Abstract
Deubiquitinating enzyme BAP1 is mutated in a hereditary cancer syndrome with increased risk of mesothelioma and uveal melanoma. Somatic BAP1 mutations occur in various malignancies. We show that mouse Bap1 gene deletion is lethal during embryogenesis, but systemic or hematopoietic-restricted deletion in adults recapitulates features of human myelodysplastic syndrome (MDS). Knock-in mice expressing BAP1 with a 3xFlag tag revealed that BAP1 interacts with HCF-1, OGT, and the polycomb group proteins ASXL1 and ASXL2 in vivo. OGT and HCF-1 levels were decreased by Bap1 deletion, indicating a critical role for BAP1 in stabilizing these epigenetic regulators. Human ASXL1 is mutated frequently in chronic myelomonocytic leukemia (CMML) so an ASXL/BAP1 complex may suppress CMML. A novel BAP1 catalytic mutation found in a MDS patient implies that BAP1 loss of function has similar consequences in mouse and man.
Somatic inactivating BAP1 mutations occur in the majority of metastatic uveal melanomas and approximately one quarter of malignant pleural mesotheliomas. Somatic mutations also have been identified in breast, lung and renal cell cancers (1–5). Recently, germline BAP1 mutations were linked to a tumor predisposition syndrome characterized by melanocytic tumors, mesothelioma, and uveal melanoma (6, 7).
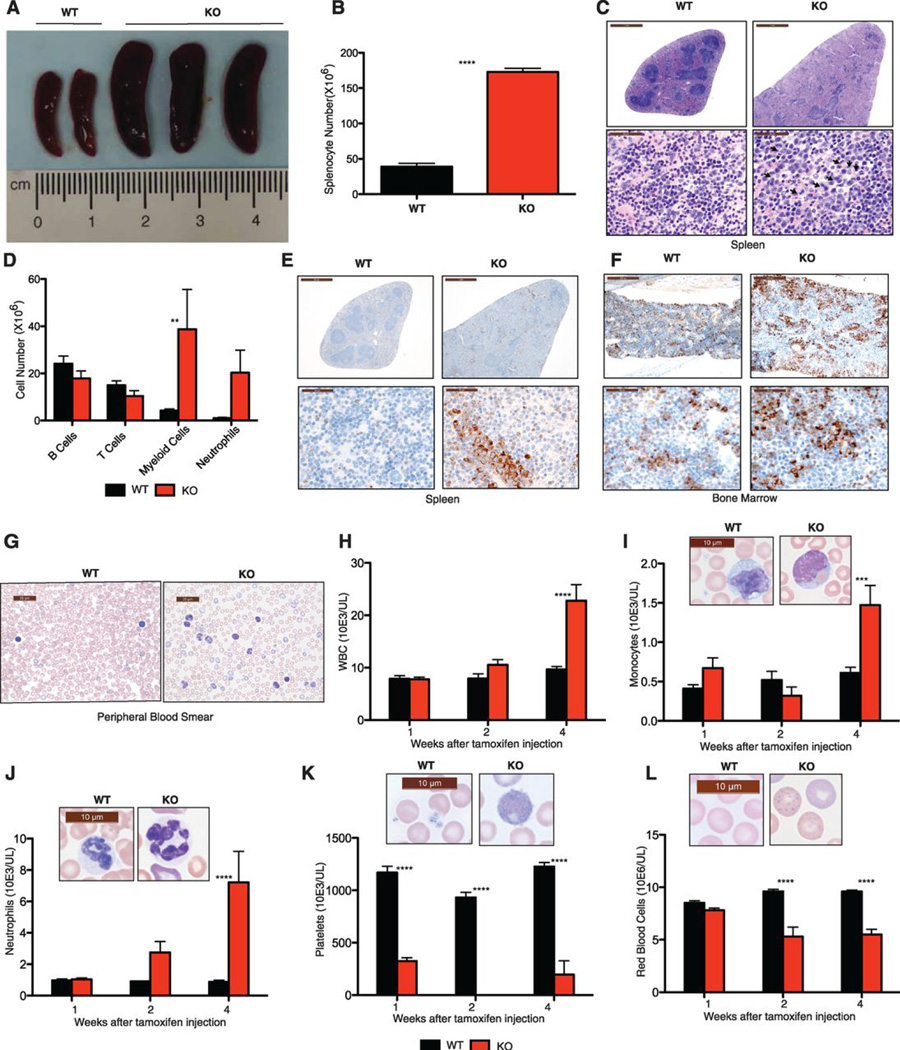
We investigated the normal physiological role of BAP1 using BAP1-deficient mice (fig. S1A). Bap1−/− embryos showed developmental retardation at E8.5 and were not detected beyond E9.5, indicating that BAP1 is essential for embryo development (fig. S1, B and C). To bypass this embryonic lethality, we bred mice that expressed the tamoxifen-inducible recombinase creERT2 ubiquitously from the Rosa26 locus (8), and had Bap1 exons 4 and 5 flanked by lox sites (fig. S1A). The floxed Bap1 exons were deleted from most adult mouse tissues at one week after completing daily tamoxifen injections for 5 days, brain being the expected exception (fig. S1D). Loss of Bap1 mRNA from hematopoietic lineages at one week after the final tamoxifen injection was confirmed by quantitative RT-PCR (fig. S1E), and BAP1 protein was no longer detected in splenocytes by western blotting (fig. S1F). Within 4 weeks of the last tamoxifen injection, 100% of the Bap1fl/fl creERT2+ mice (hereafter referred to as BAP1 KO mice) developed splenomegaly (n=12). This phenotype was never observed in Bap1+/+ creERT2+ control mice (hereafter referred to as WT mice) (Fig. 1, A and B). Histopathology, flow cytometry, and myeloperoxidase immunohistochemistry revealed that splenomegaly in the KO mice resulted from extramedullary hematopoiesis and expansion of the myeloid lineage (Fig. 1, C–E). Myeloid cells also were increased in lymph nodes (fig. S2) and bone marrow (Fig. 1F).
Fig. 1.
BAP1 deficiency results in MDS/CMML-like disease.
(A) Bap1+/+ creERT2+ (WT) and Bap1fl/fl creERT2+ (KO) spleens at 4 weeks after tamoxifen treatment.
(B) Total splenocytes at 4 weeks.
(C) Haematoxylin and eosin staining of spleens at 4 weeks.
(D) Leukocyte subsets in spleen at 4 weeks by flow cytometry. Identifying surface markers were B220 (B cells), CD3ε (T cells), CD11b (myeloid cells), and CD11b plus Gr1 (neutrophils).
