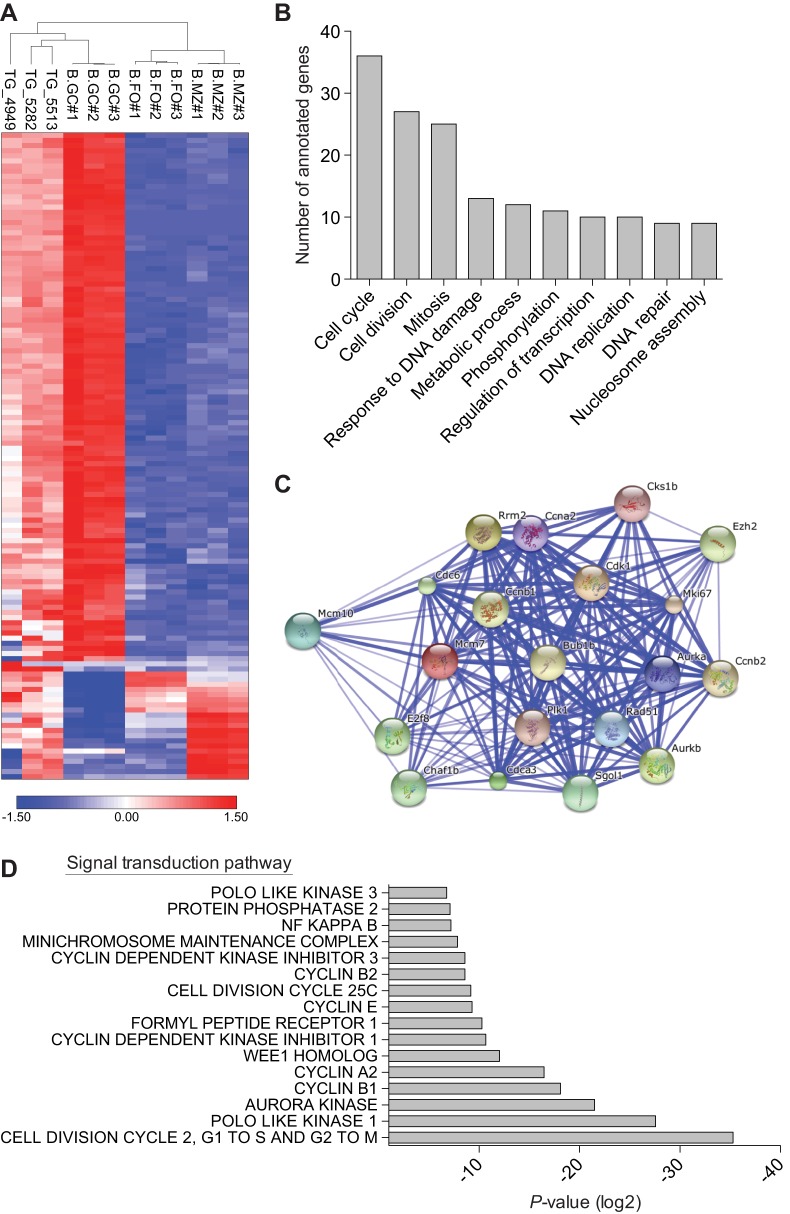
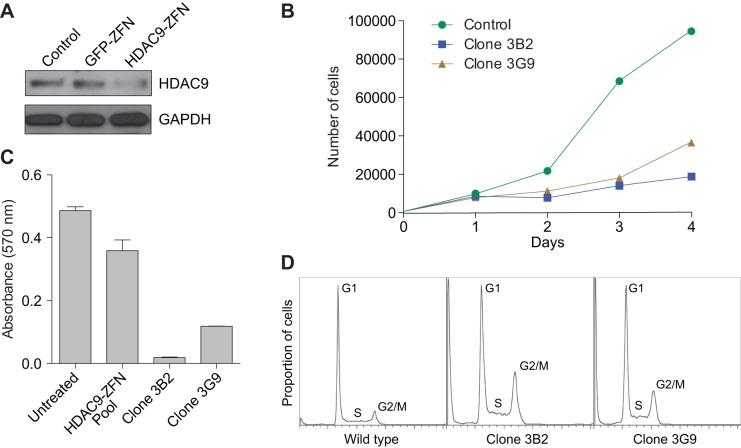
We next performed gene expression analysis, and a comparison of three representative DLBCLs from Eμ-HDAC9 mice versus normal B-cell populations indicated that Eµ-HDAC9 B-cell tumors cluster with GC B cells and separately from non-GC B cells, further supporting a GC origin, comparable with human tumors (Fig. 5A). We also compared the expression pattern of DLBCLs from Eμ-HDAC9 mice with three age- and gender-matched wild-type controls, identifying a total of 1469 upregulated and 307 downregulated transcripts (Table S2). Among the upregulated genes, pathway analysis using KEGG and gene ontology (GO) annotated databases revealed enrichment for genes involved in the cell cycle, cell division and response to DNA damage (Fig. 5B, Table 2, and Tables S3 and S4). Pathway-based hierarchical clustering (Good, 2000) for genes differentially expressed in Eμ-HDAC9 mice (Tables S5, S6 and S7) confirmed regulation of cell-cycle-related genes and pointed to modulation of MAPK/ERK pathways. Furthermore, analysis of predicted protein–protein interactions between KEGG-classified genes from HDAC9TG B-cell tumors (Table S2) showed high network connectivity (Fig. 5C). A number of differentially expressed genes from the Eμ-HDAC9-driven B-cell tumors were identified as direct targets for BCL6 and p53 that have roles in apoptosis, the cell cycle and B-cell receptor (BCR) signaling (Table S2). Additionally, signal transduction pathways revealed substantial enrichment for genes involved in G1/S and G2/M transition followed by the Polo-like kinase 1 (PLK1) pathway (Fig. 5D). Among the most upregulated genes found in HDAC9TG B-cell tumors were those encoding factors such as Plk1, Birc5, Cdk1, Aurka, Aurkb and Chek1, which are involved in proliferation and survival, G1/S and G2/M transitions, mitosis and DNA-repair/checkpoint-mediated arrest (Table 2). A role for HDAC9 in proliferation and control of the cell cycle was confirmed by zinc-finger nuclease (ZFN)-mediated gene editing of HDAC9 (Fig. 6). Monoallelic knockout of HDAC9 in the Raji Burkitt's lymphoma cell line (biallelic forms were non-viable; data not shown) led to reductions in HDAC9, consistent with a decrease in gene dosage (Fig. 6A). This resulted in growth inhibition (Fig. 6B,C) and an increase in the proportion of cells in S and G2/M phases (Fig. 6D).
Fig. 5.
Expression microarray analysis of Eµ-HDAC9 tumors. (A) Unsupervised hierarchical clustering of gene expression data from representative Eµ-HDAC9-derived (TG) lymphomas (n=3) versus normal murine mature B-cell subpopulations, including germinal center (GC), follicular (FO) and marginal zone (MZ) B cells (GSE15907). Color scale represents the range in relative expression changes (Z score) across samples, normalized by the standard deviation after log2 transformation (0, mean expression level; red, high expression; blue, low expression). (B) Functional annotation of upregulated gene expression in HDAC9TG tumors according to biological process. Values represent the number of upregulated genes for a given gene ontology (GO) term. A complete list of annotated genes for identified GO functional categories is shown in Table S5. Gene annotation was performed using Partek Genomics Suite 6.6. (C) Interactome of upregulated genes in HDAC9TG B-cell tumors. Interactions were analyzed using STRING v9.1. Thicker lines define stronger associations in the interactome. (D) Functional annotation of expression of signal transduction pathway genes upregulated in HDAC9TG tumors. Gene sets from HDAC9TG B-cell tumors were analyzed using GGA (Genomatix Genome Analyzer) and ranked according to P-value.
Table 2.
