ABSTRACT
Induction of pluripotency in differentiated cells through the exogenous expression of the transcription factors Oct4, Sox2, Klf4 and cellular Myc involves reprogramming at the epigenetic level. Histones and their metabolism governed by histone chaperones constitute an important regulator of epigenetic control. We hypothesized that histone chaperones facilitate or inhibit the course of reprogramming. For the first time, we report here that the downregulation of histone chaperone Aprataxin PNK-like factor (APLF) promotes reprogramming by augmenting the expression of E-cadherin (Cdh1), which is implicated in the mesenchymal-to-epithelial transition (MET) involved in the generation of induced pluripotent stem cells (iPSCs) from mouse embryonic fibroblasts (MEFs). Downregulation of APLF in MEFs expedites the loss of the repressive MacroH2A.1 (encoded by H2afy) histone variant from the Cdh1 promoter and enhances the incorporation of active histone H3me2K4 marks at the promoters of the pluripotency genes Nanog and Klf4, thereby accelerating the process of cellular reprogramming and increasing the efficiency of iPSC generation. We demonstrate a new histone chaperone (APLF)–MET–histone modification cohort that functions in the induction of pluripotency in fibroblasts. This regulatory axis might provide new mechanistic insights into perspectives of epigenetic regulation involved in cancer metastasis.
KEY WORDS: Histone chaperone, APLF, iPSCs, MET, MacroH2A.1, Histone modification
Summary: This study reveals, for the first time, how a histone chaperone, which is also associated with DNA repair machinery, could influence cellular transition and pluripotency and thereby regulate reprogramming.
INTRODUCTION
The discovery of Yamanaka factors that induce pluripotent stem cells through transformation of terminally differentiated fibroblasts into embryonic stem cell (ESC)-like colonies marked a major breakthrough in the field of biology and medicine (Takahashi and Yamanaka, 2006; Takahashi et al., 2007). This transformation is associated with modifications in chromatin structure and in cellular phenotype through an intricate network of transcription and epigenetic factors driving the dynamics required for the generation of induced pluripotent stem cells (iPSCs). Different epigenetic modulators – including histone-modifying enzymes such as Dot1L (Onder et al., 2012), chromatin remodelers such as Chd1 (Gaspar-Maia et al., 2009) and histone variants – have been implicated in the regulation of reprogramming (Shinagawa et al., 2014; Gaspar-Maia et al., 2013). Histone metabolism is governed by histone chaperones (Avvakumov et al., 2011) such as Asf1a, Nap1, Caf1, which also assist in different nuclear processes including repair, replication, transcription and could modulate the post-translational modification of histones (Kim and Haber, 2009; Quivy et al., 2008; Sharma and Nyborg, 2008). They influence different processes during development (Dutta et al., 2010; Majumder et al., 2015) and disease (Corpet et al., 2011). However, the role of histone chaperones in the context of the induction of pluripotency is poorly understood. Recently, histone chaperone Asf1a has been shown to be necessary for the reprogramming of human dermal fibroblasts into iPSCs (Gonzalez-Muñoz et al., 2014), whereas the function of Caf1 has been shown to retain the identity of somatic cells, thereby acting as an obstacle in reprogramming (Cheloufi et al., 2015). In this study, we observed that the expression of the newly found histone chaperone APLF was lower in embryonic stem cells (ESCs) than in mouse embryonic fibroblasts (MEFs). APLF is a recognized DNA-damage-specific histone chaperone, and it could interact with a histones H3 and H4 tetramer or could recruit variants of H2A to the damaged sites (Mehrotra et al., 2011). However, the role of APLF in reprogramming or any other aspect of developmental biology has never been considered. Thus, we focused our study on the role of APLF in reprogramming and investigated the molecular mechanism underlying how downregulation of APLF could influence the induction of pluripotency in fibroblasts. Here, we show that APLF acts as a barrier to the reprogramming process by recruiting MacroH2A.1 (encoded by H2afy) and removing histone H3 dimethylation of lysine 4 (H3me2K4) marks at the genes associated with different facets of reprogramming and that subsequent downregulation of APLF increased the pace and efficiency of generation of iPSCs.
RESULTS
APLF upregulation is associated with a decrease in expression of pluripotency factors
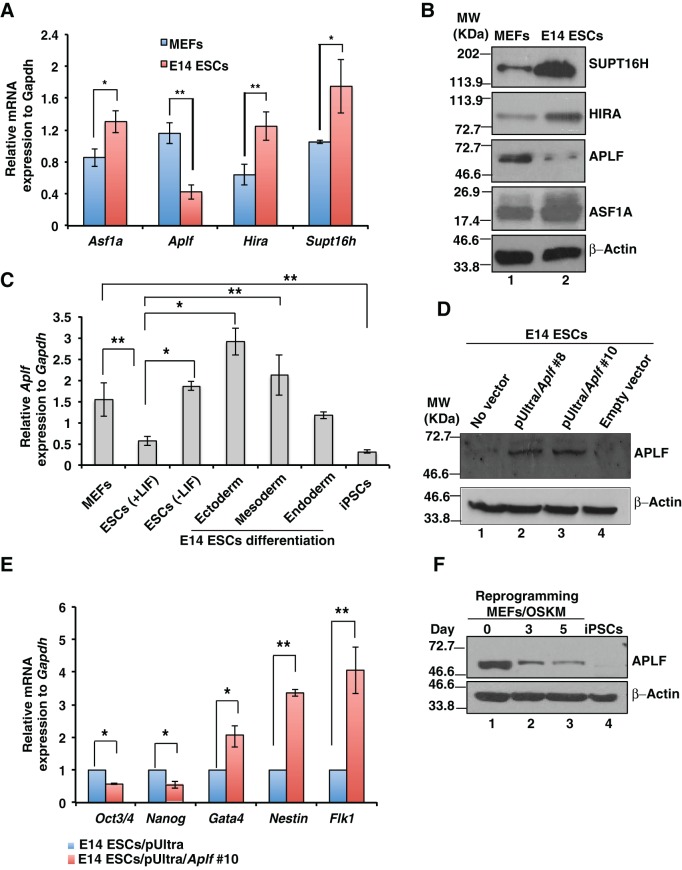
To understand the role of histone chaperones in the context of inducing pluripotency, we determined their expression in MEFs and in the mouse E14 ESC line. MEFs represent the terminally differentiated cells and constitute the point of origin for the generation of iPSCs in the present study, whereas iPSCs, the end product of the reprogramming process, closely resemble ESCs. So, we considered these two as our model systems to determine whether expressions of histone chaperones are context specific or not. Quantitative reverse-transcriptase PCR (qRT-PCR) and western blot analyses indicated that expression of histone chaperones Asf1a, Hira and Supt16h was significantly induced in E14 ESCs compared to in MEFs (Fig. 1A,B), whereas Daxx, Nap1, Caf1p150 and nucleolin expression remained unaltered (Fig. S1A). However, histone chaperone APLF was significantly downregulated in E14 ES cells, both at the mRNA and protein levels (Fig. 1A,B). Upon differentiation of ESCs either in the absence of leukemia inhibitory factor (LIF) or in the presence of factors that drive differentiations towards distinct lineages (as mentioned in the Materials and Methods), Aplf expression was induced (Fig. 1C). So, we reasoned that upregulation of APLF in ESCs might favor differentiation or decreased expression of pluripotency genes. Hence, we ectopically expressed Aplf in E14 ESCs and confirmed the overexpression by western blotting (Fig. 1D). qRT-PCR analysis demonstrated that upon overexpression of APLF, Oct3/4 (also known as Pou5f1) and Nanog expression was significantly downregulated in ESCs (Fig. 1E), whereas lineage-specific markers fetal lever kinase1 (Flk1), nestin and Gata4 were significantly upregulated (Fig. 1E). Thus, APLF acts as a negative regulator of the expression of transcription factors related to pluripotency. Next, we investigated the endogenous level of APLF present in cells during the transition from MEFs to iPSCs. MEFs were transduced with lentiviral particles expressing Oct4, Sox2, Klf4 and Myc (OSKM) under the influence of a Tet-operator (Carey et al., 2009). Western blot analysis demonstrated that concomitant with an increase in the number of days of reprogramming, the level of APLF decreased significantly until the generation of iPSCs (Fig. 1F). Therefore, we inferred that downregulation of APLF in MEFs might enhance the reprogramming process.
Fig. 1.
APLF upregulation is associated with a decrease in expression of pluripotency factors. (A,B) MEFs and feeder-free E14 ESCs were cultured for 3 days, and mRNA and protein were extracted to determine the expression of histone chaperones by performing qRT-PCR analyses and western blot analysis. (C) E14 ESCs were differentiated in the absence of LIF and towards endodermal, mesodermal and ectodermal lineages. qRT-PCR analysis showed an increase in Aplf levels in cells that had been differentiated from E14 ESCs. (D) Full-length mouse Aplf cDNA was PCR-amplified from a cDNA library derived from mRNA isolated from MEFs and was cloned into the pUltra lentiviral vector to generate lentiviral particles to transduce E14 ESCs. Western blot analysis confirmed the ectopic overexpression of APLF in clones #8 and #10. (E) E14 ESCs with empty vector and Aplf cDNA (clone 10) were analyzed for the expression of pluripotency genes Oct3/4 and Nanog, and lineage markers Gata4, Flk1 and nestin by performing qRT-PCR analysis. Error bars represent the s.e.m. for three independent experiments. (F) MEFs were transduced with lentiviral particles expressing OSKM and Tet to generate iPSCs. Cell lysates at different days of reprogramming were analyzed for the expression of APLF by western blotting. Error bars are s.e.m., n=3, *P<0.05, **P<0.01 (Student's t-test).