(E–F) Myeloperoxidase staining of spleen (E) and bone marrow (F) at 4 weeks.
(G) Peripheral blood smears at 4 weeks.
(H–L) Peripheral blood cell counts.
All graphs show the mean ± standard deviation for 5 mice of each genotype. Asterisks indicate statistically significant differences between WT and KO mice based on two-way ANOVA analysis, followed by Bonferroni post-test analysis. * P<0.05, *** P<0.001, **** P<0.0001.
Peripheral blood taken from 11 out of 12 BAP1 KO mice at 4 weeks after the final tamoxifen injection showed cytological features consistent with myelodysplasia and ineffective erythropoiesis (Fig. 1G). Total leukocyte numbers were elevated (Fig. 1H) due to monocytosis (Fig. 1I) and neutrophilia (Fig. 1J), consistent with CMML-like disease (classified as a myelodysplastic/myeloproliferative disease according to the new WHO classification of myeloid neoplasms (9)). Thrombocytopenia was detected as early as 1 week after the final tamoxifen injection (Fig. 1K) and all diseased mice developed severe, progressive anemia (Fig. 1L). We noted morphologic features of erythroid dysplasia, including increased numbers of nucleated red blood cells, anisopoikilocytosis, and prominent basophilic stippling (Fig. 1L). Hypersegmented neutrophils (Fig. 1J), bilobed granulocytes (Fig. 1I), giant platelets (Fig. 1K), hyposegmented neutrophils consistent with pseudo-Pelger-Huët anomaly, and atypical immature cells with myelomonocytic features also were observed. Mitotic figures and apoptotic cells (fig. S3) were consistent with MDS (10), while blasts were rare. Collectively, these data show that Bap1 deletion produces a myeloproliferative/myelodysplastic disorder with features of human CMML. Consistent with what is seen in patients with end-organ damage from myeloid neoplasms, the BAP1 KO heart contained microthrombi with multifocal necrosis, neutrophilic inflammation, and infiltration of myeloblastic cells (fig. S4).
Given that chronic myeloid neoplasms originate in the phenotypic hematopoietic stem cell (HSC) compartment (11), we characterized the lineage-depleted hematopoietic progenitor cell population in the BAP1 KO mice. Lineage− ScaI− c-Kit+ myeloid progenitor cells and hematopoietic stem cell-enriched lineage− ScaI+ c-Kit+ (LSK) cells were increased in BAP1 KO spleen and bone marrow as early as 2 weeks after the final tamoxifen injection (fig. S5A). Notably, given that BAP1 KO mice develop monocytosis and neutrophilia, BAP1 KO LSK cells harvested one month after tamoxifen treatment expressed higher levels of a subset of genes involved in myelopoiesis (fig. S5, B and C; (12). In methylcellulose colony forming assays, BAP1 KO LSK cells yielded fewer colonies than WT LSK cells (fig. S6, A and B). In addition, unlike cells from WT colonies, which could be replated after 10 days in culture to form new colonies, replated BAP1 KO cells did not produce well-formed colonies, and many exhibited cytoplasmic blebbing characteristic of apoptosis (fig. S6, C and D). These in vitro data suggest that BAP1 deficiency impairs HSC survival and/or self-renewal, but this may be context-dependent, such that sufficient BAP1 KO HSCs survive in vivo to reveal skewing of differentiation towards the myeloid lineage (Fig. 1 and fig. S5)
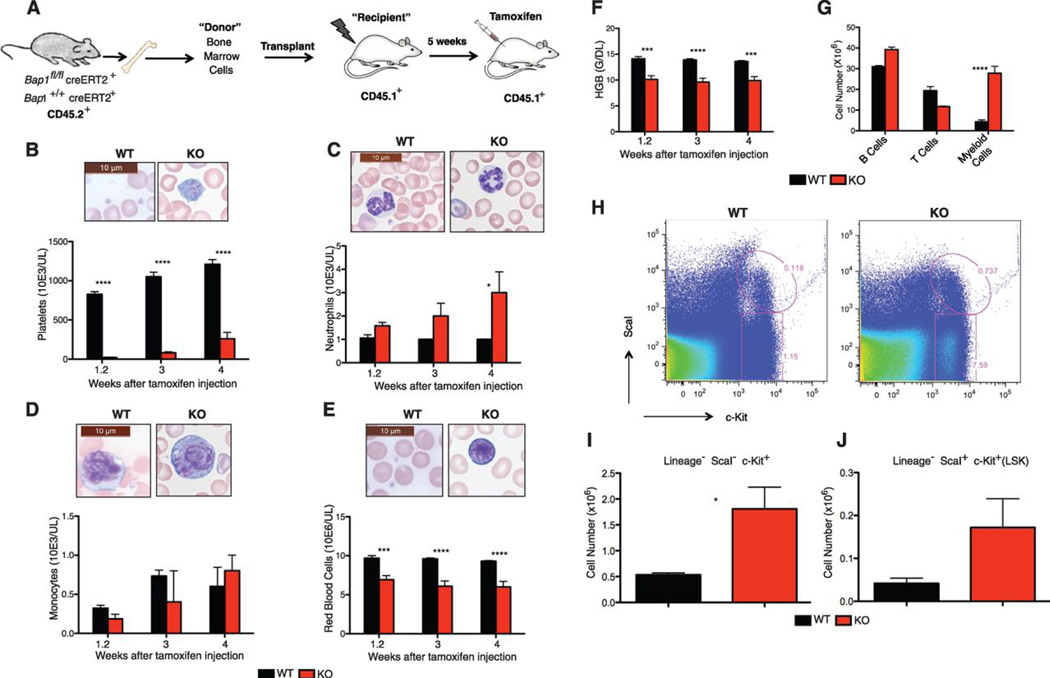
Next, we performed bone marrow transplantation studies to determine whether CMML-like disease is intrinsic to the BAP1 KO hematopoietic compartment. BAP1 KO CD45.2+ lineage− bone marrow cells harvested either 1 week (fig. S7A) or 1 month (fig. S7B) after the final tamoxifen injection were unable to reconstitute lethally irradiated congenic CD45.1+ B6.SJL recipient mice like their WT counterparts. This finding was confirmed in competitive repopulation assays where recipient mice received equal numbers of WT B6.SJL and BAP1 KO C57BL/6 bone marrow cells (fig. S8). Failure of the BAP1 KO cells to engraft might reflect an inability to home to the appropriate stem cell niche. Remarkably, however, Bap1 deletion after B6.SJL recipient mice were reconstituted with Bap1fl/fl creERT2+ bone marrow cells (Fig. 2A and fig. S9) produced features of CMML, including thrombocytopenia (Fig. 2B), neutrophilia (Fig. 2C), monocytosis (Fig. 2D), and anemia (Fig. 2, E and F) at 4 weeks. Myeloid cells were increased in spleen (Fig. 2G), and lineage− ScaI− c-Kit+ myeloid progenitor cells plus LSK cells were increased in bone marrow (Fig. 2, H–J). These data indicate that BAP1 deficiency restricted to the hematopoietic compartment is sufficient for the development of myeloid leukemia and there is no requirement for BAP1 deficiency in the bone marrow stoma.