Gene expression profiling of Eμ-HDAC9 tumors
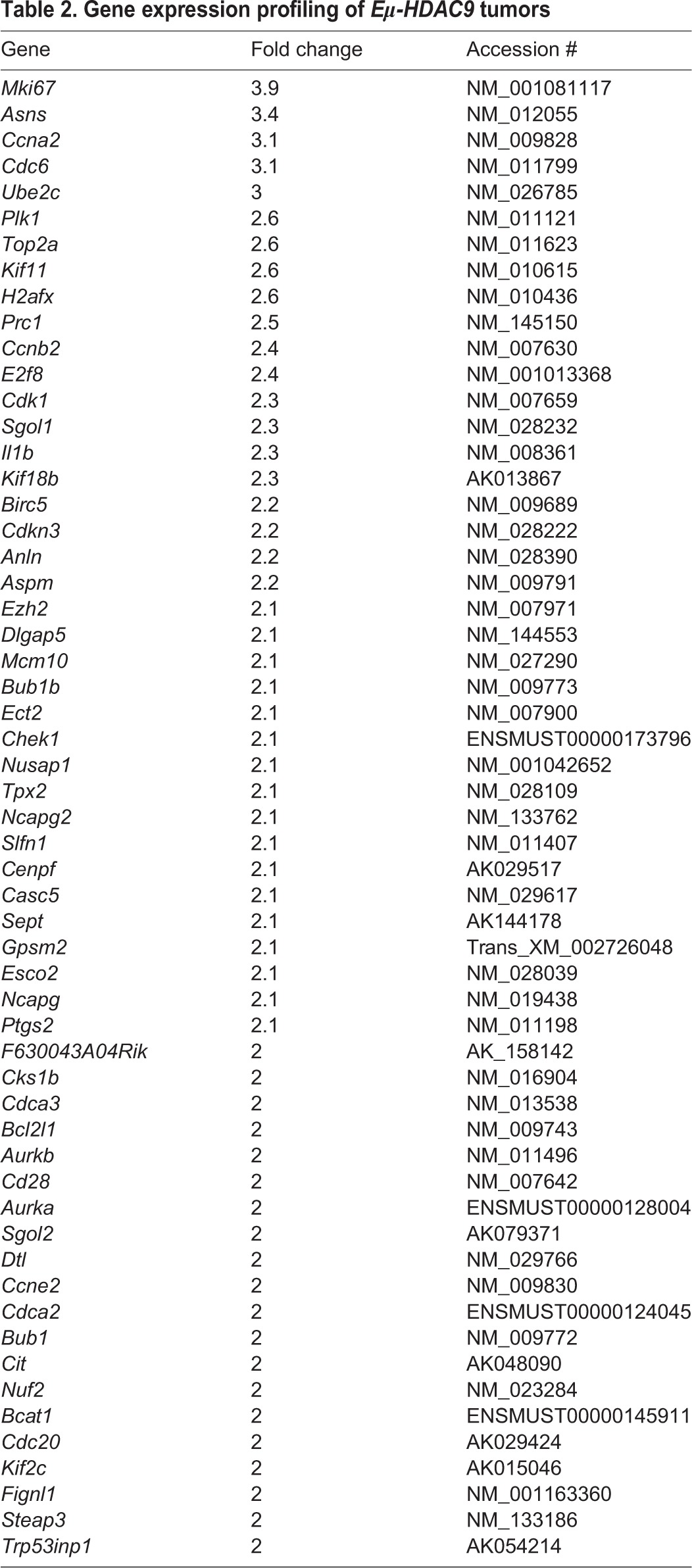
Fig. 6.
HDAC9 regulates cell cycle progression. (A) Immunoblot analysis of HDAC9 expression in untreated wild-type Raji B-cell non-Hodgkin lymphoma (B-NHL) cells (control, lane 1), and the mutants GFP ZFN-derived control (lane 2) and HDAC9 ZFN-derived cells prior to single cell sorting (lane 3). Immunoblot analysis of GAPDH expression was used as loading control. (B) Cell growth curve of wild-type Raji control and representative ZFN-generated HDAC9 mutant clones 3B2 and 3G9 obtained by single-cell sorting. (C) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay of proliferation of HDAC9 ZFN-derived clones. Shown are values for HDAC9 ZFN-derived mutant clones 3B2 and 3G9, a pool of unsorted HDAC9 ZFN mutants, and wild-type cells and untreated Raji cells. (D) Effects of HDAC9 depletion on cell cycle progression are depicted in histogram plots generated by propidium-iodide uptake.
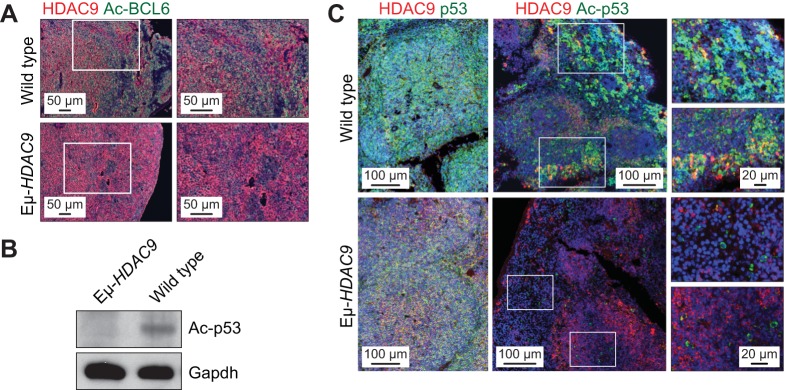
A potential role for HDAC9 in the pathogenesis of B-NHL is strengthened by its interaction with BCL6 (Petrie et al., 2003), a transcriptional repressor crucial for GC formation (Basso and Dalla-Favera, 2012) and whose deregulated expression in B cells leads to LPD and B-NHL (Cattoretti et al., 2005). BCL6 directly recruits class-II HDACs through its zinc-finger domain (Lemercier et al., 2002) and its transcriptional targets in GC B cells include TP53, thus modulating DNA-damage-induced apoptotic responses (Phan and Dalla-Favera, 2004). BCL6 and p53 function in a negative-feedback loop whereby p53 promotes BCL6 expression, which in turn suppresses the expression of TP53 (Margalit et al., 2006; Phan and Dalla-Favera, 2004). The BCL6–p53 axis is further modulated by post-translational acetylation of BCL6, which leads to its inactivation. Indicative of a potential role for HDAC9 in the acetylation of BCL6 in vivo, acetylated BCL6 was found to be abundant in normal mouse spleens but undetectable in HDAC9TG tumors (Fig. 7A). p53 is also post-translationally modified by acetylation, which is indispensable for its transcriptional activity in response to DNA damage and stress (Tang et al., 2008). Highlighting the loss of p53 tumor suppressor function as a contributory factor in the development of B-cell lymphomas, B-cell-specific disruption of TP53 leads to the development of B-NHL (Chiang et al., 2012). Consistent with this, as well as with recent research demonstrating that HDAC9 binds to the TP53 promoter to repress gene expression (Zhao et al., 2015), we found diminished levels of total p53 as well as Lys379 acetyl-p53 in Eµ-HDAC9 tumors (Fig. 7B,C).
Fig. 7.
Eµ-HDAC9 tumors display deregulated acetylation of BCL6 and p53. (A) IHC triple-immunostaining using the ABC-TSA method in mouse spleens from Eμ-HDAC9 and wild-type tumors, showing HDAC9 (red) expression in conjunction with levels of acetylated (Ac)-BCL6 (green). (B) Immunoblot analysis of Ac-p53 in Eµ-HDAC9 and wild-type spleen. GAPDH was used as a loading control. (C) ABC-TSA immunofluorescence analysis of Ac-p53 (green) and HDAC9 (red) in spleen in Eμ-HDAC9 and wild-type controls.