Downregulation of APLF enhances reprogramming of fibroblasts
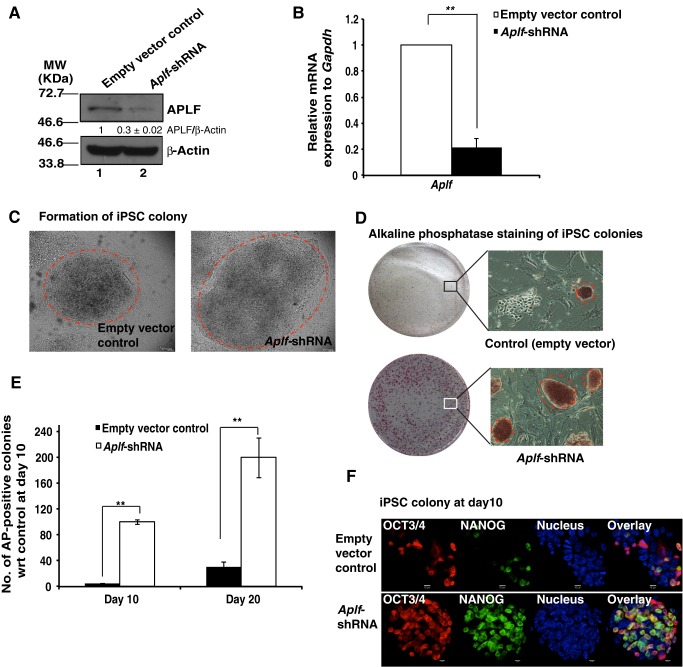
MEFs were transduced with lentiviral particles expressing small hairpin (sh)RNA against Aplf (Aplf shRNA) or an empty plko.1 vector. The Aplf-shRNA-harboring clone from Sigma-Aldrich (SHCLNG-NM_024251; TRCN0000250398) had been previously validated. The extent of knockdown of APLF in puromycin-selected MEFs was confirmed both at the protein (Fig. 2A) and mRNA levels (Fig. 2B). Western blot analysis showed that the knockdown was, on average, 70% of the total amount of APLF in control cells. To generate iPSCs, Aplf-knockdown (Aplf-kd) and control MEFs (with empty vector) were transduced with lentiviral particles expressing OSKM and Tet (Carey et al., 2009). The follow-up observation is a representation of three independent sets of experiments. On average, Aplf-kd MEFs showed the first appearance of ESC-like colonies within 6 days, whereas control MEFs required 8 days to form the first few colonies. The iPSC-like colonies were picked up at the 10th (10±2 days) and 20th (20±2 days) days after transduction for Aplf knockdown and control MEFs, respectively. No morphologic differences were observed between control and Aplf-kd iPSC-like colonies (Fig. 2C). Alkaline phosphatase staining demonstrated the enhanced number of pluripotent colonies formed from Aplf-kd MEFs (Fig. 2D,E). But, the efficiency for the derivation of colonies from Aplf-kd MEFs was enhanced to 0.2–0.25% as compared to 0.01–0.02% for control MEFs. Differential expression patterns of NANOG and OCT3/4 in the iPSC colonies were apparent in control and Aplf-kd MEFs before day 10 of reprogramming, further confirming the delay in the formation of mature iPSC colonies from control cells (Fig. 2F). The difference in time span for colony formation, colony pick-up and efficiency in generation of iPSCs was significant between control and Aplf-kd cells. These results suggest that APLF downregulation substantially improved the reprogramming process by enhancing the pace of colony formation and the number of colonies formed, and by reducing the time needed to generate the iPSC colonies.
Fig. 2.
Downregulation of APLF enhances reprogramming of fibroblasts. (A) MEFs were transduced with lentiviral particles expressing shRNA against Aplf (Aplf-shRNA) or empty plko.1 vector (empty vector control). Extent of knockdown was measured at the (A) protein and (B) mRNA level. The band intensity of blots was measured by using ImageJ software. The relative intensity of APLF to β-actin immunoreactive bands was determined, and the value for the control was normalized to 1. The intensity is the average of three independent experiments. (C) Control (with empty plko.1 vector) and Aplf-kd (expressing Aplf-shRNA) MEFs were transduced with polycistronic vector expressing OSKM under the influence of the Tet operator. Doxycycline (2 µg/ml) induced the expression of OSKM. Micrographs show iPSC colonies generated from control (left panel) and Aplf-kd MEFs (right panel). Dashed circles mark iPSC colonies. (D) Alkaline phosphatase staining of iPSC clones in control (upper panel) and Aplf-kd (lower panel) cells. Micrographs of the alkaline-phosphatase-positive clones at 10× magnification are also included. Dashed circles indicate pluripotent iPSC colonies. Similar colonies only were counted as alkaline-phosphatase-positive clones. (E) Bar graph demonstrating the number of alkaline-phosphatase-positive clones formed at day 10 and day 20 of reprogramming from control and Aplf-kd cells with respect to control set at day 10. (F) Immunofluorescence study of the expression of NANOG and OCT3/4 in control and Aplf-kd cells at day 10 of reprogramming. Error bars are s.e.m., n=3, **P<0.01 (Student's t-test).
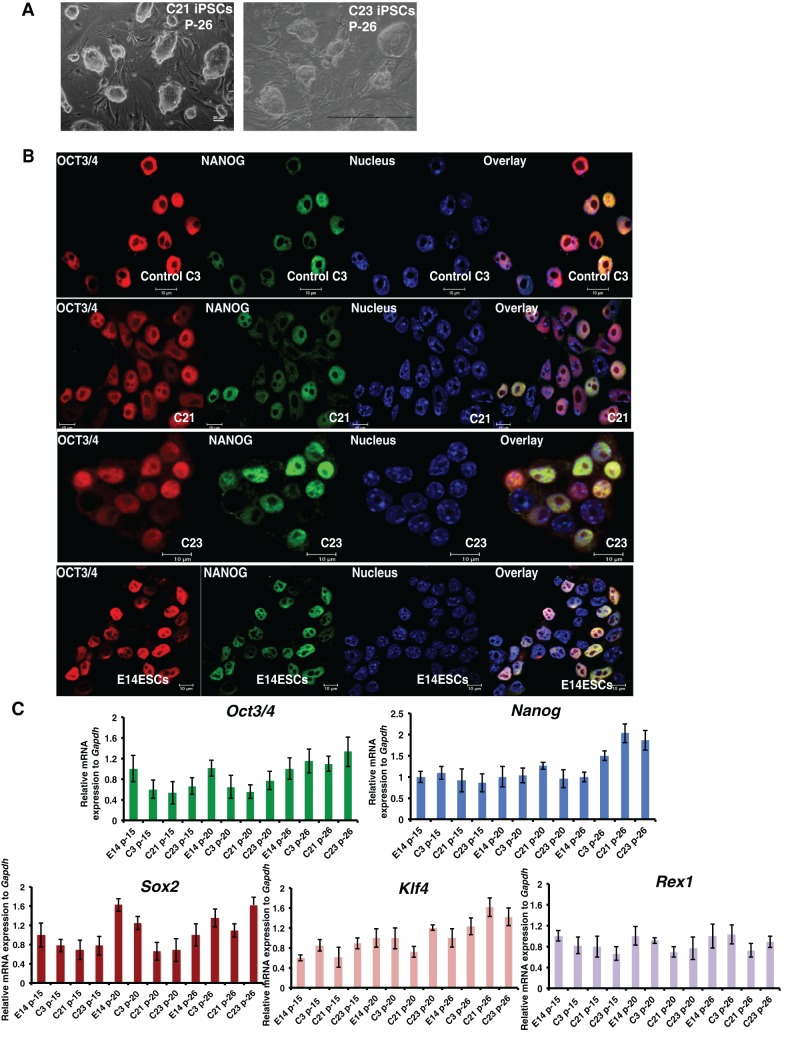
A total number of 50 iPSC colonies were derived from Aplf-kd MEFs and cultured for 15 continuous passages. Among the 50 clones, clones #21 and #23 (hereafter referred to as C21 and C23) were used for further analyses. iPSC colonies that had formed from control MEFs (empty vector) were passaged multiple times. Control clone #3, referred to as C3, was used for further analysis. Micrographs of C21 and C23 at passage 26 showed a normal ESC-like morphology of the colonies (Fig. 3A). OCT4 and NANOG expression in C21 and C23 iPSCs was similar to that in the control ESCs and iPSCs (Fig. 3B). The mRNA expression for Oct3/4, Nanog, Sox2, Klf4 and Rex1 in C21 and C23 iPSCs was similar to that in control C3 iPSCs and E14 ESCs at different passages (Fig. 3C).
Fig. 3.
iPSC colonies formed from Aplf-kd cells express pluripotency genes. (A) C21 and C23 iPSC clones at passage 26. Scale bar: 100 µm. (B) iPSCs from colonies C21 and C23 (generated from Aplf-kd MEFs), and C3 (generated from control plko.1 MEFs) were analyzed for the expression of the pluripotency proteins OCT3/4 and NANOG by performing immunofluorescence analysis. E14 ESCs were used as control. (C) qRT-PCR analysis of different pluripotency genes on samples generated from different passages (p-) of E14 ESCs and of iPSCs from C21, C23 and C3. Error bars represent s.e.m. for three independent experiments.
