ABSTRACT
Dynamic microtubule plus-ends interact with various intracellular target regions such as the cell cortex and the kinetochore. Two conserved families of microtubule plus-end-tracking proteins, the XMAP215, ch-TOG or CKAP5 family and the end-binding 1 (EB1, also known as MAPRE1) family, play pivotal roles in regulating microtubule dynamics. Here, we study the functional interplay between fission yeast Dis1, a member of the XMAP215/TOG family, and Mal3, an EB1 protein. Using an in vitro microscopy assay, we find that purified Dis1 autonomously tracks growing microtubule ends and is a bona fide microtubule polymerase. Mal3 recruits additional Dis1 to microtubule ends, explaining the synergistic enhancement of microtubule dynamicity by these proteins. A non-canonical binding motif in Dis1 mediates the interaction with Mal3. X-ray crystallography shows that this new motif interacts in an unconventional configuration with the conserved hydrophobic cavity formed within the Mal3 C-terminal region that typically interacts with the canonical SXIP motif. Selectively perturbing the Mal3–Dis1 interaction in living cells demonstrates that it is important for accurate chromosome segregation. Whereas, in some metazoans, the interaction between EB1 and the XMAP215/TOG family members requires an additional binding partner, fission yeast relies on a direct interaction, indicating evolutionary plasticity of this critical interaction module.
KEY WORDS: Crystallography, EB1, Microtubule polymerase, TIRF microscopy, XMAP215, TOG
Summary: We show that Dis1 and Mal3, fission yeast XMAP215/TOG and EB1 family members, directly interact in a non-canonical manner, thereby synergistically enhancing microtubule dynamics.
INTRODUCTION
Microtubules (MTs) are structurally polar and highly dynamic tubulin polymers that undergo spontaneous transitions from growing to shrinking phases, known as dynamic instability (Mitchison and Kirschner, 1984). Such dynamic properties of MTs play an essential role in many cellular processes including intracellular transport, cell polarity and cell division (Desai and Mitchison, 1997). MT dynamics are regulated by a cohort of evolutionarily conserved MT-associated proteins (MAPs) (Akhmanova and Steinmetz, 2008). A subclass of MAPs, MT plus-end-tracking proteins (+TIPs) have unique properties, as they can specifically interact with the dynamic ends of MTs, thereby playing a decisive role in determining the characteristics of MT plus-ends in cells (Buey et al., 2012; Duellberg et al., 2013). In particular, end-binding 1 (EB1, also known as MAPRE1) and ch-TOG [known as XMAP215 (Xenopus) and CKAP5 (Homo sapiens); hereafter denoted TOG] are important +TIPs because they can both accumulate autonomously at MT ends independently of other MAPs, although by different mechanisms (Akhmanova and Hoogenraad, 2005).
EB1 family proteins bind in a nucleotide-dependent manner to an extended region at the growing MT end, resulting in a comet-like appearance at MT plus-ends in cells (Bieling et al., 2007, 2008; Maurer et al., 2011, 2012, 2014; Mohan et al., 2013; Zanic et al., 2009; Zhang et al., 2015). EB1 family proteins can have direct and indirect effects on the MT dynamics (Galjart, 2010); several in vitro experiments have suggested that purified EB1 family proteins promote the MT growth rate and simultaneously increase the catastrophe frequency (Bieling et al., 2007; Li et al., 2012; Vitre et al., 2008; Zanic et al., 2013).
In vivo EB1 family proteins recruit several other MAPs to MT plus-ends through direct protein–protein interactions. EB1 family proteins consist of four functional regions; the N-terminal calponin homology (CH) domain required for MT binding (Hayashi and Ikura, 2003), the medial coiled-coil region involved in homo-dimerisation (De Groot et al., 2010) followed by the EB homology (EBH) domain and finally the C-terminal EEY/F motif (Duellberg et al., 2013). The EBH domain specifically binds to an SXIP motif found in a variety of +TIPs (Buey et al., 2012; Duellberg et al., 2014; Honnappa et al., 2009), whereas the EEY/F motif at the C-terminus of EB1 family proteins binds to some CAP-Gly domains found in some MAPs (Duellberg et al., 2013; Honnappa et al., 2006; Weisbrich et al., 2007). MT plus-end recruitment of other +TIPs by EB1 family proteins is responsible for the indirect effects EB1 family proteins can have on MT behaviour, and hence on a variety of MT-dependent cellular processes.
Mal3, the sole EB1 homologue in fission yeast Schizosaccharomyces pombe, is nonessential for cell division, yet mal3 deletion mutants display a variety of defects derived from abnormal MT architectures and dynamics. These include cell polarity defects during interphase (Beinhauer et al., 1997; Browning et al., 2003; Busch and Brunner, 2004; Busch et al., 2004) and chromosome segregation errors during mitosis (Asakawa et al., 2005, 2006; Asakawa and Toda, 2006; Beinhauer et al., 1997; Mana-Capelli et al., 2012). Mal3 has been shown to interact with the SXIP-motif- and CAP-Gly-domain-containing MAP Tip1, the fission yeast CLIP-170 orthologue, and the Tea2 kinesin, thereby playing a crucial role in regulation of interphase MT organisation and cell polarisation (Bieling et al., 2007; Browning et al., 2003; Busch et al., 2004). By contrast, our understanding of how Mal3 regulates mitotic progression remains poorly understood despite several earlier studies (Asakawa et al., 2006; Kerres et al., 2004). Work performed in vitro has indicated that Mal3 alone has some impact on MT dynamics (Bieling et al., 2007; des Georges et al., 2008; Katsuki et al., 2009); however, it is likely that Mal3 cooperates with other +TIPs during mitosis through direct interactions, as in interphase.
TOG proteins comprise another class of +TIPs that play pivotal roles in many MT-mediated processes (Al-Bassam and Chang, 2011; Kinoshita et al., 2002; Ohkura et al., 2001). Members of this protein family contain N-terminal TOG domains that bind soluble tubulin and a separate MT-binding site (Al-Bassam et al., 2006; Widlund et al., 2011) that, in combination, allow them to act as MT polymerases accelerating MT growth (Al-Bassam et al., 2012; Ayaz et al., 2012, 2014; Brouhard et al., 2008; Li et al., 2012; Podolski et al., 2014; Reber et al., 2013; Roostalu et al., 2015; Takeshita et al., 2013). Consequently, these TOG proteins localise to the very MT end, in contrast to EB1 family proteins that bind to an extended region (Maurer et al., 2014). In the absence of tubulin, TOG has been shown to catalyse MT depolymerisation (Brouhard et al., 2008; Roostalu et al., 2015; Shirasu-Hiza et al., 2003).
Fission yeast contains two TOG orthologues, Alp14 (also known as Mtc1) and Dis1 (Garcia et al., 2001; Nakaseko et al., 2001; Ohkura et al., 2001). These two proteins share essential functions; each single deletion mutant is viable, whereas double deletions are inviable (Aoki et al., 2006; Garcia et al., 2002; Hsu and Toda, 2011; Kakui et al., 2013; Nabeshima et al., 1995; Nakaseko et al., 2001). Whereas Alp14 has been shown to be a MT polymerase like other family members (Al-Bassam et al., 2012; Hussmann et al., 2016), a biochemical characterisation of the catalytic properties of Dis1 at MT ends has not yet been performed.
In this study, we explore the biochemical and physiological interplay between Dis1 and Mal3. Dis1 is a MT polymerase that, different from other TOG proteins, also directly binds to Mal3. In combination, the two proteins promote a synergistic change in MT dynamics. Intriguingly, Dis1 does not have a canonical EB1-binding motif. Instead, crystallographic analysis unveiled a non-canonical binding mode, whereby a new motif in Dis1 bound to the conserved hydrophobic cavity in the EBH domain present in Mal3; these structural data demonstrate similarities and differences in the interaction between EB1 and the SXIP motif versus that used by the unconventional EB1-binding motif in Dis1. Genetic studies demonstrate that the interaction between Dis1 and Mal3 is of physiological significance.
RESULTS
Fission yeast Dis1 is a MT polymerase
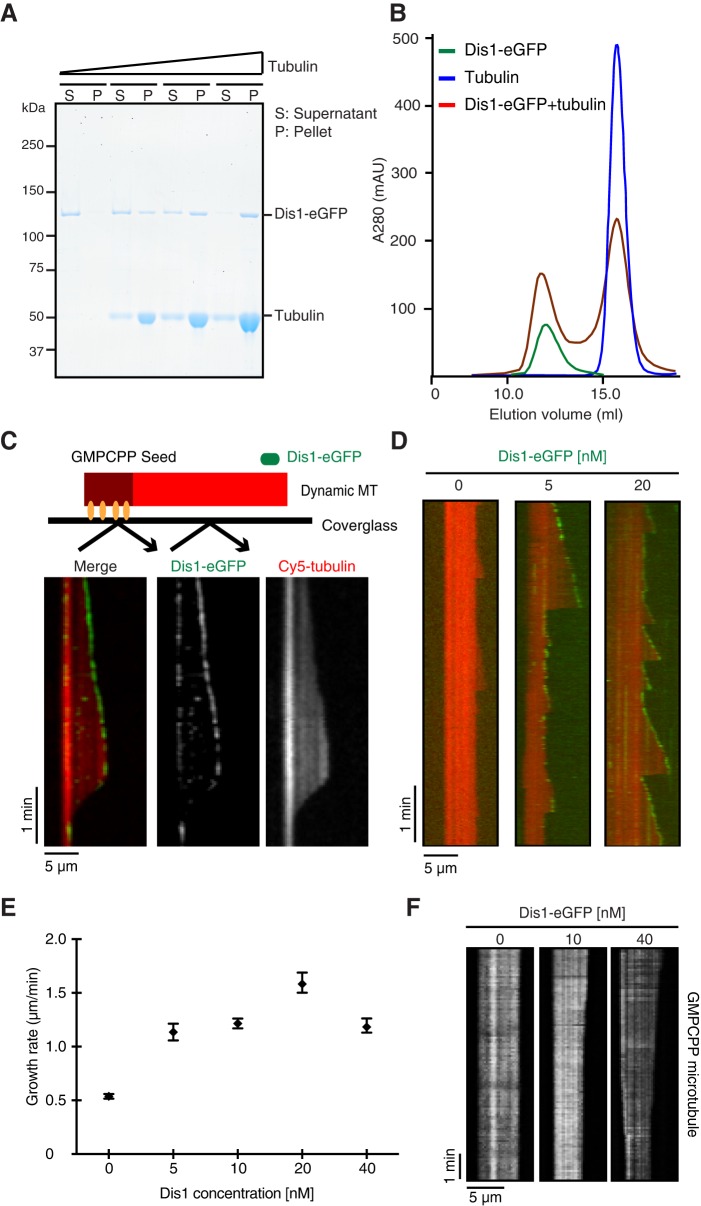
We bacterially expressed and purified recombinant full-length Dis1 protein with a hemagglutinin (HA) tag or eGFP fused to its C-terminus (Fig. S1A). It has been demonstrated that the ability of XMAP215 to accelerate MT polymerisation is dependent on its interaction with both soluble tubulin and MTs (Widlund et al., 2011). Like XMAP215, purified Dis1–eGFP co-sedimented with paclitaxel (taxol)-stabilised MTs (Fig. 1A) and also formed stable complexes with soluble tubulin dimers (Fig. 1B; Fig. S1B). These data indicate that Dis1 shares the binding characteristics of a MT polymerase.