Fig. 2.
BAP1 deficiency in hematopoietic cells is sufficient for MDS/CMML-like disease.
(A) Bone marrow cells from CD45.2+ Bap1+/+ creERT2+ or Bap1fl/fl creERT2+ mice were transplanted into CD45.1+ lethally irradiated wild-type recipients. Tamoxifen was given to recipients at 5 weeks after transplantation to induce Bap1 deletion.
(B–F) Peripheral blood cell counts (B–E) and hemoglobin levels (F) of reconstituted mice.
(G) Splenic subsets at 4 weeks after tamoxifen treatment. Identifying surface markers were B220 (B cells), CD3ε (T cells), and CD11b (myeloid cells).
(H) Flow cytometric analysis of lineage-negative bone marrow cell populations at 4 weeks after tamoxifen treatment.
(I–J) Absolute numbers of lineage− ScaI− c-Kit+ myeloid progenitors (I) and lineage ScaI+ c-Kit+ (LSK) cells (J) in bone marrow at 4 weeks after tamoxifen treatment.
All graphs show the mean ± standard deviation for 3–5 mice of each genotype. Asterisks indicate statistically significant differences between WT and KO mice based on two-way ANOVA analysis, followed by Bonferroni post-test analysis. * P<0.05, *** P<0.001, **** P<0.0001.
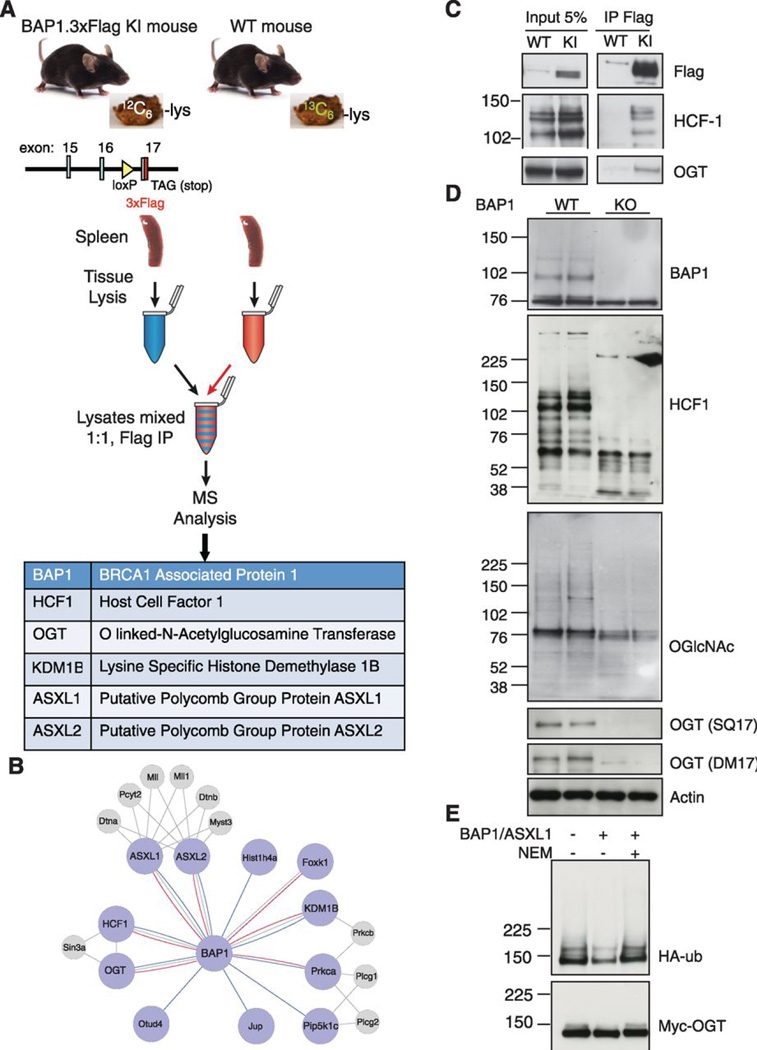
To investigate the mechanism for tumor suppression by BAP1, we characterized by mass spectrometry endogenous BAP1-interacting proteins, which we affinity purified from BAP1.3xFlag knock-in mouse spleen and brain (Fig. 3 and fig. S10A). These organs were chosen because they express BAP1 (fig. S10B) and their size means that protein yield is not a limiting factor. It is worth noting, however, that Bap1 mRNA was detected in LSK cells (fig. S1E), and BAP1.3xFlag protein was immunoblotted in lineage− bone marrow cells containing the progenitor and stem cell populations (fig. S11). Given that anti-Flag antibodies also recognize several proteins in wild-type tissues (fig. S10B), we used a subtraction strategy employing stable isotope labeling. Specifically, BAP1.3xFlag tissue lysates were mixed, prior to immunoprecipitation, in a 1:1 ratio with wild-type lysates prepared from animals fed 13C6-lysine in their chow (13). If the light version of a peptide deriving from the BAP1.3xFlag knock-in was not more abundant than its heavy wild-type counterpart by mass spectrometry, then the protein was excluded as a BAP1-interacting protein. Immunoprecipitations with tissues from an unrelated knock-in strain expressing 3xFlag.ARMC8 (fig. S10C), which is a component of the CTLH complex (14), provided a further control for specificity.
Fig. 3.
Identification and characterization of BAP1-associated proteins in mouse brain and spleen.
(A) BAP1.3xFlag knock-in (KI) mice fed regular chow and wild-type (WT) mice fed "heavy" 13C6-lysine in their chow were used to make tissue lysates for anti-Flag immunoprecipitation (IP) and mass spectrometry (MS). Top IP hits that were represented by multiple unique peptides deriving primarily from the "light" BAP1.3xFlag KI are listed.
