Abstract
During forebrain development, radial glia generate neurons through the production of intermediate progenitor cells (IPCs). The production of IPCs is a central tenet underlying the generation of the appropriate number of cortical neurons, but the transcriptional logic underpinning this process remains poorly defined. Here, we examined IPC production using mice lacking the transcription factor nuclear factor I/X (Nfix). We show that Nfix deficiency delays IPC production and prolongs the neurogenic window, resulting in an increased number of neurons in the postnatal forebrain. Loss of additional Nfi alleles (Nfib) resulted in a severe delay in IPC generation while, conversely, overexpression of NFIX led to precocious IPC generation. Mechanistically, analyses of microarray and ChIP-seq datasets, coupled with the investigation of spindle orientation during radial glial cell division, revealed that NFIX promotes the generation of IPCs via the transcriptional upregulation of inscuteable (Insc). These data thereby provide novel insights into the mechanisms controlling the timely transition of radial glia into IPCs during forebrain development.
KEY WORDS: Hippocampus, Intermediate progenitor cell, NFIX, Mouse
Summary: The Nfix and Nfib transcription factors are required for the timely transition of radial glia into intermediate progenitor cells during mouse forebrain development.
INTRODUCTION
The coordinated proliferation and lineage-specific differentiation of neural progenitor cells plays an integral role in the formation of the mammalian cerebral cortex. The primary neural progenitor cells that generate the neurons of this structure are the radial glia, which develop from neuroepithelial cells around embryonic day (E) 10.5 in rodents (Anthony et al., 2004; Mori et al., 2005). Since the large number of neurons generated during development come from a relatively small initial population of progenitor cells, the radial glial cell pool is first amplified by undergoing symmetric proliferative divisions, also known as self-expanding divisions. Subsequently, radial glial cells undergo asymmetric divisions to give rise to either a neuron that migrates directly to the cortical plate (direct neurogenesis), or, more frequently, an intermediate progenitor cell (IPC) (indirect neurogenesis) (Götz and Huttner, 2005; Huttner and Kosodo, 2005). IPCs are morphologically different from radial glia in that they are delaminated from the adherens junctional belt at the ventricular surface of the brain (Noctor et al., 2004). The majority of cortical neurons arise through the production, expansion and differentiation of IPCs (Haubensak et al., 2004; Sessa et al., 2008).
The timely generation of IPCs is required for normal neuron number in the postnatal brain. Despite the importance of IPCs, our understanding of the mechanism by which asymmetric division of radial glia is coordinated to ensure timely IPC production is limited. In the classical model of neural stem cell division, inferred largely from work in Drosophila melanogaster, large changes in spindle orientation result in the asymmetric inheritance of the apical membrane into one daughter cell and an asymmetric cell fate (Knoblich, 2008). However, the vast majority of radial glial cell cleavage planes in the mammalian telencephalon are perpendicular to the ventricular surface, and deviate only slightly from this angle. As a result, the apical membrane typically segregates into both daughter cells (Konno et al., 2008; Asami et al., 2011; Shitamukai et al., 2011). Therefore, it is unlikely that unequal segregation of the apical membrane accounts for IPC- and neuron-generating divisions in the mammalian cortex. Rather, one proposed model is that small fluctuations in cleavage plane orientation (reviewed by Matsuzaki and Shitamukai, 2015) lead to changes in cell volume and intracellular organelle inheritance to promote IPC production (Wang et al., 2009).
The argument that small fluctuations in spindle orientation promote IPC production largely comes from loss- and gain-of-function studies of the mammalian homolog of the D. melanogaster adaptor protein Inscuteable (INSC) (Konno et al., 2008; Postiglione et al., 2011; Petros et al., 2015). INSC regulates the spindle orientation of radial glia and IPC production in a gene dose-dependent manner, whereby loss of Insc reduces oblique divisions and IPC number, whereas high levels of Insc expression increase oblique divisions and IPC number (Postiglione et al., 2011; Petros et al., 2015). Currently, it remains unclear whether INSC-dependent spindle orientation directly regulates IPC production or whether another INSC-dependent mechanism might regulate the development of IPCs. However, a pivotal question that arises from these data is how Insc expression itself is regulated during cortical development to facilitate IPC development.
Transcription factors of the Nuclear factor one (Nfi) family (Nfia, Nfib, Nfix) play a crucial role in astrogliogenesis. Mice lacking Nfix exhibit markedly reduced numbers of astrocytes throughout the embryonic cerebral cortex and cerebellum (Piper et al., 2011; Heng et al., 2014). In addition to promoting astrocyte lineage progression, individual Nfi knockout mice also exhibit elevated numbers of progenitor cells and delayed expression of neuronal markers within the ammonic neuroepithelium of the presumptive hippocampus during embryonic development (Piper et al., 2010, 2014; Heng et al., 2014). From these findings we posited that NFIs could play a previously unrecognized role in the production of IPCs. Here, we use the ammonic neuroepithelium of mice lacking Nfix and Nfib as a model to investigate this hypothesis. We demonstrate that NFIs are autonomously required by radial glia for timely IPC production, and that NFIs directly activate the expression of Insc, providing novel insight into the cellular processes governing the transition of radial glial cells to IPCs during hippocampal development.