Fig. 1.
Fission yeast Dis1 is a MT polymerase. (A) The MT-binding activity of Dis1–eGFP. Dis1–eGFP concentrations were kept constant at 0.2 µM, while the tubulin concentration was varied (from left to right, 0, 1, 2 and 4 µM). Both supernatant (S) and pellet (P) were stained with Coomassie Brilliant Blue. (B) Tubulin-binding activity of Dis1–eGFP. 5 µM Dis1–eGFP (green), 10 µM tubulin dimers (blue) and their mixture (red) were fractionated on gel filtration columns. (C) Schematic of the assay (top) and dual-colour TIRF-M kymographs showing binding events between 10 nM Dis1–eGFP (green in merge) and the ends of growing MTs. (D) Dual-colour TIRF-M kymographs showing Cy5-labelled MTs growing in the absence or presence of Dis1–eGFP (green). (E) Plot of the mean growth rate. The Cy5-labelled tubulin concentration was 8 µM in C–E. Data points, black; error bars are s.e.m., n=50. (F) Kymographs showing GMPCPP-stabilised Cy5-labelled MTs. Scale bars: 5 µm (horizontal) and 1 min (vertical) (C,D,F).
To observe how Dis1–eGFP localises on dynamic MTs and how it affects their dynamic properties, we performed a total internal reflection fluorescence microscopy (TIRF-M)-based in vitro assay (Fig. 1C and Movie 1). MTs were grown from immobilised and GMPCPP-stabilised MT seeds (Materials and Methods) in the presence of Cy5-labelled tubulin and GTP (Bieling et al., 2010). Dis1–eGFP associated weakly with the MT lattice and accumulated on and tracked growing MT plus-ends in a spot-like manner (Fig. 1C), similar to other TOG orthologues (Brouhard et al., 2008; Li et al., 2012; Podolski et al., 2014); by contrast, Dis1–eGFP was not observed on shrinking MT ends, similar to Alp14, the other TOG orthologue in fission yeast (Al-Bassam et al., 2012; Garcia et al., 2001; Hussmann et al., 2016; Nakaseko et al., 2001).
Next, we measured the effect of Dis1 on the dynamics of individual MTs (Fig. 1D). MT dynamic instability parameters, such as the average growth rate, shrinkage rate and catastrophe frequency, were quantified for different Dis1 concentrations (Table S1; Fig. 1E). The strongest polymerase effect was measured for a concentration of 20 nM Dis1, where it accelerated the growth of MTs by approximately threefold, from 0.54±0.02 µm/min to 1.58±0.09 µm/min (mean±s.e.m., n=50; P<0.0001 by a Student's t-test) (Fig. 1E). By contrast, Dis1 did not have a strong impact on the shrinkage rate or catastrophe frequency (Fig. S1C). Interestingly, similar to XMAP215 and ch-TOG (Brouhard et al., 2008; Roostalu et al., 2015), Dis1 also facilitated the depolymerisation of GMPCPP-stabilised MTs in the absence of free tubulin (Fig. 1F; Fig. S1D). These data show that Dis1 is a bona fide MT polymerase.
Dis1 directly binds to Mal3 and, synergistically, they alter the dynamic properties of MTs
Previous studies have shown evidence for a functional interplay between TOG and EB1 proteins (Li et al., 2012; van der Vaart et al., 2011; Zanic et al., 2013), although there are no reports showing a direct interaction between these two +TIPs. In fission yeast, the Alp14 localisation to MT plus-ends is independent of Mal3 (Al-Bassam et al., 2012); whether this is also the case for Dis1 has not been tested. Hence, we bacterially expressed and purified recombinant full-length Mal3 proteins with or without eGFP or mCherry fused to its C-terminus (Fig. S2A). We found that GST-tagged Mal3 (GST–Mal3), but not GST alone, pulled down Dis1–eGFP (Fig. 2A). Furthermore, analytical gel filtration demonstrated that recombinant Dis1 with an HA tag (Dis1–HA) and Mal3 form a stable complex in solution (Fig. 2B; Fig. S2B). These data show that Dis1 directly binds to Mal3.
Fig. 2.
Dis1 directly binds to Mal3. (A) Binding between Dis1 and Mal3. GST or GST–Mal3 bound to glutathione beads was mixed with Dis1–eGFP. (B) Analytical gel filtration chromatography. 5 µM Dis1–HA (green), 20 µM Mal3 (blue) or the mixture of both proteins (red) was analysed. (C) TIRF-M kymographs showing binding events between 20 nM Dis1–eGFP (green) and the ends of growing Cy5-labelled MTs (red) in the absence or presence of unlabelled Mal3. The Cy5-labelled tubulin concentration was 20 µM. Snapshots are shown at the bottom. (D) Schematic of the assay (top) and triple-colour TIRF-M kymographs showing binding events between 10 nM Dis1–eGFP (green in merge) and the ends of growing Cy5-labelled MTs (blue in merge) in the presence of 200 nM Mal3–mCherry (red in merge) (bottom). (E) 10 nM Dis1–eGFP accumulates at the ends of a MT in a similar manner to 200 nM Mal3–mCherry. The Cy5-labelled tubulin concentration was 10 µM in D and E. Scale bars: 5 µm (horizontal) and 1 min (vertical) (C–E). (F) Plot of the mean growth rate. 10 nM Dis1 and/or 20 nM Mal3 were added in the presence of 8 µM tubulin. Data points, black; error bars are s.e.m., n=50.
Next, we examined the effect of Mal3 on the localisation of Dis1–eGFP on dynamic MTs using our TIRF-M assay (Fig. 2C). Whereas 20 nM Dis1–eGFP localised to growing MT ends in a spot-like manner, as with XMAP215 (Brouhard et al., 2008; Li et al., 2012; Podolski et al., 2014) (Fig. 2C and Movie 2), upon the addition of 200 nM Mal3, the amount of Dis1–eGFP at MT ends was strongly increased; Dis1–eGFP localised now to a much more elongated region at growing MT ends, rather reminiscent of the appearance of end-tracking Mal3–GFP (Fig. S2C and Movie 3) (Maurer et al., 2011). In the presence of the higher concentrations of 1 µM Mal3, Dis1–eGFP bound to the entire MT lattice (except for the GMPCPP seeds) (Fig. 2C; Movie 4), again similar in appearance to the localisation of Mal3–GFP at this high concentration (Fig. S2C).
To simultaneously visualise the localisation of Dis1, Mal3 and MTs, we used triple-colour TIRF-M. We observed that Dis1–eGFP and Mal3–mCherry colocalised to the MT end region (Fig. 2D,E; Movie 5). In control experiments, we observed that Mal3 did not directly bind to Alp14 (Fig. S2D). These results demonstrate that Mal3 recruits Dis1, but not Alp14, to the extended EB-binding region at growing MT ends.
We then examined the effect of Mal3-dependent Dis1 MT end recruitment on the dynamic properties of MTs. Compared to tubulin alone, 20 nM Mal3 mildly accelerated the growth rate by 1.6-fold (0.54±0.02 µm/min versus 0.87±0.04 µm/min; mean±s.e.m., n=50, P<0.0001 by a Student's t-test). 10 nM Dis1 accelerated the growth rate by 2.3-fold (1.22±0.05 µm/min; mean±s.e.m., n=50, P<0.0001). In the presence of both 20 nM Mal3 and 10 nM Dis1, the growth rate was even more strongly increased by 4.6 fold (to 2.48±0.08 µm/min; mean±s.e.m., n=50, P<0.0001), suggesting synergistic action of the two proteins (Table S2, Fig. 2F, Fig. S2E), reminiscent of what has been reported for the vertebrate homologues of these +TIPs (Li et al., 2012; Zanic et al., 2013). In addition, we found that Mal3 and Dis1 together substantially promoted the MT catastrophe frequency (∼3.8-fold); Mal3 alone had a modest impact (∼1.9-fold), whereas Dis1 had no effect on its own (Fig. S2F). It is of note that no synergistic effect was observed for the shrinkage rate (Fig. S2F). Therefore, the increased accumulation of Dis1 at MT ends mediated by Mal3 together with the presence of Mal3 itself strongly enhanced the dynamicity of MTs, in particular increasing the growth rate and the catastrophe frequency. Taking all these data together, we conclude that Mal3 specifically binds to Dis1 but not to Alp14, thereby enhancing MT dynamicity.