DISCUSSION
HDAC9 is highly expressed in B-NHL cell lines and patient samples (Petrie et al., 2003; Sun et al., 2011), and its locus, chromosome 7p21.1, is frequently amplified in B-NHL (Bea et al., 2005; Bentz et al., 1999; Monni et al., 1996; Rubio-Moscardo et al., 2005; Tagawa et al., 2005). In order to establish whether deregulated expression of HDAC9 in the lymphoid compartment could generate a disease phenotype, we designed a GEM model in which a human HDAC9 transgene was expressed under the control of the Eµ promoter. Our hypothesis was based on evidence of deregulation of HDAC9 expression in B-NHL cell lines and patient samples. The occurrence of B-NHL in these transgenic mice strongly indicates a link between deregulated HDAC9 expression and lymphoid neoplasia, the first time that overexpression of a histone deacetylase in mice has resulted in a cancer phenotype. This is also, to our knowledge, the first time that expression of a single epigenetic transgene has led to lymphoma – expression of mutated polycomb-group gene EZH2 fails to drive lymphomagenesis unless in a background of overexpressed BCL2 or Myc (Béguelin et al., 2013; Berg et al., 2014). This does not, however, rule out a requirement for the acquisition of additional mutations in order for progression to lymphoma in Eµ-HDAC9 mice. Our results are also in agreement with a recent study utilizing an MRL/lpr GEM model of systemic lupus erythematosus with HDAC9 deficiency (Yan et al., 2011). Here, MRL/lpr transgenic mice lacking HDAC9 displayed decreased lymphoproliferation and expression of BCL6. Although the SMZL cases studied in Eµ-HDAC9 mice were BCL6-negative, it is generally accepted that MZL in humans is derived from a post-GC B cell, with associated somatic mutations in the IgVH gene (Dunn-Walters et al., 1998; Miranda et al., 1999; Zhu et al., 1995). Moreover, progression of indolent lymphoma (SMZL) to a more aggressive lymphoma (DLBCL) is a frequent occurrence for many subtypes of indolent B-cell lymphomas and could explain the late onset of aggressive lymphomas observed in our transgenic model (Camacho et al., 2001; Freedman, 2005). The development of SMZL in Eµ-HDAC9 mice is also consistent with results obtained in mice in which TP53 was disrupted specifically in B cells, leading to the development of highly penetrant SMZL (Chiang et al., 2012).
Our results indicated that aberrant expression of HDAC9 in B cells leads to the upregulation of pathways that promote cell growth and survival, as well as impacting the activity and expression of key factors in lymphoma BCL6 and p53. The notion that transgenic expression of HDAC9 can promote lymphomagenesis, in part through deregulation of the activities of p53, is strengthened by studies suggesting that p53 might directly bind to the HDAC9 promoter and repress its expression (Akdemir et al., 2014; Wei et al., 2006). Of note, the p53-binding site in the HDAC9 promoter overlaps with a myocyte enhancer factor 2 (MEF2)-binding site, which, when bound by MEF2 family members, activates HDAC9 gene expression (Haberland et al., 2007). Activating mutations of MEF2B (which occur in 11% of DLBCL and 12% of FL) have been reported to directly upregulate expression of BCL6 (the promoter of which also contains a MEF2-binding site) in GC B cells and drives DLBCL proliferation (Ying et al., 2013). It remains to be established whether mutant MEF2B can drive HDAC9 expression in B-cell lymphomas, but recent research has identified a novel MEF2D–BCL9 fusion protein associated with high-risk acute B-cell precursor lymphoblastic leukemia (ALL) that directly upregulates HDAC9 (Suzuki et al., 2016). High expression of HDAC9 has been independently linked to poor prognosis in ALL (Moreno et al., 2010).
In recent years, numerous structurally diverse HDAC inhibitors (HDACi) have emerged as clinical candidate therapeutic agents (West and Johnstone, 2014), including the recent development of class-IIa-specific HDACi (Lobera et al., 2013). Even in the case of cutaneous T-cell lymphoma (CTCL) where HDACi have shown efficacy as single-agent targeted therapies and been approved for use in the clinic (Prince and Dickinson, 2012), the use of HDACi in rational combinations will likely maximize their therapeutic potential. It is, therefore, of interest that genes upregulated in Eµ-HDAC9 tumors, such as Cdk1, Chek1, Aurka and Aurkb, represent important clinical targets for which late-phase clinical trials are ongoing (Garrett and Collins, 2011; Lapenna and Giordano, 2009; Micel et al., 2013). The upregulation of Plk1 is also of potential clinical interest given that its high expression is a negative prognostic indicator in B-NHL (Liu et al,. 2007; Xu et al., 2013; Yim et al., 2013). PLK1 plays a crucial role at checkpoint controls during G2/M transition of the mitotic cell cycle (Barr et al., 2004) and inhibits p53 function directly by phosphorylation (Ando et al., 2004). Therefore, the Eµ-HDAC9 GEM model could serve as a valuable tool both to better understand the molecular mechanisms involved in lymphomagenesis in humans and facilitate preclinical studies of new drugs and combination therapies.
MATERIALS AND METHODS
Generation of Eμ-HDAC9 transgenic mice
FLAG-epitope-tagged full-length human HDAC9 cDNA was cloned into the pEμSR vector, placing the HDAC9-sequence-containing oligonucleotide cassette downstream of the immunoglobulin heavy chain (IgH) enhancer (Eμ) and the SRα potent promoter (Bodrug et al., 1994). The Eμ-HDAC9 transgenic fragment was isolated from the vector by enzymatic digestion using the NotI restriction sites and injected into B6CBAF1 pronuclei. Mice were backcrossed and maintained in a C57BL/6 background to generate three transgenic lines: 1468, 1469 and 1839. PCR genotyping was performed using SV40 primers: F: 5′-GGAACTGATGAATGGGAGCA-3′ and R: 5′-GCAGTGCAGCTTTTTCCTTT-3′. Mice were housed and maintained in accordance with UK Home Office regulations. Animals were monitored and analyzed from birth to 23 months of age and sacrificed if showing signs of illness. Statistical analysis was performed using Prism (GraphPad Software). Kaplan–Meier cumulative survival and the log-rank (Mantel–Cox) test were used to determine tumor-free survival and the χ2 test was used to compare B-NHL incidence in Eµ-HDAC9 mice versus wild-type controls. P<0.05 was considered statistically significant. All experimental protocols were monitored and approved by The Institute of Cancer Research Animal Welfare and Ethical Review Body, in compliance with guidelines specified by the UK Home Office Animals (Scientific Procedures) Act 1986 and the United Kingdom National Cancer Research Institute guidelines for the welfare of animals in cancer research (Workman et al., 2010). ARRIVE guidelines were applied when reporting in vivo experiments (Kilkenny et al., 2010).
Ig gene rearrangements analysis
Genomic DNA was isolated from tumor specimens and prepared using All Prep Kit (Qiagen). Primers for detection of Ig rearrangements were described previously (Ehlich et al., 1994). D-JH rearrangements of the heavy chain (JH) locus were amplified and detected in a multiplex PCR reaction using two upstream primers, DFL/DSP and DQ52, together with one reverse primer positioned downstream of JH4 (Mårtensson et al., 1997). For mutational analysis, the rearranged Ig variable heavy chain genes were amplified from genomic DNA as previously described (Cattoretti et al., 2005). V-DJH rearrangements were analyzed in separate PCR reactions using primers VHJ558a (5′-CAGGTCCAGCTGCAGCAGTCTGG-3′), VH7183b (5′-GTGAAGCCTGGAGGGTCCC-3′), VHQ52 (5′-CAGGTGCAGCTGAAACAGTCA-3′) and reverse JH4 primer (5′-TGAGGAGACGGTGACTGAGGTTCC-3′). The primers amplified all rearrangement products between JH4 and VHJ558, VH7183 or VHQ52 where four different bands would be expected by the combination of each primer set. PCR cycling conditions were: 95°C for 5 min, 95°C for 30 s, 63°C for 30 s and 72°C for 2 min, for 35 cycles. PCR products were gel-purified using the QIAQuick method (Qiagen) and directly sequenced using the ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems). Sequences were aligned to those in the international ImMunoGeneTics information system (IMGT) database.