iPSCs generated from Aplf-kd MEFs are pluripotent
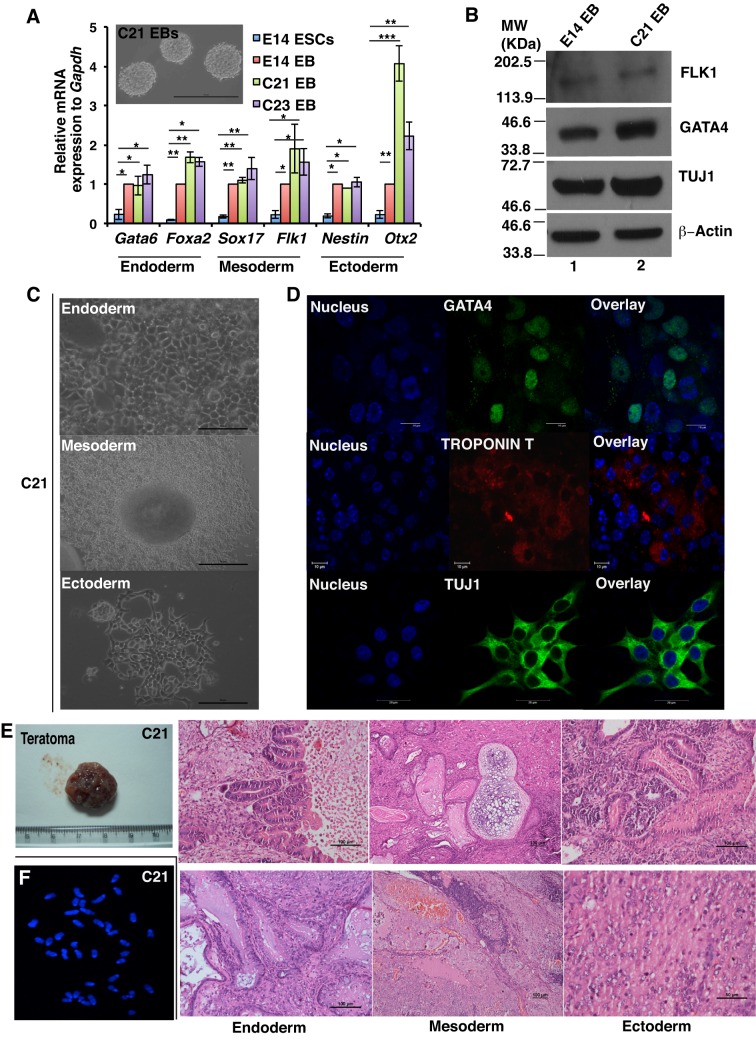
To test the pluripotent nature of iPSCs, C21 and C23 iPSC clones were differentiated to produce embryoid bodies for 5 days (Fig. 4A, inset). Embryoid bodies generated from C21 and C23 iPSCs expressed genes associated with different germ layers – namely endoderm, mesoderm and ectoderm – at the mRNA level (Fig. 4A). Western blot analysis further confirmed the expression of differentiation markers in embryoid bodies that had been generated from C21 iPSCs (Fig. 4B). Embryoid bodies formed from C21 and control C3 iPSCs were further differentiated on Matrigel to stimulate endoderm lineage (Fig. 4C; Fig. S1B, upper panel), at which stage they expressed GATA4 (Fig. 4D; Fig. S1C, upper panel). Upon differentiation into mesoderm lineage (Fig. 4C; Fig. S1B, middle panel), these cells expressed troponin T (encoded by Tnnt2) and formed beating cardiomyocytes (Fig. 4D; Movie 1, Movie 2). When differentiated into the ectoderm lineage, neural-rosette-like clusters were formed (Fig. 4C; Fig. S1A, lower panel), which expressed TUJ1 (Fig. 4D; Fig. S1C, lower panel). Consequently, these in vitro differentiation assays demonstrated that iPSCs generated from Aplf-kd MEFs were as pluripotent as control MEFs. To demonstrate pluripotency in vivo, C21 iPSCs from passage 26 were injected into SCID mice to generate teratomas (Fig. 4E). The teratomas were collected after 5 weeks for subsequent analyses (Fig. 4E). Hematoxylin and eosin (HE)-stained teratoma sections demonstrated the presence of different germ layers comprising intestinal epithelium or goblet cells (endoderm), cartilage or blood (mesoderm), and neural patches or central nervous system (ectoderm) from clone C21 (Fig. 4E). We determined the genomic stability of the C21 iPSC clone by studying the metaphase spread of the chromatin due its role in DNA repair. A normal intact chromosome number of 40 was observed in the C21 clone (Fig. 4F). These results indicate that iPSCs derived from Aplf-kd MEFs retain their developmental potential.
Fig. 4.
iPSC colonies formed from Aplf-kd cells exhibit differentiation potential in vitro and in vivo. (A) C21 and C23 iPSC clones and E14 ESCs were differentiated for 5 days to generate embryoid bodies (EBs) in a LIF-free medium. qRT-PCR analysis for the expression different lineage markers was performed. Inset, embryoid bodies formed from C21 iPSCs. (B) E14 embryoid bodies and C21 embryoid bodies were further analyzed for the expression of GATA4 (endoderm), FLK1 (mesoderm) and TUJ1 (ectoderm) by western blotting. (C) C21 embryoid bodies were differentiated into the endoderm lineage by culturing on Matrigel-coated plates in the presence of serum and activin A for 5 days. A scale-like appearance indicated the formation of endodermal lineage cells (upper panel). For the mesodermal lineage, embryoid bodies were cultured on gelatin-coated plates in the presence of 10% endothelial-specific serum for 10 days to form beating foci-like structures (middle panel). For neural differentiation, embryoid bodies were cultured in N2B27 medium for 3 days with 2 µM retinoic acid and, for the next 4 days of culture, in the absence of retinoic acid (lower panel), generating neural progenitors. (D) Immunofluorescence study for the expression of GATA4, Troponin T and TUJ1 in the cells described in C. Nucleus, Hoechst 33258, blue. (E) Around 2×106 C21 iPSCs from passage 26 were injected into SCID mice in dorsal regions. After 5 weeks, teratomas were formed on both sides. Paraffin sections of teratoma were stained with H&E. The sections demonstrated the presence of endoderm, mesoderm and ectoderm lineages. (F) Metaphase spread of chromosomes from C21 iPSCs were stained with Hoechst 33258. Error bars represent s.e.m., n=3, *P<0.05, **P<0.01, ***P<0.001 (Student's t-test). Scale bars: 100 µm (A, inset; E, endoderm, mesoderm and upper ectoderm panels); 50 µm (C; E, lower ectoderm panels); 10 µm (D, upper and middle panels); 20 µm (D, lower panels).
Aplf knockdown does not compromise the DNA repair mechanism in iPSCs
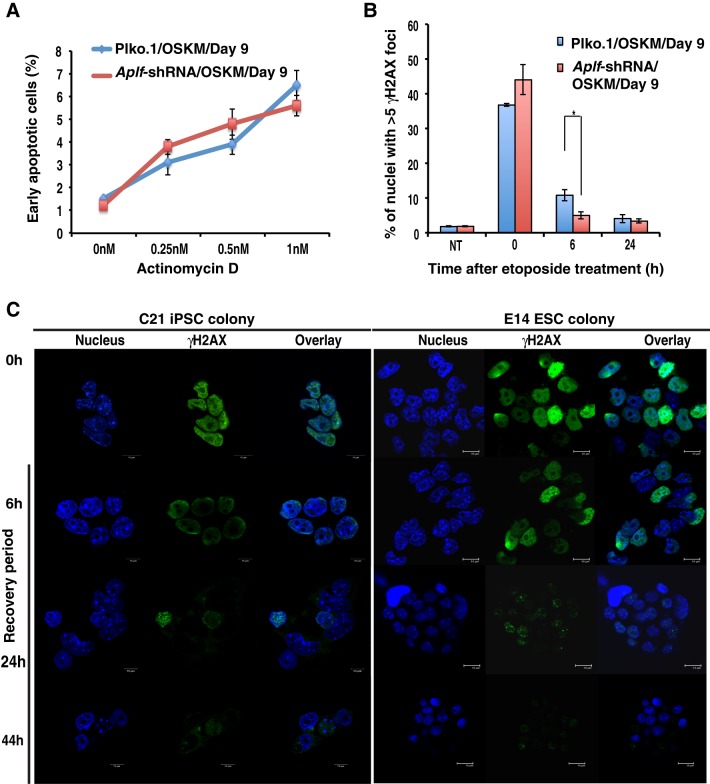
Reprogramming is often associated with the development of genomic instability (Blasco et al., 2011), which happens to be one of the main concerns in iPSC technology. APLF is an important constituent of the non-homologous end joining (NHEJ)-mediated DNA damage repair machinery (Rulten et al., 2008; Grundy et al., 2013), and its downregulation in human cells could sensitize the cells to various DNA-damaging agents (Macrae et al., 2008). In order to test whether Aplf knockdown induces DNA repair defects, the cells were first challenged with actinomycin D at different concentrations and subjected to an apoptosis assay. Actinomycin D intercalates into DNA (Sobell, 1985) and thus induces blockage during replication and transcription. We observed no significant difference in cellular apoptosis between control and Aplf-kd cells during reprogramming at day 9 (Fig. 5A). Next, we analyzed the same set of cells for their responses to the DNA-damaging agent etoposide, which induces DNA double-strand breaks (DSB) (Tichy et al., 2013). Phosphorylation of residue Ser139 of H2AX (γH2AX) is an extremely rapid and ubiquitous event after DNA DSB formation and has been extensively studied in APLF and NHEJ contexts (Fenton et al., 2013; Tong et al., 2016). An etoposide concentration profile (Shimizu et al., 2010; Tichy et al., 2013; Jamil et al., 2015) was titrated in control and Aplf-kd MEFs. Upon exposure to various concentrations of etoposide, γH2AX foci were counted at 0 h and after 24 h of recovery from etoposide treatment, which gives a measure of the DNA damage repair competency of the cells (Tichy et al., 2013). At a concentration of 10 µM, we observed a significant increase in the number of γH2AX-foci-positive cells in Aplf-kd MEFs compared to in control MEFs at the 0 h recovery time point (Fig. S1D), whereas at the higher concentration, no significant difference existed, although the number of γH2AX-foci-positive cells increased (Fig. S1D). Subsequently, we used 10 µM etoposide for the next set of experiments. During reprogramming at day 9, control and Aplf-kd cells were treated with etoposide for 4 h and were allowed to recover for 0, 6 and 24 h. Cells with ≥5 foci were considered as positive. No significant differences in γH2AX foci formation were observed in control versus Aplf-kd cells (Fig. 5B). The recovery of control cells was slower than that of the Aplf-kd cells at the 6-h time point but eventually it equalized across the two cells lines at 24 h (Fig. 5B). Next, we investigated whether the repair capacity was retained in the iPSC clones derived from Aplf-kd MEFs. We analyzed the C21 iPSCs (Fig. 5C, left panel) and E14 ESCs (Fig. 5C, right panel) in response to etoposide treatment. We observed that, within a recovery period of 24 h, the C21 iPSCs demonstrated considerable downregulation of γH2AX levels as compared to that after a recovery period of 6 h (Fig. 5C). After 44 h, the intensity of γH2AX foci was almost non-detectable in C21 iPSCs and in E14 ESCs (Fig. 5C). Western blot analysis for γH2AX levels corroborated the γH2AX staining in control and Aplf-kd cells (Fig. S1E). Thus, APLF downregulation does not impair the DNA repair capacity of cells during reprogramming as measured by γH2AX foci formation in response to DNA-DSB-inducing agents. Next, we investigated whether downregulation of APLF results in any arrest in cell proliferation.
Fig. 5.
Aplf knockdown does not compromise DNA repair competency in iPSCs. (A) Control and Aplf-kd MEFs at day 9 of reprogramming were exposed to different concentrations of actinomycin D for 16 h. Annexin-V-positive cells were counted to determine the early apoptotic cell population. (B) The same set of cells described in A were treated with 10 µM etoposide for 4 h to induce DNA damage. After 4 h, the etoposide-containing medium was replaced with normal ESC culture medium, and cells were allowed to recover. At 0 h and at different time points of recovery, cells were fixed and analyzed for the presence of γH2AX foci by immunofluorescence staining. Nuclei with ≥5 foci were counted as positive. NT, non-treated. (C) C21 iPSCs and E14 ESCs at passage 26 were grown in a 24-well plate for 16 h and treated with 10 µM etoposide in a similar manner to that described above. At 0 h and at different points of recovery, cells were fixed and analyzed for γH2AX levels by immunofluorescence. Representative colonies are shown. Nucleus, Hoechst 33258, blue. Error bars are s.e.m., n=3, *P<0.05 (Student's t-test).