RESULTS
NFIX is expressed by radial glia and IPCs during hippocampal development
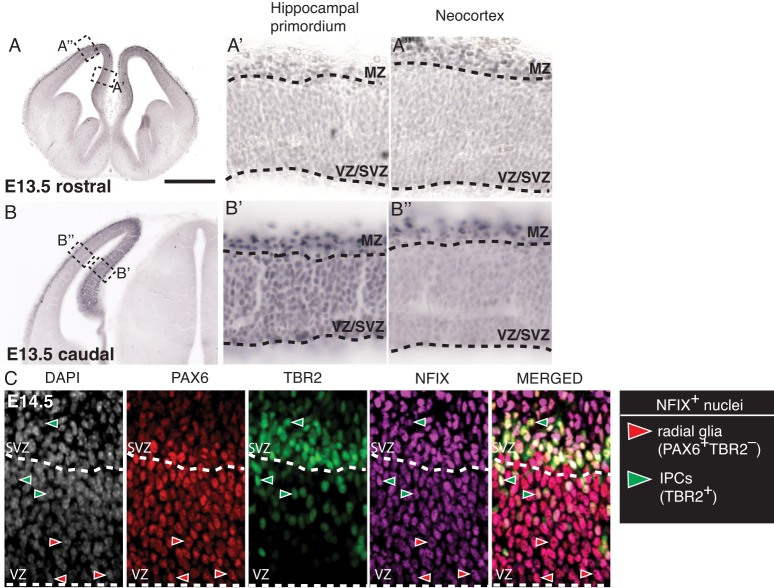
To investigate whether NFIX promotes indirect neurogenesis and IPC production during the development of the cerebral cortex, we focused predominantly on the ammonic neuroepithelium of the hippocampus. This region was chosen for two reasons; first, NFIX expression within the ventricular zone (VZ) and subventricular zone (SVZ) of the cerebral cortex exhibits a gradient, such that expression is highest in the caudomedial cortex, particularly within the presumptive hippocampal primordium (Fig. 1A,B); second, newly generated neurons from the ammonic neuroepithelium (predominantly pyramidal neurons) migrate only a short distance radially before settling in the cornu ammonis (CA) region (Altman and Bayer, 1990), rendering it a simpler model of development than the six-layered neocortex.
Fig. 1.
NFIX is expressed in hippocampal radial glia and IPCs. (A,B) NFIX immunohistochemistry at E13.5 in the mouse telencephalon. (A′,B′,A″,B″) Higher magnification of the boxed regions in A,B, with dashed lines demarcating the ventricular zone (VZ)/subventricular zone (SVZ) and marginal zone (MZ). (C) NFIX immunofluorescence within the presumptive hippocampus at E14.5. Dashed lines indicate the boundary between the VZ and SVZ. NFIX (magenta) colocalizes with both PAX6 (red) and TBR2 (green). NFIX+ radial glia and IPCs were identified as per the key to the right. Scale bar in A: A,B, 800 µm; A′,A″,B′,B″, 80 µm; C, 50 µm.
Previous studies have demonstrated that neural progenitor cells in the hippocampal neuroepithelium express NFIX during development from at least E13.5 onwards (Heng et al., 2014); however, the cell type-specific identity of these cells has not been established. The transition of radial glia to IPCs during development is demarcated by the sequential expression of PAX6 then TBR2 (EOMES) (Englund et al., 2005). We classified radial glia as PAX6+ TBR2– rather than using PAX6 expression alone as a marker for these cells, thereby excluding newborn radial glia-derived IPCs that also express low to moderate levels of PAX6 (Arai et al., 2011). Co-immunofluorescence at E14.5 (Fig. 1C) revealed that radial glia express NFIX (209/209 cells), as did the vast majority of IPCs (TBR2+ nuclei; 208/209 cells). Moreover, the expression of NFIX was maintained in radial glia and IPCs throughout the stages of the cell cycle, including mitosis (Fig. S1). This same expression pattern was also observed at E13.5 and E15.5 (Fig. S1).