Flow cytometry and cell sorting
Single-cell suspensions were obtained from dissected tissues, washed in phosphate buffered saline (PBS) supplemented with 0.2% FBS, filtered through a 45 µm cell strainer and red blood cells were lysed using ammonium chloride solution (STEMCELL Technologies). The antibody combination used for tumor analysis included: CD23, CD19, IgM, IgD, CD21 and B220. Anti-mouse conjugated antibodies were obtained from eBioscience and BioLegend. Cell sorting of splenic CD19-positive mouse B cells was carried out using immunomagnetic isolation with CD19-labeled beads (EasySep Mouse CD19 Positive Selection Kit, STEMCELL Technologies) and purity assessed using anti-mouse PE-conjugated CD19 antibody (eBioscience). Data were acquired on a FACS LSRII analyzer (BD Biosciences) and analyzed using FlowJo software (Tree Star).
Transgenic transcript detection
Total RNA from mouse bone marrow and spleen was extracted using TRIzol reagent (15596-026, Ambion) following the manufacturer's instructions. Reverse transcription was performed following standard protocol by using AccessQuick Master Mix (A1701, Promega). Real-time quantitative PCR was carried out using Fast SYBR Green Master Mix (4385612, Applied Biosystems). Primer sets used to detect mouse and human HDAC9 transcripts in B-cell subsets: F: 5′-TCTGGATGTTCACCATGGAA-3′ and R: 5′-CACTGCCAGGGAAAAAGTTC-3′. Primer sets to detect mouse Hdac9: F: 5′-GGTGATGATTCTCGGAAATTCT-3′ and R: 5′-GAAGCCAGCTCAATGACACA-3′. Abl1 mRNA expression was used as normalization control, F: 5′-CAGCGGCCAGTAGCATCTGACTT-3′ and R: 5′-GCTTCACACCATTCCCCATT-3′.
Purification of B-cell subsets
Single-cell suspensions from spleen (n=3) and bone marrow (n=5) from 8- to 12-week-old mice (transgenic and wild type) were prepared as previously described (Green et al., 2011). Bone marrow and spleen B-cell subpopulations were identified and sorted by three-color FACS method (Green et al., 2011). Anti-mouse antibodies CD43, IgM and B220 were used to obtain Pro-B, Pre-B and Mature-B-cell fractions from bone marrow. Splenic B-cell subtypes, follicular and marginal zone, were purified using anti-mouse CD19, CD21 and CD23 antibodies, whereas a combination of anti-mouse B220, IgM and CD93 was used to sort for transitional B cells. Each subset was above 90% purity. Purified cells were re-suspended in TRIzol reagent (Life Technologies) for further RNA extraction.
Histopathology and immunohistochemistry (IHC)
Tissues were freshly collected and fixed in 10% formalin for 24 h, embedded in paraffin and sectioned. Following deparaffinization and rehydration, samples were pre-treated with antigen-unmasking solution (Vector Laboratories, H-3300). After pre-treatment, tissue sections were blocked in Tris-NaCl buffer, washed in PBS and permeabilized in 0.5% Triton X-100. Slides were washed in Tris-NaCl-Tween buffer and reacted with primary antibody overnight at 4°C. Preliminary experiments were performed to determine optimal dilutions for each primary antibody used. Detection methods included: standard IHC, Vectastain Elite ABC system, Vector Laboratories or EnVision system (Dako) DAB staining and ABC-tyramide signal amplification (TSA Plus Fluorescence System, Perkin Elmer) (see below). Histopathological evaluation of HDAC9TG tumors was performed using hematoxylin and eosin (H&E)-stained sections. Immunophenotypic characterization of the HDAC9TG lymphomas was performed using antibodies against B220, PAX5, CD3, BCL6 and IRF4 as previously described (Mandelbaum et al., 2010). Human HDAC9 expression was assessed by IHC using tissue microarrays and/or individual tumor sections of 59 B-NHLs and three CHLs using a specific antibody generated against the C-terminal region of HDAC9 (Petrie et al., 2003). The B-NHL group consisted of 34 DLBCLs, nine FLs, five MZLs, six MCLs and two chronic lymphocytic leukemias/small lymphocytic lymphomas (CLLs/SLLs). Nuclear expression intensity of lymphomas was compared with that of rectal adenocarcinoma cells (positive control). If equal or higher intensity than control, HDAC9 expression was considered high (score 2+), and, if low, scored 1+.
Immunoblotting and antibody validation
Cell lysates were prepared using RIPA buffer supplemented with protease inhibitors (Roche). Rat monoclonal (IgM) antibody specific to the C-terminus of human HDAC9 (clone 45a7b5b) was developed using a synthetic peptide (DVEQPFAQEDSRTAG) conjugated to Diphtheria toxoid (Mimotopes), corresponding to unique amino acids 1046-1060. Validation was performed using Mini-PROTEAN II Multiscreen Apparatus (Bio-Rad). Other antibodies used for immunoblots included mouse monoclonal anti-FLAG M2 antibody (F1804, Sigma), β-actin loading-control antibody (BA3R) (MA5-15739, Thermo Scientific) and acetyl-p53 (Lys379) (PA5-17287, Thermo Scientific).
IHC and immunofluorescence detection methods
Single immunolabeling using ABC-TSA
Formalin-fixed paraffin-embedded tissue sections were dried for 45 min at 58°C, followed by deparaffinization and hydration in Histoclear (National Diagnostics) and a graded series of ethanol, respectively. Samples were pretreated by microwave incubation in a pH 6.0 citrate-based antigen-unmasking solution (Vector Laboratories, H-3300) followed by 2×5 min washes in PBS and permeabilization in 0.5% Triton X-100 (in PBS) for 20 min at room temperature (RT). Samples were washed in PBS prior to blocking for 30 min at RT in Tris-NaCl (TNB) blocking buffer with subsequent incubation in the monoclonal anti-HDAC9 antibody (clone 45a7b5b, 1/100 dilution) overnight at 4°C. Primary antibody was followed by biotinylated rabbit anti-rat secondary antibody for 30 min at RT. Endogenous peroxidase activity was inactivated by incubation with 3% hydrogen peroxide in methanol for 15 min at RT. After washing in Tris–NaCl–Tween-20 (TNT) buffer, samples were incubated in streptavidin (SA)-horseradish peroxidase (HRP) (Vectastain ABC Elite Kit, Vector Laboratories, PK-6104) for 45 min at RT. Following this, FITC fluorophore tyramide (Perkin Elmer, NEL741) was added for 10 min at RT, which results in the deposition of numerous fluorophore labels adjacent to the HRP. This fluorescent signal was then converted to a chromogenic signal by the addition of HRP-labeled anti-FITC (Perkin Elmer, NEF710), 1/200 dilution in TNB for 45 min at RT, 3×5 min washes in TNT and incubation in chromogen 3-amino-9-ethylcarbazole (AEC) (BUF019B, AbD Serotec). Hematoxylin QS (Vector Laboratories, H-3404) was utilized for nuclear counterstaining and the sections mounted in Vectamount aqueous mounting medium (Vector Laboratories, H-5501). Images were collected on a Zeiss Axioskop 2 light microscope utilizing Axiovision 4.4 software.
Double/triple labeling with ABC-TSA
The following immunolabeling reactions were applied using ABC-TSA method as described elsewhere (Asson-Batres and Smith, 2006). Preliminary controls showed no antibody carryover when one primary antibody was a rabbit polyclonal and the other was a rat monoclonal antibody, or when both primary antibodies were made in rabbits. Images were collected on Zeiss LSM700 confocal microscope using Zen 2009 software with sequential collection to prevent any spectral crosstalk.