The C-terminal tail region of Dis1 is the primary site for Mal3 binding
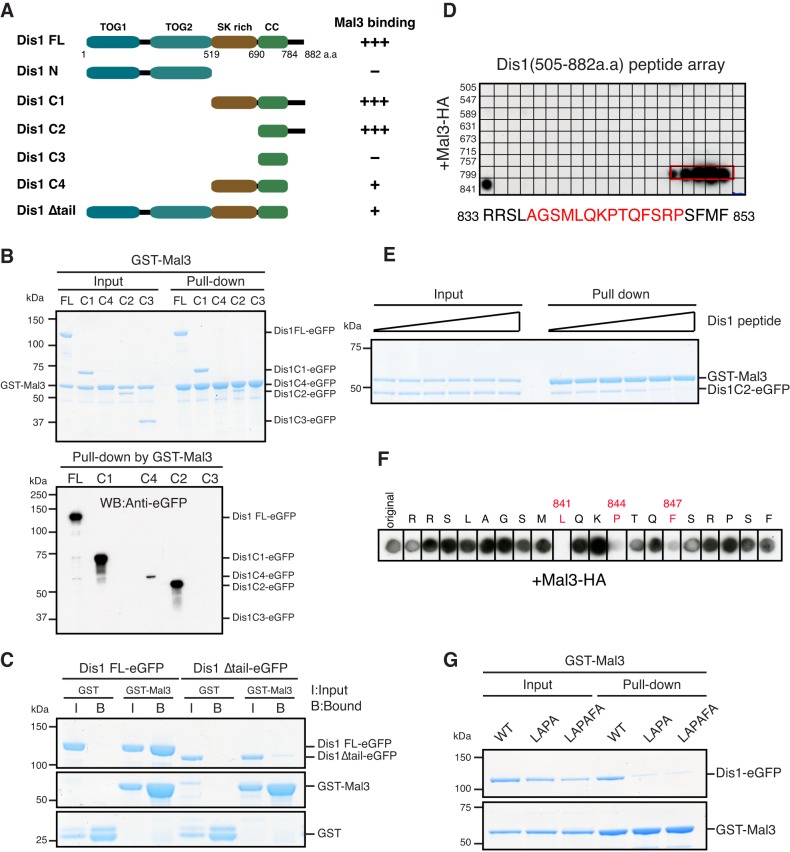
Although Dis1 directly binds to Mal3 (Fig. 2), there is no SXIP motif or CAP-Gly domain within the Dis1 protein. Therefore, we decided to experimentally identify the interaction site of Dis1 for Mal3 binding. Using GST pulldown assays with several truncated Dis1 constructs (Fig. 3A), we found that the C-terminal region of Dis1 (Dis1 C2, amino acids 690–882) is the minimal region that interacts with Mal3 as efficiently as full-length Dis1 (Dis1 FL); the preceding coiled-coil region (Dis1 C3, amino acids 690–784) alone did not bind to Mal3 (Fig. 3A,B). This data indicated that the C-terminal tail of Dis1 (amino acids 784–882) is necessary for Mal3 binding. To test this possibility, we constructed the C-terminally truncated Dis1 that lacked this region (Dis1Δtail) and compared the binding to Mal3 between Dis1 FL and Dis1Δtail. We found that the binding of Dis1Δtail to Mal3 was substantially reduced compared to that of Dis1 FL (Fig. 3A,C).
Fig. 3.
The C-terminal tail region of Dis1 is the primary binding site for Mal3. (A) A schematic representation and a summary of their binding to Mal3. TOG1, TOG2, TOG homology domains; SK rich, rich in serine and lysine residues; CC, coiled-coil. (B) Binding between various truncated Dis1 proteins and Mal3. GST–Mal3 bound to glutathione beads was mixed with truncated Dis1–eGFP. The vertical line found between the last two lanes on the right was introduced unintentionally during scanning of the original data. (C) Binding between the full-length or tail-less Dis1 protein and Mal3. (D) Peptide array analysis. The arrays cover from 505th to 882nd amino acid residues of Dis1. The arrays were incubated with a solution containing Mal3–HA protein, followed by immunoblotting against an anti-HA antibody. Amino acid sequences corresponding to bound residues (red) are shown on the bottom. (E) Binding between the tail region of Dis1 and Mal3. GST–Mal3 bound to the glutathione beads were pre-incubated with various concentrations of the Dis1 peptide (amino acids 833–852) (from left to right, 0, 0.5, 2.5, 5.0, 50, 100 µM) for 30 min before incubation with 0.5 µM Dis1 C2–eGFP for an additional hour. (F) Alanine-scanning mutagenesis analysis. Each position in the Dis1 peptide was replaced with alanine. Amino acid residues shown in red were sensitive to alterations. (G) Binding between Mal3 and the wild-type (WT) or mutated Dis1 protein. Dis1–eGFP proteins were LAPA (L841A and P844A) or LAPAFA (L841A, P844A and F847A).
Consistent with these pulldown assays, TIRF-M assays demonstrated that Dis1 constructs containing the C-terminal tail could track the MT plus-end in the presence of Mal3 (Fig. S3A,B). Importantly, although Dis1 FL could track the MT end regardless of the presence or absence of Mal3, Dis1 C2 (amino acids 690–882), which lacked the N-terminal part of Dis1 required for its autonomous end-tracking ability, tracked ends only in the presence of Mal3 (Fig. S3B). Notably, Dis1 C2 in the presence of Mal3 tracked a more extended region at MT ends compared to Dis1-FL alone, which showed a spot-like appearance on the MT ends. These data strongly suggest that the C-terminal tail region is the primary site for Mal3 binding.
Three amino acid residues in the C-terminus of Dis1 are crucial for binding to Mal3
To identify the Mal3-binding site within the C-terminal tail region of Dis1, we analysed the Dis1–Mal3 interaction by means of a tiling peptide array. A membrane was spotted with 20-residue peptides covering the C-terminal region of Dis1 (amino acids 505–882) with a two-residue start increment per spot, and then probed with Mal3–HA protein followed by immunoblotting. Remarkably, a single region consisting of 21 amino acid residues (amino acids 833–853) bound to Mal3 (Fig. 3D). To verify this interaction, we synthesised the corresponding peptide (Dis1 peptide) and conducted a competition experiment in the GST pulldown assay. Confirming the importance of the identified peptide for the Dis1–Mal3 interaction, an excess amount of the Dis1 peptide effectively abolished the binding between Mal3 and Dis1 C2 (amino acids 690–882) (Fig. 3E).
To identify the amino acid residues that are most crucial for the interaction between Dis1 and Mal3, we further performed alanine-scanning mutagenesis against each amino acid and found that the Dis1 residues L841, P844 and F847 were indispensable residues for Mal3 binding (Fig. 3F). To further test the importance of these key residues, we mutated them in full-length Dis1; we constructed two Dis1 replacement mutants (LAPA, L841A and P844A; and LAPAFA, L841A, P844A and F847A) and compared their binding to Mal3. Both Dis1 mutant proteins bound more weakly to Mal3 in comparison to wild-type Dis1 (Fig. 3G), confirming that the three amino acid residues L841, P844 and F847 in the Dis1 C-terminal region are crucial for the interaction with Mal3. Given that the identified binding motif differs from the canonical SXIP motif, Dis1 likely binds to Mal3 in a new and unconventional manner.
The coiled-coil and EBH domains of Mal3 are necessary and sufficient for binding to Dis1
To identify in turn the Dis1-binding region within Mal3, several truncated Mal3 constructs were made and used for Dis1-binding GST pulldown assays. We found that the C-terminal half (Mal3 C1, amino acids 144–308) is responsible for binding to Dis1 (Fig. 4A,B). This C-terminal region consists of the parallel coiled-coil region (amino acids 144–194) followed by the EBH domain (amino acids 197–247) (Slep et al., 2005) (Fig. 4A). We produced fragments corresponding to these two regions (Mal3 C2 and Mal3 C3, respectively) and found that neither bound to Dis1 (Fig. 4C). However, a Mal3 construct that consists of the C-terminal part of the coiled-coil and the EBH domains (Mal3 C4, amino acids 174–247) was sufficient to bind to Dis1 (Fig. 4A,D). Introduction of the LAPA mutations (L841A and P844) to the Dis1 C2 construct abrogated the binding of Mal3 C4 (Fig. 4D). Taken together, the dimerised EBH domain within Mal3 is necessary and sufficient for the binding to Dis1.
Fig. 4.
The coiled-coil and EBH domains of Mal3 are necessary and sufficient for binding to Dis1. (A) A schematic representation of various truncated Mal3 proteins and a summary of their binding to Dis1. CH, calponin-homology domain; CC, coiled-coil; EBH, EB homology. (B,C) Binding between various truncated Mal3 proteins and Dis1. Dis1 C1 (amino acids 520–882) tagged with eGFP was mixed with various GST-tagged truncated Mal3 proteins (B, FL, N, C1 or C, C2, C3), which were bound to the glutathione beads. (D) Binding between the dimerised EBH domain of Mal3 protein and Dis1. EBH-domain-containing Mal3 protein (amino acids 174–247) was mixed with wild-type (WT) or mutated Dis1-C2–eGFP (LAPA; L841A, P844A) and pulled down by GFP trap beads.
Direct interaction between Dis1 and Mal3 is essential for the synergistic impact on MT dynamics
We next examined the localisation of 10 nM Dis1-LAPA–eGFP in the presence or absence of 20 nM Mal3 on dynamic MTs using TIRF-M assays. As shown in Fig. 5A, the localisation patterns of Dis1-LAPA to the MT tip were indistinguishable between in the presence and in the absence of Mal3; they displayed thin spot-like appearances rather than the thick comets seen in wild-type Dis1–eGFP in the presence of Mal3. This firmly established that Dis1-LAPA fails to interact with Mal3 not only in solution but also on MTs.
Fig. 5.
Physical binding between Dis1 and Mal3 is required for the synergistic impact on MT dynamics. (A) TIRF-M kymographs showing Dis1–eGFP (10 nM, green in merge) in the presence or absence of 20 nM Mal3. The Cy5-labelled tubulin (red) concentration was 8 µM. Scale bars: 5 µm (horizontal) and 1 min (vertical). (B,C) Plot of the mean growth rate (B) or catastrophe frequency (C). 10 nM Dis1-eGFP [wild-type (WT) or LAPA] and/or 20 nM Mal3 were added in the presence of 8 µM tubulin. Data points, black; error bars are s.e.m. n=50.
We then asked whether Dis1 and Mal3 in combination are capable of altering the dynamic properties of MTs without their direct interaction. To this end, we quantified various parameters of the MT dynamics. Remarkably, no additive or synergistic impact was observed between Dis1-LAPA and Mal3 (Fig. 5B; Table S3); the growth rate was 1.08±0.09 µm/min (mean±s.e.m., n=50), which was very similar to that of Dis1-LAPA alone (1.02±0.04 µm/min, mean±s.e.m.), whereas the catastrophe frequency was almost identical to that of Mal3 alone (0.22±0.02 µm/min versus 0.23±0.01 µm/min; mean±s.e.m., n=50). It is of note that authentic activities of Dis1-LAPA on its own towards the MT dynamics are indistinguishable to those of wild-type Dis1 (Fig. 5B; Table S3). Collectively, these results illuminate the marked differences in the TOG and EB1 families between fission yeast and vertebrates; whereas fission yeast Dis1 (the TOG protein) and Mal3 (the EB1 protein) need to interact directly in order to exert a synergistic impact on the MT dynamics, vertebrate orthologues are poised to do so without physical binding.