Delayed IPC development in Nfix−/− mice
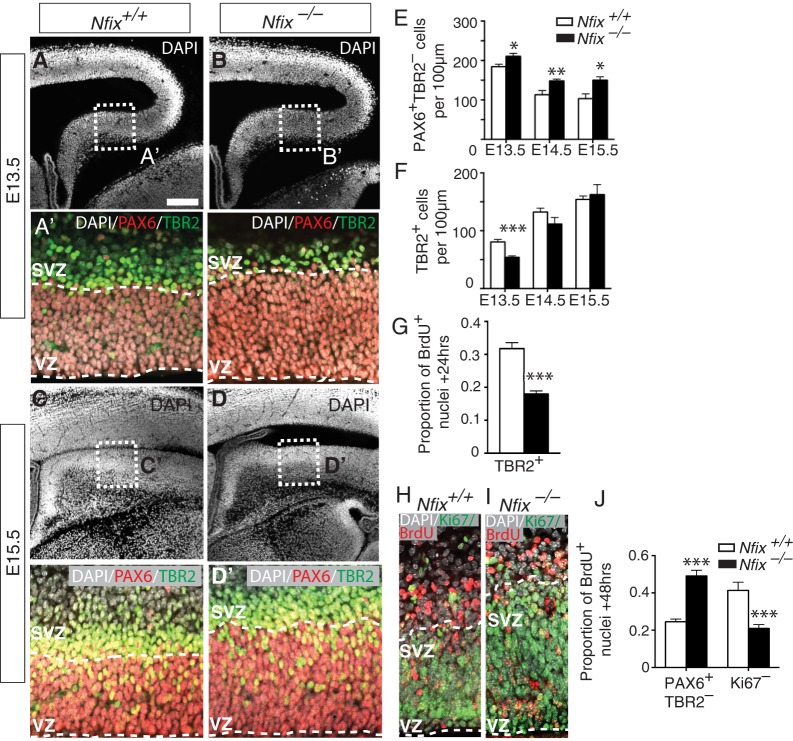
We have previously reported elevated numbers of PAX6+ cells (indicative of increased numbers of radial glial cells) and delayed neuronal differentiation in the hippocampus of Nfix−/− mice at E16.5 (Heng et al., 2014). It remained unclear, however, when these developmental defects first become apparent and which stage of the lineage transition from a stem cell to a mature neuron is affected in these mice. Using PAX6 and TBR2 colabeling, we investigated how the numbers of radial glial cells in the ammonic neuroepithelium of Nfix−/− mice change from the onset of hippocampal neurogenesis at E13.5 to the peak of neurogenesis at E15.5. Relative to controls, we found significantly more radial glia (PAX6+ TBR2–) in Nfix−/− mice at E13.5, E14.5 and E15.5 (Fig. 2A-E). Moreover, the magnitude of the change was smallest at E13.5 (P=0.022) and progressively increased at E14.5 (P=0.0096) and E15.5 (P=0.014), from which we infer that the lack of Nfix culminates in an ongoing (rather than a temporally restricted) delay in the transition of radial glia into IPCs. In support of this, we found fewer IPCs at E13.5 in Nfix−/− mice than in controls (P=0.00013) (Fig. 2F), demonstrating that radial glia generate IPCs less efficiently in these mice. By E14.5 and E15.5, the total number of IPCs in Nfix−/− mice was comparable to that in wild type (Fig. 2F), probably because the increased size of the radial glial pool resulted in an increase in the absolute number of IPC-generating divisions. However, measured as a proportion of all cell types in the VZ/SVZ, the number of IPCs remained reduced at E15.5 in Nfix−/− mice (data not shown).
Fig. 2.
Increased numbers of radial glia and delayed IPC generation in the hippocampus of Nfix−/− mice from E13.5-E15.5. (A-D) DAPI staining (white) in wild-type and Nfix−/− mice at E13.5 and E15.5. (A′-D′) Higher magnification of the boxed regions in A-D, showing DAPI (white), PAX6 (red) and TBR2 (green) staining, with dashed lines demarcating the VZ/SVZ. (E,F) Cell counts of (E) radial glia and (F) IPCs from E13.5-E15.5 in wild-type and Nfix−/− mice. Mean±s.e.m. of seven, eight and five embryos at E13.5, E14.5 and E15.5, respectively. *P<0.05, **P<0.01, ***P<0.001, ANOVA. (G) Cell counts of the proportion of total BrdU+ cells expressing TBR2 in wild-type and Nfix−/− mice at E14.5 following a BrdU chase at E13.5. Mean±s.e.m. of five embryos. ***P<0.001, t-test. (H,I) DAPI (white), BrdU (red) and Ki67 (green) staining in wild-type and Nfix−/− mice at E15.5 following a BrdU chase at E13.5. (J) Cell counts reveal the proportion of BrdU+ cells that were PAX6+ TBR2– or Ki67– in wild-type and Nfix−/− mice. Mean±s.e.m. of five embryos ***P<0.001, ANOVA. Scale bar in A: A-D, 220 µm; A′-D′, 50 µm; H,I, 40 µm.
Collectively, these data suggest that Nfix-deficient radial glial cells undergo increased self-expanding divisions and reduced IPC-generating divisions during early hippocampal development. To examine this further, we performed birth-dating experiments by injecting time-mated dams with the DNA analog BrdU at E13.5 before sacrificing embryos 24 h later, thereby labeling all proliferating cells (predominantly radial glia) that were in S-phase at the time of injection, as well as their progeny. We found that 24 h later (at E14.5), there were substantially fewer IPCs generated by BrdU-labeled radial glia in Nfix−/− mice than in controls (Fig. 2G; P=0.0001).
The impaired IPC development observed in Nfix−/− mice could potentially occur as a result of increased direct neurogenesis from radial glia, similar to that observed in Elp3 conditional knockout mice (Laguesse et al., 2015). To determine whether this was the case we performed a 48 h BrdU chase experiment (labeling with BrdU from E13.5). There were significantly fewer BrdU+ cells that had exited the cell cycle (BrdU+ Ki67–) in mutant mice relative to controls at E15.5 (P=0.00016; Fig. 2H-J). This argues that the impaired IPC development in Nfix−/− mice is not offset by an increase in direct neurogenesis. Indeed, a significantly greater number of BrdU+ cells remained as PAX6+ TBR2− radial glia at E15.5 (P=2.2×10−5). These data, coupled with the expansion of the radial glial population from E13.5-E15.5, support the hypothesis that Nfix-deficient radial glia undergo more self-expanding divisions at the expense of IPC development during this stage of hippocampal development.