APLF downregulation does not induce cellular arrest
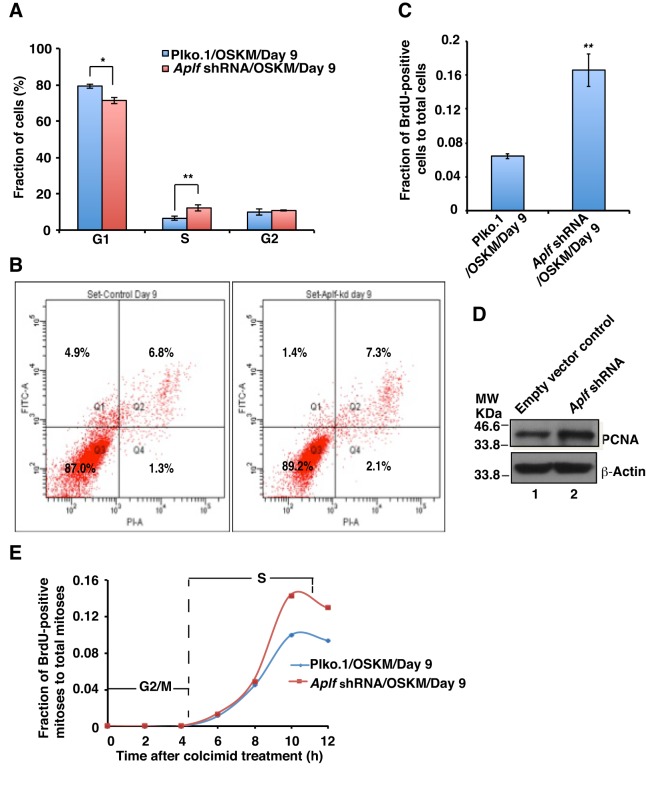
To investigate the inhibitory role of APLF in reprogramming, we determined the effect of Aplf knockdown at the cellular level. Cell cycle analyses of control and Aplf-kd MEFs, on day 9 of reprogramming, were performed by using flow cytometry after staining with propidium iodide. We observed a significant decrease in the G1 cell population and an increase in the S-phase cell population in Aplf-kd cells compared to control, but no significant difference was observed in G2/M phase cells (Fig. 6A). Accumulation of cells in S-phase indicates either increased proliferation to complete the cell cycle more quickly or arrest with impaired repair, consequently leading to cell death by apoptosis. Annexin-V and FITC staining followed by flow cytometry analysis revealed no apoptosis in either the control (Fig. 6B, left panel) or Aplf-kd (Fig. 6B, right panel) cell populations (4.9% vs 1.4%, respectively) at day 9 of reprogramming, thus ruling out the possibility of apoptosis induced as a result of a defective DNA damage response due to Aplf-knockdown.
Fig. 6.
Aplf downregulation does not induce cellular arrest. (A) Cell cycle analysis. Control and Aplf-kd cells at day 9 were stained with propidium iodide and analyzed using flow cytometry. (B) Apoptosis assay. Control (left panel) and Aplf-kd (right panel) cells at day 9 were stained with Annexin-V–FITC and propidium iodide, followed by FACS analysis. Q1 represents FITC-labeled early apoptotic cells. (C) BrdU incorporation into control and Aplf-kd cells during reprogramming. Cells were pulsed with BrdU and analyzed for BrdU and Hoechst 33258 staining by immunofluorescence analysis. BrdU-positive nuclei were counted and expressed as the fraction of total nuclei present. ∼1000 cells were counted for each set. (D) The same set of cells as described in A,B were analyzed for the expression of PCNA by western blotting. (E) Determination of cell cycle phase length. Control and Aplf-kd cells were treated with BrdU in a similar manner as described in C and treated with colcemid (100 ng/ml) for 12 h in ESC culture medium. At each interval of 2 h, cells were fixed to analyze the BrdU staining and observed using fluorescence microscopy to enumerate the BrdU-positive mitoses. Plot represents the fraction of cells undergoing BrdU-positive mitoses (of the total mitoses) over time. For each time point, 30–40 mitoses were analyzed. Error bars are s.e.m., n=3, *P<0.05, **P<0.01 (Student's t-test).
Next, to investigate the possibility of proliferation over cellular arrest, we measured the incorporation of 5-bromo-2′-deoxyuridine (BrdU, a synthetic thymidine analogue) by control and Aplf-kd cells during reprogramming. Proliferating cells can incorporate BrdU in S-phase of the cell cycle (Kimura et al., 2016). Day 9 cells were pulsed with BrdU, and the level of BrdU incorporation was determined by immunofluorescence analysis (Becker et al., 2006). The Aplf-kd cell population demonstrated a significant increase in BrdU-positive cells (expressed as BrdU-positive nuclei relative to the total number of nuclei) compared to control cells (Fig. 6C; Fig. S2A). Further, expression of an S-phase-specific proliferation marker, proliferating cell nuclear antigen (PCNA) (Kurki et al., 1986), was enhanced in Aplf-kd cells in comparison to in control cells (Fig. 6D). But, are these highly proliferating cells more prone to DNA damage? To answer this, we simply co-stained the BrdU-incorporated cells for γH2AX. The extent of γH2AX foci formation was very low among BrdU-positive cells and similar in control and Aplf-kd cells (Fig. S2B). Next, we asked whether the enhanced proliferation reduced the length of cell cycle phases. To determine that, we analyzed BrdU incorporation in cells that had been arrested at mitosis by treatment with colcemid. Cells in G2/M phase are the first to enter mitosis but do not incorporate BrdU; however, cells in the late S-phase acquire BrdU and continue to do so until the time they reach the peak value shown in Fig. 6E, representing the fraction of BrdU-positive mitoses relative to the total mitoses versus time. The time spanning from exposure to colcemid until the appearance of BrdU-positive mitoses is the length of G2/M phase. Both control and Aplf-kd cells had a ∼7-h S-phase and a ∼4-h G2/M phase (Fig. 6E; Fig. S2C). The plot showed no variation in duration of phases of cell cycle, except for the increased percentage of BrdU-positive mitoses in Aplf-kd cells (Fig. 6E); essentially Aplf knockdown in MEFs did not induce any cellular arrest.
Next, we investigated the molecular mechanism involved in the regulation of reprogramming by APLF.
APLF regulates genes that are implicated in MET during the generation of iPSCs from fibroblasts
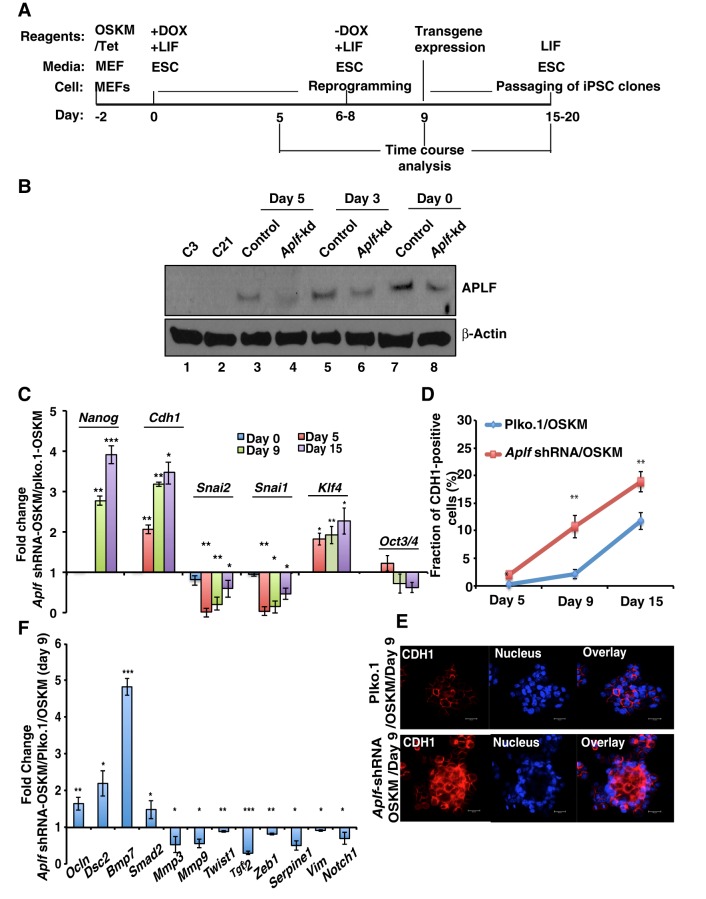
Having shown a new inhibitory role for APLF in pluripotency, we examined the possible mechanisms that could be altered through reduced expression of APLF. During the course of iPSC generation (Fig. 7A), we observed that timing for the formation of colonies significantly varied in control and Aplf-kd cells and so determined the level of APLF during the stages of reprogramming. As expected, reduction of APLF levels in Aplf-kd cells over time was faster than that observed for control cells (Fig. 7B). At day 5, the amount of APLF had been reduced considerably compared to that seen at day 0, and it can be recalled here that the average time required to form the first few colonies from Aplf-kd MEFs was around 6 days, whereas in control cells it was around 8 days. As the timing in appearance of iPSC colonies was affected, we focused on genes implicated in the initiation phase along with pluripotency factors that are activated during the maturation phase. During the generation of iPSCs from fibroblasts, cells undergo a morphological change from a mesenchymal-to-epithelial phenotype, which constitutes one of the vital steps in the initiation phase of reprogramming. This phenomenon is associated with the upregulation of epithelial cell markers and downregulation of mesenchymal cell markers (Li et al., 2010). In the time course analysis mesenchymal-to-epithelial transition (MET) and epithelial-to-mesenchymal (EMT) genes showed differential expression patterns in control and Aplf-kd MEFs. Expression of Cdh1, which is upregulated during MET, was significantly higher in Aplf-kd cells compared to the control cells within 5 days (Fig. 7C), whereas the EMT markers Snai2 and Snai1 were significantly downregulated in Aplf-kd cells compared to in control MEFs (Fig. 7C). Fluorescence-activated cell sorting (FACS) analysis demonstrated that the fraction of CDH1-positive cells in the Aplf-kd population was significantly larger than in the control cell population at any given point of time during reprogramming (Fig. 7D,E).
Fig. 7.