Crystallographic analysis of the Mal3–Dis1 binding interface
Previous crystal structures of the EB1 C-terminal domain showed that the highly conserved surface of the EBH domain comprises a hydrophobic cavity, which is important for the binding to SXIP-containing +TIPs (Honnappa et al., 2005, 2009; Slep et al., 2005). A crystal structure of the corresponding Mal3 EBH domain has not been reported yet. To obtain structural insight into the basis of Mal3–Dis1 interaction, we crystallised Mal3 C4 (amino acids 174–247) alone and in complex with the minimal Mal3-binding Dis1 peptide (amino acids 833–852) (Fig. 3E), and solved their structures (Fig. 6A; Fig. S4A,B and Table S4). Like the human EBH domain (Honnappa et al., 2005; Slep et al., 2005), the homodimerised EBH domain of Mal3 also displayed a hydrophobic cavity, demonstrating that this structural feature is conserved (Fig. 6B). The Dis1 peptide bound to this homodimerised EBH domain (Fig. 6A; Fig. S4A,B), and its binding was predominantly driven by hydrophobic interactions between L841, P844 and F847 in the Dis1 peptide and the conserved hydrophobic cavity on the outside edge of the EBH domain of Mal3 (Fig. 6B). These structural data are fully consistent with the results of alanine scanning and pull-down assay using the mutant Dis1 protein with the disrupted binding motif (Fig. 3F,G).
Fig. 6.
Molecular details of the Mal3–Dis1 interaction. (A) Ribbon diagram of the complex between the coiled-coil and EBH domains of Mal3 (residues 174–247 forming a homodimer as depicted in blue and pink) and the interacting Dis1 peptide (residues 833–852 as depicted in green). A second peptide binds on the other side of the dimer, but has been omitted from the figure for the sake of clarity. (B) Details of the key interacting residues. Surface conservation plot of the EBH domain of Mal3 from low (blue) to high (purple) similarity. (C,D) Close-up view of the interaction seen between the Mal3-EBH dimer (residues 174–247 depicted in blue and pink) and the Dis1 peptide (residues 833–852 depicted in green) (C) or the human EB1-EBH dimer (residues 191–260 depicted in yellow and green) and the SXIP motif containing the MACF peptide (residues 5468–5497 depicted in orange) (D). (E) Cartoon showing the comparison of the interaction interfaces between Mal3–Dis1 (left) and EB1–MCAF (right). A,A′ and B,B′ represent the EBH domain derived from two dimerised Mal3 molecules.
Next, we carried out a detailed comparison of the interaction interface between Mal3–Dis1 and EB1–MACF (microtubule actin cross-linking factor), an EB1 interactor containing the canonical SXIP motif (Honnappa et al., 2009) (Fig. 6C,D; Fig. S4C,D). We found that the S839MLQKP844 residues in the Dis1 peptide are functionally equivalent to the S5477LIPTP5482 motif in the MACF peptide, especially the L841 residue in the Dis1 peptide and the corresponding I5479 residue in the MACF peptide, which fit into the conserved hydrophobic cavity formed by the EBH domain. Intriguingly, however, we noted a clear difference between the individual interaction modes of the two peptides: whereas the binding interface for EB1–MCAF was provided predominantly by one EB1 monomer, the interface for the Dis1 peptide spans two Mal3 monomers (Fig. 6E). This structural data explains the necessity of dimerisation of the EBH domain for Dis1 binding through the preceding coiled-coil of Mal3 (Fig. 4A,C). These findings indicate that the binding between Dis1 and Mal3 is indeed non-canonical, although the hydrophobic cavity is conserved between Mal3 and EB1.
Physiological significance of the binding between Dis1 and Mal3
Given that Dis1 and Mal3 directly interact in vitro, we next examined their in vivo interaction with GFP trap pulldown from fission yeast lysates. We found that Dis1–2GFP and Mal3–5FLAG indeed formed a complex (Fig. 7A). We then created a mutant strain in which wild-type dis1+-2GFP was replaced by dis1-LAPA-2GFP (L841A and P844A). Consistent with the in vitro data (Fig. 3F,G), the pulldown assay clearly indicated that Dis1-LAPA–2GFP did not bind to Mal3–5FLAG (Fig. 7A), demonstrating that the identified Mal3-binding site in Dis1 is relevant in the context of living cells.
Fig. 7.
Dis1 interacts with Mal3 in vivo and its interaction is crucial for Dis1 function. (A) Interaction between Dis1 and Mal3. Whole-cell extracts (1.0 mg) were pulled down with GFP trap beads. Immunoblotting was performed with anti-GFP and anti-FLAG antibodies. Input: 15 µg. WT, wild type. (B) Summary of genetic interaction. Solid lines, synthetically lethal; dotted line, viable. (C) Tetrad dissection. Five representative tetrads are shown [the parental ditype (1), tetratypes (2–4) and the non-parental ditype (5)]. Double mutants were inviable (red circles). (D) Spot assay. Indicated strains were serially diluted (tenfold each), spotted on YES plates and incubated for 4 days or at 26°C for 4 days in the presence of 10 µg/ml TBZ (right). (E) Mitotic progression and MT dynamics in dis1-LAPA cells. Mitotic progression of wild-type (left, n=38) and dis1-LAPA (right, n=31) cells containing a tubulin marker (mCherry–Atb2) was followed in live cells under fluorescence microscopy at room temperature (23°C). Phase I (red), the initial stage of spindle elongation; phase II (yellow), the pre-anaphase stage with constant spindle length; Phase III (blue), anaphase B (Nabeshima et al., 1998). Results are mean±s.d.; n>30 cells; *P<0.01; n.s., not significant (two-tailed unpaired Student's t-tests). (F) Minichromosome loss assay. Indicated strains carrying the minichromosome Ch16 (Niwa et al., 1989) were grown on rich YE plates (lacking adenine) and incubated at 25°C for 6 days. The percentage of red and/or sectored colonies is shown at the bottom (n≥1,000).
In order to explore the functional significance of the direct binding between Dis1 and Mal3, we examined genetic interactions. It has been previously shown that the double mutant between Δdis1 and Δalp14 was synthetically lethal (Garcia et al., 2002; Nakaseko et al., 2001). Interestingly, whereas the Δmal3 mutant displayed synthetic lethality with Δalp14, Δmal3Δdis1 was viable (Kerres et al., 2004). This result implies that Dis1 and Mal3 act in the same genetic pathway, which shares essentiality with Alp14 (Fig. 7B). Moreover, dis1-LAPA mutants also showed synthetic lethal interaction with Δalp14 (Fig. 7B,C). It is noteworthy that, unlike Δdis1 cells, which displayed cold sensitivity (Ohkura et al., 1988), dis1-LAPA cells were capable of forming colonies at low temperature (Fig. 7D). This result indicates that failure of interaction between Dis1 and Mal3 did not abolish all the cellular functions of Dis1 (Garcia et al., 2002; George and Walworth, 2015; Sanchez-Perez et al., 2005); instead by forming a complex with Mal3, Dis1 plays an essential role in cell division, which can be compensated for by Alp14.
Although a strain containing dis1-LAPA cells grew apparently normally, these cells displayed hyper-sensitivity to the MT-destabilising drug thiabendazole (TBZ) (Fig. 7D), which slows down growth in mutants with defects in MT regulation and chromosome segregation (Garcia et al., 2001; Toyoda et al., 2002; Umesono et al., 1983); note that Δmal3 and Δdis1 cells were also individually hypersensitive to TBZ (Fig. 7D) as previously shown (Aoki et al., 2006; Beinhauer et al., 1997). We then examined MT integrity and mitotic progression in the dis1-LAPA mutant. No noticeable differences in MT structures and intensities were observed; however, a modest yet reproducible increase in the rate of MT elongation during anaphase B (phase III, Nabeshima et al., 1998) was detected (Fig. 7E). Currently the reason for this faster anaphase B progression is unknown; nonetheless, this result uncovered that a direct interaction between Dis1 and Mal3 plays a role in the proper MT dynamics in late mitosis.
Mutations in genes required for chromosome transmission fidelity can be identified by a minichromosome loss assay (Fleig et al., 1996; Niwa et al., 1989; Takahashi et al., 1994). It was previously observed that both mal3 and dis1 mutants display a minichromosome loss phenotype (Aoki et al., 2006; Beinhauer et al., 1997). Although it was not as severe as in Δdis1 cells (6.9%), a strain containing Dis1-LAPA lost minichromosomes at an elevated rate compared to wild-type cells (2.0% versus <0.1%) (Fig. 7F). Taken together, these results indicate that the physical interaction between Dis1 and Mal3 is indeed necessary for accurate chromosome segregation.
DISCUSSION
Our study of the biochemical and physiological interplay between fission yeast Dis1 and Mal3 has provided five major results. First, Dis1 is a bona fide MT polymerase like other TOG proteins. Second, Dis1 and Mal3 synergistically control MT dynamics. Third, this is explained by a direct interaction between Dis1 and Mal3 involving a new EB1-binding motif. Fourth, the binding mode of this motif in Dis1 to the conserved hydrophobic pocket in the EBH domain of Mal3 is unconventional. Fifth and, finally, the Dis1–Mal3 interaction is of physiological significance.
Our in vitro reconstitution experiments showed that Dis1 is a MT polymerase that autonomously localises to the growing MT plus-ends, thereby accelerating the MT growth rate by about threefold. This behaviour is very similar to that of Alp14, the other TOG orthologue in fission yeast (Al-Bassam et al., 2012; Garcia et al., 2001; Nakaseko et al., 2001). As for other TOG proteins, such as frog XMAP215, budding yeast Stu2 or fission yeast Alp14 (Al-Bassam et al., 2006, 2012; Hussmann et al., 2016; Widlund et al., 2011), the N-terminal TOG domains of Dis1 are likely to bind tubulin dimers, whereas the central Ser-Lys-rich (SK-rich) domain of Dis1 is probably responsible for binding to the MT lattice, which is consistent with our results with truncated Dis1–eGFP constructs (Fig. S3B) and also with the detailed domain analysis previously performed (Nakaseko et al., 1996).