Nfix-deficient radial glia have a longer S-phase
We next posited that Nfix-deficient radial glial cells should display cellular characteristics associated with fewer IPC-generating divisions. Interestingly, a recent study revealed that radial glia committed to neurogenic divisions have a shorter S-phase (1.8 h) than radial glia during self-expanding divisions (8 h) (Arai et al., 2011). To further assess if NFIX promotes IPC production, we measured S-phase duration as a proxy for the frequency of neurogenic divisions. We hypothesized that the population of Nfix-deficient radial glia would exhibit a longer average S-phase due to the reduction in neurogenic divisions. To test this, we performed consecutive injections of the S-phase markers EdU and BrdU at E14.5 to determine the relative rate of progression through S-phase, as well as cell cycle duration (see Materials and Methods, Fig. 3A-C) (Martynoga et al., 2005). We identified radial glia as TBR2− nuclei in the VZ, defined as the region apical to the thick band of TBR2+ cells (Fig. 3C). Consistent with our hypothesis, Nfix−/− radial glia had a significantly longer S-phase than wild-type radial glia (P=0.025; Fig. 3D). Total cell cycle duration in Nfix−/− radial glia was not significantly different (P=0.051; Fig. 3E), nor was G1/G2/M phase length (P=0.11; Fig. 3F). Thus, the increased S-phase duration further indicates that Nfix−/− radial glia undergo proportionally fewer IPC-generating divisions during the period of peak neurogenesis.
Fig. 3.
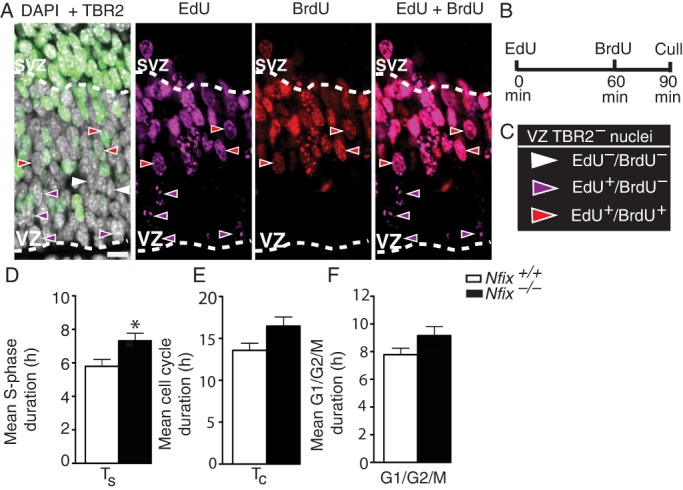
Nfix−/− radial glia undergo proportionally fewer neurogenic divisions. (A) Wild-type hippocampus showing DAPI (white), TBR2 (green), EdU (magenta) and BrdU (red) staining at E14.5, with dashed lines demarcating the VZ/SVZ. (B) Pregnant dams were injected with EdU, followed by BrdU 60 min later, and sacrificed at 90 min. (C) Radial glia were identified as cells with TBR2– nuclei in the VZ. (D-F) Quantification of cell cycle kinetics for (D) mean S-phase duration (Ts), (E) mean total cell cycle duration (Tc) and (F) mean G1/G2/M phase length in radial glia of wild-type and Nfix−/− mice. Mean±s.e.m. of eight embryos *P<0.05, t-test. Scale bar: 22.5 µm.
Overexpression of NFIX in vivo promotes IPC and neuron generation
If loss of NFIX impairs IPC generation, then NFIX overexpression should result in an increased rate of IPC and neuronal differentiation. To investigate this, we used in utero electroporation to overexpress an HA-tagged mouse NFIX construct containing a bicistronic GFP reporter (NFIX pCAGIG) (Heng et al., 2014) or vector-only control (pCAGIG) in the presumptive hippocampus of wild-type CD1 mice at E12.5 (Fig. 4A,B). At E14.5 we found that NFIX overexpression led to a significantly higher percentage of electroporated cells becoming IPCs (P=0.037) and neurons (P=0.016), while fewer cells remained as radial glia (P=1.8×10−5), as compared with electroporated cells in the control condition (Fig. 4C-E). Importantly, co-staining for the neuronal marker TBR1 revealed that all neurons were located basal to the band of TBR2+ cells, demonstrating that NFIX overexpression did not induce gross neuronal migration errors (data not shown). Together, these data are supportive of a role for NFIX in promoting the commitment of radial glia into IPCs.
Fig. 4.