(B) The mouse BAP1 interactome. Large nodes represent BAP1-interacting proteins identified in brain (blue lines) or spleen (red lines). Gray lines indicate known interactions from the STRING database. Smaller gray nodes represent proteins from the STRING database that are connected to two or more proteins from our dataset and include manually crated annotations.
(C) Anti-Flag immunoprecipitations (IP) from WT and BAP1.3xFlag splenocytes.
(D) Western blot analysis of WT and BAP1 KO splenocytes at 3 weeks after tamoxifen treatment.
(E) Myc-tagged OGT was affinity purified from HEK293T cells co-transfected with HA-tagged ubiquitin and then incubated with BAP1/ASXL1 purified from Sf9 cells. Where indicated, BAP1 protease activity was inactivated with N-ethylmaleimide (NEM) prior to adding OGT.
Captured proteins were washed briefly under low stringency conditions, such that silver staining revealed no differential bands between wild-type versus BAP1.3xFlag spleen (fig. S10D). After subtracting out the wild-type background, however, known BAP1-interacting proteins were revealed (15, 16), including Host Cell Factor-1 (HCF-1), O-linked N-acetylglucosamine transferase (OGT), the polycomb group proteins ASXL1 and ASXL2 (Fig. 3A), the lysine demethylase KDM1B, and the transcriptional regulator FOXK1. The BAP1 network defined by these experiments is represented in Fig. 3B and it also includes select interactors from the STRING database or manually curated annotations if they connected to two or more proteins within our experimental dataset. Immunoprecipitation and western blotting of BAP1.3xFlag mouse spleen validated the interaction of OGT and HCF-1 with BAP1 (Fig. 3C), but we lacked quality antibodies to assess the other endogenous interactions in this manner. Notably, ASXL1 is a co-factor that is essential for BAP1 enzymatic activity (17) and is mutated frequently in MDS and CMML (18, 19). Given the development of MDS in our BAP1 KO mice, it is tempting to speculate that the BAP1-ASXL1 axis has a critical role in suppressing CMML.
Deubiquitylation of the conserved epigenetic regulator HCF-1 by BAP1 prevents HCF-1 proteasomal degradation (20) and, consistent with this notion, BAP1 KO splenocytes contained far less HCF-1 than their WT counterparts (Fig. 3D). OGT also was decreased in BAP1 KO splenocytes (Fig. 3D) and, being the sole cellular enzyme responsible for O-GlcNAcylation, the cells showed a corresponding decrease in O-GlcNAcylated proteins. To investigate whether BAP1 can deubiquitylate and stabilize OGT directly, rather than decreased OGT being secondary to a reduction in HCF-1, we affinity purified ubiquitylated and Myc-tagged human OGT from HEK293T cells, and then incubated it with human BAP1 and human ASXL1 residues 2–365 co-purified from Sf9 cells. Consistent with OGT being a BAP1 substrate, ubiquitylation on OGT was reduced by the BAP1/ASXL1 complex, and this deubiquitylation was blocked when BAP1/ASXL1 were pre-treated with the cysteine protease inhibitor N-ethylmaleimide (NEM) (Fig. 3E). NEM-mediated inactivation of BAP1 catalytic activity was confirmed in a Ub-AMC cleavage assay (fig. S12A). In contrast to the situation in splenocytes, BAP1 deficiency in mouse embryo fibroblasts (MEFs) did not alter steady-state OGT protein abundance but, in keeping with OGT being a BAP1 substrate, increased OGT protein turnover was revealed when translation was inhibited with cycloheximide (fig. S12B). Given that O-GlcNAcylation of HCF-1 by OGT is necessary for proteolytic maturation of HCF-1 (21, 22), our data argue that BAP1 not only stabilizes HCF-1, but also is necessary for HCF-1 activation via OGT stabilization.
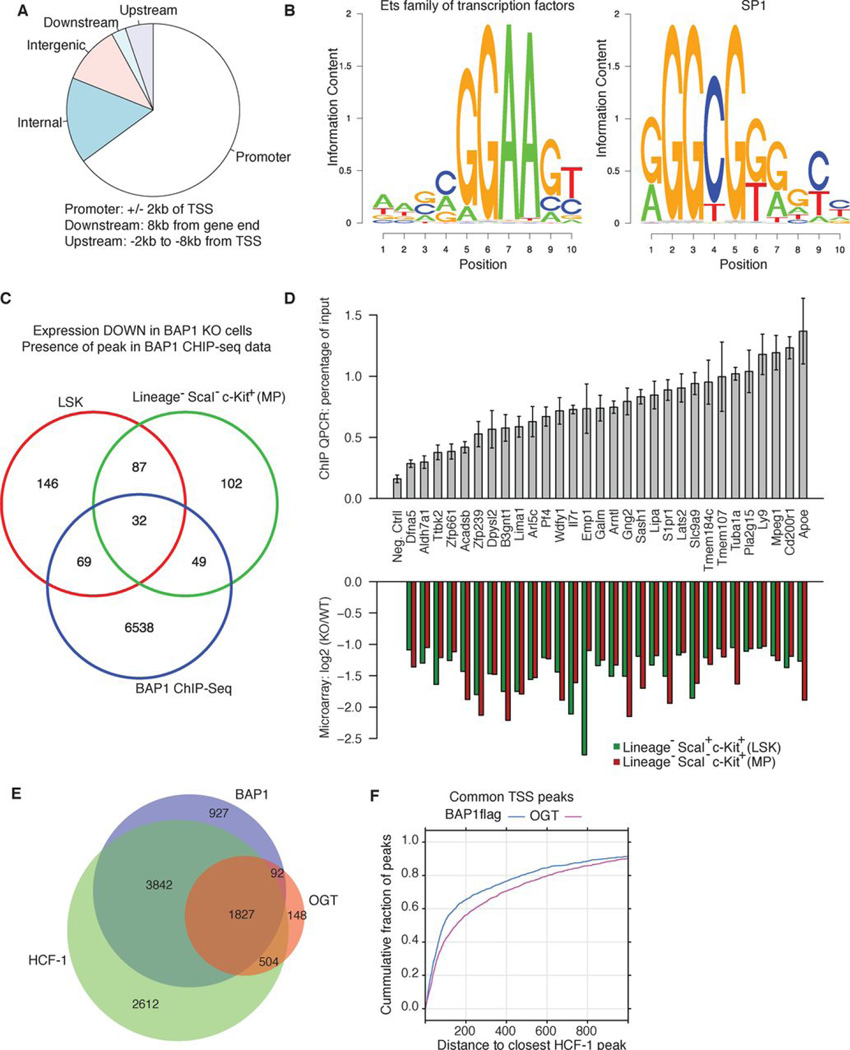
HCF-1 is a pleiotropic transcriptional coregulator (23), so BAP1 probably regulates gene expression via HCF-1 stabilization. To identify potentially critical BAP1-regulated genes, we determined what genes are: (i) dysregulated in BAP1 KO cells by microarray analyses, and (ii) normally have BAP1 bound indirectly to their promoters (via yet to be characterized DNA binding proteins) as judged by chromatin immunoprecipitation and DNA sequencing (ChIP-seq). Low chromatin yields from hematopoietic stem/progenitor cell populations precluded ChIP-seq studies, so we used BAP1.3xFlag knock-in bone marrow-derived macrophages (BMDMs) as a surrogate to identify genome-wide BAP1 binding sites. A total of 9,128 significant BAP1 peaks were identified after ChIP with anti-Flag antibodies (MACS analysis software with stringent criteria: p-value < 1×10−10 and fold-enrichment >5). Of these sites, 5,926 (65%) were located near the transcription start sites of 5,731 genes (Fig. 4A). Motif enrichment analysis of the BAP1-binding sites revealed that the top two occurring motifs are most similar to known binding sites of the Ets (CGGAAG) family of transcription factors and SP1 (GGGCGGGG) (Fig. 4B). Gene expression datasets were generated with WT and BAP1 KO lineage− ScaI− c-Kit+ myeloid progenitor cells, as well as LSK cells. Among the 5,731 genes with BAP1 peaks by ChIP-seq, 32 were down-regulated significantly in BAP1 KO cells (Fig. 4C), while 18 were up-regulated (data not shown). We validated BAP1 localization to 30/32 of the down-regulated genes by ChIP-qPCR (Fig. 4D). Several of the genes identified impact the immune system, including Il7r, which is particularly interesting as a candidate BAP1 target gene because it is a known regulator of hematopoietic cell survival, and IL7R expression is decreased in MDS patients (24).
