Abstract
Purpose
Systemic lupus erythematosus (SLE) is a chronic inflammatory female-predominant autoimmune disease that can affect the central nervous system and exhibit neuropsychiatric symptoms. In SLE patients without neuropsychiatric symptoms (non-NPSLE), recent diffusion tensor imaging studies showed white matter abnormalities in their brains. The present study investigated the entire brain white matter structural connectivity in non-NPSLE patients by using probabilistic tractography and connectivity-based analyses.
Methods
Whole-brain structural networks of 29 non-NPSLE patients and 29 healthy controls (HCs) were examined. The structural networks were constructed with interregional probabilistic connectivity. Graph theory analysis was performed to investigate the topological properties, and network-based statistic was employed to assess the alterations of the interregional connections among non-NPSLE patients and controls.
Results
Compared with HCs, non-NPSLE patients demonstrated significantly decreased global and local network efficiencies and showed increased characteristic path length. This finding suggests that the global integration and local specialization were impaired. Moreover, the regional properties (nodal efficiency and degree) in the frontal, occipital, and cingulum regions of the non-NPSLE patients were significantly changed and negatively correlated with the disease activity index. The distribution pattern of the hubs measured by nodal degree was altered in the patient group. Finally, the non-NPSLE group exhibited decreased structural connectivity in the left median cingulate-centered component and increased connectivity in the left precuneus-centered component and right middle temporal lobe-centered component.
Conclusion
This study reveals an altered topological organization of white matter networks in non-NPSLE patients. Furthermore, this research provides new insights into the structural disruptions underlying the functional and neurocognitive deficits in non-NPSLE patients.
Keywords: Non-neuropsychiatric systemic lupus erythematosus, Diffusion tensor imaging, White matter, Probabilistic tractography, Graph theory
Highlights
-
•
The global integration and local specialization of the white matter networks were disrupted in non-NPSLE patients.
-
•
The brain regions with significantly altered nodal properties were mainly involved in language, visual, and DMN systems.
-
•
The properties of some identified regions presented negative correlations with the SLE disease activity index scores.
-
•
The distribution pattern of the hubs measured by nodal degree was altered in the non-NPSLE patients.
-
•
The non-NPSLE group exhibited a subnetwork of decreased connectivity and a subnetwork of increased connectivity.
1. Introduction
Systemic lupus erythematosus (SLE) is a chronic and female-predominant inflammatory autoimmune disease affecting multiple organs and systems (Stojanovich et al., 2007). The causal etiologies of SLE include vasculopathy, autoantibodies, inflammatory mediators, and cytokines. When the pathological etiologies involve the central nervous system, the disease evolves to neuropsychiatric systemic lupus erythematosus (NPSLE), which is the most common complication of SLE. NPSLE has been reported to occur in approximately 75% of SLE patients with diverse clinical symptoms, such as mood disorders, headaches, seizures, and cognitive dysfunctions (Bertsias and Boumpas, 2010). However, the neuropsychiatric pathology and neuronal mechanism of NPSLE have not been elucidated.
SLE subjects without neuropsychiatric symptoms have been referred to as non-NPSLE patients (Liang et al., 1999). Although non-NPSLE patients do not manifest neuropsychiatric symptoms, neuroimaging studies have demonstrated some abnormalities in their brains. With regard to brain functions, functional magnetic resonance imaging (fMRI) researches reported some disruptions in several functional systems, such as working memory (Hou et al., 2013, Mackay et al., 2011), attention (Hou et al., 2013), learning memory (Shapira-Lichter et al., 2013), and language processing functional systems (DiFrancesco et al., 2007) in non-NPSLE patient groups. With regard to brain structure, the volume and thickness of gray matter show no abnormality in a non-NPSLE group (Jung et al., 2010b). Recent research with diffusion tensor imaging (DTI) indicated some alterations in the white matter integrity in non-NPSLE patients (Schmidt-Wilcke et al., 2014). These findings suggest that the cerebral function and structure of non-NPSLE patients have already been altered compared with those of healthy people, and the disruption or abnormality of the white matter plays a relatively critical role. Structural alterations demonstrate causal relationships with the functional alterations to some extent (Wang et al., 2015b). Thus, investigation of the abnormality of the white matter structure may enhance understanding of the potential mechanisms of neuronal pathogenesis at the early stages of SLE.
The abnormalities in the white matter structure can be investigated by DTI analysis with tract-based spatial statistics (TBSS) (Smith et al., 2006). Application of TBSS-based analysis has shown that SLE patients exhibit a reduction in their white matter integrity in the inferior fronto-occipital fasciculus (Ercan et al., 2015, Schmidt-Wilcke et al., 2014), internal capsule (Emmer et al., 2010), and inferior and superior longitudinal fasciculi (Ercan et al., 2015). Notably, the TBSS approach is a kind of voxel-wise statistical analysis of diffusion-related metrics on fractional anisotropy (FA) skeletons, which consist of voxels with large FA values and represent the centers of fiber bundles. Actually, the brain is composed of highly interconnected distinct regions integrating massive neural information (Gong et al., 2009, Iturria-Medina et al., 2008, Tymofiyeva et al., 2014). Sporns et al. have proposed a conceptual framework to model the entire brain structural connectivity as a network, which was referred to as the human connectome (Sporns, 2011, Sporns et al., 2005). The connectome framework can explore extensive white matter alterations underlying the key organizational abnormalities associated with a disease, and this framework has been applied to investigate topological disruptions in several pathological situations, such as Alzheimer's disease (Lo et al., 2010), schizophrenia (Skudlarski et al., 2010, Zalesky et al., 2011), epilepsy (Bernhardt et al., 2011), and multiple sclerosis (Shu et al., 2011). However, alteration in the topological organization of the white matter network is rarely reported for non-NPSLE patients (Xu et al., 2016).
Analysis of the topological organization of the white matter network generally consists of two main steps: mapping of large-scale white matter network through DTI-based tractography and analysis of the network through graph theoretical methods (Bullmore and Sporns, 2009, He and Evans, 2010). The DTI-based tractography constructs the 3D curves of the maximal diffusion coherence and facilitates the noninvasive estimation of the heterogeneous fiber bundles of the white matter (Hagmann et al., 2008). On the basis of the identified fiber bundles, the structural connections among the various brain regions can be estimated to construct the white matter network (Sporns et al., 2005). Graph theoretical analysis is a mathematical framework used to represent complex graphs, and this framework also provides a powerful approach to describe the topological features, such as the network integration, segregation, centrality, small-world properties, and highly connected hubs (He and Evans, 2010). The graph theory can detect subtle white matter network alterations, which may be disregarded by traditional techniques (Drakesmith et al., 2015). The network-based statistic method (Zalesky et al., 2010a) provides a complementary way to characterize the interregional connective features within the networks and facilitates a multiple hypothesis testing at the network level. The network connections can be constructed by deterministic and probabilistic tractography methods. However, deterministic tractography suffers from the crossing fiber problem, that is, this tracking method cannot determine whether the two tracts are crossing or kissing within a voxel (Basser et al., 2000, Mori and van Zijl, 2002). Probabilistic tractography method has been found to outperform deterministic tractography in tracking white matter tracts with fiber crossing (Behrens et al., 2007).
In the present study, DTI probabilistic tractography and connectivity-based analyses were performed to investigate the topological organization of the white matter networks in non-NPSLE patients and healthy individuals. This research aims to detect whether the patients exhibit changes in the aforementioned topological features, and if so, whether these alterations demonstrate correlations with the clinical characteristics of the disease.
2. Materials and methods
2.1. Participants
A total of 58 participants, comprising 29 non-NPSLE patients and 29 age-matched healthy controls (HCs) were included in the following network analyses and statistical comparisons. The participants were recruited from the Medical Imaging Center of Nanfang Hospital, and each participant provided written informed consent prior to the study. The research protocol was approved by the Research Ethics Committee of Nanfang Hospital of the Southern Medical University. All the SLE patients were diagnosed and classified by an experienced rheumatologist in accordance with the American College of Rheumatology (Hochberg, 1997, Liang et al., 1999). The exclusion criteria of participants were as follows: (1) male; (2) left–handedness; (3) the history of neuropsychiatric manifestations regardless of attribution, alcohol abuse, drug abuse, head injury and central nervous system infection; (4) suffering from hypertension, diabetes, stroke, epilepsy, Alzheimer's disease, and cardiovascular disorders; (5) white matter hyperintensities observed in the FLAIR images. All the included participants were then submitted to a standardized clinical examination, including medical history, physical examination and standard laboratory assessment. After that, a complete neurological examination, cognitive and psychiatric charts, the Beck Depression Inventory (BDI), and the Hospital Anxiety and Depression Scale (HADS) were applied, and the patients with no nervous system involvement were identified as non-NPSLE patients. After the above screening, 33 non-NPSLE patients and 33 healthy controls were accepted in the data preprocessing. We inspected the quality control results of the preprocessing, and found that spatial mismatches existed between the T1-wighted images and DTI images of several participants after registration. Hence, participants with inaccurate registration were further excluded from the two groups, and 29 non-NPSLE patients and 29 healthy controls were retained in the final network analysis. Data on age, gender, disease duration, disease activity index, cumulative damage, anti-Sm antibodies, serum assay indices and medication treatment of the participants were collected by standard clinical examination. Among which, the disease duration was defined as the interval time between the diagnosis of SLE and the MRI acquisition time. The disease activity index was determined using the systemic lupus disease activity index (SLEDAI). The cumulative damage was assessed on the basis of the damage index score of the Systemic Lupus International Collaborating Clinics (SLICC). Detailed clinical information and demographic data from non-NPSLE patients and HCs are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of all participants in this study.