Double immunolabeling
Formalin-fixed paraffin-embedded tissue sections were dried for 45 min at 58°C followed by deparaffinization and hydration in Histoclear (National Diagnostics) and a graded series of ethanol, respectively. Samples were pre-treated by microwave incubation in a pH 6.0 citrate-based antigen-unmasking solution (Vector Laboratories, H-3300) followed by 2×5 min washes in PBS and permeabilization in 0.5% Triton X-100 in PBS for 20 min at RT. Samples were then washed in PBS prior to blocking for 30 min at RT in TNB blocking buffer with subsequent incubation in the monoclonal anti-HDAC9 antibody (clone 45a7b5b, 1/100 dilution) overnight at 4°C. Primary antibody was followed by biotinylated rabbit anti-rat secondary antibody (Vectastain ABC Elite Kit, Vector Laboratories, PK-6104) for 30 min at RT. Endogenous peroxidase activity was quenched by incubation in 3% hydrogen peroxide (in methanol) for 15 min at RT. After washing in TNT buffer, samples were incubated in SA-HRP (Vectastain, Vector Laboratories ABC Elite Kit, PK-6104) for 45 min at RT. Next, a Cy3 fluorophore tyramide (Perkin Elmer, NEL741) was added (1/50 dilution prepared in the supplied amplification buffer) for 10 min at RT, which results in the deposition of numerous fluorophore labels adjacent to the HRP. Slides were washed 3×5 min in TNT buffer, any remaining HRP was deactivated by incubating in 3% hydrogen peroxide (in methanol) for 15 min at RT. A second primary antibody was added sequentially [anti-acetylated p53 (Lys379) Thermo Scientific, cat. # 17287, 1/100 dilution] and the protocol was repeated with the FITC fluorophore tyramide (Perkin Elmer, NEL741) utilized to detect acetylated p53. Slides were counterstained with To-pro 3 iodide (Life Technologies, T-3605) and mounted in Vectashield (Vector Laboratories, cat. # H-1000) mounting medium. Negative control slides were prepared by excluding the primary antibody, by excluding the conjugated secondary antibody-fluorophore and by excluding TSA reagents; negative controls showed no immunoreactivity. Single-labeling experiments carried out to observe the patterns of staining of each primary antibody validated the double-immunostaining results.
Triple immunolabeling
First, the primary antibodies monoclonal rat anti-HDAC9 (clone 7b5b, 1/100 dilution) and rabbit polyclonal anti-acetylated lysine (Millipore 06-933, 1/500 dilution) were added together and left overnight at 4°C. Following this, secondary anti-rat biotinylated antibody (1/200 dilution) was added and the TSA protocol was carried out using Cy3 fluorophore tyramide (Perkin Elmer, NEL753). Next, HRP was inactivated before the secondary antibody against acetylated lysine was added (anti-rabbit biotinylated) and the TSA protocol followed using FITC fluorophore tyramide (Perkin Elmer, NEL753). The HRP was once again inactivated with 3% hydrogen peroxide (in methanol) for 15 min at RT and rabbit polyclonal anti-BCL6 antibody (ab19011) (1/1000 dilution) was added (overnight at 4°C). Following this, the TSA protocol was applied using Cy5 fluorophore tyramide (Perkin Elmer, NEL745). Secondary-antibody controls were performed to detect any non-specific background staining. Single-staining controls were carried out as in the same-species double-labeling experiments.
IHC of human tumors
The EnVision detection system HRP/DAB+ (Dako) was used as previously described (Kim et al., 2009). Briefly, tissue sections were deparaffinized in xylene and rehydrated in ethanol following treatment in pre-heated target retrieval solution. Following washes, serum-free blocking solution was applied for 40 min at RT. In-house anti-HDAC9 monoclonal antibody was used overnight at 4°C then treated with polymer/HRP and DAB. After washes, the slides were counterstained with hematoxylin, dried and mounted with Permount. Photomicrographs were captured using an Olympus BX41 dual head light microscope equipped with an Olympus Q-Color 5 digital camera (Olympus America).
Antibodies IHC/IF
Primary antibodies included polyclonal rabbit anti-Ac-p53 (Lys379) (Thermo Scientific), monoclonal rat anti-HDAC9 antibody (clone 45a7b5b), monoclonal mouse anti-FLAG M2 (Sigma), polyclonal rabbit anti-acetylated lysine (Millipore 06-933), polyclonal rabbit anti-BCL6 (ab19011), anti-CD45R(B220) (ab64100) and anti-CD3 (ab5690). Secondary antibodies included: biotinylated rabbit anti-rat secondary antibody (Vector Laboratories, PK-6104), biotinylated horse anti-mouse secondary (Vector Laboratories, PK6102) and biotinylated goat anti-rabbit secondary (Vector Laboratories, PK6101). Fluorochromes and chromogens included SA-HRP (Vector Laboratories, PK-6102), FITC fluorophore tyramide (Perkin Elmer, NEL741, NEL753), FITC-HRP (Perkin Elmer, NEF710), Cy3 fluorophore tyramide (Perkin Elmer, NEL741, NEL753), Cy5 fluorophore tyramide (Perkin Elmer, NEL745) and AEC (AbD Serotec, BUF019B).
High-density SNP array analysis
Genome-wide DNA profiles were obtained from high-molecular-weight genomic DNA of DLBCL patients using the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix) following the manufacturer's instructions. Image data analysis and quality control for the hybridized samples were performed using the Affymetrix Genotyping Console 3.0.1 software, and only samples passing the Affymetrix recommended contrast QC and SNP call rates threshold (in the Birdseed v2.0 algorithm) were considered for analysis. Affymetrix CEL files and corresponding SNP genotype call files generated by the Affymetrix Genotyping Console tool were then analyzed using the dCHIP software. Model-based expression was performed using the perfect-match/mismatch (PM/MM) model to summarize signal intensities for each probe set. Probe intensity data for each array were normalized using a diploid reference set of three normal (non-tumor) DNA samples that had been processed and hybridized in the same experiment as the tumor samples. The standard invariant-set normalization approach in dCHIP was implemented by a karyotype-guided normalization method as previously described (Mullighan et al., 2007; Pounds et al., 2009). To identify regions of amplification and deletion, the circular binary segmentation algorithm was applied to the SNP array data as described (Mullighan et al., 2007). The following criteria were used to obtain candidate genomic regions (gains or loss): (1) mean log2 ratios of ≥0.2 or ≤−0.2; (2) ≥8 SNP markers within a segment. The results of the CBS algorithm were then compared to those of dCHIP. To exclude calls of genomic gains or loss arising from inherited genomic copy number variants (CNVs), the dCHIPSNP algorithm was also applied to 130 normal DNAs from an independent study as well as to 230 normal DNAs from the HapMap project; alterations identified in the pool of reference samples were excluded. In addition, CNVs were excluded if present in the Database of Genomic Variants (http://projects.tcag.ca/cgi-bin/variation/gbrowse/hg18/).
Gene expression analysis
Affymetrix GeneChip Mouse Gene 1.0 ST hybridizations were performed using biotin end-labeled cDNA prepared from CD19-positive B cells isolated from tumors. Unsupervised hierarchical clustering was performed on gene expression data from representative HDAC9TG-derived lymphomas versus normal murine mature B-cell subpopulations, including GC and non-GC (follicular and marginal zone) B cells (GeneChip Mouse Gene 1.0 ST Arrays). Data from normal B-cell subsets were obtained from the Immunological Genome Project (GSE15907) (www.immgen.org). Only probes with minimal expression level equal to 10 and minimal standard deviation of 1.5 (log2 transformed) were considered. The hierarchical clustering algorithm is based on the average-linkage, Pearson correlation.
Gene data sets were also analyzed for interactions and pathways using: GGA (Genomatix Genome Analyzer, https://mygga.genomatix.de), the Search Tool for the Retrieval of Interacting Genes/Proteins STRING v9.1 (http://string-db.org) to develop interactomes or networks (Franceschini et al., 2013) and the KEGG pathway database (Kanehisa et al., 2004). GO (Ashburner et al., 2000) clustering was performed with csbl.go (Ovaska et al., 2008). Partek Genomics Suite 6.6 was additionally used for data set analysis and comparisons.