APLF regulates genes implicated in MET during the generation of iPSCs from fibroblasts. (A) Schematic representation for the time frame (not to scale) for the generation of iPSCs. Dox, doxycycline. (B) Western blot analysis for the expression of APLF in control and Aplf-kd MEFs at different stages of reprogramming. (C) qRT-PCR analysis was performed from samples collected at different days of reprogramming, and fold changes in gene expression were calculated in Aplf-kd cells relative to those in control cells. (D) CDH1-positive cells were determined by FACS analysis at different stages of reprogramming. (E) Immunofluorescence study of the expression of CDH1 in control and Aplf-kd MEFs at day 9. Nucleus, Hoechst 33258. (F) The same set of samples as described in E were analyzed for the expression of EMT-specific genes by performing qRT-PCR. Bar graphs represent the fold change in expression relative to in cells without shRNA. Error bars are s.e.m., n=3, *P<0.05, **P<0.01, ***P<0.001 (Student's t-test). Scale bar: 20 µm.
In the case of pluripotency genes, Nanog expression was evidently induced more than 2.5-fold in Aplf-kd cells as compared to the control cells, only on day 9 after induction with OSKM (Fig. 7C). Klf4 was induced around twofold more on day 9 of induction in Aplf-kd cells (Fig. 7C). However, no significant difference was observed in Oct3/4 expression in control and Aplf-kd cells (Fig. 7C). On appearance of iPSC colonies at day 6–8, doxycycline-supplemented ESC medium was replaced with normal ESC medium for further culturing of these cells (Fig. 7A). In the absence of doxycycline, we would not expect any noticeable contribution from ectopic or viral genes toward the expression of different pluripotency factors. The viral transgene expression of OSKM using the E2A-Myc primer was almost undetectable (Fig. S1F) (Carey et al., 2009); therefore, expression of pluripotency factors is solely endogenously derived from the reprogrammed cells.
qRT-PCR analyses demonstrated that in addition to the EMT markers mentioned above, other classic EMT markers Twist1, Zeb1, Mmp3, Mmp9 and Serpine1 were significantly downregulated in Aplf-kd cells (Fig. 7F), whereas genes associated with induction of MET, including Ocln and Dsc2, were significantly upregulated (Fig. 7F). Interestingly, expression of few signaling molecules were altered in Aplf-kd cells in comparison to the control cells. Notch1, implicated in EMT in cancer cells, was downregulated, whereas Smad2, implicated in MET, was upregulated in Aplf-kd cells (Fig. 7F). The analysis showed upregulation of Bmp7 and downregulation of Tgfb2 in Aplf-kd cells compared to control cells (Fig. 7F), which suggests an auxiliary role of MET in the generation of iPSCs from Aplf-kd MEFs (Samavarchi-Tehrani et al., 2010). Induction of MET improved the efficiency in the generation of iPSCs (Li et al., 2010).
We next questioned how APLF could regulate these genes within the context of cellular reprogramming.
Expedited loss of MacroH2A.1 variants from the E-cadherin promoter in APLF-depleted fibroblasts
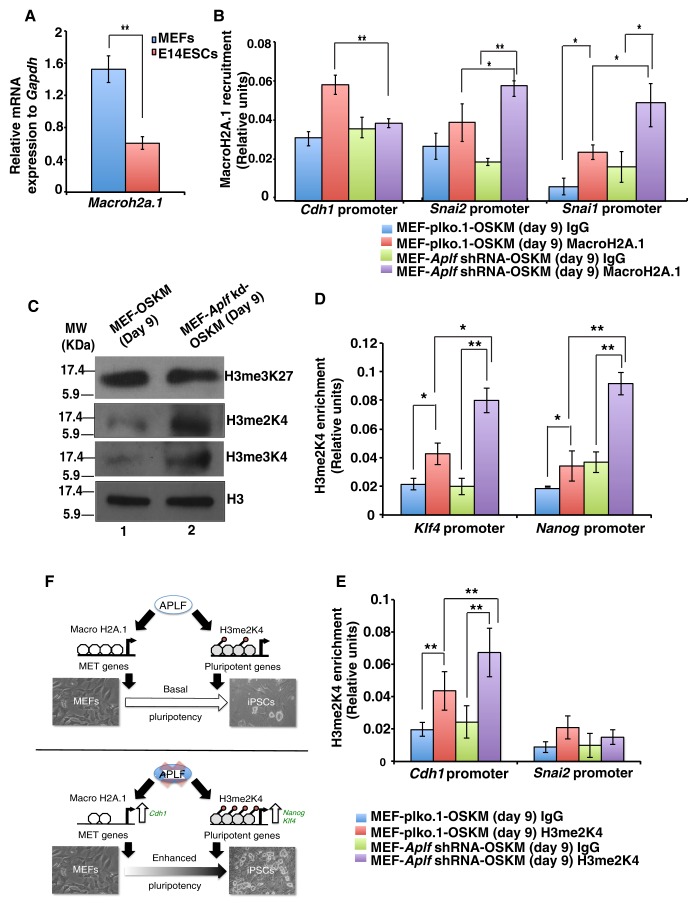
It is known that histone chaperones can recruit, exchange or replace different histone variants in a replication-dependent or -independent manner (Avvakumov et al., 2011). MacroH2A.1 is a repressive mark that is associated with inactive chromatin, and earlier studies have demonstrated that isoforms of the histone variant MacroH2A can act as a barrier to the reprogramming of fibroblasts to iPSCs (Gaspar-Maia et al., 2013). APLF can recruit the MacroH2A.1 histone variant in response to DNA damage induced by lasers (Mehrotra et al., 2011). We studied whether knockdown of APLF could suppress MacroH2A.1 incorporation at the chromatin level and hence whether this might be responsible for the more efficient reprogramming of MEFs. qRT-PCR analyses showed that the gene encoding the MacroH2A.1 variant was significantly downregulated in E14 ESCs as compared to in MEFs (Fig. 8A). Additionally, in MEFs, MacroH2A.1 physically interacted with APLF (Fig. S3A). Because of its known role as a repressor of chromatin, we hypothesized that an expedited loss of MacroH2A.1 from the MET genes in Aplf-kd MEFs could enhance reprogramming. Chromatin immunoprecipitation (ChIP) analyses of cells after 9 days of reprogramming showed that the level of MacroH2A.1 was significantly downregulated at the Cdh1 promoter in Aplf-kd versus control cells (Fig. 8B). At the promoters of the EMT markers Snai1 and Snai2, a significant enrichment in MacroH2A.1 levels was observed in Aplf-kd cells (Fig. 8B). The recruitment pattern of MacroH2A.1 further validates the differences in the expression of EMT and MET genes noted above (Fig. 7C). To gain further insight into the molecular mechanism, we analyzed the recruitment of MacroH2A.1 at the pluripotent and lineage-specific genes as well. No significant difference was observed at the Nanog or Klf4 promoters and Oct3/4 proximal enhancer (Fig. S3B), or at the promoters of developmental genes such as Gata4 and nestin in control and Aplf-kd cells (Fig. S3C). In sharp contrast, a significant difference was observed in the pattern of MacroH2A.1 recruitment at the α-smooth muscle actin (Acta2; herein α-Sma) promoter (Fig. S3C), a gene frequently used as a fibroblast marker. Enrichment of MacroH2A.1 at the α-Sma promoter in Aplf-kd cells indicates reduced expression of α-Sma with a loss of fibroblast character or phenotype.
Fig. 8.
Expedited loss of MacroH2A.1 variants and increased levels of H3me2K4 in APLF-depleted fibroblasts enhances reprogramming. (A) qRT-PCR analysis for the gene expression of Macroh2A.1 in MEFs and E14 ESCs. (B) ChIP analysis was performed with control (MEF-plko.1-OSKM) and Aplf-kd (MEF-Aplf shRNA-OSKM) cells at day 9 of reprogramming. The plots represent the level of MacroH2A.1 variant at different promoters. IgG was used as the negative control. (C) The same sets of cells as described in B were analyzed for the presence of different histone modifications. (D,E) The same sets of cells in C were analyzed for enrichment of H3me2K4 at different loci by performing ChIP analyses. (F) Proposed model for the regulation of reprogramming by APLF. Error bars are s.e.m., n=3, *P<0.05, **P<0.01 (Student's t-test).
With no significant alteration in the MacroH2A.1 levels, we wondered which mechanism might be responsible for the upregulation of pluripotency genes in Aplf-deficient MEFs that could contribute to the enhanced expression of Nanog and Klf4 (Fig. 7C).
Downregulation of APLF favors enrichment of H3me2K4 marks at pluripotency genes during reprogramming
Recently, histone chaperone ASF1A has been reported to be involved in the acetylation of histone H3 residue K56, which in turn activates pluripotency genes (Gonzalez-Muñoz et al., 2014). Moreover, there are other instances in which histone chaperones mediate change in the histone modification pattern in the context of development (Dutta et al., 2010). Bivalent chromatin comprising histone H3 methylated at residues K4 and K27 constitutes the signature of pluripotent ESCs. We therefore determined the status of different histone modification marks related to pluripotency in control and Aplf-kd MEFs during reprogramming. Western blot analyses of the bivalent chromatin marks demonstrated that active histone methylation marks of histone H3 trimethylation at K4 and dimethylation at K4 (H3me2K4) were enhanced in Aplf-kd OSKM-expressing cells (Fig. 8C). We reasoned that an increased level of active histone modification marks might contribute towards the induction in expression of pluripotency genes in Aplf-kd cells. A similar pattern was observed in control and Aplf-kd MEFs (Fig. S3D). But, upon downregulation of APLF, no change in the levels of histone H3 trimethylation at K27 was observed (Fig. 8C; Fig. S3D). Dimethylated-H3me2K4 is one of the early marks associated with reprogramming, and enrichment of this mark at different loci is inhibited by the recruitment of the MacroH2A.1 variant (Barrero et al., 2013). We determined the incorporation of the H3me2K4 level within the pluripotency genes. ChIP analysis demonstrated the enrichment of the H3me2K4 mark at the promoter regions of Nanog and Klf4 in control and Aplf-kd iPSCs at day 9 (Fig. 8D). Enrichment of H3me2K4 was significantly more in Aplf-kd cells at these promoters than at those of the control cells (Fig. 8D); however, a similar enrichment was not observed in the Oct3/4 region (Fig. S3E). This result could be further correlated with the expression pattern of these transcription factors in Aplf-kd cells during time course analysis shown previously (Fig. 7C).