| Characteristics | HC (n = 29 females) |
Non-NPSLE (n = 29 females) |
p value |
|---|---|---|---|
| Age | 30.97 ± 9.76 | 29.50 ± 9.91 | 0.569 |
| Duration of illness (months) | N.A | 39.67 ± 53.80 | N.A |
| SLEDAI | N.A | 10.40 ± 6.77 | N.A |
| SLICC | N.A | 0.67 ± 0.61 | N.A |
| Anti-SM | N.A | 31.00 ± 49.85 | N.A |
| C3(g/l) | N.A | 0.58 ± 0.28 | N.A |
| C4(g/l) | N.A | 0.11 ± 0.07 | N.A |
| CH50(g/l) | N.A | 31.01 ± 20.56 | N.A |
| Current medication treatment | |||
| Prednisone | N.A | 29 / 29 | N.A |
| Dose of prednisone (mg/day) | N.A | 38.83 ± 21.81 | N.A |
| Cyclophosphamide | N.A | 19 / 29 | N.A |
| Azathioprine | N.A | 10 / 29 | N.A |
| Methotrexate | N.A | 16 / 29 | N.A |
| Hydroxychloroquine | N.A | 11 / 29 | N.A |
HC = healthy control; non-NPSLE = non-neuropsychiatric systemic lupus erythematosus; SLICC = Systemic Lupus International Collaborating; SLEDAI = Systemic Lupus Erythematosus Disease Activity Index; Serum assay indexes: C3, C4, and CH50. All participants were female and were matched for age. Values were represented as mean ± SD. With regard to the current medication treatment, all the patients were treated with prednisone and the dose of the prednisone were shown. Some patients were also treated with other drugs, such as cyclophosphamide, azathioprine, methotrexate and hydroxychloroquine. Values were represented as (number of patients treated with other drugs)/(the total patient number). All the participants were right-handedness.
2.2. Image acquisition
Images were acquired by a Philips 3.0-T Achieva MRI scanner with an 8-channel head coil. The T1-weighted and sagittal 3D data were acquired through a spoiled gradient echo sequence which covered the entire brain (176 slices; slice thickness = 1.0 mm; repetition time [TR]/echo time [TE] = 9 ms/4 ms; acquisition matrix = 256 × 256; field of view [FOV] = 256 × 256 mm2). The diffusion tensor images were acquired through a single-shot spin-echo EPI sequence (75 axial contiguous slices; slice thickness = 2.0 mm; TR/TE = 9737 ms/88 ms; acquisition matrix = 112 × 112; FOV = 224 × 224 mm2; 32 diffusion orientations with b-value = 1000 s/mm2; one b0 image without diffusion weighting).
2.3. Data preprocessing
The data preprocessing consisted of the following steps: (1) the non-brain tissues from the head images were deleted by employing the brain extraction tool (BET) (Smith, 2002); (2) the eddy current distortions and head motion artifacts were corrected by applying an affine alignment of each diffusion-weighted image to the non-diffusion b0 image (Andersson and Sotiropoulos, 2016) and adjusting the gradient orientations to accommodate the slight rotations because of head movement (Leemans and Jones, 2009); (3) the diffusion tensor models were estimated by solving the Stejskal and Tanner equation (Stejskal and Tanner, 1965); and (4) the probabilistic distribution of the fiber diffusion parameters at each voxel was estimated by Markov Chain Monte Carlo sampling (Behrens et al., 2007). All the preprocessing steps were accomplished using the PANDA standard pipeline (Cui et al., 2013), which was an integration analysis toolbox comprising the Diffusion Toolkit (Wang et al., 2007), FMRIB Software Library (FSL) (Smith et al., 2004), MRIcron (Rorden et al., 2007) and Pipeline System for Octave and Matlab (PSOM) (Bellec et al., 2012).
2.4. Network construction
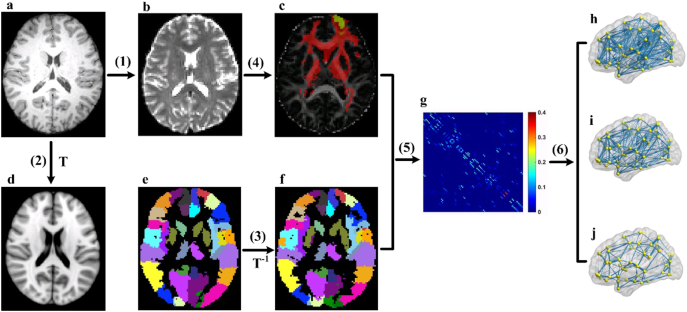
The construction of the structural network requires the following basic elements: nodes and connection edges. Fig. 1 shows the detailed procedure of the white matter network construction. The main steps are described in the following sections.
Fig. 1.
Flowchart for the construction of the white matter structural network. (1) Register the T1-weighted image (a) to the non-diffusion image (b) in DTI native space for each subject. (2) Register the resultant T1 image to the ICBM152 T1 template (d) in the MNI space, resulting in a nonlinear transformation (T). (3) Apply the inverse transformation (T− 1) to the AAL90 template (e) in the MNI space, resulting in a subject-specific parcellation (f) in the DTI native space. (4) For each region, estimate the connectivity probability with other brain regions by using probabilistic tractography (c). In c, the yellow–red colors represent the connected probability from the right superior frontal gyrus (marked as green) to the other regions. (5) Construct the weighted network (g) for each subject by computing the connection probability between each pair of regions. (6) Threshold the individual matrix under a range of sparsity from 5% to 25% with an interval of 0.5%: sparsity = 25% (h); sparsity = 15% (i); sparsity = 5% (j). The visualization of the sparse networks was represented at lateral view.
2.4.1. Brain parcellation
For the network construction, the brain parcellation framework in PANDA was used to segment the entire cerebrum into 90 regions of interest (45 regions for each hemisphere) (Gong et al., 2009). Each anatomical region represented a node in the structural network. In particular, the structural T1-weighted image of each subject was first coregistered to the corresponding non-diffusion image in the DTI native space by using an affine transformation. The registered structural image was then nonlinearly transformed in the ICBM152 T1 template in the Montreal Neurological Institute (MNI) space. Finally, the automated anatomical labeling (AAL) atlas (Tzourio-Mazoyer et al., 2002) was wrapped from the standard space to each native space by inversing the nonlinear transformation from the last step. During implementation, the nearest-neighbor interpolation method was applied to preserve the discrete labeling values.
2.4.2. Tractography
The probabilistic tracking method in the FMRIB Diffusion Toolbox (FDT) was used for the whole-brain tractography. The probabilistic tracking model presents advantages with regard to the tracking sensitivity of non-dominant fiber populations but exerts no significant influence on the results of the dominant pathways (Behrens et al., 2007). In tractography, 5000 streamlines were sampled in each voxel in accordance with the default configuration in the software. For each sampled fiber, a pathway direction was calculated from the distribution of the voxel-wise principal diffusion direction, which was estimated in the preprocessing stage, and then the pathway proceeded to a new position with a default distance of 0.5 mm along the diffusion direction. Finally, a new sample direction was derived from the distribution at the new location. The procedure was iterated until the fibers reached the surface of the brain or satisfied the termination conditions. When the model extended to the regional level, 5000 × n streamlines were sampled for a seed region, where n was the number of voxels in that region. The connective probability from the seed region to the target region was the number of the fibers that traversed the target region divided by the number of fibers originating from the seed region. The connection probability between the seed and target regions was asymmetric because it relied on the source locations (Gong et al., 2009). Thus, the unidirectional probabilistic connectivity between the two regions was set as the average of the two probabilities from one region to another.
2.4.3. Connection definition
The connection edges in the structural network were defined as the above connectivity probability Pij between regions i and j. Thus, a 90 × 90 symmetric weighted matrix was obtained for each subject to represent the constructed network. Given the nature of the probabilistic tractography, the resultant network usually contains a large number of false positive connections. This finding contradicts the sparse connectivity pattern of the human brain connectome, and the spurious connections can influence the subsequent inferences of the network topological parameters. To remove the spurious connections, the weakest connection weights under a given sparse threshold in the probability matrix were discarded for each subject. To exclude the bias of a single sparse threshold, a sparsity ranging from 5% to 25% with an interval of 0.5% was used, and the statistic comparison of the topological metrics was performed across this sparsity range.