Zinc-finger nuclease (ZFN) knockout of HDAC9
ZFNs targeting human HDAC9 sequence were obtained from Sigma-Aldrich (CompoZr Knockout Zinc Finger Nucleases, CKOZFND9935-1KT). The ZFN binding-cutting site (in lowercase) was 5′-CTCTGGTCCCAGTTCACCaaacaATGGGCCAACTGGAAGTG-3′. Delivery of ZFN was performed following the manufacturer's protocol (Sigma-Aldrich). The human Burkitt's lymphoma cell line Raji (ATCC CCL-86) was used for the study. Cells were maintained in RPMI medium 1640 (1×) (Gibco) supplemented with 10% FBS (Sigma-Aldrich) and grown in a 5% CO2 incubator at 37°C. Cells were transfected by nucleofection (electroporation) using Amaxa Cell Line Nucleofector Kit V (Lonza), and grown for 48 h followed by single-cell sorting in a 96-well format using BD FACS Aria (BD Biosciences). After 3-4 weeks, single-cell-derived clones were screened and analyzed using CEL-I assay (SURVEYOR mutation detection assay) following the manufacturer's instructions (Transgenomic). Genomic DNA was obtained by high-throughput HotSHOT DNA preparation method in 96-well plates. ZFN mutant clones were confirmed by sequencing.
Acknowledgements
We thank Mel Greaves, Daniel Catovsky, David Gonzalez de Castro, Laura Pasqualucci and Riccardo Dalla-Favera for their support and advice. We acknowledge Dr Jerry Adams (Walter and Eliza Hall Institute, Melbourne, Australia) for his gift of the pEµSR vector.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
V.S.G. conceived the study, performed all experiments unless otherwise indicated, analyzed the data, performed statistical analyses, and co-wrote and co-edited the manuscript. G.B., L.H. and C.H.K. performed immunohistochemistry and immunofluorescence experiments. G.B. and F.V. carried out histopathological analysis of tumor samples. J.Z. and S.S. performed genetic and expression microarray analysis of tumor samples. A.Z. conceived the study and analyzed the data. K.P. conceived the study, analyzed the data, and co-wrote, co-edited and submitted the final version of the manuscript.
Funding
V.S.G. was supported by funding from Worldwide Cancer Research (formerly Association for International Cancer Research) (11-0301). S.S. was supported by Cancer Research UK (A12747), and K.P. and L.H. were supported by a Bloodwise (formerly Leukaemia and Lymphoma Research UK) Specialist Programme Grant (11046).
Data availability
Microarray expression profiles of HDAC9TG mouse tumors and wild-type counterparts have been deposited The Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE89954 and GSE89955. The GEO Series accession number for this study is GSE89956.
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.023366.supplemental
References
- Akdemir K. C., Jain A. K., Allton K., Aronow B., Xu X., Cooney A. J., Li W. and Barton M. C. (2014). Genome-wide profiling reveals stimulus-specific functions of p53 during differentiation and DNA damage of human embryonic stem cells. Nucleic Acids Res. 42, 205-223. 10.1093/nar/gkt866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K., Ozaki T., Yamamoto H., Furuya K., Hosoda M., Hayashi S., Fukuzawa M. and Nakagawara A. (2004). Polo-like kinase 1 (Plk1) inhibits p53 function by physical interaction and phosphorylation. J. Biol. Chem. 279, 25549-25561. 10.1074/jbc.M314182200 [DOI] [PubMed] [Google Scholar]
- Ansell S. M. (2015). Non-hodgkin lymphoma: diagnosis and treatment. Mayo Clin. Proc. 90, 1152-1163. 10.1016/j.mayocp.2015.04.025 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T. et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25-29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asson-Batres M. A. and Smith W. B. (2006). Localization of retinaldehyde dehydrogenases and retinoid binding proteins to sustentacular cells, glia, Bowman's gland cells, and stroma: potential sites of retinoic acid synthesis in the postnatal rat olfactory organ. J. Comp. Neurol. 496, 149-171. 10.1002/cne.20904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F. A., Sillje H. H. and Nigg E. A. (2004). Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429-440. 10.1038/nrm1401 [DOI] [PubMed] [Google Scholar]
- Basso K. and Dalla-Favera R. (2012). Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev. 247, 172-183. 10.1111/j.1600-065X.2012.01112.x [DOI] [PubMed] [Google Scholar]
- Basso K., Saito M., Sumazin P., Margolin A. A., Wang K., Lim W.-K., Kitagawa Y., Schneider C., Alvarez M. J., Califano A. et al. (2010). Integrated biochemical and computational approach identifies BCL6 direct target genes controlling multiple pathways in normal germinal center B cells. Blood 115, 975-984. 10.1182/blood-2009-06-227017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bea S., Zettl A., Wright G., Salaverria I., Jehn P., Moreno V., Burek C., Ott G., Puig X., Yang L. et al. (2005). Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood 106, 3183-3190. 10.1182/blood-2005-04-1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béguelin W., Popovic R., Teater M., Jiang Y., Bunting K. L., Rosen M., Shen H., Yang S. N., Wang L., Ezponda T. et al. (2013). EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677-692. 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier U. H., Wang L., Han R., Akimova T., Liu Y. and Hancock W. W. (2012). Histone deacetylases 6 and 9 and sirtuin-1 control Foxp3+ regulatory T cell function through shared and isoform-specific mechanisms. Sci. Signal. 5, ra45 10.1126/scisignal.2002873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellenguez C., Bevan S., Gschwendtner A., Spencer C. C. A., Burgess A. I., Pirinen M., Jackson C. A., Traylor M., Strange A., Su Z. et al. (2012). Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat. Genet. 44, 328-333. 10.1038/ng.1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz M., Werner C. A., Dohner H., Joos S., Barth T. F., Siebert R., Schroder M., Stilgenbauer S., Fischer K., Moller P. et al. (1996). High incidence of chromosomal imbalances and gene amplifications in the classical follicular variant of follicle center lymphoma. Blood 88, 1437-1444. [PubMed] [Google Scholar]
- Bentz M., Stilgenbauer S., Lichter P. and Dohner H. (1999). Interphase FISH in chronic lymphoproliferative disorders and comparative genomic hybridisation in the study of lymphomas. Haematologica 84 Suppl. EHA-4, 102-106. [PubMed] [Google Scholar]
- Berg T., Thoene S., Yap D., Wee T., Schoeler N., Rosten P., Lim E., Bilenky M., Mungall A. J., Oellerich T. et al. (2014). A transgenic mouse model demonstrating the oncogenic role of mutations in the polycomb-group gene EZH2 in lymphomagenesis. Blood 123, 3914-3924. 10.1182/blood-2012-12-473439 [DOI] [PubMed] [Google Scholar]
- Bodrug S. E., Warner B. J., Bath M. L., Lindeman G. J., Harris A. W. and Adams J. M. (1994). Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 13, 2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho F. I., Mollejo M., Mateo M.-S., Algara P., Navas C., Hernández J.-M., Santoja C., Solé F., Sánchez-Beato M. and Piris M. A. (2001). Progression to large B-cell lymphoma in splenic marginal zone lymphoma: a description of a series of 12 cases. Am. J. Surg. Pathol. 25, 1268-1276. 10.1097/00000478-200110000-00007 [DOI] [PubMed] [Google Scholar]
- Cattoretti G., Pasqualucci L., Ballon G., Tam W., Nandula S. V., Shen Q., Mo T., Murty V. V. and Dalla-Favera R. (2005). Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 7, 445-455. 10.1016/j.ccr.2005.03.037 [DOI] [PubMed] [Google Scholar]
- Chatterjee T. K., Basford J. E., Knoll E., Tong W. S., Blanco V., Blomkalns A. L., Rudich S., Lentsch A. B., Hui D. Y. and Weintraub N. L. (2014). HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high-fat feeding. Diabetes 63, 176-187. 10.2337/db13-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y. J., Difilippantonio M. J., Tessarollo L., Morse H. C. and Hodes R. J. (2012). Exon 1 disruption alters tissue-specific expression of mouse p53 and results in selective development of B cell lymphomas. PLoS ONE 7, e49305 10.1371/journal.pone.0049305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoeten E. F., Wang L., Sai H., Dillmann W. H. and Hancock W. W. (2010). Inhibition of HDAC9 increases T regulatory cell function and prevents colitis in mice. Gastroenterology 138, 583-594. 10.1053/j.gastro.2009.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn-Walters D. K., Boursier L., Spencer J. O. and Isaacson P. G. (1998). Analysis of immunoglobulin genes in splenic marginal zone lymphoma suggests ongoing mutation. Hum. Pathol. 29, 585-593. 10.1016/S0046-8177(98)80007-5 [DOI] [PubMed] [Google Scholar]
- Ehlich A., Martin V., Müller W. and Rajewsky K. (1994). Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4, 573-583. 10.1016/S0960-9822(00)00129-9 [DOI] [PubMed] [Google Scholar]
- Falini B., Fizzotti M., Pucciarini A., Bigerna B., Marafioti T., Gambacorta M., Pacini R., Alunni C., Natali-Tanci L., Ugolini B. et al. (2000). A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood 95, 2084-2092. [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Ervik M., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D. M., Forman D. and Bray F. (2013). GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer. [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C. et al. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808-D815. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman A. S. (2005). Biology and management of histologic transformation of indolent lymphoma. Hematol. Am. Soc. Hematol. Educ. Program 1, 314-320. 10.1182/asheducation-2005.1.314 [DOI] [PubMed] [Google Scholar]
- Garrett M. D. and Collins I. (2011). Anticancer therapy with checkpoint inhibitors: what, where and when? Trends Pharmacol. Sci. 32, 308-316. 10.1016/j.tips.2011.02.014 [DOI] [PubMed] [Google Scholar]
- Good P. (2000). Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. New York: Springer-Verlag.