Cdh1 induction is involved in the early stages of reprogramming and, because H3me2K4 is also associated with early marks involved in this transition, we analyzed the incorporation of H3me2K4 at the Cdh1 promoter. A significant increase in H3me2K4 levels was present at the Cdh1 promoter in Aplf-kd cells in contrast to that at the Snai2 promoter (Fig. 8E).
On consideration of the developmental genes, interestingly, the H3me2K4 level at the α-Sma promoter in Aplf-kd cells exhibited a significant reduction in comparison to that in control cells (Fig. S3F). Effectively, presence of the H3me2K4 mark (Fig. S3F) and reduced incorporation of MacroH2A.1 (Fig. S3C) at the α-Sma promoter, even at day 9 post OSKM infection, implies a consistent expression of α-Sma in control cells and, hence, its function as a barrier to reprogramming. As a result, Aplf-kd cells tend to lose MEF characteristics sooner than the control cells and consequently transform into iPSCs more quickly.
We have demonstrated that downregulation of APLF in MEFs could facilitate the reprogramming process through reducing the recruitment of the MacroH2A.1 histone variant to the Cdh1 promoter and inducing the incorporation of H3me2K4 marks at the Nanog and Klf4 promoters (Fig. 8F).
DISCUSSION
The efficiency of the generation of iPSCs to date remains very poor. In this regard, understanding and exploiting epigenetic mechanisms involved in remodeling represents a good option to improve the process of cellular reprogramming. Histone chaperones are associated with diverse aspects of histone metabolism and function in different molecular mechanisms. We have demonstrated that an accelerated loss of APLF in Aplf-kd cells during reprogramming could enhance the rate and efficiency of inducing pluripotency (Figs 1F, 7B). Timely variation in APLF levels could account for this difference in kinetics and efficiency. We ectopically overexpressed APLF in these cells and tried to generate iPSCs but after three consecutive viral transductions and selections, cell survival was very poor and we failed to successfully accomplish the experiment. Aplf-knockout MEFs would have served as the best option to categorically study the effect of ectopically expressed APLF at different stages of reprogramming.
Mechanistically, two predominant barriers to the reprogramming of MEFs constitute (1) MET and (2) setting the stage for induction of the pluripotency network. Here, an enforced reduction of APLF levels in MEFs potentiated the cells to overcome the first barrier through enhanced expression of Cdh1 due to the loss of repressive MacroH2A.1 occupancy at the Cdh1 promoter. This was reflected in the faster reduction in cell size and gain in epithelial morphology of Aplf-kd cells over control cells that provided the impetus to attain induced pluripotency (Fig. S4). Additionally, a proliferation-independent network mediated by Nanog has been associated with accelerated reprogramming (Hanna et al., 2009). Our findings also demonstrated a significant increase in Nanog expression, which is necessary to establish the pluripotent network, in Aplf-kd cells during reprogramming. Although we observed an increase in the proliferative rate of Aplf-kd cells without alteration in the length of the cell cycle phases, these did not conform to the accelerated kinetics or efficiency of reprogramming as evident from earlier reports. Increased proliferation could only expand the pool of cells available for generation of pluripotency and, in later stages, might result in altered fates (Smith et al., 2010). Also, higher proliferation did not drive the reprogrammed cells towards an epithelial state, given a small fraction of highly proliferating fibroblasts could not achieve the epithelial fate (Smith et al., 2010; Liang and Zhang, 2013). An increased rate of proliferation due to Myc overexpression has been associated with a reduction in reprogramming efficiency in mouse iPSCs (Xu et al., 2013). Essentially, only cell cycle acceleration could not increase the efficiency of generation of iPSCs. So, APLF could modulate both rate and efficiency of induction of reprogramming by regulating MET and the early establishment of the pluripotency network.
To fix the instability arising as a result of DNA metabolism, a certain level of APLF should be present in the cells. But, a 70% of knockdown of APLF at the protein level failed to reveal any issue with the stability of iPSCs. APLF, along with its interacting partner PARP3, basically promotes NHEJ (Rulten et al., 2008). Even in the absence of Aplf, overexpression of XRCC4 and DNA ligase IV leads to NHEJ repair (Rulten et al., 2008). So, APLF accelerates NHEJ by acting as a scaffold in the recruitment of the Ku complexes (Rulten et al., 2008; Fenton et al., 2013; Grundy et al., 2013). Quite expectedly, here, in presence of the 30% remaining APLF protein in Aplf-kd cells, no significant alteration in repair capacity was demonstrated in comparison to controls.
We questioned, mechanistically, how APLF affects the other histone modifications, including active histone H3 acetylation at K9 (H3acK9) and repressive histone H3 trimethylation at K9 (H3me3K9). Downregulation of APLF enhanced the levels of H3acK9 and reduced the H3me3K9 levels in cells undergoing reprogramming at day 9 (Fig. S3G). We reasoned that, indirectly, APLF might alter the activity of different histone lysine acetyl transferases (KATs) or histone lysine methyl transferases (KMTs) and thus regulate the modification pattern as well. We analyzed the expression pattern of different KATs and KMTs in Aplf-kd and control cells. A significant variation in the expression pattern of different enzymes, including KATs, KMTs and histone deacetylases (HDACs), was observed (Fig. S3H). Downregulation of the histone H3 K9 methylases Suv39h1, Ehmt1 and Ehmt2 is associated with induction of pluripotency (Papp and Plath, 2013). Increases in the dimethylation marks can be co-related with an induction in Kmt2e expression in Aplf-kd cells (Black et al., 2012), whereas the induction of Kat2a and Kat2b (Jin et al., 2011) can be linked to the increase in H3acK9 levels in Aplf-kd cells. APLF interaction with Ku complexes could mask the KAT2A acetyl transferase activity (Shirodkar et al., 2013; Barlev et al., 1998). Thus, we speculate that downregulation of APLF might restrict the shielding of KAT2A that is reflected by enriched levels of H3acK9 (Fig. S3G). Direct proof of this mechanism will present a new theory in the context of pluripotency.
Population studies and single-cell analyses have indicated the role of genes involved in proliferation, epigenetic modification, MET, pluripotency and MEF markers in the reprogramming of fibroblasts to iPSCs (Buganim et al., 2013). Understanding the role of APLF in reprogramming could lead to explanation of important rate-limiting factors and potentially reveal a new function for a histone chaperone in the context of reprogramming. Acquisition of mutations that might be present in parent cells constitutes one of the concerns regarding iPSC technology (Blasco et al., 2011). But, on exposure to different challenges, Aplf-kd cells show a repair capacity equivalent to that of control cells. Even a metaphase spread of chromosomes of iPSCs at passage 26 demonstrated a normal karyotype (Fig. 4F). Because chromosomally abnormal metaphases, determined by karyotyping analysis, closely correlate with failure in chimerism or germline transmission (Longo et al., 1997; Nagy et al., 1993), we expect that iPSCs derived from Aplf-kd MEFs would retain their full developmental potential. However, improper NHEJ might lead to translocations or nucleotide-level defects. In Aplf−/− mice, the number of ionizing-radiation-induced translocations is significantly reduced (Tong et al., 2016). We therefore predict that 70% knockdown of APLF in fibroblasts under normal conditions might not induce any significant translocation in the iPSCs. Nucleotide-level defects are an outcome of improper nuclease activities of the NHEJ components APLF and Artemis; however, we anticipate that with 30% of APLF still present in the cell, and with no effect on Artemis levels (data not shown), there would be no defect in nucleotide end joining because either of the nucleases present can fulfil this function, and the effects would be small (Pannunzio et al., 2014).
APLF stands out as a unique factor that is related to both DNA repair machinery and histone metabolism. This study demonstrates how its dual nature could be exploited to both improve and understand the highly fascinating phenomenon of reprogramming. It would be further interesting to unravel whether APLF is involved in a similar manner in the induction of pluripotency in human fibroblasts.
MATERIALS AND METHODS
Cell culture
HEK293T (a gift from Dr Soumen Paul, University of Kansas Medical Center) and MEFs were cultured in standard medium, as reported previously (Dutta et al., 2011). E14 ESCs were cultured in feeder-free conditions in ES culture medium (Dutta et al., 2011) and were a gift from Dr. Jay L Vivian (University of Kansas Medical Center).
Animal experiments
All animal-related experiments were performed at the host institute (Rajiv Gandhi Centre for Biotechnology) and according to the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) guidelines and institutional protocol approved by Institutional Animal Ethics Committee (#IAEC/160/DSD/2012 and #IAEC/265/DSD/2014). C57Bl/6J mice (6–8–week–old females and 12–15–week–old males) with an average weight of 21 g were used for the isolation of MEFs. 12–15–week–old NOD/SCID mice (n=3) were used for the generation of teratoma.
Isolation of mouse embryonic fibroblasts
Pregnant C57Bl/6J mice at embryonic day 13.5 post coitum were killed. Uterine horns were removed, and embryos collected, finely minced and incubated with 0.25% trypsin for 30 min to complete the digestion. Trypsin was inactivated by adding MEF medium, and suspensions were centrifuged at a low speed, and supernatant was collected and cultured in MEF medium (Dutta et al., 2011).
Generation of iPSCs
Induced pluripotent stem cells were generated using a lentiviral-based polycistronic vector (FUW-TetO-OSKM; Addgene; 20321) carrying OSKM under the control of a doxycycline-inducible TetO operator (Tet) (Carey et al., 2009; Dutta et al., 2011). MEFs were infected with concentrated lentiviral particles expressing OSKM and Tet. After 30 h of infection, cells were trypsinized, and 3×105 MEFs were plated onto 6-well plates. From here, MEFs were maintained in ESC culture medium in the presence of 2 µg/ml doxycycline until the formation of ESC-like colonies. After the formation of colonies, they were manually picked up, trypsinized and cultured on a feeder layer in ES culture medium without doxycycline. MEFs were inactivated with mitomycin C at a concentration of 10 µg/ml for 2 h for the feeder layer. Next, cells were trypsinized, and ∼5×105 cells were seeded in 6-well plates as a feeder layer for the passaging of iPSCs. The generation of iPSCs was repeated three times for statistical analysis of the data.
RNA interference
An shRNA in a plko.1 vector targeting mouse Aplf was purchased from Sigma (SHCLNG-NM_024251; TRCN0000250398). Stable transfection using CaCl2 was performed in HEK293T cells to produce lentiviral particles harboring shRNA against Aplf, as detailed in previous reports (Majumder et al., 2015). Another set of MEFs was transduced with lentiviral particles encoding the empty plko.1 vector and were treated as the control cells for the generation of iPSCs.
Alkaline phosphatase staining
iPSCs were fixed with 4% paraformaldehyde for 2 min and stained with an alkaline phosphatase detection kit (Millipore; SCR004) according to manufacturer's instructions.