2.5. Network measures
To characterize the topological organization of the white matter structural connectomes, the following graph theoretical quantitative measures were assessed: network strength (Sp), global efficiency (Eglob), local efficiency (Eloc) (Latora and Marchiori, 2001), cluster coefficient (Cp), characteristic path length (Lp), normalized cluster coefficient (gamma, γ), normalized characteristic path length (lambda, λ), and small-worldness (sigma, σ) (Bassett and Bullmore, 2006, Bullmore and Sporns, 2009, Sporns and Honey, 2006). For regional properties, we considered the nodal efficiency (Enodal) and nodal degree (Dnodal) (Rubinov and Sporns, 2010). See the supplementary materials for detailed definitions and interpretations of these network metrics. All the network measures were calculated using the GRETNA package (Wang et al., 2015a). In addition, network hubs refer to essential brain regions interacting with several other regions and playing a critical role in functional integration. A brain region was identified as a network hub if its nodal measurement (efficiency or degree) was at least 1SD larger than the mean of whole network.
2.6. Network-based statistic analysis
The network-based statistic (NBS) analysis method (Zalesky et al., 2010a) was used to localize the interregional connections with significant differences between the non-NPSLE and HC groups. In the NBS framework, a two-sample one-tailed t-test at each connection was first performed independently. A primary threshold was then applied to the statistic values to generate a set of supra-threshold connections before identifying the maximal connected subnetwork components. To estimate the significance of each subnetwork, we employed nonparametric permutation test for 10,000 times. An empirical null distribution of the maximal subnetwork size was then obtained. Finally, a corrected p value was calculated for each original subnetwork component observed from this null distribution. The age of the subjects was considered as nuisance covariates in the regression analyses. The significant subnetwork results were visualized using the BrainNet Viewer package (Xia et al., 2013).
2.7. Statistical analysis
2.7.1. Between-group differences
The between-group age difference was tested through a two-sample t-test. For each global network metric, the between-group difference in the area under the metric curve (AUC) value was calculated. The significance of the difference was evaluated by employing a nonparametric permutation test (10,000 permutations). In the permutation test, the influence of age was regarded as the nuisance covariate and was thus removed by multiple linear regression analysis. For the regional comparisons, an additional false discovery rate (FDR) correction (Genovese et al., 2002) was applied to correct the false positive error caused by the multiple comparisons.
2.7.2. Correlations between network measures and clinical variables
For the brain regions with significantly different nodal efficiency or nodal degree, the correlations between the above network metrics and the clinical variables (i.e., SLEDAI, SLICC, disease duration, dose of prednisone, anti-Sm antibodies, and serum assay indexes C3, C4, and CH50) in the non-NPSLE group were evaluated by multiple linear regression analyses. In the regression analyses, the regional parameters were considered as dependent variables, and the clinical variables were considered as independent variables. Age was also considered as a nuisance covariate and was thus regressed out.
2.8. Effect of high-resolution parcellation
Previous studies reported that the results of brain network topological parameters are related to the resolution of the network (Wang et al., 2009, Zalesky et al., 2010b). Hence, a reproducibility analysis at a relatively high resolution was necessary. We randomly subdivided the coarse parcellation template of 90 nodes into 1024 regions (Hagmann et al., 2008, Zalesky et al., 2010b). The connection edge among the regions was defined in a same manner as that in the low-resolution case. With this procedure, a symmetric 1024 × 1024 network matrix was obtained for each subject. Similarly, the network analysis of the global and regional measurements was performed at high-resolution to investigate the differences between the non-NPSLE and HC groups. The network sparsity ranged from 1.0% to 2.5% with an interval of 0.1%.
3. Results
3.1. Alterations of global network measures
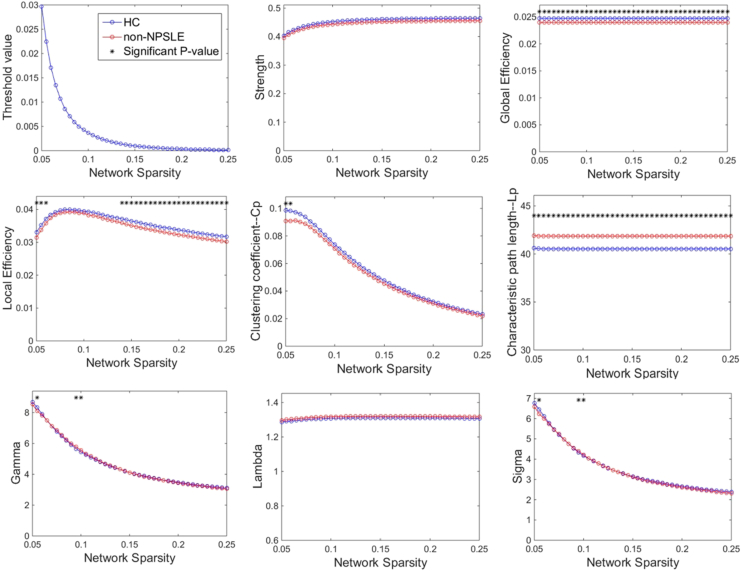
Fig. 2 compares the global network measurements under varying sparsity levels between the non-NPSLE patients and HCs. Within the scope of the applied network sparsity, the structural networks of both the patients and controls exhibited small-world characteristics. Compared with the controls, the patient group exhibited a significant decrease in their global and local efficiencies, and their characteristic path length was significantly increased within a wide range of sparsity. In addition, significant differences were observed in γ and σ at several thresholds. With regard to the AUC value of each metric over the entire range of the thresholds, the patients presented a significant decrease in their global efficiency (p = 0.0262), local efficiency (p = 0.0208) and an increase in their characteristic path length (p = 0.0289), but they did not demonstrate significant differences among the other metrics (Table 2).
Fig. 2.
Differences in global topological properties of the networks between the non-NPSLE patients and HCs across the sparsity range. The data points marked with an asterisk indicate a significant group difference (p < 0.05) under the sparsity. Both controls and patients showed a small-world property of networks characterized by γ ≫ 1 and λ ≈ 1. However, significant between-group differences were observed for most of the sparsity in global efficiency, local efficiency, and characteristic path length.
Table 2.
Comparisons of AUC values of the global network metrics between the HC and non-NPSLE groups.
| AUC value | HC | Non-NPSLE | p value |
|---|---|---|---|
| Strength (× e-03) | 91 ± 4.5 | 89 ± 5.6 | 0.0653 |
| E_Global (× e-04) | 49 ± 2.3 | 48 ± 3.1 | 0.0262* |
| E_local (× e-04) | 72 ± 3.9 | 70 ± 4.3 | 0.0208* |
| Cp (× e-03) | 10.8 ± 1.2 | 10.2 ± 1.1 | 0.1018 |
| Lp | 8.11 ± 0.39 | 8.37 ± 0.55 | 0.0289* |
| Gamma | 0.93 ± 0.02 | 0.93 ± 0.02 | 0.4190 |
| Lambda | 0.26 ± 0.005 | 0.26 ± 0.005 | 0.2488 |
| Sigma | 0.72 ± 0.02 | 0.71 ± 0.02 | 0.3040 |
The values were the AUC values of the global metrics, represented as mean ± SD. The between-group comparisons of the network measures were performed by using nonparametric permutation test. *p < 0.05 was considered significant.
3.2. Alterations of regional network measures
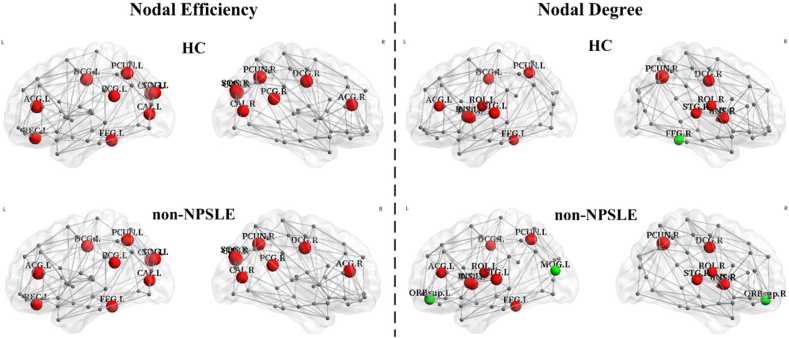
3.2.1. Hub identification
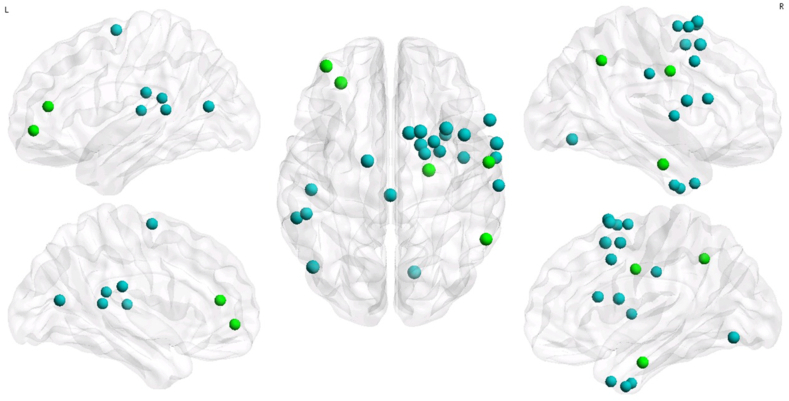
By using the nodal efficiency, the patient and control groups had been identified with same 16 hub regions for each group. By using the nodal degree, 17 and 15 hub regions were identified for the patient and control groups, respectively. The bilateral orbital part of the superior frontal gyrus (ORBsup.L/R) and the left middle occipital gyrus (MOG.L) were identified as hubs in the patient group but not in the control group. The right fusiform gyrus (FFG.R) was identified as a hub in the control group only. The visualization and detailed results are presented in Fig. 3.