- Green M. R., Monti S., Dalla-Favera R., Pasqualucci L., Walsh N. C., Schmidt-Supprian M., Kutok J. L., Rodig S. J., Neuberg D. S., Rajewsky K. et al. (2011). Signatures of murine B-cell development implicate Yy1 as a regulator of the germinal center-specific program. Proc. Natl. Acad. Sci. USA 108, 2873-2878. 10.1073/pnas.1019537108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover N. S. and Park S. I. (2015). Novel targeted agents in Hodgkin and non-Hodgkin lymphoma therapy. Pharmaceuticals 8, 607-636. 10.3390/ph8030607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Arnold M. A., McAnally J., Phan D., Kim Y. and Olson E. N. (2007). Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol. Cell. Biol. 27, 518-525. 10.1128/MCB.01415-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M., Montgomery R. L. and Olson E. N. (2009). The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat. Rev. Genet. 10, 32-42. 10.1038/nrg2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haery L., Thompson R. C. and Gilmore T. D. (2015). Histone acetyltransferases and histone deacetylases in B- and T-cell development, physiology and malignancy. Genes Cancer 6, 184-213. 10.18632/genesandcancer.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassler M. R., Schiefer A.-I. and Egger G. (2013). Combating the epigenome: epigenetic drugs against non-Hodgkin's lymphoma. Epigenomics 5, 397-415. 10.2217/epi.13.39 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Kawashima S., Okuno Y. and Hattori M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, D277-D280. 10.1093/nar/gkh063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilkenny C., Browne W. J., Cuthill I. C., Emerson M. and Altman D. G. (2010). Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 10.1371/journal.pbio.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. E., Singh R. R., Cho-Vega J. H., Drakos E., Davuluri Y., Khokhar F. A., Fayad L., Medeiros L. J. and Vega F. (2009). Sonic hedgehog signaling proteins and ATP-binding cassette G2 are aberrantly expressed in diffuse large B-cell lymphoma. Mod. Pathol. 22, 1312-1320. 10.1038/modpathol.2009.98 [DOI] [PubMed] [Google Scholar]
- Klein U., Tu Y., Stolovitzky G. A., Keller J. L., Haddad J. Jr., Miljkovic V., Cattoretti G., Califano A. and Dalla-Favera R. (2003). Transcriptional analysis of the B cell germinal center reaction. Proc. Natl. Acad. Sci. USA 100, 2639-2644. 10.1073/pnas.0437996100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang B., Alrahbeni T. M., Clair D. S., Blackwood D. H., International Schizophrenia C., McCaig C. D. and Shen S. (2012). HDAC9 is implicated in schizophrenia and expressed specifically in post-mitotic neurons but not in adult neural stem cells. Am. J. Stem Cells 1, 31-41. [PMC free article] [PubMed] [Google Scholar]
- Lapenna S. and Giordano A. (2009). Cell cycle kinases as therapeutic targets for cancer. Nat. Rev. Drug Discov. 8, 547-566. 10.1038/nrd2907 [DOI] [PubMed] [Google Scholar]
- Lemercier C., Brocard M.-P., Puvion-Dutilleul F., Kao H.-Y., Albagli O. and Khochbin S. (2002). Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor. J. Biol. Chem. 277, 22045-22052. 10.1074/jbc.M201736200 [DOI] [PubMed] [Google Scholar]
- Lemoine M. and Younes A. (2010). Histone deacetylase inhibitors in the treatment of lymphoma. Discov. Med. 10, 462-470. [PubMed] [Google Scholar]
- Liu L., Zhang M. and Zou P. (2007). Expression of PLK1 and survivin in diffuse large B-cell lymphoma. Leuk. Lymphoma 48, 2179-2183. 10.1080/10428190701615918 [DOI] [PubMed] [Google Scholar]
- Lobera M., Madauss K. P., Pohlhaus D. T., Wright Q. G., Trocha M., Schmidt D. R., Baloglu E., Trump R. P., Head M. S., Hofmann G. A. et al. (2013). Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat. Chem. Biol. 9, 319-325. 10.1038/nchembio.1223 [DOI] [PubMed] [Google Scholar]
- Mandelbaum J., Bhagat G., Tang H., Mo T., Brahmachary M., Shen Q., Chadburn A., Rajewsky K., Tarakhovsky A., Pasqualucci L. et al. (2010). BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell 18, 568-579. 10.1016/j.ccr.2010.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit O., Amram H., Amariglio N., Simon A. J., Shaklai S., Granot G., Minsky N., Shimoni A., Harmelin A., Givol D. et al. (2006). BCL6 is regulated by p53 through a response element frequently disrupted in B-cell non-Hodgkin lymphoma. Blood 107, 1599-1607. 10.1182/blood-2005-04-1629 [DOI] [PubMed] [Google Scholar]
- Martelli M., Ferreri A. J. M., Agostinelli C., Di Rocco A., Pfreundschuh M. and Pileri S. A. (2013). Diffuse large B-cell lymphoma. Crit. Rev. Oncol. Hematol. 87, 146-171. 10.1016/j.critrevonc.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Mårtensson I.-L., Melchers F. and Winkler T. H. (1997). A transgenic marker for mouse B lymphoid precursors. J. Exp. Med. 185, 653-661. 10.1084/jem.185.4.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Kettmann R. and Dequiedt F. (2009). Class IIa histone deacetylases: conducting development and differentiation. Int. J. Dev. Biol. 53, 291-301. 10.1387/ijdb.082698mm [DOI] [PubMed] [Google Scholar]
- Micel L. N., Tentler J. J., Smith P. G. and Eckhardt G. S. (2013). Role of ubiquitin ligases and the proteasome in oncogenesis: novel targets for anticancer therapies. J. Clin. Oncol. 31, 1231-1238. 10.1200/JCO.2012.44.0958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milde T., Oehme I., Korshunov A., Kopp-Schneider A., Remke M., Northcott P., Deubzer H. E., Lodrini M., Taylor M. D., von Deimling A. et al. (2010). HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin. Cancer Res. 16, 3240-3252. 10.1158/1078-0432.CCR-10-0395 [DOI] [PubMed] [Google Scholar]
- Miles R. R., Crockett D. K., Lim M. S. and Elenitoba-Johnson K. S. J. (2005). Analysis of BCL6-interacting proteins by tandem mass spectrometry. Mol. Cell. Proteomics 4, 1898-1909. 10.1074/mcp.M500112-MCP200 [DOI] [PubMed] [Google Scholar]
- Miranda R. N., Cousar J. B., Hammer R. D., Collins R. D. and Vnencak-Jones C. L. (1999). Somatic mutation analysis of IgH variable regions reveals that tumor cells of most parafollicular (monocytoid) B-cell lymphoma, splenic marginal zone B-cell lymphoma, and some hairy cell leukemia are composed of memory B lymphocytes. Hum. Pathol. 30, 306-312. 10.1016/S0046-8177(99)90010-2 [DOI] [PubMed] [Google Scholar]
- Monni O., Joensuu H., Franssila K. and Knuutila S. (1996). DNA copy number changes in diffuse large B-cell lymphoma--comparative genomic hybridization study. Blood 87, 5269-5278. [PubMed] [Google Scholar]