The vertebrate proteins Xenopus XMAP215 and human EB1 have previously been reported to synergistically increase the MT polymerisation rate in vitro, although these two proteins do not interact directly with each other (Zanic et al., 2013). Synergy was proposed to result from the ability of EB1 to potentially alter the curvature of the MT end structure allosterically, allowing XMAP215 to promote MT growth more efficiently (Zanic et al., 2013). Here, we have also observed a synergistic enhancement of the MT growth rate by fission yeast Mal3 and Dis1, even if to a lesser extent. For the fission yeast proteins, synergy is likely to arise from the enhanced accumulation of Dis1 at MT ends as a consequence of its recruitment by Mal3 through direct interaction, as our TIRF microscopy experiments have demonstrated. Consistent with this notion, the Dis1 mutant protein with specific defects in binding to Mal3 (Dis1-LAPA) failed to exhibit additive or synergistic effects on the MT dynamics in combination with Mal3, although Dis1-LAPA on its own still retained normal enzymatic activities (Fig. 5B). In this context, it is interesting to note that in budding yeast, an interaction between Bim1 (an EB1 protein) and Stu2 (a TOG protein) is known; however their mode of interaction is unclear (Wolyniak et al., 2006). This suggests that fungi and vertebrates have developed similar yet distinct regulatory systems by which to regulate the MT dynamics changed mediated by the TOG and EB1 families.
In human cells, the protein SLAIN2 acts as a linker mediating an indirect interaction between ch-TOG and EB1 (van der Vaart et al., 2011). The effect of all three human proteins together on MT dynamics has not been investigated yet in vitro. However, the indirect interaction between XMAP215 and EB1 proteins is probably conserved in metazoa, as the unrelated Drosophila protein Sentin plays a similar role for Drosophila Msps (a TOG protein) and EB1, mediating their indirect interaction (Gluszek et al., 2015; Mohan et al., 2013; Moriwaki and Goshima, 2016; Li et al., 2011, 2012). A mild synergistic effect on the growth rate of MTs exerted by a combination of these three Drosophila proteins has also been observed, possibly as a consequence of their tripartite interaction (Li et al., 2012). Taken together, it is possible that members of the TOG and EB1 families act collaboratively at the MT plus-ends in many, if not all, eukaryotes, having a synergistic impact on MT dynamics. Their interaction can either be indirect as in metazoans, or direct as in fission yeast, demonstrating evolutionary plasticity of this regulatory module (Fig. 8A,B).
Fig. 8.
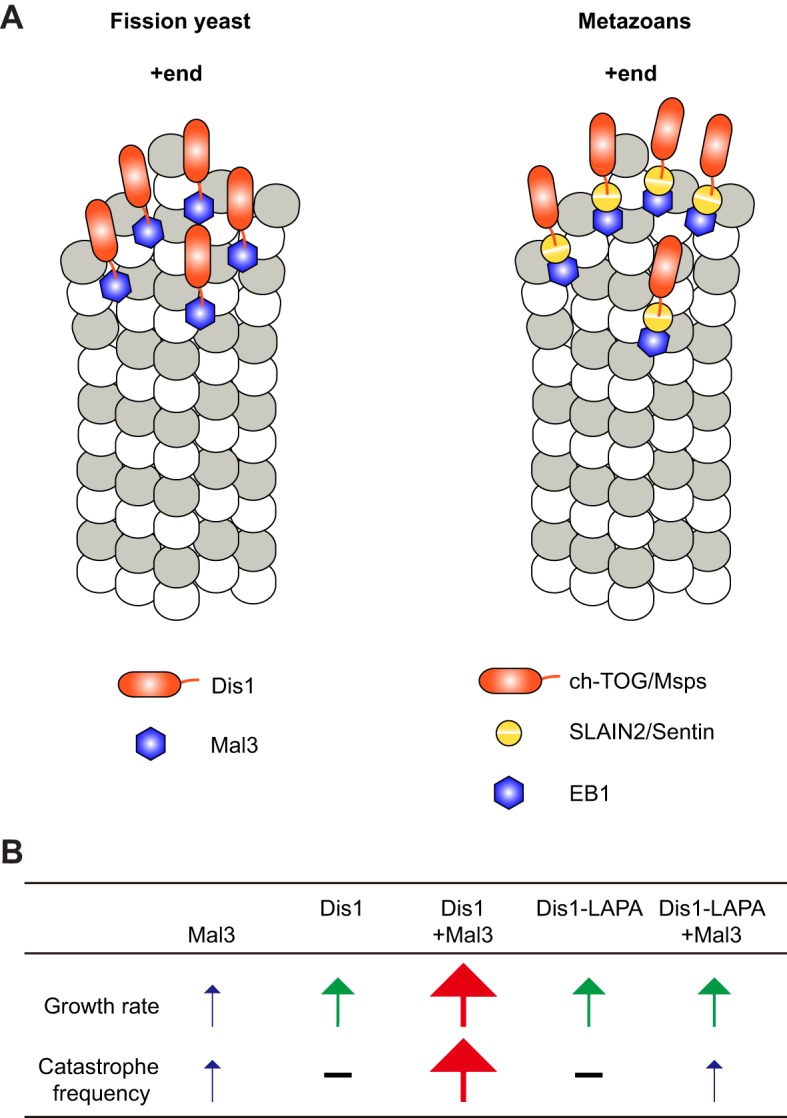
Interplay between Dis1 and Mal3 and comparison to metazoans. (A) In fission yeast, Dis1 (orange) binds directly to Mal3 (blue), thereby enhancing the MT growth rate and the catastrophe frequency in a synergistic manner (left). By contrast, in metazoans including humans and Drosophila, ch-TOG and Msps (orange) interact with EB1 (blue) through SLAIN2 and Sentin (yellow), respectively (right), thereby accelerating MT dynamics. Dimer formation of EB1/Mal3 is not shown for simplicity. (B) Summary of individual parameters representing MT dynamics in the presence of Dis1 (wild-type or LAPA) and/or Mal3. –, not altered compared to control samples (tubulin only); blue arrow, increased by up to twofold; green arrow, increased by ∼threefold; red arrow, increased by more than fourfold.
Here, we have solved the first X-ray structure of the EBH domain of Mal3, the protein originally used to reconstitute autonomous MT end tracking by an EB protein and its recruitment of other +TIPs to growing MT ends (Bieling et al., 2007). Our crystallographic analysis shows the two hydrophobic pockets that are formed by the C-terminal part of the Mal3 EBH domains and to which typically recruited +TIPs bind through their canonical SXIP motif. This pocket is conserved, in agreement with the observation of the SXIP motif containing proteins being recruited by Mal3 in vitro (Bieling et al., 2007). The polypeptide fold, however, is unconventional, because a hydrophobic pocket is formed from helices of two Mal3 polypeptides (trans-configuration), whereas in EB1 a hydrophobic pocket is formed by the corresponding helices of a single EB1 polypeptide (cis-configuration).
The interaction of the non-canonical Dis1 peptide with the hydrophobic pocket is also unconventional compared to the binding of SXIP-motif-containing peptides. Although both peptides interact strongly with the hydrophobic cavity of the EBH domain, their exact binding interface varies (Fig. 6E). Owing to these differences, the binding interface in Mal3–Dis1 is at 90° to the linker between the two helices in the EB monomer, whereas in EB1–MACF (containing the canonical SXIP motif) it is on the same side (Fig. 6C–E; Fig. S4C,D). Hence, the SXIP motif is not be the sole motif that can interact with the EBH domain of EB1. In fact, previous comprehensive proteomic analysis of EB1-binding proteins identified a fairly large number of interactors that do not contain the canonical SXIP sequence (Jiang et al., 2012; Tamura et al., 2015). Therefore, it is likely that non-canonical EB1 interaction motifs similar to the one we discovered in Dis1 will exist in proteins of other organisms than fission yeast, likely also including humans.
Previous work has shown that Dis1 localises to the mitotic kinetochore through interaction with the Ndc80 (also known as Hec1) outer kinetochore component and that it plays a crucial role in proper kinetochore–MT attachment (Hsu and Toda, 2011). It is, therefore, likely that Dis1 and Mal3 interact at the kinetochore–MT interface upon kinetochore capture by the MT plus-ends. Interestingly, Alp14 also localises to the mitotic kinetochore, yet in a different manner; Alp7 (a TACC family protein) binds to Ndc80, thereby recruiting the Alp7–Alp14 complex to the kinetochore (Sato et al., 2004; Tang et al., 2015). The result that Δalp14 and Mal3-binding defective dis1-LAPA display synthetic lethality indicates that these two fission yeast TOG members in concert establish and maintain kinetochore–MT attachment by exploiting distinct binding partners, Mal3 for Dis1 and Alp7 for Alp14.
Intriguingly, the interaction between TOG and the Ndc80 complex is conserved in budding yeast and humans (Miller et al., 2016). Importantly, TOG orthologues of these two organisms (Stu2 and ch-TOG) regulate kinetochore–MT attachment in a tension-sensitive manner. Intriguingly, in vitro work showed that the TOG proteins play this crucial role without altering MT dynamics (Miller et al., 2016). However, in cells undergoing mitosis, the fine-tuning of MT dynamics and the state of tension should temporally be coupled. We envisage that the EB1 proteins might be important proteins promoting proper kinetochore–MT attachment in response to the state of tension. Given these circumstances, in vitro studies examining addition of the EB1 proteins (and SLAIN2) to TOG and the Ndc80 complex would be of great interest to explore this possibility.
In conclusion, our work provides new insight into the unconventional mode through which Mal3 binds with Dis1, which is likely to indicate the possibility of further modes of interactions for other uncharacterised EB1 interactors. From an evolutionary point of view, the interaction between EB1 and TOG proteins is probably ubiquitous; intriguingly, however, the mechanism by which they interact varies among different organisms, illuminating diversification and remarkable adaptability through evolution.
MATERIALS AND METHODS
Protein expression and purification
dis1 cDNA optimised for E. coli codons was synthesised (GeneArt Gene Synthesis) and subcloned into the pETM-11 vector (Maurer et al., 2011). The purification of the various Dis1–eGFP constructs was performed as described previously (Maurer et al., 2011) except that Dis1 buffer [50 mM Tris-HCl pH 7.5, 300 mM KCl, 10% (v/v) glycerol, 1 mM EDTA and 1 mM DTT] containing EDTA-free protease inhibitors (Roche) and IgG Sepharose 6 Fast Flow (GE Healthcare) were used. In addition, TEV protease (for Dis1–eGFP) or Precision protease (for Dis1–HA) was used. The purification of the Mal3, Mal3–eGFP and Mal3–mCherry was performed as described previously (Maurer et al., 2011).