Fig. 3.
Constructed structural networks and the corresponding distributions of hubs for the HC group (upper half) and non-NPSLE patients (lower half). The hubs were identified using nodal efficiency (left half) and nodal degree (right half). The hubs are shown in red and green with the node sizes representing their nodal property values. The red node represents the same node between the two groups, and the green node represents the different node between the two groups. The connections among the regions were constructed by averaging the probabilistic connections of all the subjects in the HC group at a threshold sparsity of 1%. The regions and connections were mapped onto the cortical surfaces at a lateral view and visualized by using the BrainNet Viewer software.
3.2.2. Between-group regional differences
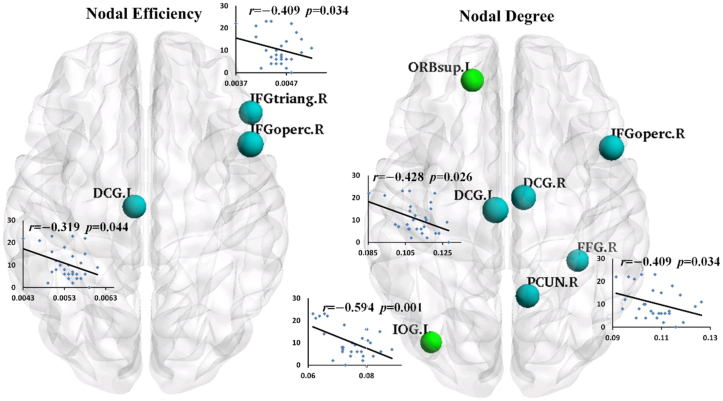
The non-NPSLE patient group showed a significant reduction in the nodal efficiency of the right opercular and triangular parts of their inferior frontal gyrus (IFGoperc.R and IFGtriang.R) and the left median cingulate and paracingulate gyri (DCG.L) (p < 0.01, uncorrected). The nodal degree decreased in the right inferior frontal gyrus (IFGoperc.R), bilateral median cingulate and paracingulate gyri (bilateral DCG), right fusiform gyrus (FFG.R), and right precuneus (PCUN.R) but increased in the left superior frontal gyrus (ORBsup.L) and the left inferior occipital gyrus (IOG.L) (p < 0.01, uncorrected). None of these regions survived after FDR correction. (Fig. 4 and Table 3).
Fig. 4.
The visualization of the brain regions with significant between-group differences in nodal efficiency (left half) and nodal degree (right half) at p < 0.01 (uncorrected). The blue nodes indicate decreased regional property in patients by comparison with HCs, and the green nodes indicate increased regional property in patients by comparison with HCs. The node sizes represent the significance of the between-group differences among the regional properties. These regions were mapped onto the cortical surface at the axial view, and were located according to the centroid stereotaxic coordinates. The nodal degrees of the DCG.L, IOG.L and FFG.R were significantly negatively correlated with SLEDAI values (DCG.L: r = − 0.428, p = 0.026; IOG.L: r = − 0.594, p = 0.001; FFG.R: r = − 0.409, p = 0.034), and the nodal efficiency values of the IFGtriang.R and DCG.L were significantly negatively correlated with SLEDAI values (IFGtriang.R: r = − 0.409, p = 0.034; DCG.L: r = − 0.391, p = 0.044). The corresponding scatter diagrams were overlaid translucently on the brain surface.
Table 3.
Brain regions with significant between-group differences in nodal efficiency and nodal degree respectively.
| Metric | Regions | Category | HC | Non-NPSLE | p value (uncorrected) | T value (p value) of clinical correlations with SLEDAI |
|---|---|---|---|---|---|---|
| Nodal efficiency |
IFGoperc.R | Frontal | 0.471 ± 0.029 (× e-02) | 0.447 ± 0.032(× e-02) | 0.0052 ↓ | —— |
| IFGtriang.R | Frontal | 0.477 ± 0.031(× e-02) | 0.456 ± 0.031(× e-02) | 0.0069 ↓ | − 0.409 (0.034) | |
| DCG.L | Paralimbic | 0.569 ± 0.045(× e-02) | 0.537 ± 0.039(× e-02) | 0.0070 ↓ | − 0.391 (0.044) | |
| Nodal degree |
ORBsup.L | Frontal | 0.105 ± 0.011 | 0.113 ± 0.007 | 0.0061 ↑ | —— |
| IFGoperc.R | Frontal | 0.093 ± 0.009 | 0.087 ± 0.009 | 0.0038 ↓ | —— | |
| DCG.L | Paralimbic | 0.125 ± 0.019 | 0.111 ± 0.009 | 0.0026 ↓ | − 0.428 (0.026) | |
| DCG.R | Paralimbic | 0.131 ± 0.013 | 0.123 ± 0.012 | 0.0041 ↓ | —— | |
| IOG.L | Occipital | 0.069 ± 0.007 | 0.075 ± 0.007 | 0.0072 ↑ | − 0.594 (0.001) | |
| FFG.R | Occipital | 0.115 ± 0.016 | 0.107 ± 0.008 | 0.0066 ↓ | − 0.409 (0.034) | |
| PCUN.R | Parietal | 0.153 ± 0.018 | 0.143 ± 0.013 | 0.0055 ↓ | —— | |
The values of the nodal efficiency and nodal degree were measured as the AUC values across the sparsity range. The between-group comparisons of the regional properties were performed by using nonparametric permutation test (p < 0.01, uncorrected). The age effect was removed in these comparisons. ——, indicating that the nodal property has no significant correlations with the SLEDAI scores.
3.2.3. Correlations between network measures and clinical variables
The results of the multiple linear regression analyses indicated that the global and local network efficiencies in the non-NPSLE patients presented a significant negative correlation with the SLEDAI values (Eglob: r = − 0.407, p = 0.035; Eloc: r = − 0.402, p = 0.038). The nodal degrees of the DCG.L, IOG.L, and FFG.R were also significantly negatively correlated with the SLEDAI values (DCG.L: r = − 0.428, p = 0.026; IOG.L: r = − 0.594, p = 0.001; FFG.R: r = − 0.409, p = 0.034). Similarly, the nodal efficiencies of IFGtriang.R and DCG.L were significantly negatively correlated with the SLEDAI values (IFGtriang.R: r = − 0.409, p = 0.034; DCG.L: r = − 0.391, p = 0.044). The correlation results are shown in Fig. 4 and Table 3. No significant correlation was observed between the regional metrics and the other non-NPSLE clinical variables.
3.3. Whole-brain mapping of connectivity alterations
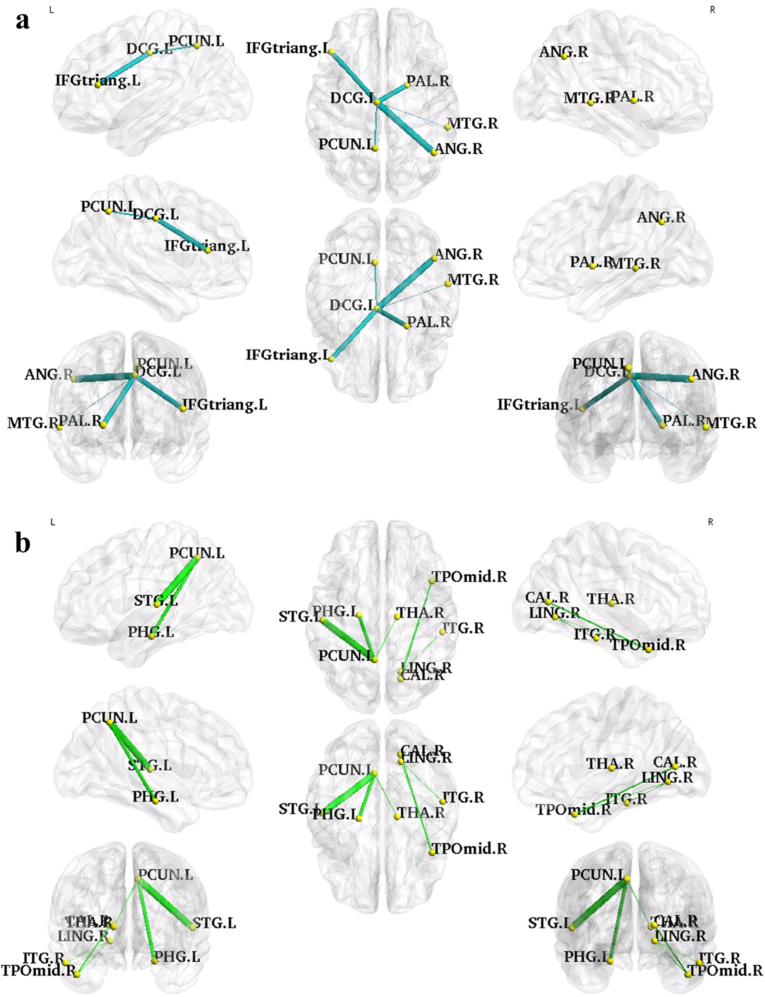
Compared with the HCs, the patient group presented a significantly decreased subnetwork component and two significantly increased subnetwork components in the NBS analysis results (p < 0.01, NBS corrected, Table 4). The decreased subnetwork was a DCG.L-centered component consisting of five edges connecting the DCG.L to five different regions, particularly IFGtriang.L, right angular gyrus (ANG.R), PCUN.L, left pallidum (PAL.L), and right middle temporal gyrus (MTG.R). The increased subnetwork contained a PCUN.L-centered component and a right middle temporal pole (TPOmid.R)-centered component. The other regions that linked with these two center regions were the left parahippocampal gyrus (PHG.L), right thalamus (THA.R), left superior temporal gyrus (STG.L), right calcarine (CAL.R), right lingual gyrus (LING.R), and right inferior temporal gyrus (ITG.R). The visualization of the connectivity alterations is shown in Fig. 5.