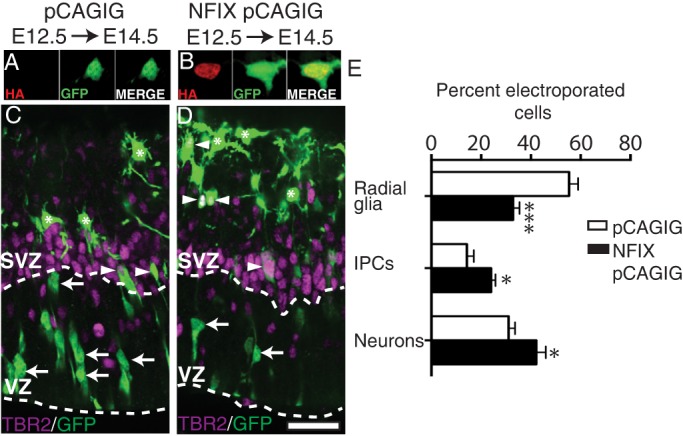
Overexpression of NFIX promotes IPC and neuron generation in vivo. (A,B) Cortical neurons from E14.5 hippocampi expressing (A) the empty vector control pCAGIG or (B) an HA-tagged mouse NFIX isoform (NFIX pCAGIG), with HA in red and GFP in green. (C,D) pCAGIG (C) and NFIX pCAGIG (D) were targeted to the hippocampal primordium of wild-type CD1 mice at E12.5 using in utero electroporation. Electroporated brains were collected and analyzed at E14.5. Image shows GFP in green and TBR2 in magenta, with dashed lines demarcating the VZ/SVZ. Radial glia were identified as TBR2– nuclei in the VZ (arrows), IPCs were identified as any cell expressing TBR2 (arrowheads) and neurons were identified as TBR2– nuclei basal to the SVZ (asterisks). (E) Quantification of the percentage of electroporated (GFP-positive) cells that are radial glia, IPCs or neurons in pCAGIG and NFIX pCAGIG hippocampi. Mean±s.e.m. of five (pCAGIG) and seven (NFIX pCAGIG) embryos. *P<0.05, ***P<0.001, ANOVA. Scale bar in D: A,B, 24 µm; C,D, 40 µm.
Loss of four Nfi alleles results in a more severe IPC phenotype
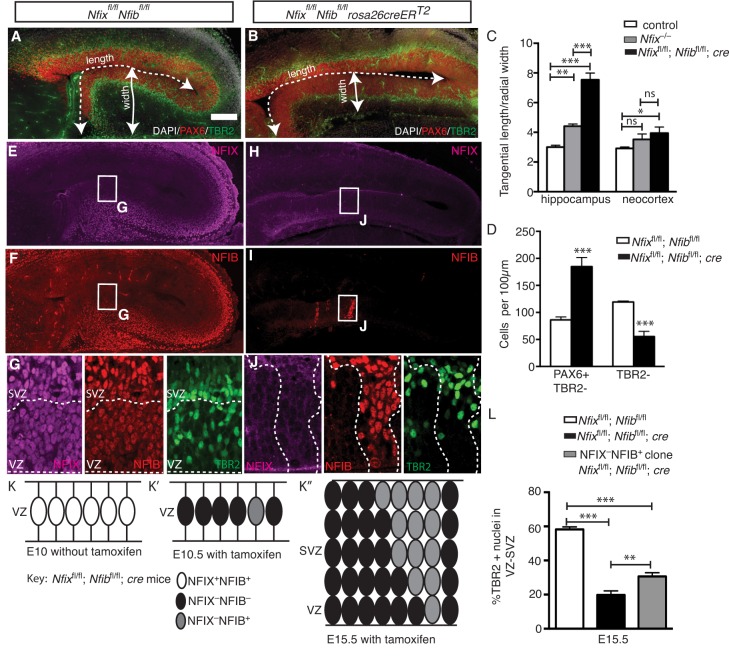
We have thus far demonstrated that Nfix-deficient radial glia generate IPCs with reduced efficiency. Despite this, Nfix−/− mice lack the gross morphological abnormalities often associated with mouse models in which radial glial populations are expanded and there is decreased neurogenesis (Chenn and Walsh, 2002; Farkas et al., 2008). For example, these phenotypes are usually associated with increased tangential length of the cortex and reduced radial thickness. We therefore questioned whether the loss of additional Nfi alleles would result in a more severe phenotype. We removed four Nfi alleles by generating conditional floxed Nfix and Nfib mice, which were then crossed to a mouse line expressing a tamoxifen-activated form of Cre recombinase under the control of the ubiquitous Rosa26 promoter. Time-mated dams were then injected with tamoxifen (two sequential injections, at E10.5 and the other at E11.5) to delete Nfix and Nfib, and embryos were analyzed at E15.5. We found that Cre-expressing Nfixfl/fl; Nfibfl/fl (hereafter Nfixfl/fl; Nfibfl/fl; cre) embryos were almost entirely devoid of NFIX and NFIB immunoreactivity (Fig. 5H,I), whereas cre-negative animals retained NFIX and NFIB expression (Fig. 5E,F). In Nfixfl/fl; Nfibfl/fl; cre animals, the tangential length to radial width ratio of the presumptive hippocampus was markedly increased (251%) compared with control animals (P<0.0001), as was that of the neocortex (P=0.025) (Fig. 5A-C). The length/width ratio of the Nfixfl/fl; Nfibfl/fl; cre hippocampus was also significantly larger than in Nfix−/− mice (P=0.0008), suggestive of a more severe phenotype than in Nfix−/− animals (Fig. 5C). Furthermore, cellular analysis revealed that, compared with controls, Nfixfl/fl; Nfibfl/fl; cre animals had a greater increase in radial glial cell number (PAX6+ TBR2−; P=0.0006; Fig. 5D) than Nfix−/− mice (Fig. 2E) at E15.5. Moreover, Nfixfl/fl; Nfibfl/fl; cre animals had a 50% reduction in IPC number at E15.5 (P=0.0002; Fig. 5D), whereas in Nfix−/− mice the IPC number did not differ significantly from that of wild type at this age (Fig. 2F). Together, these data demonstrate that the loss of additional Nfi alleles results in morphological and cellular phenotypes associated with reduced IPC production.
Fig. 5.