Immunofluorescence
Immunostaining was performed as described previously (Majumder et al., 2015). Cells were incubated with primary antibodies overnight (see Tables S2, S4) and labeled with fluorescence-conjugated secondary antibodies (Alexa-Fluor-568, rabbit Alexa-Fluor-488 and mouse Alexa-Fluor-488). Cells were co-stained with Hoechst dye 33258 (1 µg/ml) for nuclear staining. Images were captured using confocal microscopy.
Cell cycle analysis and apoptosis assay
Approximately 1×106 iPSCs were washed with PBS and fixed in 70% ethanol. Subsequently, cells were treated with RNaseA and incubated with propidium iodide, followed by analysis on a BD FACS AviaTM II instrument. For the apoptosis assay, cells were stained with Annexin-V–FITC and propidium iodide followed by flow cytometry analysis, as per the manufacturer's protocol (Sigma; APOAF-20TST).
FACS analysis
Cells were trypsinized with TrypLE and fixed in 2% paraformaldehyde. The pellet was suspended in the FACS buffer [DPBS with 0.8% (v/v) FBS and 0.263 mM EDTA] and incubated with an anti-E-cadherin (CDH1) primary antibody (ab76055) for 1 h (see Tables S2, S4), followed by incubation with a fluorescence-conjugated secondary antibody (Alexa-Fluor-488). Subsequently, cells were centrifuged and resuspended in FACS buffer, filtered and analyzed by performing FACS.
BrdU incorporation assay
For the BrdU (Sigma, #B5002) incorporation assay, OSKM-transduced control and Aplf-kd MEFs (at day 9) were allowed to incorporate 10 µM BrdU for 30 min. Cells were fixed in ethanol and 50 mM glycine (pH 2.0) for 20 min at −20°C, which was followed by blocking and overnight incubation with primary anti-BrdU antibody (BD Biosciences #347580, see Tables S2, S4) and visualized using fluorescence-conjugated secondary antibodies using fluorescence microscopy (Becker et al., 2006). Nuclei were stained with Hoechst dye 33258. A minimum of 1000 cells was counted in different fields for three biological replicates for each sample.
To determine the cell cycle phase duration, the BrdU-labeled mitotic chromosomes that had accumulated in the presence of the mitotic inhibitor colcemid were counted (Becker et al., 2006) in OSKM-transduced control and Aplf-kd MEFs at day 9. In the presence of 100 ng/ml colcemid, samples were collected at every 2-h interval up to 12 h, processed as mentioned above and incubated overnight with a primary anti-BrdU antibody (see Tables S2, S4). BrdU-incorporated mitoses (condensed chromatin) were analyzed using fluorescence microscopy. 30–40 total mitoses were calculated for each time point for a sample.
To determine the extent of γH2AX foci formation in the same set of BrdU-incorporated cells at day 9, cells were fixed as mentioned above with an additional step of permeabilization with 0.2% Tween-20 followed by overnight incubation with primary anti-γH2AX and anti-BrdU antibodies (see Table S2) and subsequently stained with Hoechst 33258 and visualized by using fluorescence-conjugated secondary antibodies and confocal microscopy.
qRT-PCR
Total RNA was isolated using Qiagen RNeasy Kit (#74106) according to the manufacturer's protocol. cDNA was prepared, and Sybr Green mastermix was used for qRT-PCR analysis (Dutta et al., 2010). Primers used are listed in Table S1.
Western blotting
Cell pellets were suspended and lysed in RIPA buffer (10 mM Tris-HCl pH 7.6, 1% Triton X-100, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 5 mM EDTA, 1 mM sodium orthovanadate, 1 mM PMSF and 10 mg/ml aprotinin), and the protein concentrations were determined by using the Bradford reagent (Bio-Rad; 500-0006) (Majumder et al., 2015), resolved on 10% or 12% SDS-PAGE gels. Antibodies used are listed in Tables S2, S4.
Embryoid body formation and in vitro differentiation
iPSCs and E14 ESCs were trypsinized into a single-cell suspension, and 4×103 cells were cultured in ultra-low attachment dishes without any growth factors for 5 days to stimulate embryoid body formation (Dutta et al., 2011). For endodermal differentiation, embryoid bodies were cultured in Matrigel-coated plates with 0.5% ESC-qualified FBS and 50 ng/ml of activin A for five days (Li et al., 2009). On the appearance of scale-like cells, cultures were examined for the expression of GATA4 by immunofluorescence analysis. For mesodermal differentiation, embryoid bodies were cultured in 0.1%-gelatin-coated plates with Dulbecco's modified Eagle's medium (DMEM) and endothelial-specific FBS (Stem Cell Technologies; 06907) for 10 days (Li et al., 2009). After 10 days, beating foci were seen, and the cells were examined for the expression of troponin T using immunofluorescent staining. For neural differentiation, 72-h-old embryoid bodies were cultured in N2B27 medium (Majumder et al., 2015) in 0.1%-gelatin-coated plates and treated with 2 µM retinoic acid for another 3 days (Jiang et al., 2007). From day 4, embryoid bodies were maintained in retinoic-acid-free medium and cultured for another 3–5 days. Upon observation of neural-rosette-like structures, the cells were subsequently analyzed for TUJ1 expression by using immunofluorescence.
Ectopic expression of Aplf
Mouse Aplf full-length cDNA (accession number: NM_001170489) was amplified from MEFs by performing PCR by using a superscript reverse transcriptase kit (Invitrogen; 18080044). The amplified Aplf cDNA was cloned into XbaI and BamHI sites in the pUltra lentiviral vector, a gift from Malcolm Moore (Dept. of Cell Biology, Sloan-Kettering Institute, Memorial Sloan-Kettering Cancer Center, USA; Addgene; #24129; Lou et al., 2012). Lentiviral particles were generated as described above. E14 ESCs were transfected with these particles and grown for 72 h to analyse the expression of APLF by western blotting and qRT-PCR analyses.
Teratoma formation
C21 iPSCs (2×106 cells) suspended in Iscove's modified Dulbecco's medium (IMDM) were injected subcutaneously at a dorsal flank region (two sites) of NOD/SCID mice (Takahashi and Yamanaka, 2006). After 5 weeks, teratomas were harvested from both dorsal regions and fixed in 4% paraformaldehyde overnight. Fixed teratoma samples were embedded in paraffin, and hematoxylin and eosin was used to stain the sections for further analysis.
DNA damage and recovery assay
Control and Aplf-kd cells were treated with 10 µM etoposide for 4 h and then harvested immediately for the 0-h time point analysis. To remove etoposide from the medium, cells were thoroughly washed with 1× PBS and allowed to recover in ESC medium with LIF for different time points. Recovered cells were analyzed for the presence of γH2AX foci by using confocal microscopy (Tichy et al., 2013). ∼2×104 C21 iPSCs and E14 ESCs were plated in 24-well plates and cultured for another 24 h by adding 105 units of LIF/ml (Millipore; ESG1106) and then analyzed for the presence of γH2AX foci following the same protocol mentioned above.
ChIP analysis
Real-time PCR-based quantitative ChIP analysis was performed as described previously (Dutta et al., 2011). Briefly, ∼1×106 cells per immunoprecipitation were trypsinized, cross-linked with 1% formaldehyde and sonicated to generate chromatin fragments. Antibodies were used to immunoprecipitate protein–DNA cross-linked fragments. Precipitated complexes were eluted and reverse crosslinked. Enrichment of chromatin fragments was measured by performing qRT-PCR analysis using Sybr Green fluorescence relative to a standard curve of input chromatin. Primer sequences are listed in Table S3.
Metaphase spread of chromosomes and Hoechst staining
C21 iPSCs were cultured for 24 h and then treated with colcemid (10 µg/ml) to induce metaphase arrest. Cells were treated with 0.075 M KCl, and fixed and lysed by dropping on the edge of slides (Karyotyping ES cells - Andras Nagy's Laboratory, Samuel Lunenfeld Research Institute, Canada; http://sunnybrook.ca/uploads/sri_tf_form_karyotyping_es_cells.pdf). Chromosomes were stained with Hoechst dye 33258 and observed under a fluorescence microscope.
Co-immunoprecipitation
Protein lysates were prepared in RIPA buffer (Majumder et al., 2015), and concentrations determined by using Bradford reagent. Lysates were immunoprecipitated with primary antibodies (see Tables S2, S4). Immune complexes were adsorbed to protein-A–Sepharose and resolved on 10% PAGE gels.
Statistical analysis
Student's two-tailed, unpaired t-test was used to determine statistical significance. P-values less than 0.05 were considered to be significant.
Acknowledgements
We thank Dr K. B. Harikumar for help with the teratoma study. We sincerely thank Prof. Parimal Karmakar for providing reagents for the BrdU-related experiments. We also thank Prof. C. C. Kartha, Dr S. Sreeja, Dr Tessy and Thomas Maliekkel for sharing reagents.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
K.M.S., S.J., A. Mukherjee and A. Majumder performed the experiments. J.M.T. and M.R.P. provided inputs and shared reagents. D.D. conceived the idea, performed experiments, analyzed data, co-wrote the manuscript with A. Mukherjee and J.M.T.