Table 4.
The NBS results: the connected sub-networks that show decreased (left half) and increased (right half) connections in non-NPSLE patients compared with the HCs (p < 0.01, NBS corrected).
| HC > non-NPSLE |
HC < non-NPSLE |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Connection |
HC (× e-04) |
Non-NPSLE (× e-04) |
T-stat | Connection |
HC (× e-04) |
Non-NPSLE (× e-04) |
T-stat | ||
| Region A | Region B | Region A | Region B | ||||||
| DCG.L | IFGtriang.L | 10.75 ± 11.33 | 2.671 ± 3.312 | 3.63 | PCUN.L | PHG.L | 2.912 ± 2.965 | 11.53 ± 10.47 | 4.20 |
| DCG.L | ANG.R | 2.291 ± 2.853 | 0.296 ± 0.566 | 3.68 | PCUN.L | THA.R | 6.429 ± 5.741 | 12.26 ± 6.963 | 3.59 |
| DCG.L | PCUN.L | 908.8 ± 170.8 | 775.5 ± 106.2 | 3.53 | PCUN.L | STG.L | 9.910 ± 7.802 | 27.73 ± 18.24 | 4.96 |
| DCG.L | PAL.L | 0.439 ± 0.555 | 0.056 ± 0.065 | 3.63 | TPOmid.R | CAL.R | 0.196 ± 0.365 | 1.039 ± 1.148 | 3.75 |
| DCG.L | MTG.R | 0.972 ± 1.137 | 0.195 ± 0.229 | 3.50 | TPOmid.R | LING.R | 0.102 ± 0.365 | 1.076 ± 1.410 | 3.54 |
| ITG.R | LING.R | 2.320 ± 3.625 | 8.152 ± 7.905 | 3.53 | |||||
Fig. 5.
The visualization of the connected subnetworks that showed significant decreased (a, blue lines) or increased (b, green lines) structural connections in non-NPSLE groups as compared with those of the HC group (p < 0.01, NBS corrected). The edge width represents the significance of the between-group difference in the probabilistic connection strength. The nodes and edges were mapped onto the brain surfaces at omnibearing views, and visualized by using the BrainNet Viewer software.
3.4. Effects of high-resolution parcellation
With regard to the global network measurements, the patient group demonstrated a decreased tendency in their global and local network efficiencies, and their characteristic path length exhibited an increased tendency. Notably, these alterations were insignificant (p > 0.05). With regard to the regional network measurements, the regions with decreased nodal degree in the non-NPSLE group were mainly involved in the cortex of the bilateral superior frontal gyrus (SFGdor.L/R), right middle and inferior frontal gyrus (ORBsup.R, MFG.R), bilateral supplementary motor areas (SMA.L/R), DCG.L/R, FFG.R, MTG.L, and THA.L/R (p < 0.01, uncorrected). The regions with increased nodal degree in the non-NPSLE group were mainly involved in the areas of MFG.L, IFGtriang.L, right hippocampus (HIP.R), parahippocampal gyrus (PHG.R), and right supramarginal gyrus (SMG.R) (p < 0.01, uncorrected). The visualization of the results is shown in Fig. 6. Under the same significance level, no significant between-group difference on nodal efficiency was identified in any brain region. Overall, the high-resolution analysis produced some results consistent with the low-resolution analysis, for example, the decreased nodal degrees in FFG and DCG. On the other hand, the global properties and several regional alterations (e.g., altered degree in SMA, THA, and HIP) identified by high-resolution analysis were different from those of low-resolution analysis. This inconsistency may be attributed to the dependence of the topological properties on the spatial regional scales (Zalesky et al., 2010b).
Fig. 6.
The distribution of the regions with significant between-group differences in the nodal degree of high resolution (H-1024) networks. The regions with decreased and increased efficiency in non-NPSLE patients are shown in blue and green nodes, respectively (p < 0.01, uncorrected). The node size represents the significance of the between-group differences in the nodal degree. The nodes were mapped onto the brain surfaces at lateral and axial views, and visualized by using the BrainNet Viewer software.
4. Discussion
In this study, the topological alterations of the white matter networks between the non-NPSLE patients and HCs were investigated using probabilistic tractography and connectivity-based analysis methods. The main findings were presented as follows. First, compared with the controls, the non-NPSLE patients showed significantly decreased global and local network efficiencies but presented increased characteristic path length. Second, the patients demonstrated significantly changed regional properties in their frontal, occipital, and cingulum regions, and the changes were negatively correlated with the disease activity index. Third, the distribution pattern of the hubs measured by nodal degree was altered in the patient group. Fourth, the patients exhibited a subnetwork of decreased connectivity and a subnetwork of increased connectivity. Overall, these findings revealed the alteration of the topological organization in non-NPSLE disease from a network perspective.
4.1. Disrupted global network topological organization
The white matter networks in the patients showed small-world properties across the sparsity range. This result showed that the basic brain organization was conserved in non-NPSLE disease, indicating that the small-world network can tolerate structural alteration to some extent (He et al., 2009, Supekar et al., 2009).
However, compared with the HCs, the non-NPSLE patients exhibited significantly decreased global and local efficiencies and significantly increased characteristic path length, as well as a tendency to achieve decreased connection strength. First, the decreased global efficiency reflected the reduction in effective interaction and neural information transmission across remote cortical regions. The decreased local efficiency reflects the decreased local specialization which mediates the modularized information processing among the neighboring cortical regions (Latora and Marchiori, 2001). The alterations in the network efficiencies in the patients reflected a disruption in the topological organization because of a disease. These findings were consistent with the insufficient effective integration among the distributed functional cortical regions reported in a previous SLE fMRI study (Mikdashi, 2016). In addition, the increased short path represented prolonged optimal interactions among the neurons, which are essential for functional cognitive processes within and across the brain regions (Sporns and Zwi, 2004). The degeneration of the fiber tracts is generally considered the cause of the increase in the shortest path length in the patient group. Previous DTI studies reported the degeneration of the fiber tracts, and this condition was reflected by the significantly decreased FA values and increased MD values in several brain fiber tracts of NPSLE and non-NPSLE patients (Emmer et al., 2010, Ercan et al., 2015, Jung et al., 2010a, Schmidt-Wilcke et al., 2014).
4.2. Altered regional network topological organization
With regard to the regional network properties, the non-NPSLE patients showed significantly decreased nodal efficiencies in their regions of IFGoperc.R, IFGtriang.R, and DCG.L. They also exhibited significantly increased nodal degrees in their regions of ORBsup.L and IOG.L (p < 0.01, uncorrected). On the contrary, they exhibited significantly decreased nodal degrees in their regions of IFGoperc.R, DCG.L/R, FFG.R, and PCUN.R (p < 0.01, uncorrected). These regions can be categorized into language, visual, and default-mode network (DMN) systems.
4.2.1. Language system
Decreased regional properties were observed in the inferior frontal regions. The inferior frontal regions were considered to be the pivotal components for the language processing. Previous TBSS-based DTI analysis observed the decrease in the white matter integrity in the prefrontal fiber tracts in the non-NPSLE group (Schmidt-Wilcke et al., 2014). Morphological studies reported the decreased cortical thickness in clusters of the frontal lobe (Appenzeller et al., 2007, Jung et al., 2010b). Several fMRI studies also observed abnormal neuronal activity in the prefrontal regions of SLE patients (Fitzgibbon et al., 2008, Hou et al., 2013, Mackay et al., 2011). The alterations of the efficiency and degree of the structural connections to and from the identified regions can influence the functional interactions mediated by these structural pathways (Honey et al., 2010). Notably, in the current results, the IFGoperc and IFGtriang comprising the Broca's area were the essential regions for language processing. By performing a verb generation functional task, DiFrancesco et al. (DiFrancesco et al., 2007) found that deficits in the language ability were among the earliest signs of developmental neurologic disorders even in non-NPSLE patients.