The EBH domain (amino acids 174–247) of Mal3 was purified by the same procedures as described above except that buffer A (50 mM Tris-HCl pH7.5, 500 mM NaCl and 10 mM β-mercaptoethanol) containing 30 mM imidazole was used for cell suspension and column washing. In addition, His-tag purification resin (Roche) and buffer A supplemented with 500 mM imidazole were used for affinity purification and elution, respectively. The cleavage of the oligo-histidine tag was performed by incubation with TEV protease as described earlier. Mal3 proteins were further purified by gel filtration using Superose200 10/300 columns equilibrated with buffer B (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM DTT).
Porcine brain tubulin was purified and cycled tubulin fractions were labelled with Cy5 (Lumiprobe) or EZ-link NHS Biotin (Thermo Scientific) by standard methods as described previously (Bieling et al., 2007). The protein concentration was determined by Bradford assay (Bio-Rad).
MT co-sedimentation and binding assays
The MT co-sedimentation assay was performed as described previously (Maurer et al., 2011). 0.2 µM Dis1–eGFP was incubated with taxol-stabilised MTs at concentrations from 0 to 4 µM in BRB80 supplemented with 85 mM KCl and 85 mM CH3COOK in the presence of 20 µM taxol.
Analytical gel filtration
5 µM Dis1–eGFP and/or 10 µM tubulin (or 5 µM Dis1–HA and/or 20 µM Mal3) was incubated in gel filtration buffer (25 mM K-HEPES pH 7.5, 200 mM KCl, 1 mM EGTA, 1 mM MgCl2) before loading on a Superose 6 10/300 (GE Healthcare) equilibrated with gel filtration buffer. The absorbance of the eluted protein was measure at 280 nm.
TIRF microscopy and image analysis
TIRF-microscopy-based dynamic MT assays were performed as previously described (Bieling et al., 2010). TIRF assay buffer consisted of BRB80 supplemented with 85 mM KCl, 1 mM GTP, 10 mM β-mercaptoethanol, 0.1% Brij-35, 0.1% methylcellulose (Sigma-Aldrich) and an oxygen scavenger system [glucose, glucose oxidase (Serva) and catalase (Sigma-Aldrich)]. For simultaneous dual (or triple)-colour time-lapse imaging of the Cy5 and GFP (and mCherry) channel, imaging was performed at 1-s intervals with 100 ms exposure time, using a ×100 objective lens at 30±1°C on a custom TIRF microscope equipped with a Cascade II, cooled charge-coupled device camera (Photometrics), illuminating the sample with 488 nm and 640 nm (and 561 nm) lasers. Image analysis was performed as described previously (Duellberg et al., 2014).
GST pulldown assay
For domain analysis of the Dis1 protein, 1 µM of GST or GST–Mal3 and 0.5 µM of several truncated Dis1–eGFP proteins were incubated in pulldown buffer (BRB80 supplemented with 85 mM KCl, 0.01% Brij-35 and 1 mM DTT). Then, 20 µl of glutathione–Sepharose beads (GE Healthcare) were added and after incubation for 1 h at 4°C, beads were washed and eluted with the buffer containing 10 mM reduced glutathione. For domain analysis of Mal3, 0.5 µM Dis1 C1-eGFP and 1 µM of several truncated GST–Mal3 proteins were used and the same procedures were followed.
For the peptide competition assay, 1 µM of purified GST–Mal3 bound to the glutathione beads were pre-incubated with various concentration of the Dis1 peptide (amino acids 833–852; 0, 0.5, 2.5, 5.0, 50, 100 µM) in pulldown buffer for 30 min, followed by further incubation with 0.5 µM Dis1 C2-eGFP for 1 h.
Peptide array assay
Peptide array assays were performed as previously described (Hsu and Toda, 2011).
The blocked membrane was incubated with 1 µg/ml Mal3–HA protein and bound Mal3–HA was detected by immunoblotting with an anti-HA antibody (diluted 1:1000, 16B12, Covance).
X-ray crystallography
X-ray data were collected on beamlines I04 and I03 of the Diamond Light Source. Data were integrated and scaled using the Xia2 pipeline. The unliganded Mal3 EBH (hereafter Mal3) domain was solved using ab initio molecular replacement as implemented in the ARCIMBOLDO-LIGHT package (Sammito et al., 2015). 20-residue α-helical segments were used as a search model. A strong (30.2% origin) native Patterson peak at 0.155, 0.5, 0.5 in the native data suggested the presence of translational pseudo-symmetry, and the appropriate correction was applied in Phaser. The preliminary poly-alanine model was then assigned sequence in Buccaneer, and manually rebuilt and refined with Coot (Emsley and Cowtan, 2004) and Refmac5 (Murshudov et al., 2011). The final model was refined with anisotropic temperature factors applied to all atoms. The Mal3–Dis1 structure was solved by molecular replacement in Phaser (McCoy et al., 2007) using the apo-Mal3 structure as a search model. Model building and refinement were carried out as for the apo-structure. Clear density could be seen for the bound Dis1 peptides in an unbiased difference map at both homodimer interfaces and was built in the final stages of refinement. All data collection and refinement statistics are provided in Table S4.
Crystallisation and structure solution
Crystals were grown by sitting-drop vapour diffusion. The Mal3174-247 proteins at 7 mg/ml were mixed with crystallisation solution comprising 0.2 M NaCl, 30% MPD and 0.1 M sodium acetate at pH 4.6. Crystals grew to full size after 5 days and were flash-cooled in liquid nitrogen with no additional cryoprotection. To obtain the Mal3 EBH domain-Dis1 complex, the Mal3174-247 protein was incubated with the Dis1 peptide (residues 833-852) in a 1:5 molar ration for 1 h at 4°C before crystallisation. Crystals were grown by sitting-drop vapour diffusion. The protein complex at 7 mg/ml was mixed with the crystallisation solution consisting of 0.2 M MgCl2, 0.1 M HEPES pH 7, 20% PEG 6K. Crystals grew to full size after a week and were flash-cooled in liquid nitrogen.
Immunoprecipitation
Immunoprecipitation was performed as previously described (Hsu and Toda, 2011) except that immunoprecipitation buffer [50 mM Na-HEPES, pH7.5, 150 mM NaCl, 1 mM EDTA, 0.1% Tween20, 1 mM DTT, protease inhibitor cocktail (Sigma-Aldrich)], GFP-Trap A and an anti-FLAG antibody (diluted 1:2000, Anti-FLAG M2, Sigma-Aldrich) or an anti-GFP antibody (diluted 1:2000, Anti-eGFP 632569, Clontech) was used.
Fission yeast strains, media, genetic methods, minichromosome loss assay and cell biology
Fission yeast strains used in this study are listed in Table S5. Standard fission yeast methodologies were followed (Moreno et al., 1991). Spot assays were carried out by spotting 5–10 μl of cells at a concentration of 2×106 cells/ml after 10-fold serial dilutions onto rich YE5S plates with or without a drug (10 µg/ml TBZ). C-terminal tagging and gene disruption were performed using PCR-generated fragments as described previously (Bähler et al., 1998; Sato et al., 2005). The minichromosome loss assay was carried out as described previously (Niwa et al., 1989; Tange et al., 2016).
Fluorescence microscope images were obtained using the DeltaVision microscope system (Applied Precision, Inc.) with a cooled CCD camera CoolSNAP.HQ (Photometrics). Live cells were imaged in a glass-bottomed culture dish (MatTek Corporation) coated with soybean lectin.
Statistical data analysis
In all the experiments in which statistical data are presented, the number of experiments and sample size are described in the main text, tables and figure legends. Results are expressed as the mean±s.e.m. or mean±s.d. Statistical evaluation was performed using Prism (GraphPad). All P-values are from two-tailed unpaired Student t-tests. We followed this key for asterisk placeholders for P-values in the figures: *P<0.01, n.s. not significant.
Note added in proof
After acceptance of this manuscript, a new paper on the interaction between budding yeast Kar9 and Bim1 (the yeast EB1) was published (Manatschal et al., 2016). In this work, it is shown that the amino acid sequence containing TRLRPPTPLS binds to Bim1. We notice that ‘TRLRPPTPLS’ is very similar to ‘SMLQKPTQFS’ included in the Dis1 peptide. It is possible that Kar9 binds to Bim1 in a non-canonical manner analogous to the binding between Dis1 and Mal3.
Acknowledgements
We are grateful to Hirofumi Takada for his contribution to the initial part of this study, N. O'Reilly for peptide synthesis, A. Purkiss for assistance with X-ray data collection, Yasuto Murayama for plasmid and Yasutaka Kakui, all members of the Cell Regulation Laboratory and MT Cytoskeleton Laboratory for discussion and technical advice.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Y.M., S.P.M., M.R.S., T.S. and T.T. designed the experiments. M.Y. performed most experiments. S.P.M. performed initial protein purifications and TIRF-microscopy experiments, and S.Z. and M.R.S. performed crystallisation and X-ray data acquisition and analysis. M.Y. analysed dis1-LAPA mutants under microscopy. Y.M., M.R.S., T.S. and T.T. wrote the manuscript with input from S.P.M., M.Y. and S.Z.
Funding
This work was supported by Cancer Research UK and the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001155, FC001163, FC001184), the UK Medical Research Council (FC001155, FC001163, FC001184), and the Wellcome Trust (FC001155, FC001163, FC001184), the Japan Society for the Promotion of Science (JSPS) [KAKENHI Scientific Research (A) (16H02503 to T.T.), a Challenging Exploratory Research grant (16K14672 to T.T.), Scientific Research (C) (16K07694 to M.Y.)], the Naito Foundation (T.T.) and a Marie Curie fellowship from the European Commission (PIEF-GA-2009-253043 to S.P.M.). Deposited in PMC for immediate release.