- Moreno D. A., Scrideli C. A., Cortez M. A. A., de Paula Queiroz R., Valera E. T., da Silva Silveira V., Yunes J. A., Brandalise S. R. and Tone L. G. (2010). Differential expression of HDAC3, HDAC7 and HDAC9 is associated with prognosis and survival in childhood acute lymphoblastic leukaemia. Br. J. Haematol. 150, 665-673. 10.1111/j.1365-2141.2010.08301.x [DOI] [PubMed] [Google Scholar]
- Mullighan C. G., Goorha S., Radtke I., Miller C. B., Coustan-Smith E., Dalton J. D., Girtman K., Mathew S., Ma J., Pounds S. B. et al. (2007). Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446, 758-764. 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Ovaska K., Laakso M. and Hautaniemi S. (2008). Fast gene ontology based clustering for microarray experiments. BioData Min. 1, 11 10.1186/1756-0381-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra M. (2015). Class IIa HDACs - new insights into their functions in physiology and pathology. FEBS J. 282, 1736-1744. 10.1111/febs.13061 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L. and Dalla-Favera R. (2015). The genetic landscape of diffuse large B-cell lymphoma. Semin. Hematol. 52, 67-76. 10.1053/j.seminhematol.2015.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Dominguez-Sola D., Chiarenza A., Fabbri G., Grunn A., Trifonov V., Kasper L. H., Lerach S., Tang H., Ma J. et al. (2011). Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471, 189-195. 10.1038/nature09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K., Guidez F., Howell L., Healy L., Waxman S., Greaves M. and Zelent A. (2003). The histone deacetylase 9 gene encodes multiple protein isoforms. J. Biol. Chem. 278, 16059-16072. 10.1074/jbc.M212935200 [DOI] [PubMed] [Google Scholar]
- Phan R. T. and Dalla-Favera R. (2004). The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635-639. 10.1038/nature03147 [DOI] [PubMed] [Google Scholar]
- Pounds S., Cheng C., Mullighan C., Raimondi S. C., Shurtleff S. and Downing J. R. (2009). Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics 25, 315-321. 10.1093/bioinformatics/btn624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince H. M. and Dickinson M. (2012). Romidepsin for cutaneous T-cell lymphoma. Clin. Cancer Res. 18, 3509-3515. 10.1158/1078-0432.CCR-11-3144 [DOI] [PubMed] [Google Scholar]
- Rubio-Moscardo F., Climent J., Siebert R., Piris M. A., Martín-Subero J. I., Nieländer I., Garcia-Conde J., Dyer M. J. S., Terol M. J., Pinkel D. et al. (2005). Mantle-cell lymphoma genotypes identified with CGH to BAC microarrays define a leukemic subgroup of disease and predict patient outcome. Blood 105, 4445-4454. 10.1182/blood-2004-10-3907 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D. and Jemal A. (2015). Cancer statistics, 2015. CA Cancer J. Clin. 65, 5-29. 10.3322/caac.21254 [DOI] [PubMed] [Google Scholar]
- Sun J. Y., Xu L., Tseng H., Ciccarelli B., Fulciniti M., Hunter Z. R., Maghsoudi K., Hatjiharissi E., Zhou Y., Yang G. et al. (2011). Histone deacetylase inhibitors demonstrate significant preclinical activity as single agents, and in combination with bortezomib in Waldenström's macroglobulinemia. Clin. Lymphoma Myeloma Leukemia 11, 152-156. 10.3816/CLML.2011.n.036 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Okuno Y., Kawashima N., Muramatsu H., Okuno T., Wang X., Kataoka S., Sekiya Y., Hamada M., Murakami N. et al. (2016). MEF2D-BCL9 fusion gene is associated with high-risk acute B-cell precursor lymphoblastic leukemia in adolescents. J. Clin. Oncol. 34, 3451-3459. 10.1200/JCO.2016.66.5547 [DOI] [PubMed] [Google Scholar]
- Tagawa H., Suguro M., Tsuzuki S., Matsuo K., Karnan S., Ohshima K., Okamoto M., Morishima Y., Nakamura S. and Seto M. (2005). Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood 106, 1770-1777. 10.1182/blood-2005-02-0542 [DOI] [PubMed] [Google Scholar]
- Tang Y., Zhao W., Chen Y., Zhao Y. and Gu W. (2008). Acetylation is indispensable for p53 activation. Cell 133, 612-626. 10.1016/j.cell.2008.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R., de Zoeten E. F., Özkaynak E., Chen C., Wang L., Porrett P. M., Li B., Turka L. A., Olson E. N., Greene M. I. et al. (2007). Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 13, 1299-1307. 10.1038/nm1652 [DOI] [PubMed] [Google Scholar]
- Wei C.-L., Wu Q., Vega V. B., Chiu K. P., Ng P., Zhang T., Shahab A., Yong H. C., Fu Y. T., Weng Z. et al. (2006). A global map of p53 transcription-factor binding sites in the human genome. Cell 124, 207-219. 10.1016/j.cell.2005.10.043 [DOI] [PubMed] [Google Scholar]
- West A. C. and Johnstone R. W. (2014). New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124, 30-39. 10.1172/JCI69738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt O., Deubzer H. E., Milde T. and Oehme I. (2009). HDAC family: what are the cancer relevant targets? Cancer Lett. 277, 8-21. 10.1016/j.canlet.2008.08.016 [DOI] [PubMed] [Google Scholar]
- Workman P., Aboagye E. O., Balkwill F., Balmain A., Bruder G., Chaplin D. J., Double J. A., Everitt J., Farningham D. A. H., Glennie M. J. et al. (2010). Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer 102, 1555-1577. 10.1038/sj.bjc.6605642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Shen C., Wang T. and Quan J. (2013). Structural basis for the inhibition of Polo-like kinase 1. Nat. Struct. Mol. Biol. 20, 1047-1053. 10.1038/nsmb.2623 [DOI] [PubMed] [Google Scholar]
- Yan K., Cao Q., Reilly C. M., Young N. L., Garcia B. A. and Mishra N. (2011). Histone deacetylase 9 deficiency protects against effector T cell-mediated systemic autoimmunity. J. Biol. Chem. 286, 28833-28843. 10.1074/jbc.M111.233932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.-J. and Seto E. (2008). The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9, 206-218. 10.1038/nrm2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim H. (2013). Current clinical trials with polo-like kinase 1 inhibitors in solid tumors. Anticancer Drugs Biol. 24, 999-1006. 10.1097/CAD.0000000000000007 [DOI] [PubMed] [Google Scholar]
- Ying C. Y., Dominguez-Sola D., Fabi M., Lorenz I. C., Hussein S., Bansal M., Califano A., Pasqualucci L., Basso K. and Dalla-Favera R. (2013). MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat. Immunol. 14, 1084-1092. 10.1038/ni.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. X., Wang Y. S., Cai Q. Q., Wang J. Q. and Yao W. T. (2015). Up-regulation of HDAC9 promotes cell proliferation through suppressing p53 transcription in osteosarcoma. Int. J. Clin. Exp. Med. 8, 11818-11823. [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Oscier D. G. and Stevenson F. K. (1995). Splenic lymphoma with villous lymphocytes involves B cells with extensively mutated Ig heavy chain variable region genes. Blood 85, 1603-1607. [PubMed] [Google Scholar]