4.2.2. Visual system
Altered nodal degree was observed in several occipital regions (FFG and IOG), which are essential in visual processing. In the DTI studies, Emmer et al. found a reduction in the white matter tract integrity in the inferior fronto-occipital fasciculi of SLE patients (Emmer et al., 2010). Previous morphological studies have revealed alterations in the gray matter volume in the occipital lobe and lower white matter volume in the inferior frontal-occipital fasciculi in SLE patients (Appenzeller et al., 2007, Jung et al., 2010b, Mak et al., 2016). In the fMRI study, DeFrancesco et al. (DiFrancesco et al., 2013) found a significant increase in the neural activation of visual association regions (bilateral fusiform gyrus and inferior occipital lobes) in non-NPSLE patients when they performed a visual stimuli paradigm to evaluate the visuoconstructional ability. The increased functional cortical activity observed in the regions showed that the increased recruitment of neural activation was needed to accomplish the functional tasks. The alterations in the regional properties in anatomical connectivity were suggested to be associated with the compensation in the functional connectivity. These results provided evidence for the abnormality in the visual system of non-NPSLE patients.
4.2.3. Default-mode network system
Decreased regional efficiency and degree in several key components of the DMN regions (DCG and PCUN) were observed in non-NPSLE patients. Morphological analysis indicated that the middle cingulate cortex presented relatively low gray and white matter volumes in SLE patients (Mak et al., 2016). Many previous fMRI studies revealed abnormal neuronal activities in the cingulate gyrus and precuneus regions when non-NPSLE patients performed working memory functional tasks (DiFrancesco et al., 2013, Mackay et al., 2011). These results supported the current findings with regard to the abnormalities in the core components of the DMN system of the non-NPSLE group. The DMN was suggested to play an important role in cognitive dysfunctions. In addition, the DMN regions were demonstrated to possess the highest structure–function connectivity agreement within the entire brain (Horn et al., 2014). Thus, the structural disruption of the regional topological features in DMN was assumed to be the underlying cause of the decrease in the cognitive function of non-NPSLE and NPSLE patients (Mackay et al., 2011, Ren et al., 2012).
4.3. Clinical relevance of network metrics
Both the global and local efficiencies of the networks were significantly negatively correlated with the SLEDAI scores of the non-NPSLE patients. This result suggested that the disease state of SLE can affect the topological organization of the brain network, even in the early stages before the appearance of neuropsychiatric manifestations. Considering that the decrease in network efficiency reflects the disruption of topological organization, high SLE disease activity index may be associated with severe disruption of the topological organization. Furthermore, the regional properties in the right inferior frontal gyrus, left median cingulum, right fusiform, and right inferior occipital were negatively correlated with the SLEDAI values. This result indicated that the alterations in these regions play a key role in the pathogenesis of SLE. Many previous studies found that both the pathological white matter foci volume and the frontal-parietal functional connectivity strength were correlated with the SLEDAI score (Hou et al., 2013, Podrazilova et al., 2008, Xu et al., 2010). No other serum or clinical marker (e.g., C3, C4, anti-Sm antibodies, SLICC, etc.) showed correlation with the cerebral involvement indicated by either functional or structural method. These results suggested that SLEDAI score is the sole correlated indicator of the investigated biomarkers, which revealed the extent of the involvement of the brain nervous system in SLE patients. During the clinical treatment of the disease, doctors should focus on the level of the SLEDAI to prevent its increase, thereby preventing further damage on the patient's brain.
4.4. Altered hub distribution pattern
Compared with the HC group, the patient group exhibited additional hubs of ORBsup.L/R and MOG.L, as well as absence of the FFG.R hub identified by nodal degree, although the hub distribution identified by nodal efficiency was similar between the two groups. The additional hubs were consistent with the significantly increased nodal degree in the ORBsup and occipital region of the patients, and the absence of a hub was consistent with the significantly decreased nodal degree in the FFG region of the patients. The alterations in the hub distribution in non-NPSLE patients may be attributed to the undergoing changes in the brain network during the course of the disease. These changes can affect the channels for the optimal neural information transmission. As a result, some brain regions become more or less integral, and this condition is manifested by the increased or decreased nodal degree. The increased degree in the ORBsup and MOG.L regions of the non-NPSLE patients suggested increased reliance on these hubs because of the reconstruction of the local topology around them, whereas the decreased degree in the FFG.R region suggested a decreased reliance on it.
4.5. Altered structural network connectivity
By using NBS analysis, the two subnetwork components with altered connectivity were observed in non-NPSLE patients (p < 0.01, NBS corrected). The first subnetwork with decreased connections was mainly composed of a left median cingulate-centered component. The second subnetwork with increased connections was mainly composed of a left precuneus-centered component and a middle temporal lobe-centered component. The decreased structural connections may reflected a disrupted myelination mechanism, which was caused by the illness, in the connected fiber tracts (Cao et al., 2013, Pavuluri et al., 2009, Silk et al., 2009). The increased white matter connections in the subnetwork components may reflect the compensatory mechanisms of fiber connectivity (Jacobs et al., 2015, Riederer et al., 2016) or decreased fiber crossing and branching along these tracts (Cao et al., 2013, Tamm et al., 2012). The center regions of these subnetwork components belonged to the DMN (Raichle et al., 2001) and were considered to be the core hubs in a healthy connectome. Abnormality in the connections in the DMN was associated with faulty self-processing and psychosis (Whitfield-Gabrieli and Ford, 2012). Thus, the altered structural connectivity in these subnetwork components can confer a particular risk of dysfunction. As expected, Lin et al. demonstrated that the ReHo values of several DMN regions were significantly altered in non-NPSLE patients (Lin et al., 2011). The decreased and increased probabilistic connections among the identified subnetworks in the current study might be the structural foundation of the ReHo alterations.
4.6. Limitations
Several issues were found and needed to be addressed. First, the results of the regional topological alterations did not survive after the FDR correction, and they were considered as exploratory conclusions. The number of patients should be increased in future research to improve the statistical power. Second, the alterations in the global properties were not significant in high spatial resolution, and the significantly altered regions identified by the nodal properties showed discrepancies among the different resolutions. Such discrepancies may have been caused by the reliance of the topological attributes on the nodal scale (Zalesky et al., 2010b). Third, analysis was only performed on the white matter anatomical connectivity. A combination of the structural network analysis and functional network analysis can provide various perspectives with regard to the connectivity disruptions in non-NPSLE patients. Finally, only the non-NPSLE patients were focused, excluding the NPSLE patients. Previous studies found that differences between the non-NPSLE and NPSLE groups were present when the white matter integrity was investigated (Jung et al., 2012, Schmidt-Wilcke et al., 2014). Therefore, further systematic study on NPSLE, non-NPSLE, and controls can benefit the exploration of the pathogenesis of SLE.
5. Conclusion
Probabilistic tractography and connectivity-based analyses were performed to investigate the alterations in the structural networks of non-NPSLE patients relative to healthy people. The non-NPSLE patients exhibited impaired global integration and local specialization. The brain regions with significantly altered nodal properties were mainly involved in language, visual, and DMN systems. Some of these regions presented negative correlations with the SLE disease activity index scores. The brain hub distribution, which was identified by the nodal degree metric, exhibited changes in the non-NPSLE patient group. Two subnetwork components with abnormal connectivity were identified by network-based statistics. These findings demonstrated the alterations in the white matter connectome in SLE even without neuropsychiatric symptoms, and thus enhanced the understanding of the structural disruptions influencing the functional and neurocognitive deficits in non-NPSLE. Despite the limitations, the results provided insights into the neural structural bases of non-NPSLE and suggested that the structural network analysis method might be a promising approach to reflect the neurobiological mechanism in SLE individuals.
Acknowledgements
This work was supported by the National Natural Science Funds of China [grant number 81371539 and 61671228], the National Key Technology R&D Program [grant number 2015BAI01B03] and the Provincial Natural Science Funds of Guangdong, China [grant number 2016A030310380].
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.nicl.2016.12.021.
Contributor Information
Yikai Xu, Email: yikai.xu@163.com.
Yanqiu Feng, Email: foree@163.com.