Data availability
Atomic coordinates and structure factors of Mal3 174–247 domain and the Mal3 174–247 domain interacting with Dis1 peptide have been deposited in the Protein Data Bank under accession codes 5M97 and 5M9E, respectively.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.197533.supplemental
References
- Akhmanova A. and Hoogenraad C. C. (2005). Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 17, 47-54. 10.1016/j.ceb.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Akhmanova A. and Steinmetz M. O. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309-322. 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Al-Bassam J. and Chang F. (2011). Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 21, 604-614. 10.1016/j.tcb.2011.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S. C. and Hyman A. (2006). Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 172, 1009-1022. 10.1083/jcb.200511010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., Kim H., Flor-Parra I., Lal N., Velji H. and Chang F. (2012). Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol. Biol. Cell 23, 2878-2890. 10.1091/mbc.E12-03-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Nakaseko Y., Kinoshita K., Goshima G. and Yanagida M. (2006). Cdc2 phosphorylation of the fission yeast Dis1 ensures accurate chromosome segregation. Curr. Biol. 16, 1627-1635. 10.1016/j.cub.2006.06.065 [DOI] [PubMed] [Google Scholar]
- Asakawa K. and Toda T. (2006). Cooperation of EB1-Mal3 and the Bub1 spindle checkpoint. Cell Cycle 5, 27-30. 10.4161/cc.5.1.2262 [DOI] [PubMed] [Google Scholar]
- Asakawa K., Toya M., Sato M., Kanai M., Kume M., Goshima T., Garcia M. A., Hirata D. and Toda T. (2005). Mal3, the fission yeast EB1 homolog. cooperates with Bub1 spindle checkpoint to prevent monopolar attachment. EMBO Rep. 6, 1194-1120. 10.1038/sj.embor.7400540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K., Kume K., Kanai M., Goshima T., Miyahara K., Dhut S., Tee W. W., Hirata D. and Toda T. (2006). The V260I mutation in fission yeast α-tubulin Atb2 affects microtubule dynamics and EB1-Mal3 localization and activates the Bub1 branch of the spindle checkpoint. Mol. Biol. Cell 17, 1422-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Ye X., Huddleston P., Brautigam C. A. and Rice L. M. (2012). A TOG:αβ-tubulin complex structure reveals conformation-based mechanisms for a microtubule polymerase. Science 337, 857-860. 10.1126/science.1221698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz P., Munyoki S., Geyer E. A., Piedra F.-A., Vu E. S., Bromberg R., Otwinowski Z., Grishin N. V., Brautigam C. A. and Rice L. M. (2014). A tethered delivery mechanism explains the catalytic action of a microtubule polymerase. eLife 3, e03069 10.7554/eLife.03069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu J.-Q., Longtine M. S., Shah N. G., McKenzie A. III, Steever A. B., Wach A., Philippsen P. and Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951. [DOI] [PubMed] [Google Scholar]
- Beinhauer J. D., Hagan I. M., Hegemann J. H. and Fleig U. (1997). Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 139, 717-728. 10.1083/jcb.139.3.717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Laan L., Schek H., Munteanu E. L., Sandblad L., Dogterom M., Brunner D. and Surrey T. (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100-1105. 10.1038/nature06386 [DOI] [PubMed] [Google Scholar]
- Bieling P., Kandels-Lewis S., Telley I. A., van Dijk J., Janke C. and Surrey T. (2008). CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J. Cell Biol. 183, 1223-1233. 10.1083/jcb.200809190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P., Telley I. A., Hentrich C., Piehler J. and Surrey T. (2010). Fluorescence microscopy assays on chemically functionalized surfaces for quantitative imaging of microtubule, motor, and +TIP dynamics. Methods Cell Biol. 95, 555-580. 10.1016/S0091-679X(10)95028-0 [DOI] [PubMed] [Google Scholar]
- Brouhard G. J., Stear J. H., Noetzel T. L., Al-Bassam J., Kinoshita K., Harrison S. C., Howard J. and Hyman A. A. (2008). XMAP215 is a processive microtubule polymerase. Cell 132, 79-88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning H., Hackney D. D. and Nurse P. (2003). Targeted movement of cell end factors in fission yeast. Nat. Cell Biol. 5, 812-818. 10.1038/ncb1034 [DOI] [PubMed] [Google Scholar]
- Buey R. M., Sen I., Kortt O., Mohan R., Gfeller D., Veprintsev D., Kretzschmar I., Scheuermann J., Neri D., Zoete V. et al. (2012). Sequence determinants of a microtubule tip localization signal (MtLS). J. Biol. Chem. 287, 28227-28242. 10.1074/jbc.M112.373928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K. E. and Brunner D. (2004). The microtubule plus end-tracking proteins mal3p and tip1p cooperate for cell-end targeting of interphase microtubules. Curr. Biol. 14, 548-559. 10.1016/j.cub.2004.03.029 [DOI] [PubMed] [Google Scholar]
- Busch K. E., Hayles J., Nurse P. and Brunner D. (2004). Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev. Cell 6, 831-843. 10.1016/j.devcel.2004.05.008 [DOI] [PubMed] [Google Scholar]
- De Groot C. O., Jelesarov I., Damberger F. F., Bjelic S., Scharer M. A., Bhavesh N. S., Grigoriev I., Buey R. M., Wuthrich K., Capitani G. et al. (2010). Molecular insights into mammalian end-binding protein heterodimerization. J. Biol. Chem. 285, 5802-5814. 10.1074/jbc.M109.068130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A., Katsuki M., Drummond D. R., Osei M., Cross R. A. and Amos L. A. (2008). Mal3, the Schizosaccharomyces pombe homolog of EB1, changes the microtubule lattice. Nat. Struct. Mol. Biol. 15, 1102-1108. 10.1038/nsmb.1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A. and Mitchison T. J. (1997). Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13, 83-117. 10.1146/annurev.cellbio.13.1.83 [DOI] [PubMed] [Google Scholar]
- Duellberg C., Fourniol F. J., Maurer S. P., Roostalu J. and Surrey T. (2013). End-binding proteins and Ase1/PRC1 define local functionality of structurally distinct parts of the microtubule cytoskeleton. Trends Cell Biol. 23, 54-63. 10.1016/j.tcb.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Duellberg C., Trokter M., Jha R., Sen I., Steinmetz M. O. and Surrey T. (2014). Reconstitution of a hierarchical +TIP interaction network controlling microtubule end tracking of dynein. Nat. Cell Biol. 16, 804-811. 10.1038/ncb2999 [DOI] [PubMed] [Google Scholar]
- Emsley P. and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126-2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fleig U., Sen-Gupta M. and Hegemann J. H. (1996). Fission yeast mal2+ is required for chromosome segregation. Mol. Cell. Biol. 16, 6169-6177. 10.1128/MCB.16.11.6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galjart N. (2010). Plus-end-tracking proteins and their interactions at microtubule ends. Curr. Biol. 20, R528-R537. 10.1016/j.cub.2010.05.022 [DOI] [PubMed] [Google Scholar]
- Garcia M. A., Vardy L., Koonrugsa N. and Toda T. (2001). Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 20, 3389-3401. 10.1093/emboj/20.13.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. A., Koonrugsa N. and Toda T. (2002). Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr. Biol. 12, 610-621. 10.1016/S0960-9822(02)00761-3 [DOI] [PubMed] [Google Scholar]
- George A. A. and Walworth N. C. (2015). Escape from mitotic arrest: an unexpected connection between microtubule dynamics and epigenetic regulation of centromeric chromatin in Schizosaccharomyces pombe. Genetics 201, 1467-1478. 10.1534/genetics.115.181792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluszek A. A., Cullen C. F., Li W., Battaglia R. A., Radford S. J., Costa M. F., McKim K. S., Goshima G. and Ohkura H. (2015). The microtubule catastrophe promoter Sentin delays stable kinetochore-microtubule attachment in oocytes. J. Cell Biol. 211, 1113-1120. 10.1083/jcb.201507006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I. and Ikura M. (2003). Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278, 36430-36434. 10.1074/jbc.M305773200 [DOI] [PubMed] [Google Scholar]
- Honnappa S., John C. M., Kostrewa D., Winkler F. K. and Steinmetz M. O. (2005). Structural insights into the EB1-APC interaction. EMBO J. 24, 261-269. 10.1038/sj.emboj.7600590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S., Okhrimenko O., Jaussi R., Jawhari H., Jelesarov I., Winkler F. K. and Steinmetz M. O. (2006). Key interaction modes of dynamic +TIP networks. Mol. Cell 23, 663-671. 10.1016/j.molcel.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Honnappa S., Gouveia S. M., Weisbrich A., Damberger F. F., Bhavesh N. S., Jawhari H., Grigoriev I., van Rijssel F. J. A., Buey R. M., Lawera A. et al. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell 138, 366-376. 10.1016/j.cell.2009.04.065 [DOI] [PubMed] [Google Scholar]
- Hsu K.-S. and Toda T. (2011). Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr. Biol. 21, 214-220. 10.1016/j.cub.2010.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann F., Drummond D. R., Peet D. R., Martin D. S. and Cross R. A. (2016). Alp7/TACC-Alp14/TOG generates long-lived, fast-growing MTs by an unconventional mechanism. Sci. Rep. 6, 20653 10.1038/srep20653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Toedt G., Montenegro Gouveia S., Davey N. E., Hua S., van der Vaart B., Grigoriev I., Larsen J., Pedersen L. B., Bezstarosti K. et al. (2012). A proteome-wide screen for mammalian SxIP motif-containing microtubule plus-end tracking proteins. Curr. Biol. 22, 1800-1807. 10.1016/j.cub.2012.07.047 [DOI] [PubMed] [Google Scholar]
- Kakui Y., Sato M., Okada N., Toda T. and Yamamoto M. (2013). Microtubules and Alp7-Alp14 (TACC-TOG) reposition chromosomes before meiotic segregation. Nat. Cell Biol. 15, 786-796. 10.1038/ncb2782 [DOI] [PubMed] [Google Scholar]
- Katsuki M., Drummond D. R., Osei M. and Cross R. A. (2009). MAL3 masks catastrophe events in Schizosaccharomyces pombe microtubules by inhibiting shrinkage and promoting rescue. J. Biol. Chem. 284, 29246-29250. 10.1074/jbc.C109.052159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerres A., Vietmeier-Decker C., Ortiz J., Karig I., Beuter C., Hegemann J., Lechner J. and Fleig U. (2004). The fission yeast kinetochore component Spc7 associates with the EB1 family member Mal3 and is required for kinetochore-spindle association. Mol. Biol. Cell 15, 5255-5267. 10.1091/mbc.E04-06-0443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K., Habermann B. and Hyman A. A. (2002). XMAP215: a key component of the dynamic microtubule cytoskeleton. Trends Cell Biol. 12, 267-273. 10.1016/S0962-8924(02)02295-X [DOI] [PubMed] [Google Scholar]