Loss of four Nfi alleles results in a more severe IPC phenotype. (A,B) DAPI staining (white) and PAX6 (red) and TBR2 (green) expression in Nfixfl/fl; Nfibfl/fl and Nfixfl/fl; Nfibfl/fl; cre mice at E15.5. (C) Quantification of tangential length/radial width of the hippocampus and neocortex in control, Nfix−/− and Nfixfl/fl; Nfibfl/fl; cre mice. Mean±s.e.m. from ten control, five Nfix−/− and five Nfixfl/fl; Nfibfl/fl; cre embryos, *P<0.05, **P<0.01, ***P<0.001; ns, not significant; ANOVA. (D) Cell counts of radial glia and IPCs in E15.5 Nfixfl/fl; Nfibfl/fl and Nfixfl/fl; Nfibfl/fl; cre mice. Mean±s.e.m. from five embryos. ***P<0.001, t-test. (E-J) NFIX (E,H, magenta) and NFIB (F,I, red) expression in Nfixfl/fl; Nfibfl/fl and Nfixfl/fl; Nfibfl/fl; cre mice at E15.5. (G,J) Boxed regions shown at higher magnification. Dashed lines (G) delineate the VZ and SVZ or (J) demarcate the border of the NFIB+ clone. (K) Schematic representing stochastic deletion of the Nfibfl/fl allele in Nfixfl/fl; Nfibfl/fl; cre mice. Radial glia express NFIX and NFIB (white ovals). (K′) Upon administration of tamoxifen, Nfib and Nfix are deleted in most radial glia (black ovals), but some cells do not delete Nfib fully (gray ovals). (K″) Incomplete recombination at the Nfib locus results in Nfixfl/fl; Nfibfl/fl; cre hippocampi being sparsely patterned with NFIX– NFIB+ clones. (L) Quantification of total VZ/SVZ nuclei that were TBR2+ in Nfixfl/fl; Nfibfl/fl mice (white bar) and Nfixfl/fl; Nfibfl/fl cre mice (black bar), including NFIX– NFIB+ cells in Nfixfl/fl; Nfibfl/fl; cre mice (gray bar). Mean±s.e.m. from five embryos. **P<0.01, ***P<0.001, ANOVA. Scale bar in A: A,B,E,H,F,I, 100 µm; G,J, 25 µm.
During development, the cell-cell interactions between different progenitor populations is important for regulating the balance between the self-expansion of radial glia or their commitment to IPC and neuronal differentiation (Namihira et al., 2009). Owing to the ubiquitous expression of NFIs by radial glia during cortical development, it is possible that NFIs promote the generation of IPCs by regulating the expression of extrinsic signaling molecules that affect the fate of neighboring radial glial cells (non-cell-autonomous effect), rather than the host cell itself (cell-autonomous effect). Distinguishing between these scenarios could provide important insight into the mechanisms through which NFIs drive the generation of IPCs from radial glia. To analyze these possibilities, we took advantage of the different DNA recombination efficiencies of the floxed Nfix and Nfib alleles (Fig. S2). The reduced recombination efficiency of the Nfib floxed allele resulted in a stochastic effect whereby, in random areas of the cortex, both Nfix alleles had recombined after administration of tamoxifen whereas both Nfib alleles had not. The result was that, at E15.5, Nfixfl/fl; Nfibfl/fl; cre cortices were sparsely patterned with NFIX– NFIB+ clones (Fig. 5E-K). We took advantage of this serendipitous occurrence to determine whether NFIs were cell-autonomously required by radial glia for IPC development. Were this the case, then NFIX– NFIB+ clones should show increased numbers of IPCs compared with the cells surrounding the clone. Conversely, a non-cell-autonomous requirement for NFI would predict that the number of IPCs within NFIX– NFIB+ clones would be similar to that in the surrounding cells. We stained Nfixfl/fl; Nfibfl/fl; cre hippocampi for NFIB, NFIX and TBR2, and analyzed the proportion of cells within NFIX– NFIB+ clones that were IPCs (TBR2+). Within these NFIX– NFIB+ clones there was a greater proportion of IPCs compared with immediately adjacent NFIX– NFIB– regions (P=0.0029; Fig. 5J-L). These data, together with our results from overexpressing NFIX through in utero electroporation (Fig. 4), support a model in which NFIs are autonomously required by radial glia for normal IPC development.