Appendix A. Supplementary data
Supplementary material
References
- Andersson J.L., Sotiropoulos S.N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage. 2016;125:1063–1078. doi: 10.1016/j.neuroimage.2015.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller S., Bonilha L., Rio P.A., Min Li L., Costallat L.T., Cendes F. Longitudinal analysis of gray and white matter loss in patients with systemic lupus erythematosus. NeuroImage. 2007;34:694–701. doi: 10.1016/j.neuroimage.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Basser P.J., Pajevic S., Pierpaoli C., Duda J., Aldroubi A. In vivo fiber tractography using DT-MRI data. Magn. Reson. Med. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Bassett D.S., Bullmore E. Small-world brain networks. Neuroscientist. 2006;12:512–523. doi: 10.1177/1073858406293182. [DOI] [PubMed] [Google Scholar]
- Behrens T.E., Berg H.J., Jbabdi S., Rushworth M.F., Woolrich M.W. Probabilistic diffusion tractography with multiple fibre orientations: what can we gain? NeuroImage. 2007;34:144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellec P., Lavoie-Courchesne S., Dickinson P., Lerch J.P., Zijdenbos A.P., Evans A.C. The pipeline system for octave and Matlab (PSOM): a lightweight scripting framework and execution engine for scientific workflows. Front Neuroinform. 2012;6:7. doi: 10.3389/fninf.2012.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt B.C., Chen Z., He Y., Evans A.C., Bernasconi N. Graph-theoretical analysis reveals disrupted small-world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cereb. Cortex. 2011;21:2147–2157. doi: 10.1093/cercor/bhq291. [DOI] [PubMed] [Google Scholar]
- Bertsias G.K., Boumpas D.T. Pathogenesis, diagnosis and management of neuropsychiatric SLE manifestations. Nat. Rev. Rheumatol. 2010;6:358–367. doi: 10.1038/nrrheum.2010.62. [DOI] [PubMed] [Google Scholar]
- Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Cao Q., Shu N., An L., Wang P., Sun L., Xia M.R., Wang J.H., Gong G.L., Zang Y.F., Wang Y.F., He Y. Probabilistic diffusion tractography and graph theory analysis reveal abnormal white matter structural connectivity networks in drug-naive boys with attention deficit/hyperactivity disorder. J. Neurosci. 2013;33:10676–10687. doi: 10.1523/JNEUROSCI.4793-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Zhong S., Xu P., He Y., Gong G. PANDA: a pipeline toolbox for analyzing brain diffusion images. Front. Hum. Neurosci. 2013;7:42. doi: 10.3389/fnhum.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFrancesco M.W., Holland S.K., Ris M.D., Adler C.M., Nelson S., DelBello M.P., Altaye M., Brunner H.I. Functional magnetic resonance imaging assessment of cognitive function in childhood-onset systemic lupus erythematosus: a pilot study. Arthritis Rheum. 2007;56:4151–4163. doi: 10.1002/art.23132. [DOI] [PubMed] [Google Scholar]
- DiFrancesco M.W., Gitelman D.R., Klein-Gitelman M.S., Sagcal-Gironella A.C., Zelko F., Beebe D., Parrish T., Hummel J., Ying J., Brunner H.I. Functional neuronal network activity differs with cognitive dysfunction in childhood-onset systemic lupus erythematosus. Arthritis Res Ther. 2013;15 doi: 10.1186/ar4197. (R40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakesmith M., Caeyenberghs K., Dutt A., Zammit S., Evans C.J., Reichenberg A., Lewis G., David A.S., Jones D.K. Schizophrenia-like topological changes in the structural connectome of individuals with subclinical psychotic experiences. Hum. Brain Mapp. 2015;36:2629–2643. doi: 10.1002/hbm.22796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer B.J., Veer I.M., Steup-Beekman G.M., Huizinga T.W., van der Grond J., van Buchem M.A. Tract-based spatial statistics on diffusion tensor imaging in systemic lupus erythematosus reveals localized involvement of white matter tracts. Arthritis Rheum. 2010;62:3716–3721. doi: 10.1002/art.27717. [DOI] [PubMed] [Google Scholar]
- Ercan E., Ingo C., Tritanon O., Magro-Checa C., Smith A., Smith S., Huizinga T., van Buchem M.A., Ronen I. A multimodal MRI approach to identify and characterize microstructural brain changes in neuropsychiatric systemic lupus erythematosus. Neuroimage Clin. 2015;8:337–344. doi: 10.1016/j.nicl.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon B.M., Fairhall S.L., Kirk I.J., Kalev-Zylinska M., Pui K., Dalbeth N., Keelan S., Robinson E., During M., McQueen F.M. Functional MRI in NPSLE patients reveals increased parietal and frontal brain activation during a working memory task compared with controls. Rheumatology (Oxford) 2008;47:50–53. doi: 10.1093/rheumatology/kem287. [DOI] [PubMed] [Google Scholar]
- Genovese C.R., Lazar N.A., Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 2002;15:870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Gong G., He Y., Concha L., Lebel C., Gross D.W., Evans A.C., Beaulieu C. Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb. Cortex. 2009;19:524–536. doi: 10.1093/cercor/bhn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P., Cammoun L., Gigandet X., Meuli R., Honey C.J., Wedeen V.J., Sporns O. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060159. (e159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Evans A. Graph theoretical modeling of brain connectivity. Curr. Opin. Neurol. 2010;23:341–350. doi: 10.1097/WCO.0b013e32833aa567. [DOI] [PubMed] [Google Scholar]
- He Y., Dagher A., Chen Z., Charil A., Zijdenbos A., Worsley K., Evans A. Impaired small-world efficiency in structural cortical networks in multiple sclerosis associated with white matter lesion load. Brain. 2009;132:3366–3379. doi: 10.1093/brain/awp089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg M.C. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- Honey C.J., Thivierge J.P., Sporns O. Can structure predict function in the human brain? NeuroImage. 2010;52:766–776. doi: 10.1016/j.neuroimage.2010.01.071. [DOI] [PubMed] [Google Scholar]
- Horn A., Ostwald D., Reisert M., Blankenburg F. The structural-functional connectome and the default mode network of the human brain. NeuroImage. 2014;102(Pt 1):142–151. doi: 10.1016/j.neuroimage.2013.09.069. [DOI] [PubMed] [Google Scholar]
- Hou J., Lin Y., Zhang W., Song L., Wu W., Wang J., Zhou D., Zou Q., Fang Y., He M., Li H. Abnormalities of frontal-parietal resting-state functional connectivity are related to disease activity in patients with systemic lupus erythematosus. PLoS One. 2013;8 doi: 10.1371/journal.pone.0074530. (e74530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturria-Medina Y., Sotero R.C., Canales-Rodriguez E.J., Aleman-Gomez Y., Melie-Garcia L. Studying the human brain anatomical network via diffusion-weighted MRI and graph theory. NeuroImage. 2008;40:1064–1076. doi: 10.1016/j.neuroimage.2007.10.060. [DOI] [PubMed] [Google Scholar]
- Jacobs H.I., Gronenschild E.H., Evers E.A., Ramakers I.H., Hofman P.A., Backes W.H., Jolles J., Verhey F.R., Van Boxtel M.P. Visuospatial processing in early Alzheimer's disease: a multimodal neuroimaging study. Cortex. 2015;64:394–406. doi: 10.1016/j.cortex.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Jung R.E., Caprihan A., Chavez R.S., Flores R.A., Sharrar J., Qualls C.R., Sibbitt W., Roldan C.A. Diffusion tensor imaging in neuropsychiatric systemic lupus erythematosus. BMC Neurol. 2010;10:65. doi: 10.1186/1471-2377-10-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung R.E., Segall J.M., Grazioplene R.G., Qualls C., Sibbitt W.L., Jr., Roldan C.A. Cortical thickness and subcortical graymatter reductions in neuropsychiatric systemic lupus erythematosus. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung R.E., Chavez R.S., Flores R.A., Qualls C., Sibbitt W.L., Jr., Roldan C.A. White matter correlates of neuropsychological dysfunction in systemic lupus erythematosus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0028373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V., Marchiori M. Efficient behavior of small-world networks. Phys. Rev. Lett. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Leemans A., Jones D.K. The B-matrix must be rotated when correcting for subject motion in DTI data. Magn. Reson. Med. 2009;61:1336–1349. doi: 10.1002/mrm.21890. [DOI] [PubMed] [Google Scholar]