Fig. 4.
Identification of BAP1-regulated genes.
(A) Characterization of BAP1 binding sites identified by anti-Flag ChIP-seq analysis of BAP1.3xFlag BMDMs.
(B) De novo motif enrichment analysis of BAP1 binding sites identified similarities to Ets (CGGAAG) and SP1 (GGGCGGGG) transcription factor binding sites.
(C) Venn diagram of genes with BAP1 recruited to their promoter in BAP1.3xFlag BMDMs (blue circle), and genes with reduced expression in BAP1 KO lineage− ScaI− c-Kit+ myeloid progenitors (MP; green circle) or BAP1 KO LSK cells (red circle).
(D) Putative BAP1 target genes identified in (C) were validated by ChIP-qPCR on BAP1.3xFlag BMDMs (top panel). Negative control (Neg. Ctrl) primers amplify a region in a gene desert and therefore binding of transcription factors is not expected. Bars show the mean ± standard deviation of triplicate wells, the chromatin deriving from BMDMs pooled from multiple KI mice. The lower panel shows microarray expression data for these genes in WT versus BAP1 KO MP or LSK cells.
(E) Venn diagram showing the overlap between promoters occupied by BAP1.3xFlag, HCF-1, or OGT based on ChIP-seq analyses.
(F) Distances separating BAP1 peaks and OGT peaks from HCF-1 peaks by ChIP-seq.
Next we performed additional ChIP-seq studies for HCF-1 and OGT using WT BMDMs, the goal being to determine which promoters occupied by BAP1 also contain HCF1 and/or OGT. Most HCF-1 and OGT peaks (57.3% and 52.4%, respectively) were located near transcriptional start sites (fig. S13). Strikingly, 85% of promoters occupied by BAP1 also contained HCF-1 (Fig. 4E). Fewer OGT sites were identified but, of these, most were found in genes containing BAP1 and/or HCF-1 sites (Fig. 4E). Within the 1827 promoters occupied by BAP, OGT, and HCF-1, more than 65% of BAP1 peaks are within 200bp of an HCF-1 peak, while 70% of OGT peaks are within 400bp of an HCF1 peak (Fig. 4F). The close proximity of the BAP1, OGT, and HCF-1 peaks (Fig. 4F) fits with their being recruited to promoters as a complex.
Given that BAP1 KO mice develop MDS, we investigated whether BAP1 mutations occur in human MDS by full length resequencing of BAP1 in 32 paired tumor/normal samples from patients with de novo MDS. We identified a patient with a frameshift mutation that causes premature termination within the UCH catalytic domain of BAP1.
Analysis of matched normal DNA did not identify the frameshift allele, consistent with somatic acquisition of the frameshift BAP1 mutation by the MDS clone (fig. S14). Of note, the patient with the somatic BAP1 mutation presented with refractory cytopenias and multi-lineage dysplasia (RCMD), similar to the multi-lineage dysplasia and cytopenias seen in our murine model. Mutational profiling revealed that this patient was wild-type for known MDS mutations, including TET2, ASXL1, EZH2, NRAS, C-KIT, FLT3, IDH1, and IDH2, and had del(20)(q11.2q13.3) as the sole cytogenetic abnormality on metaphase chromosome analysis (data not shown). Interestingly, in a separate microarray dataset (25), BAP1 mRNA expression was reduced significantly in CD34+ cells from MDS patients compared to healthy controls (fig. S15), which is in keeping with BAP1 being a tumor suppressor. To mimic BAP1 haploinsufficiency, we characterized heterozygous Bap1fl/+ creERT2+ mice after tamoxifen treatment. Loss of one copy of Bap1 caused very mild, but progressive hematological defects (fig. S16), which is significant because the frameshift mutation identified in the patient is heterozygous.
In summary, our results identify a new and potent tumor suppressor function for BAP1 in myeloid neoplasia. The BAP1 ortholog in Drosophila called Calypso suppresses hox gene expression (17), but we did not see increased Hox gene expression in BAP1 KO cells (fig. S17). Divergent epigenetic functions for fly and vertebrate BAP1 might reflect that the HCF-1 binding motif conserved in vertebrate BAP1 (16, 20) is absent from Calypso. We propose that BAP1 forms a core complex with HCF-1 and OGT that can differentially recruit additional histone-modifying enzymes to regulate gene expression and thereby preserve normal hematopoiesis. It will be interesting to determine if Bap1 deficiency restricted to non-hematopoietic mouse tissues also promotes tumor development.