Funding
Department of Biotechnology, Ministry of Science and Technology, India (BT/PR5754/MED/31/167/2012) supported the work. K.M.S. is also funded by Department of Biotechnology, Ministry of Science and Technology (DBT/JRF/13/AL/452). J.M.T. is supported by National Institutes of Health (NIH HD072489 and NIH OD012206). The work was also supported by the intramural funding from the Rajiv Gandhi Centre for Biotechnology. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.194035.supplemental
References
- Avvakumov N., Nourani A. and Côté J. (2011). Histone chaperones: modulators of chromatin marks. Mol. Cell 41, 502-514. 10.1016/j.molcel.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Barlev N. A., Poltoratsky V., Owen-Hughes T., Ying C., Liu L., Workman J. L. and Berger S. L. (1998). Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol. 18, 1349-1358. 10.1128/MCB.18.3.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero M. J., Sese B., Kuebler B., Bilic J., Boue S., Martí M. and Izpisua Belmonte J. C. (2013). Macrohistone variants preserve cell identity by preventing the gain of H3K4me2 during reprogramming to pluripotency. Cell Rep. 3, 1005-1011. 10.1016/j.celrep.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Becker K. A., Ghule P. N., Therrien J. A., Lian J. B., Stein J. L., van Wijnen A. J. and Stein G. S. (2006). Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell Physiol. 209, 883-893. 10.1002/jcp.20776 [DOI] [PubMed] [Google Scholar]
- Black J. C., Van Rechem C. and Whetstine J. R. (2012). Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491-507. 10.1016/j.molcel.2012.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M. A., Serrano M. and Fernandez-Capetillo O. (2011). Genomic instability in iPS: time for a break. EMBO J. 30, 991-993. 10.1038/emboj.2011.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Faddah D. A. and Jaenish R. (2013). Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 14, 427-439. 10.1038/nrg3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B. W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M. and Jaenisch R. (2009). Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA 106, 157-162. 10.1073/pnas.0811426106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S., Elling U., Hopfgartner B., Jung Y. L., Murn J., Ninova M., Hubmann M., Badeaux A. I., Euong Ang C., Tenen D. et al. (2015). The histone chaperone CAF-1 safeguards somatic cell identity. Nature 528, 218-224. 10.1038/nature15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet A., De Koning L., Toedling J., Savignoni A., Berger F., Lemaître C., O'Sullivan R. J., Karlseder J., Barillot E., Asselain B. et al. (2011). Asf1b, the necessary Asf1 isoform for proliferation, is predictive of outcome in breast cancer. EMBO J. 30, 480-493. 10.1038/emboj.2010.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Ray S., Home P., Saha B., Wang S., Sheibani N., Tawfik O., Cheng N. and Paul S. (2010). Regulation of angiogenesis by histone chaperone HIRA-mediated incorporation of lysine 56-acetylated histone H3.3 at chromatin domains of endothelial genes. J. Biol. Chem. 285, 41567-41577. 10.1074/jbc.M110.190025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., Ray S., Home P., Larson M., Wolfe M. W. and Paul S. (2011). Self-renewal versus lineage commitment of embryonic stem cells: protein kinase C signaling shifts the balance. Stem Cells 29, 618-628. 10.1002/stem.605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A. L., Shirodkar P., Macrae C. J., Meng L. and Koch C. A. (2013). The PARP3- and ATM-dependent phosphorylation of APLF facilitates DNA double-strand break repair. Nucleic Acids Res. 41, 4080-4092. 10.1093/nar/gkt134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M. J., Heidersbach A., Ramalho-Santos J., McManus M. T., Plath K., Meshorer E. et al. (2009). Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460, 863-868. 10.1038/nature08212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar-Maia A., Qadeer Z. A., Hasson D., Ratnakumar K., Leu N. A., Leroy G., Liu S., Costanzi C., Valle-Garcia D., Schaniel C. et al. (2013). MacroH2A histone variants act as a barrier upon reprogramming towards pluripotency. Nat. Commun. 24, 565 10.1038/ncomms2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Muñoz E., Arboleda-Estudillo Y., Out H. H. and Cibelli J. B. (2014). Cell reprogramming. Histone chaperone ASF1A is required for maintenance of pluripotency and cellular reprogramming. Science 345, 822-825. 10.1126/science.1254745 [DOI] [PubMed] [Google Scholar]
- Grundy G. J., Rulten S. L., Zeng Z., Arribas-Bosacoma R., Iles N., Manley K., Oliver A. and Caldecott K. W. (2013). APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 32, 112-125. 10.1038/emboj.2012.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Saha K., Pando B., van Zon J., Lengner C. J., Creyghton M. P., van Oudenaarden A. and Jaenisch R. (2009). Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 462, 595-601. 10.1038/nature08592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamil S., Lam I., Majd M., Tsai S.-H. and Duronio V. (2015). Etoposide induces cell death via mitochondrial-dependent actions of p53. Cancer Cell Int. 15, 79 10.1186/s12935-015-0231-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Sun B., Wang W., Zhang Z., Gao F., Shi G., Cui B., Kong X., He Z., Ding X. et al. (2007). Activation of paternally expressed imprinted genes in newly derived germline-competent mouse parthenogenetic embryonic stem cell lines. Cell Res. 17, 792-803. 10.1038/cr.2007.70 [DOI] [PubMed] [Google Scholar]
- Jin Q., Yu L.-R., Wang L., Zhang Z., Kasper L. H., Lee J.-E., Wang C., Brindle P. K., Dent S. Y. R. and Ge K. (2011). Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249-262. 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-A. and Haber J. E. (2009). Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc. Natl. Acad. Sci. USA 106, 1151-1156. 10.1073/pnas.0812578106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Nakajima-Koyama M., Lee J. and Nishida E. (2016). Transient expression of WNT2 promotes somatic cell reprogramming by inducing β-catenin nuclear accumulation. Stem Cell Rep. 6, 834-843. 10.1016/j.stemcr.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurki P., Vanderlaan M., Dolbeare F., Gray J. and Tan E. M. (1986). Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp. Cell. Res. 66, 209-219. 10.1016/0014-4827(86)90520-3 [DOI] [PubMed] [Google Scholar]
- Li C., Yu H., Ma Y., Shi G., Jiang J., Gu J., Yang Y., Jin S., Wei Z., Jiang H. et al. (2009). Germline-competent mouse-induced pluripotent stem cell lines generated on human fibroblasts without exogenous leukemia inhibitory factor. PLoS ONE 4, e6724 10.1371/journal.pone.0006724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. et al. (2010). A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51-63. 10.1016/j.stem.2010.04.014 [DOI] [PubMed] [Google Scholar]
- Liang G. and Zhang Y. (2013). Embryonic stem cell and induced pluripotent stem cell: an epigenetic perspective. Cell Res. 23, 49-69. 10.1038/cr.2012.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo L., Bygrave A., Grosveld F. G. and Pandolfi P. P. (1997). The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res. 6, 321-328. 10.1023/A:1018418914106 [DOI] [PubMed] [Google Scholar]
- Lou E., Fujisawa S., Morozov A., Barlas A., Romin Y., Dogan Y., Gholami S., Moreira A. L., Manova-Todorova K. and Moore M. A. (2012). Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS ONE 7, e33093 10.1371/journal.pone.0033093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae C. J., McCulloch R. D., Ylanko J., Durocher D. and Koch C. A. (2008). APLF (C2orf13) facilitates nonhomologous end-joining and undergoes ATM-dependent hyperphosphorylation following ionizing radiation. DNA Repair 7, 292-302. 10.1016/j.dnarep.2007.10.008 [DOI] [PubMed] [Google Scholar]
- Majumder A., Syed K. M., Joseph S., Scambler P. J. and Dutta D. (2015). Histone chaperone HIRA in regulation of transcription factor RUNX1. J. Biol. Chem. 290, 13053-13063. 10.1074/jbc.m114.615492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra P. V., Ahel D., Ryan D. P., Weston R., Wiechens N., Kraehenbuehl R., Owen-Hughes T. and Ahel I. (2011). DNA repair factor APLF is a histone chaperone. Mol. Cell 41, 46-55. 10.1016/j.molcel.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Rossant J., Nagy R., Abramow-Newerly W. and Roder J. C. (1993). Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci USA 90, 8424-8428. 10.1073/pnas.90.18.8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T. T., Kara N., Cherry A., Sinha A. U., Zhu N., Bernt K. M., Cahan P., Marcarci B. O., Unternaehrer J., Gupta P. B. et al. (2012). Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598-602. 10.1038/nature10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannunzio N. R., Li S., Watanabe G. and Lieber M. R. (2014). Non-homologous end joining often uses microhomology: implications for alternative end joining. DNA Repair 17, 74-80. 10.1016/j.dnarep.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B. and Plath K. (2013). Epigenetics of reprogramming to induced pluripotency. Cell 152, 1324-1343. 10.1016/j.cell.2013.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy J. P., Gérard A., Cook A. J., Roche D. and Almouzni G. (2008). The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat. Struct. Mol. Biol. 15, 972-979. 10.1038/nsmb.1470 [DOI] [PubMed] [Google Scholar]
- Rulten S. L., Cortes-Ledesma F., Guo L., Iles N. J. and Caldecott K. W. (2008). APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 28, 4620-4628. 10.1128/MCB.02243-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.-k., Beyer T. A., Datti A., Woltjen K., Nagy A. and Wrana J. L. (2010). Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64-77. 10.1016/j.stem.2010.04.015 [DOI] [PubMed] [Google Scholar]
- Sharma N. and Nyborg J. K. (2008). The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc. Natl. Acad. Sci. USA 105, 7959-7963. 10.1073/pnas.0800534105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S., Konishi A., Nishida Y., Mizuta T., Nishina H., Yamamoto A. and Tsujimoto Y. (2010). Involvement of JNK in the regulation of autophagic cell death. Oncogene 29, 2070-2082. 10.1038/onc.2009.487 [DOI] [PubMed] [Google Scholar]
- Shinagawa T., Takagi T., Tsukamoto D., Tomaru C., Huynh L. M., Sivaraman P., Kumarevel T., Inoue K., Nakato R., Katou Y. et al. (2014). Histone variants enriched in oocytes enhance reprogramming to induced pluripotent stem cells. Cell Stem Cell 14, 217-227. 10.1016/j.stem.2013.12.015 [DOI] [PubMed] [Google Scholar]
- Shirodkar P., Fenton A. L., Meng L. and Koch C. A. (2013). Identification and functional characterization of a Ku-binding motif in aprataxin polynucleotide kinase/phosphatase-like factor (APLF). J. Biol. Chem. 288, 19604-19613. 10.1074/jbc.M112.440388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. D., Nachman I., Regev A. and Meissner A. (2010). Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotechnol. 28, 521-526. 10.1038/nbt.1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H. M. (1985). Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA 82, 5328-5331. 10.1073/pnas.82.16.5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K. and Yamanaka S. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861-872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Tichy E. D., Stephan Z. A., Osterburg A., Noel G. and Stambrook P. J. (2013). Mouse embryonic stem cells undergo charontosis, a novel programmed cell death pathway dependent upon cathepsins, p53, and EndoG, in response to etoposide treatment. Stem Cell Res. 10, 428-441. 10.1016/j.scr.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong K. I., Ota K., Komuro A., Ueda T., Ito A., Anne Koch C. and Okada H. (2016). Attenuated DNA damage repair delays therapy-related myeloid neoplasms in a mouse model. Cell Death Dis. 7, e2401 10.1038/cddis.2016.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Wei X., Wang M., Zhang R., Fu Y., Xing M., Hua Q. and Xie X. (2013). Proliferation rate of somatic cells affects reprogramming efficiency. J. Biol. Chem. 288, 9767-9778. 10.1074/jbc.M112.403881 [DOI] [PMC free article] [PubMed] [Google Scholar]