- Liang M., Corzillius M., Bae S., Lew R., Fortin P., Gordon C., Isenberg D., Alarcon G., Straaton K., Denburg S. The American College of Rheumatology nomenclature and case definitions for neuropsychiatric lupus syndromes. Arthritis Rheum. 1999;42:599–608. doi: 10.1002/1529-0131(199904)42:4<599::AID-ANR2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Lin Y., Zou Q.H., Wang J., Wang Y., Zhou D.Q., Zhang R.H., Zhang Y.W., Lii H.T., Fang Y.F. Localization of cerebral functional deficits in patients with non-neuropsychiatric systemic lupus erythematosus. Hum. Brain Mapp. 2011;32:1847–1855. doi: 10.1002/hbm.21158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo C.Y., Wang P.N., Chou K.H., Wang J., He Y., Lin C.P. Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J. Neurosci. 2010;30:16876–16885. doi: 10.1523/JNEUROSCI.4136-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay M., Bussa M.P., Aranow C., Ulug A.M., Volpe B.T., Huerta P.T., Argyelan M., Mandel A., Hirsch J., Diamond B., Eidelberg D. Differences in regional brain activation patterns assessed by functional magnetic resonance imaging in patients with systemic lupus erythematosus stratified by disease duration. Mol. Med. 2011;17:1349–1356. doi: 10.2119/molmed.2011.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A., Ho R.C., Tng H.Y., Koh H.L., Chong J.S., Zhou J. Early cerebral volume reductions and their associations with reduced lupus disease activity in patients with newly-diagnosed systemic lupus erythematosus. Sci. Rep. 2016;6:22231. doi: 10.1038/srep22231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikdashi J.A. Altered functional neuronal activity in neuropsychiatric lupus: a systematic review of the fMRI investigations. Semin. Arthritis Rheum. 2016;45:455–462. doi: 10.1016/j.semarthrit.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Mori S., van Zijl P.C. Fiber tracking: principles and strategies - a technical review. NMR Biomed. 2002;15:468–480. doi: 10.1002/nbm.781. [DOI] [PubMed] [Google Scholar]
- Pavuluri M.N., Yang S., Kamineni K., Passarotti A.M., Srinivasan G., Harral E.M., Sweeney J.A., Zhou X.J. Diffusion tensor imaging study of white matter fiber tracts in pediatric bipolar disorder and attention-deficit/hyperactivity disorder. Biol. Psychiatry. 2009;65:586–593. doi: 10.1016/j.biopsych.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podrazilova L., Peterova V., Olejarova M., Tegzova D., Krasensky J., Seidl Z., Kozelek P., Dostal C. Magnetic resonance volumetry of pathological brain foci in patients with systemic lupus erythematosus. Clin. Exp. Rheumatol. 2008;26:604–610. [PubMed] [Google Scholar]
- Raichle M.E., MacLeod A.M., Snyder A.Z., Powers W.J., Gusnard D.A., Shulman G.L. A default mode of brain function. Proc. Natl. Acad. Sci. U. S. A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Ho R.C., Mak A. Dysfunctional cortico-basal ganglia-thalamic circuit and altered hippocampal-amygdala activity on cognitive set-shifting in non-neuropsychiatric systemic lupus erythematosus. Arthritis Rheum. 2012;64:4048–4059. doi: 10.1002/art.34660. [DOI] [PubMed] [Google Scholar]
- Riederer J.W., Shott M.E., Deguzman M., Pryor T.L., Frank G.K. Understanding neuronal architecture in obesity through analysis of white matter connection strength. Front. Hum. Neurosci. 2016;10:271. doi: 10.3389/fnhum.2016.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden C., Karnath H.O., Bonilha L. Improving lesion-symptom mapping. J. Cogn. Neurosci. 2007;19:1081–1088. doi: 10.1162/jocn.2007.19.7.1081. [DOI] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. NeuroImage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Schmidt-Wilcke T., Cagnoli P., Wang P., Schultz T., Lotz A., McCune W.J., Sundgren P.C. Diminished white matter integrity in patients with systemic lupus erythematosus. Neuroimage Clin. 2014;5:291–297. doi: 10.1016/j.nicl.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira-Lichter I., Vakil E., Litinsky I., Oren N., Glikmann-Johnston Y., Caspi D., Hendler T., Paran D. Learning and memory-related brain activity dynamics are altered in systemic lupus erythematosus: a functional magnetic resonance imaging study. Lupus. 2013;22:562–573. doi: 10.1177/0961203313480399. [DOI] [PubMed] [Google Scholar]
- Shu N., Liu Y., Li K., Duan Y., Wang J., Yu C., Dong H., Ye J., He Y. Diffusion tensor tractography reveals disrupted topological efficiency in white matter structural networks in multiple sclerosis. Cereb. Cortex. 2011;21:2565–2577. doi: 10.1093/cercor/bhr039. [DOI] [PubMed] [Google Scholar]
- Silk T.J., Vance A., Rinehart N., Bradshaw J.L., Cunnington R. White-matter abnormalities in attention deficit hyperactivity disorder: a diffusion tensor imaging study. Hum. Brain Mapp. 2009;30:2757–2765. doi: 10.1002/hbm.20703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skudlarski P., Jagannathan K., Anderson K., Stevens M.C., Calhoun V.D., Skudlarska B.A., Pearlson G. Brain connectivity is not only lower but different in schizophrenia: a combined anatomical and functional approach. Biol. Psychiatry. 2010;68:61–69. doi: 10.1016/j.biopsych.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M. Fast robust automated brain extraction. Hum. Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E., Johansen-Berg H., Bannister P.R., De Luca M., Drobnjak I., Flitney D.E., Niazy R.K., Saunders J., Vickers J., Zhang Y., De Stefano N., Brady J.M., Matthews P.M. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl. 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Jenkinson M., Johansen-Berg H., Rueckert D., Nichols T.E., Mackay C.E., Watkins K.E., Ciccarelli O., Cader M.Z., Matthews P.M., Behrens T.E. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. NeuroImage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Sporns O. The human connectome: a complex network. Ann. N. Y. Acad. Sci. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Sporns O., Honey C.J. Small worlds inside big brains. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19219–19220. doi: 10.1073/pnas.0609523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O., Zwi J.D. The small world of the cerebral cortex. Neuroinformatics. 2004;2:145–162. doi: 10.1385/NI:2:2:145. [DOI] [PubMed] [Google Scholar]
- Sporns O., Tononi G., Kotter R. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stejskal E.O., Tanner J.E. Spin diffusion measurements: spin echoes in the presence of a time-dependent field gradient. J. Chem. Phys. 1965;42:288. [Google Scholar]
- Stojanovich L., Zandman-Goddard G., Pavlovich S., Sikanich N. Psychiatric manifestations in systemic lupus erythematosus. Autoimmun. Rev. 2007;6:421–426. doi: 10.1016/j.autrev.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Supekar K., Musen M.Menon., V. Development of large-scale functional brain networks in children. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm L., Barnea-Goraly N., Reiss A.L. Diffusion tensor imaging reveals white matter abnormalities in attention-deficit/hyperactivity disorder. Psychiatry Res. 2012;202:150–154. doi: 10.1016/j.pscychresns.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymofiyeva O., Hess C.P., Xu D., Barkovich A.J. Structural MRI connectome in development: challenges of the changing brain. Br. J. Radiol. 2014;87:20140086. doi: 10.1259/bjr.20140086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio-Mazoyer N., Landeau B., Papathanassiou D., Crivello F., Etard O., Delcroix N., Mazoyer B., Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage. 2002;15:273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- Wang R., Benner T., Sorensen A.G., Wedeen V.J. Diffusion toolkit: a software package for diffusion imaging data processing and tractography. Proc. Int. Soc. Magn. Reson. Med. 2007;15:3720. [Google Scholar]
- Wang J., Wang L., Zang Y., Yang H., Tang H., Gong Q., Chen Z., Zhu C., He Y. Parcellation-dependent small-world brain functional networks: a resting-state fMRI study. Hum. Brain Mapp. 2009;30:1511–1523. doi: 10.1002/hbm.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wang X., Xia M., Liao X., Evans A., He Y. GRETNA: a graph theoretical network analysis toolbox for imaging connectomics. Front. Hum. Neurosci. 2015;9:386. doi: 10.3389/fnhum.2015.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Dai Z., Gong G., Zhou C., He Y. Understanding structural-functional relationships in the human brain: a large-scale network perspective. Neuroscientist. 2015;21:290–305. doi: 10.1177/1073858414537560. [DOI] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S., Ford J.M. Default mode network activity and connectivity in psychopathology. Annu. Rev. Clin. Psychol. 2012;8:49–76. doi: 10.1146/annurev-clinpsy-032511-143049. [DOI] [PubMed] [Google Scholar]
- Xia M., Wang J., He Y. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Cheng Y., Chai P., Lu Z., Li H., Luo C., Li X., Li L., Zhou Q., Chen B., Cao J., Xu X., Shan B., Xu L., Wen J. White-matter volume reduction and the protective effect of immunosuppressive therapy in systemic lupus erythematosus patients with normal appearance by conventional magnetic resonance imaging. J. Rheumatol. 2010;37:974–986. doi: 10.3899/jrheum.090967. [DOI] [PubMed] [Google Scholar]
- Xu X., Hui E.S., Mok M.Y., Jian J., Lau C.S., Mak H.K. Structural brain network reorganization in patients with neuropsychiatric systemic lupus erythematosus. AJNR Am. J. Neuroradiol. 2016 doi: 10.3174/ajnr.A4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A., Fornito A., Bullmore E.T. Network-based statistic: identifying differences in brain networks. NeuroImage. 2010;53:1197–1207. doi: 10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Zalesky A., Fornito A., Harding I.H., Cocchi L., Yucel M., Pantelis C., Bullmore E.T. Whole-brain anatomical networks: does the choice of nodes matter? NeuroImage. 2010;50:970–983. doi: 10.1016/j.neuroimage.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Zalesky A., Fornito A., Seal M.L., Cocchi L., Westin C.F., Bullmore E.T., Egan G.F., Pantelis C. Disrupted axonal fiber connectivity in schizophrenia. Biol. Psychiatry. 2011;69:80–89. doi: 10.1016/j.biopsych.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material