Materials and Methods
Mass Spectrometry
Flash frozen BAP1.3xFlag, 3xFlag.ARMC8, or 13C6-lysine-labeled wild-type (Cambridge Isotope Laboratories) spleens or brains were disrupted in lysis buffer [50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, complete protease inhibitor cocktail (Roche)] using a PowerGen homogenizer (3 × 20 s on ice). Samples were left on ice for 30 min, and then soluble lysates were recovered by centrifugation (16000 × g for 10 min at 4°C). For immunoprecipitation (IP), equal amounts of protein from a “light” (either BAP1.3xFlag or ARMC8.3xFlag) and “heavy” wild-type lysate were mixed and incubated with M2 anti-Flag resin (Sigma) for 1 h at 4°C. The resin was washed 3 times with lysis buffer and bound proteins were eluted at room temperature for 30 min in 0.2 mg/mL 3xFlag peptide (Sigma), 50 mM Tris-HCl pH 7.4, 150 mM NaCl, and 1 mM EDTA. Proteins were resolved by SDS-PAGE and stained with Simply Blue Safe Stain. Input lysate mixtures were also resolved to evaluate mixing ratios.
LC-MS/MS analyses followed previous descriptions (1), with minor modifications. Brain IPs were cut into 16 gel regions and digested with trypsin. Spleen IPs were divided into 15 regions and digested with Lys-C protease. Digested peptides were extracted and dried prior to resuspension in 2% acetonitrile/0.1% formic acid. Samples were injected onto a 0.1 × 100 mm column packed with 1.7 µm BEH-130 C18 using a NanoAcquity UPLC (Waters) and introduced to a LTQ-Orbitrap XL mass spectrometer (ThermoFisher Scientific) through an ADVANCE electrospray ionization source. The Orbitrap was operated in data dependent top8 mode, with a full MS scan collected in the FTMS at 60000 resolution and MS/MS collected in the ion trap on the top 8 most abundant species.
Raw spectral data were converted to mzXML file format and corrected monoisotopic masses corresponding to each selected precursor determined using in house software tools as described previously (2). MS/MS spectra were searched with Mascot (3) using a 50 ppm precursor ion tolerance and full trypsin/LysC specificity against a concatenated target-decoy database containing marine proteins extracted from the Uniprot database (Version 2010_12) and common contaminants. Lysine (13C6) and oxidized methionine were considered as variable modifications. Peptide spectral matches (PSMs) were filtered to a 1% FDR using linear discriminant analysis. For BAP1.3xFlag samples, area under curve (AUC) measurements, and the corresponding SILAC ratios for “light” and “heavy” peptides, were determined using the VistaQuant algorithm. Chromatographic peak pairs were filtered using a heuristic score cutoff of 83 as described previously (4). PSMs were consolidated to remove redundant observations of chromatographic peak pairs. Pairs where either the “light” or “heavy” chromatographic peak area remained undetermined after consolidation had missing area values scaled proportionally to the signal-to-noise ratio of the identified species. After removing common laboratory contaminants (e.g. keratins, digestive enzymes), data from each tissue-bait pair were combined and normalized to the median ratio (log2) of all peak observations where both “light” and “heavy” AUC values were observed within each tissue-bait set. Normalized AUC values were summed for each protein identified as determined by Mascot and an overall ratio (log2) determined. Proteins were mapped to genes based upon the Unipart database and putative BAP1 interactors selected based on at least 8 observed AUC pairs and a log2 normalized protein ratio > 3 (light BAP1:heavy WT) in either tissue. In order to remove nonspecific relationships, putative BAP1 interactors were compared to a similar analysis of ARMC8 interactors. Genes appearing in the BAP1 list that also appeared in the ARMC8 analysis after filtering to at least 4 AUC observations and a log2 ratio > 2 (ARMC8:WT) were eliminated from consideration. Select integrators were imported from the STRING database (Version 9.0) (5) and included within the BAP1 network if the gene connected to two or more genes identified within the SILAC experiment.
Mice
Mutant mouse strains were generated using C57BL/6 ES cells. Mice with loxP sites flanking Bap1 exons 4 and 5 were generated by genOway, who crossed the mice to a Flp recombinase deleter strain to remove the neo selection cassette (fig. S1A). These Bap1fl/fl mice were crossed to the inducible general deleter C567BL/6 NTac-Gt(ROSA)26Sortm9(Cre/ESR1)Arte or the constitutive general deleter C57BL/6-Gt(ROSA)26Sortm16(Cre)Arte (Taconic). BAP1.3xFlag mice had a 3xFlag sequence inserted ahead of the Bap1 translation termination codon in exon 17 (fig. S10A). 3xFlag.ARMC8 mice had the 3xFlag sequence inserted after the Armc8 translation initiation codon in exon 1 and were generated by genOway (fig. S10C). The neo selection cassette was removed from both knock-in strains. CreERT2+ mice aged 6–8 wks were injected intraperitoneally with 40 mg/kg tamoxifen dissolved in sunflower oil daily for 5 days. The Genentech Institutional Animal Care and Use Committee approved all protocols.
Genotyping
Bap1 genotyping primers 5' CCA TCA GTG ACT ACT GGG GAG CAA C, 5' ACA GAT GGC TGG GCA CAT CTG, and 5' GAA CCC TCC GTT GCA TAG TGT TG amplified 234 bp WT, 350 bp floxed, and 503 bp KO DNA fragments. Cre primers 5′-GCT AAA CAT GCT TCA TCG TCG GTC and 5′-CCA GAC CAG GCC AGG TAT CTC TG amplified a 582 bp DNA fragment. BAP1.3xFlag genotyping primers 5' CTT CAT ATC CAT GCT GGC TCA G and 5' TGG GCT CTC GTT GAA CTA CTC amplified 441 bp WT and 567 bp KI DNA fragments. Armc8 genotyping primers 5' GAT TCA CCA ATT ATG TTC CCT GGA AG and 5' CTT AAT TTC AGG CAT CAG TAT GTT CCT G amplified 307 bp WT and 404 bp KI fragments.