Insc is a target for transcriptional activation by NFIs during hippocampal development
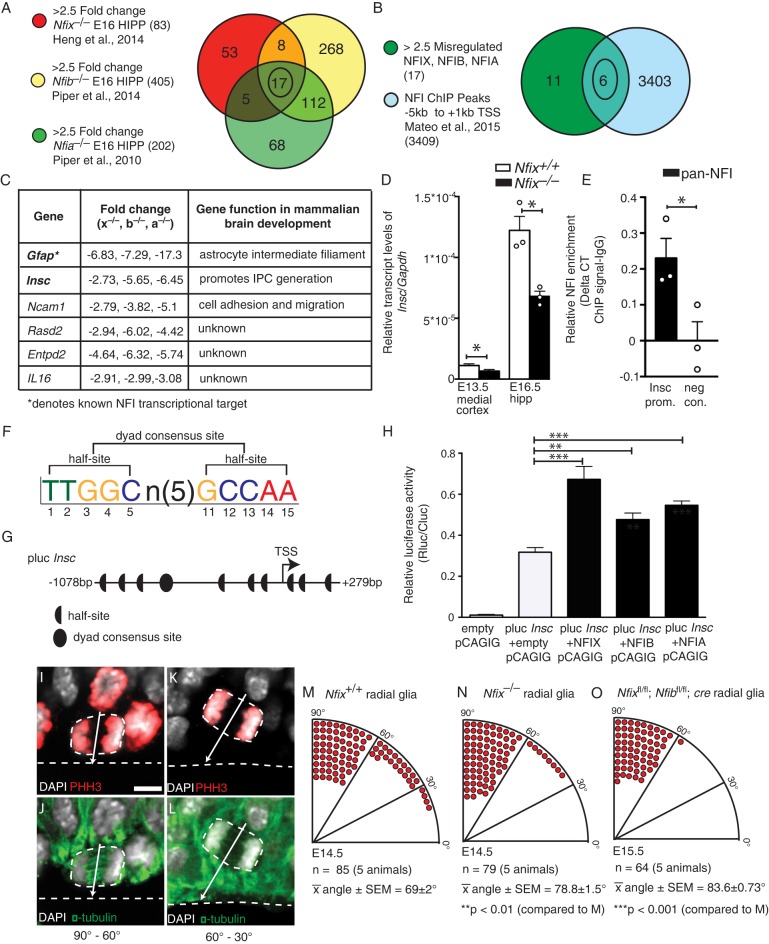
What are the transcriptional mechanisms through which NFIs promote IPC production? To address this question, we analyzed existing microarray datasets that examined alterations in gene expression within the entire hippocampal primordium of E16.5 Nfix−/− (Heng et al., 2014), Nfib−/− (Piper et al., 2014) and Nfia−/− (Piper et al., 2010) mice. In these datasets there were 668, 1893 and 1099 misregulated genes relative to wild-type controls, respectively (±1.5-fold change, P<0.05). To focus our search, we further filtered these data using a number of stringent parameters. First, we refined our analysis to only include genes misregulated in all three microarray datasets. We took this approach because it is likely that NFIs promote IPC production through a common transcriptional mechanism; this is supported by previous data highlighting the functional similarity of NFIs in the development of other cortical phenotypes (Vidovic et al., 2015), the additive effects of deleting NFIX and NFIB on IPC development (Fig. 5), and our analysis of the microarray datasets, which revealed that a highly significant proportion of genes are commonly misregulated across each of the three datasets (Table S1). Next, we only included genes with a greater than ±2.5-fold change (Fig. 6A, Table S2). Finally, we used a recently derived chromatin immunoprecipitation and sequencing (ChIP-seq) dataset (performed using a pan-NFI antibody on embryonic stem cell-derived neural stem cell cultures) to identify which of these remaining genes had a promoter region bound by an NFI protein (Fig. 6B) (Mateo et al., 2015). After this final filter, only six candidate genes remained in the analysis pipeline (Fig. 6C). Crucially, one of these was Gfap, an established target of NFI transcriptional activation in vitro (Namihira et al., 2009) and in vivo (Heng et al., 2014), highlighting the effectiveness of our filters. Ncam1, which regulates neural cell adhesion and migration, was also misregulated, as were three genes of unknown function in central nervous system development (Rasd2, Entpd2, Il16).
Fig. 6.
Insc is a target for transcriptional activation by NFIs during cortical development. (A) Venn diagram of genes misregulated (>2.5-fold) in published Nfix−/− (red), Nfib−/− (yellow), Nfia−/− (green) E16 hippocampal microarrays. (B) Venn diagram of misregulated genes common to all three mutants in A (green) overlapped with NFI ChIP peaks from −5 kb to +1 kb of the gene transcriptional start site (TSS) (blue). (C) The six genes identified in B. (D) qPCR of Insc expression in the E13.5 medial cortex (hippocampal primordium and medial neocortex) and E16.5 hippocampus from wild-type and Nfix−/− mice. Mean±s.e.m. from five (E13.5) and three (E16.5) embryos. *P≤0.05, t-test, Mann–Whitney test. (E) ChIP-PCR showing enrichment of pan-NFI antibody (relative to IgG) within the Insc promoter, and no enrichment (relative to IgG) with a negative control primer located within a gene desert. *P≤0.05, Mann–Whitney test. (F) NFI half-site and dyad consensus site sequence. (G) The cloned region of the Insc promoter showing NFI half-sites and the dyad consensus site. (H) Relative luciferase activity (Rluc/Cluc) after cotransfection of NFIX, NFIB or NFIA with the Insc luciferase construct. Mean±s.e.m. from five experiments. **P<0.01, ***P<0.001, ANOVA. (I-L) Representative images of wild-type radial glia stained with DAPI (white) and for α-tubulin (green) and PHH3 (red), revealing vertical (90°-60°; I,J) or oblique (60°-30°; K,L) cleavage planes. (M-O) The number of radial glia undergoing vertical (90°-60°), oblique (60°-30°) or horizontal (30°-0°) cleavage in wild-type (M), Nfix−/− (N) and Nfixfl/fl; Nfibfl/fl; cre (O) radial glia. Mean±s.e.m. from five embryos. t-tests. Scale bar: 5.5 µm.