- Li W., Miki T., Watanabe T., Kakeno M., Sugiyama I., Kaibuchi K. and Goshima G. (2011). EB1 promotes microtubule dynamics by recruiting Sentin in Drosophila cells. J. Cell Biol. 193, 973-983. 10.1083/jcb.201101108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moriwaki T., Tani T., Watanabe T., Kaibuchi K. and Goshima G. (2012). Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J. Cell Biol. 199, 849-862. 10.1083/jcb.201206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S., McLean J. R., Chen C.-T., Gould K. L. and McCollum D. (2012). The kinesin-14 Klp2 is negatively regulated by the SIN for proper spindle elongation and telophase nuclear positioning. Mol. Biol. Cell 23, 4592-4600. 10.1091/mbc.E12-07-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manatschal C., Farcas A. M., Degen M. S., Bayer M., Kumar A., Landgraf C., Volkmer R., Barral Y. and Steinmetz M. O. (2016). Molecular basis of Kar9-Bim1 complex function during mating and spindle positioning. Mol. Biol Cell. doi:10.1091/mbc.E16-07-0552 10.1091/mbc.E16-07-0552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer S. P., Bieling P., Cope J., Hoenger A. and Surrey T. (2011). GTPγS microtubules mimic the growing microtubule end structure recognized by end-binding proteins (EBs). Proc. Natl. Acad. Sci. USA 108, 3988-3993. 10.1073/pnas.1014758108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer S. P., Fourniol F. J., Bohner G., Moores C. A. and Surrey T. (2012). EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell 149, 371-382. 10.1016/j.cell.2012.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer S. P., Cade N. I., Bohner G., Gustafsson N., Boutant E. and Surrey T. (2014). EB1 accelerates two conformational transitions important for microtubule maturation and dynamics. Curr. Biol. 24, 372-384. 10.1016/j.cub.2013.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C. and Read R. J. (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658-674. 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M. P., Asbury C. L. and Biggins S. (2016). A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell 165, 1428-1439. 10.1016/j.cell.2016.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T. and Kirschner M. W. W. (1984). Dynamic instability of microtubule growth. Nature 312, 237-242. 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Mohan R., Katrukha E. A., Doodhi H., Smal I., Meijering E., Kapitein L. C., Steinmetz M. O. and Akhmanova A. (2013). End-binding proteins sensitize microtubules to the action of microtubule-targeting agents. Proc. Natl. Acad. Sci. USA 110, 8900-8905. 10.1073/pnas.1300395110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A. and Nurse P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823. 10.1016/0076-6879(91)94059-L [DOI] [PubMed] [Google Scholar]
- Moriwaki T. and Goshima G. (2016). Five factors can reconstitute all three phases of microtubule polymerization dynamics. J. Cell Biol. 215, 305-307. 10.1083/jcb.201604118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F. and Vagin A. A. (2011). REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355-367. 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Kurooka H., Takeuchi M., Kinoshita K., Nakaseko Y. and Yanagida M. (1995). p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev. 9, 1572-1585. 10.1101/gad.9.13.1572 [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa T., Straight A. F., Murray A., Chikashige Y., Yamashita Y. M., Hiraoka Y. and Yanagida M. (1998). Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell 9, 3211-3225. 10.1091/mbc.9.11.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y., Nabeshima K., Kinoshita K. and Yanagida M. (1996). Dissection of fission yeast microtubule associating protein p93dis1: regions implicated in regulated localization and microtubule interaction. Genes Cells 1, 633-644. 10.1046/j.1365-2443.1996.00253.x [DOI] [PubMed] [Google Scholar]
- Nakaseko Y., Goshima G., Morishita J. and Yanagida M. (2001). M phase-specific kinetochore proteins in fission yeast microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr. Biol. 11, 537-549. 10.1016/S0960-9822(01)00155-5 [DOI] [PubMed] [Google Scholar]
- Niwa O., Matsumoto T., Chikashige Y. and Yanagida M. (1989). Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 8, 3045-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Adachi Y., Kinoshita N., Niwa O., Toda T. and Yanagida M. (1988). Cold-sensitive and caffeine supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J. 7, 1465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Garcia M. A. and Toda T. (2001). Dis1/TOG universal microtubule adaptors - one MAP for all? J. Cell Sci. 114, 3805-3812. [DOI] [PubMed] [Google Scholar]
- Podolski M., Mahamdeh M. and Howard J. (2014). Stu2, the budding yeast XMAP215/Dis1 homolog, promotes assembly of yeast microtubules by increasing growth rate and decreasing catastrophe frequency. J. Biol. Chem. 289, 28087-28093. 10.1074/jbc.M114.584300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber S. B., Baumgart J., Widlund P. O., Pozniakovsky A., Howard J., Hyman A. A. and Jülicher F. (2013). XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat. Cell Biol. 15, 1116-1122. 10.1038/ncb2834 [DOI] [PubMed] [Google Scholar]
- Roostalu J., Cade N. I. and Surrey T. (2015). Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 17, 1422-1434. 10.1038/ncb3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammito M., Millán C., Frieske D., Rodríguez-Freire E., Borges R. J. and Usón I. (2015). ARCIMBOLDO_LITE: single-workstation implementation and use. Acta Crystallogr. D Biol. Crystallogr. 71, 1921-1930. 10.1107/S1399004715010846 [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S. J., Crawley K., Karig I., Buck V., Meadows J. C., Franco-Sanchez A., Fleig U., Toda T. and Millar J. B. A. (2005). The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24, 2931-2943. 10.1038/sj.emboj.7600761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Vardy L., Garcia M. A., Koonrugsa N. and Toda T. (2004). Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol. Biol. Cell 15, 1609-1622. 10.1091/mbc.E03-11-0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Dhut S. and Toda T. (2005). New drug-resistant cassettes for gene disruption and epitope tagging in Schizosaccharomyces pombe. Yeast 22, 583-591. 10.1002/yea.1233 [DOI] [PubMed] [Google Scholar]
- Shirasu-Hiza M., Coughlin P. and Mitchison T. (2003). Identification of XMAP215 as a microtubule-destabilizing factor in Xenopus egg extract by biochemical purification. J. Cell Biol. 161, 349-358. 10.1083/jcb.200211095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep K. C., Rogers S. L., Elliott S. L., Ohkura H., Kolodziej P. A. and Vale R. D. (2005). Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J. Cell Biol. 168, 587-598. 10.1083/jcb.200410114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamada H. and Yanagida M. (1994). Fission yeast minichromosome loss mutants mis cause lethal aneuploidy and replication abnormality. Mol. Biol. Cell 5, 1145-1158. 10.1091/mbc.5.10.1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita N., Mania D., Herrero S., Ishitsuka Y., Nienhaus G. U., Podolski M., Howard J. and Fischer R. (2013). The cell-end marker TeaA and the microtubule polymerase AlpA contribute to microtubule guidance at the hyphal tip cortex of Aspergillus nidulans to provide polarity maintenance. J. Cell Sci. 126, 5400-5411. 10.1242/jcs.129841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N., Simon J. E., Nayak A., Shenoy R., Hiroi N., Boilot V., Funahashi A. and Draviam V. M. (2015). A proteomic study of mitotic phase-specific interactors of EB1 reveals a role for SXIP-mediated protein interactions in anaphase onset. Biol. Open 4, 155-169. 10.1242/bio.201410413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N. H. and Toda T. (2015). Alp7/TACC recruits kinesin-8-PP1 to the Ndc80 kinetochore protein for timely mitotic progression and chromosome movement. J. Cell Sci. 128, 354-363. 10.1242/jcs.160036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y., Chikashige Y., Takahata S., Kawakami K., Higashi M., Mori C., Kojidani T., Hirano Y., Asakawa H., Murakami Y. et al. (2016). Inner nuclear membrane protein Lem2 augments heterochromatin formation in response to nutritional conditions. Genes Cells 21, 812-832. 10.1111/gtc.12385 [DOI] [PubMed] [Google Scholar]
- Toyoda Y., Furuya K., Goshima G., Nagao K., Takahashi K. and Yanagida M. (2002). Requirement of chromatid cohesion proteins Rad21/Scc1 and Mis4/Scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12, 347-358. 10.1016/S0960-9822(02)00692-9 [DOI] [PubMed] [Google Scholar]
- Umesono K., Toda T., Hayashi S. and Yanagida M. (1983). Two cell division cycle genes NDA2 and NDA3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol. 168, 271-284. 10.1016/S0022-2836(83)80018-7 [DOI] [PubMed] [Google Scholar]
- van der Vaart B., Manatschal C., Grigoriev I., Olieric V., Gouveia S. M., Bjelić S., Demmers J., Vorobjev I., Hoogenraad C. C., Steinmetz M. O. et al. (2011). SLAIN2 links microtubule plus end-tracking proteins and controls microtubule growth in interphase. J. Cell Biol. 193, 1083-1099. 10.1083/jcb.201012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B., Coquelle F. M., Heichette C., Garnier C., Chrétien D. and Arnal I. (2008). EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat. Cell Biol. 10, 415-421. 10.1038/ncb1703 [DOI] [PubMed] [Google Scholar]
- Weisbrich A., Honnappa S., Jaussi R., Okhrimenko O., Frey D., Jelesarov I., Akhmanova A. and Steinmetz M. O. (2007). Structure-function relationship of CAP-Gly domains. Nat. Struct. Mol. Biol. 14, 959-967. 10.1038/nsmb1291 [DOI] [PubMed] [Google Scholar]
- Widlund P. O., Stear J. H., Pozniakovsky A., Zanic M., Reber S., Brouhard G. J., Hyman A. A. and Howard J. (2011). XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. Proc. Natl. Acad. Sci. USA 108, 2741-2746. 10.1073/pnas.1016498108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K., Hwang E., You L. and Huffaker T. C. (2006). The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell 17, 2789-2798. 10.1091/mbc.E05-09-0892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M., Stear J. H., Hyman A. A. and Howard J. (2009). EB1 recognizes the nucleotide state of tubulin in the microtubule lattice. PLoS ONE 4, e7585 10.1371/journal.pone.0007585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M., Widlund P. O., Hyman A. A. and Howard J. (2013). Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nat. Cell Biol. 15, 688-693. 10.1038/ncb2744 [DOI] [PubMed] [Google Scholar]
- Zhang R., Alushin G. M., Brown A. and Nogales E. (2015). Mechanistic origin of microtubule dynamic instability and Its modulation by EB proteins. Cell 162, 849-859. 10.1016/j.cell.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]