Flow cytometry
Anti-CD49b (DX5), anti-Gr-1 (RB6–8C5), anti-CD11b (M1/70), anti-B220 (RA3–6B2), anti-CD4 (GK1.5), anti-CD3e (145–2C11), anti-TER-119 (TER-119), and anti-NK1.1 (PK136) antibodies from BD Biosciences were used for negative sorting of lineage− cells. Anti-c-kit (2B8), anti-Sca-1 (D7), anti-CD45.1 (A20), anti-F4/80 (BM8), anti-Ly-6G (1A8), anti-CD34 (8G12), anti-FcγR (2.4G2) and anti-CD16/CD32 were from BD Biosciences. Anti-CD45.2 (104) was from eBioscience. Flow cytometric analyses were done in a LSR II (BD Biosciences) and cell sorting with a FACSAria (BD Biosciences). Data were analyzed with Flow software (Version 9.3). Propidium iodide staining excluded dead cells from analyses and sorts.
Hematology
Blood was obtained from the retro-orbital sinus under anesthesia and collected in tubes containing K3-EDTA. Total peripheral blood cell counts and red blood cell indices were measured using a Cell-Dyn 3700 hematology analyzer (Abbott Laboratories). Blood smears were prepared, air-dried, fixed in methanol and stained with a Modified Wright-Giemsa stain using an automated slide stainer (Leica Autostainer XL).
In vitro colony forming assays
Colony formation assays were done using cytokine supplemented methylcellulose media (Methocult M3434 and M3534, STEMCELL Technologies) according to manufacturer’s instructions. LSK cells were sorted from WT and KO bone marrow at 1 and 3 weeks after tamoxifen treatment and seeded at 1000 cells/3.5 cm plate. Colonies formed in culture were counted after 1 week. Representative colonies from the plates were isolated for cytospins and stained with Wright-Giemsa stain after 10 days in culture. Remaining cells were resuspended in tissue culture DMEM media, counted, and replated (10000 cells/ plate) in M3434 and M3534 media for another 10 days.
Bone marrow transplants
Recipient animals received 2 doses of 525 Rads from a 137Cs source separated by a 4 h interval, and then donor bone marrow cells or lineage-depleted progenitors were injected into the tail vein. Lineage− cells were isolated with a lineage cell depletion kit (Miltenyi Biotec). Reconstituted mice were given water containing 0.11 mg/mL polymyxin B and 1.1 mg/mL neomycin for two weeks and then switched to regular water.
Western Blotting
Whole cell lysates were prepared in NP-40 buffer (1% NP-40, 120 mM NaCl, 50 mM Tris, pH 7.4, 1 mM EDTA, pH 7.4, 20mM NEM, protease and phosphatase inhibitors (Roche)). Anti-BAP1 and anti-HCF1 antibodies were from Bethyl laboratories. Anti-OGT (SQ17 and DM17), anti-actin, and anti-O-GlcNAc antibodies were from Sigma-Aldrich. Anti-HA and anti-myc were from Cell Signaling Technology.
In vitro deubiquitination (DUB) assay
Myc-Flag-human OGT and HA-ubiquitin (ub) plasmids were transfected into HEK293T cells at a ratio of 1:1.5 with Fugene 6. After 48 hours, cells were lysed in NP-40 buffer (1% NP-40, 120 mM NaCl, 50 mM Tris, pH 7.4, 1 mM EDTA, pH 7.4, 20mM cysteine protease inhibitor N-ethylmaleimide (NEM), protease and phosphatase inhibitors (Roche)). Proteins were denatured with 1% SDS and then diluted to 0.05% SDS prior to anti-flag immunoprecipitation. Flag-tagged human BAP1 (amino acids 2–729) was co-expressed with His-tagged human ASXL1 (amino acids 2–365) in Sf9 cells and affinity purified through anti-Flag M2 affinity resin (Sigma) and a Superdex S200 size-exclusion column (GE) in 300 mM NaCl, 50 mM Tris 8.0, and 2 mM β-mercaptoethanol (Buffer A). The BAP1/ASXL1 concentration was calculated by measuring the absorbance at 280 nm, using an extinction coefficient 78,000 M−1 cm−1. To mimic catalytically inactive BAP1, purified BAP1/ASXL1 was pre-treated with 20 mM NEM for 15 minutes at room temperature. Ub-AMC cleavage assays were performed by adding 625 nM BAP1/ASXL1 +/− NEM to 2 mM ub-AMC (Sigma Aldrich) in Buffer A. Reactions were monitored continuously on a Wallac Victor 3V Plate 1420 (Perkin Elmer) plate reader using a 340 nm (12 nm band pass) excitation filter and a 460 nm (60 nm band pass) emission filter. BAP1/ASXL1 was added to HA-ub-conjugated Myc-Flag-OGT for one hour at 37°C. The reaction was terminated with SDS loading dye and boiled for 10 minutes.
Microarray analyses
RNA from Bap1+/+ or Bap1−/− myeloid progenitors (lineage− ScaI− c-Kit+) and LSK cells (lineage− ScaI+ c-Kit+) was isolated using the RNeasy Mini kit (Qiagen) and hybridized to the Mouse v2 Whole Genome OneArray® (Phalanx Biotech). RNA quality was determined utilizing an Agilent 2100 Bioanalyzer (Agilent Technologies) and a NanoDrop spectrophotometer (Thermo Scientific). Only RNA with a RIN of > 7.0, and absorbance ratios A260/A280 > 1.8 and A260/A230 > 1.6 was utilized for further experimentation. RNA was converted to double-stranded cDNA and amplified using in vitro transcription that included amino-ally UTP, and the aRNA product was subsequently conjugated with Cy5™ NHS ester (GEH Lifesciences). Fragmented aRNA was hybridized at 42°C overnight using the HybBag mixing system with 1X OneArray® Hybridization Buffer (Phalanx Biotech), 10 mg/ml sheared salmon sperm DNA (Promega), at a concentration of 25 mg/ml labeled target. After hybridization, the arrays were washed according to the OneArray® protocol.
Raw intensity signals for each microarray were captured using a Molecular Dynamics™ Axon 4100A scanner, measured using GenePixPro™ Software, and stored in GPR format. Expression summary values for all probes were obtained by doing background correction and quantile normalization with the ‘limma’ package from Bioconductor. Statistical analyses of differentially expressed genes were performed using linear models and empirical Bayes moderated statistics as implemented in the ‘limma’ package. Significant genes were defined as those with Benjamini-Hochberg adjusted p-values < 0.1 and fold changes > 2. Gene set enrichment analysis was carried out by using aggregate gene-level t-statistics to calculate the gene-set effect statistics (6), as implemeted in the ‘GSEAlm’ package from Bioconductor. All gene expression data set described in this study will be deposited into Gene Expression Omnibus (GEO) database.
Chromatin immunoprecipitation (ChIP)
Active Motif Epigenetic Services performed ChIP assays. Cells fixed with 1% formaldehyde for 15 min were quenched with 0.125 M lysine, and then swollen in lysis buffer for 30 min on ice. Chromatin was isolated after sonication to shear the DNA to an average length of 300–500 bp. Input genomic DNA was prepared by treating aliquots of chromatin with RNase, proteinase K and heat for de-crosslinking, followed by ethanol precipitation. DNA was quantified in a NanoDrop spectrophotometer. Extrapolation to the original chromatin volume allowed quantitation of the total chromatin yield.
An aliquot of chromatin was precleared with protein G-agarose (Invitrogen). Anti-Flag ChIP was performed with M2 Flag resin (Sigma) at 4°C overnight. Protein G-agarose was used to capture immune complexes involving free antibody. Washed beads were eluted with SDS sample buffer. Complexes were treated with RNase and proteinase K, crosslink’s were reversed at 65°C overnight, and ChIP DNA was extracted with phenol-chloroform prior to ethanol precipitation.
Quantitative PCRs (qPCR) were performed in triplicate on specific genomic regions using SYBR Green Supermix (Bio-Rad). Data were normalized for primer efficiency by carrying out qPCR on input DNA with each primer pair.
ChIP-sequencing
ChIP and input DNAs were prepared for amplification by converting overhangs into phosphorylated blunt ends and adding an adenine to the 3’ ends. Illumina adaptors were added and the library was size-selected (175–225 bp) on an agarose gel. The adaptor-ligated libraries were amplified for 18 cycles. The resulting DNA libraries were purified, quantified, and tested by qPCR at the same specific genomic regions as the original ChIP DNA to assess quality of the amplification reactions. DNA libraries then were sequenced on the Illumina Genome Analyzer II.
Reads were filtered to keep only those with > 70% of bases having Phred quality score of 20 or better. These filtered reads were mapped to Mus musculus genome revision 9 (mm9) by GSNAP (7), allowing maximum of two mismatches. Mean fragment length was estimated using a correlation method implemented in the ‘chipseq’ package from Bioconductor (8). Reads were then extended according to the estimated fragment length, and per base coverage was calculated using the extended reads. WIG files were generated from the coverage for visualization purposes. ChIP-seq enrichment regions for BAP1.3xFlag ChIP were called using MACS 1.4 (9) with a p-value threshold of 1×10−10 and fold enrichment threshold of 5. Mapping enrichment regions to different regions of the genome was performed with Genentech in-house software tools using R/Bioconductor. De novo motif discovery analysis was carried out using HOMER software (10).
BAP1 sequence analysis
Peripheral blood and/or bone marrow was collected from 32 patients with clinicopathologically-confirmed MDS after obtaining informed consent. Matched normal DNA isolated from buccal swabs was available for all patients. All DNA samples were whole genome amplified using F29 polymerase, and then M13-appended BAP1-specific primers were used to amplify and sequence all BAP1 coding exons. PCR products amplified from genomic DNA (20 ng) underwent magnetic bead purification (SPRI, Agencourt Bioscience) and bidirectional sequencing in an ABI 3730 (Agencourt Bioscience) as described (11). Sequence traces were analyzed for missense and nonsense mutations using Mutation Surveyor (Softgenetics, Inc.), and all traces were reviewed manually and with Mutation Surveyor for frameshift mutations. Mutations in BAP1 were annotated according to the predicted effects on BAP1 coding sequence. Non-synonymous mutations were compared to published SNP data (http://www.ncbi.nlm.nih.gov/projects/SNP) such that previously annotated SNPs were not considered pathogenic mutations. Resequencing of non-amplified source germline and tumor DNA validated candidate somatic mutations.
Supplementary Material
Acknowledgments
We thank members of the Dixit and Martin laboratories for advice and discussions, and core laboratories for technical assistance.
References
- 1.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jensen DE, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene. 1998;16:1097. doi: 10.1038/sj.onc.1201861. [DOI] [PubMed] [Google Scholar]
- 3.Jensen DE, Rauscher FJ., 3rd Defining biochemical functions for the BRCA1 tumor suppressor protein: analysis of the BRCA1 binding protein BAP1. Cancer Lett. 1999;(143 Suppl 1):S13. doi: 10.1016/s0304-3835(99)90004-6. [DOI] [PubMed] [Google Scholar]
- 4.Guo G, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44:17. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 5.Abdel-Rahman MH, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet. 2011;48:856. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Testa JR, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet. 2011;43:1022. doi: 10.1038/ng.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner T, et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat Genet. 2011;43:1018. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seibler J, et al. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 10.Beachy SH, Aplan PD. Mouse models of myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24:361. doi: 10.1016/j.hoc.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corey SJ, et al. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007;7:118. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- 12.Klinakis A, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kruger M, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi N, et al. RanBPM, Muskelin, p48EMLP, p44CTLH, and the armadillo-repeat proteins ARMC8alpha and ARMC8beta are components of the CTLH complex. Gene. 2007;396:236. doi: 10.1016/j.gene.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 15.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem. 2009;284:34179. doi: 10.1074/jbc.M109.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheuermann JC, et al. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelsi-Boyer V, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 19.Abdel-Wahab O, et al. Concomitant analysis of EZH2 and ASXL1 mutations in myelofibrosis, chronic myelomonocytic leukemia and blast-phase myeloproliferative neoplasms. Leukemia. 2011;25:1200. doi: 10.1038/leu.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misaghi S, et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol Cell Biol. 2009;29:2181. doi: 10.1128/MCB.01517-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capotosti F, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 22.Daou S, et al. Crosstalk between O-GlcNAcylation and proteolytic cleavage regulates the host cell factor-1 maturation pathway. Proc Natl Acad Sci U S A. 2011;108:2747. doi: 10.1073/pnas.1013822108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristie TM, Liang Y, Vogel JL. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochimica et biophysica acta. 2010;1799:257. doi: 10.1016/j.bbagrm.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pellagatti A, et al. Gene expression profiling in the myelodysplastic syndromes using cDNA microarray technology. Br J Haematol. 2004;125:576. doi: 10.1111/j.1365-2141.2004.04958.x. [DOI] [PubMed] [Google Scholar]
- 25.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44:53. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Phu L, et al. Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.003756. M110003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas W, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 4.Bakalarski CE, et al. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7:4756. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research. 2011;39:D561. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang Z, Gentleman R. Extensions to gene set enrichment. Bioinformatics. 2007;23:306. doi: 10.1093/bioinformatics/btl599. [DOI] [PubMed] [Google Scholar]
- 7.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26:1351. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.