Significantly, the final gene in the analysis pipeline was the mammalian homolog of Drosophila insc. Because Insc promotes IPC production during mouse cortical development (Postiglione et al., 2011), we inferred that NFIs might activate Insc expression, and that the downregulation of Insc in Nfi null mice (Fig. 6C) contributes to the IPC phenotype observed within these lines.
NFIs activate Insc promoter-driven transcriptional activity
To investigate whether NFIs activate transcription of Insc, we first validated the microarray results from Nfix−/− E16.5 hippocampi (Heng et al., 2014). Using qPCR we determined that there was a ∼50% reduction in hippocampal Insc mRNA compared with that in wild-type littermates at E16.5 (P=0.05; Fig. 6D). Importantly, Insc was also downregulated in the medial cortex (hippocampal primordium and medial neocortex) of Nfix−/− mice at E13.5, when the IPC phenotype of these mice first becomes apparent (P=0.044).
Next, we verified the capacity of NFI proteins to physically bind to the Insc promoter by performing an in vivo ChIP assay on tissue isolated from E14.5 forebrains using a pan-NFI antibody. The ChIP assay revealed enrichment of NFI protein binding at a region in the Insc promoter that corresponds to both the NFI dyad consensus site (P=0.05; Fig. 6E) and the NFI ChIP peak detected by Mateo et al. (2015) in vitro.
Finally, we asked whether NFIs could active Insc promoter-driven transcriptional activity. A region of the Insc promoter, which included the dyad consensus site, was cloned upstream of the luciferase gene (Fig. 6F,G). Co-transfection of the Insc luciferase construct with an NFIX expression plasmid in Neuro2a cells revealed that NFIX strongly activated Insc promoter-driven transcriptional activity (P=1.8×10−6; Fig. 6H). Likewise, NFIA (P=0.0001) and NFIB (P=0.0026) also enhanced Insc promoter-driven transcriptional activity (Fig. 6H).
NFI-deficient radial glia phenocopy the cleavage plane defects of Insc knockout mice
If the downregulation of Insc contributes to the impaired generation of IPCs in Nfix−/− mice, then we would anticipate that the spindle orientation and cleavage plane deficits seen within Insc conditional knockout mice (Postiglione et al., 2011) would be recapitulated within the radial glia of Nfix−/− mice. We assessed cleavage plane orientation of the dividing progenitor cell relative to the ventricular surface (Fig. 6I-L). At E14.5 in wild-type mice, 71% of radial glia divided with a vertical cleavage plane (the angle between the cleavage plane and the ventricular surface was between 90° and 60°), whereas 23.5% of radial glia divided with an oblique division plane (between 60° and 30°; Fig. 6M). Consistent with previous reports (Postiglione et al., 2011; Insolera et al., 2014), mitotic radial glia with a horizontal cleavage plane (between 30° and 0°) were rare, occurring in only 4.7% of cells (Fig. 6M). In Nfix−/− mice, however, the vast majority of radial glia divided with a vertical cleavage plane (89.9%), and the proportion of oblique divisions was greatly reduced (10.1%) (Fig. 6N). Moreover, no radial glial cell was observed to divide with a horizontal cleavage plane in Nfix−/− mice. Similarly, in Nfixfl/fl; Nfibfl/fl; cre radial glia at E15.5, 98.4% of radial glia divided with vertical cleavage plane, 1.6% with an oblique cleavage plane, and no cells were observed dividing horizontally (Fig. 6O). This phenotype is reminiscent of the cleavage plane phenotype of Insc conditional knockout mice, where there are substantially fewer oblique divisions, and horizontal cleavage planes are not seen (Insolera et al., 2014). Thus, the data support a model whereby downregulation of Insc in NFI-deficient mice results in fewer oblique divisions in radial glia.
In vivo rescue of IPC number in Nfix−/− radial glia through INSC overexpression
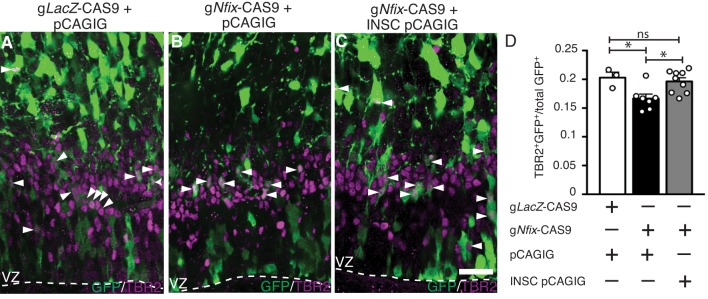
If NFIX promotes IPC development, in part or wholly through activating Insc expression, then overexpressing INSC in Nfix−/− radial glia should help restore IPC generation to wild-type levels. To test this hypothesis, we used the CRISPR/Cas system to delete Nfix from radial glia within the ammonic neuroepithelium of embryonic CD1 wild-type mice. After electroporation at E12.5 with plasmids encoding Nfix-CAS9 and pCAGIG (knockout condition) or lacZ-CAS9 and pCAGIG (control condition), embryos were assessed for NFIX expression. In the knockout condition, only 8% of electroporated (GFP+) cells retained NFIX immunoreactivity compared with 96% in the control (Fig. S3). We then assessed TBR2 expression, and found proportionally fewer TBR2+ GFP+ cells in the knockout compared with the control condition (P=0.03), reminiscent of our findings using Nfix−/− mice (Fig. 7A-D). Finally, for the rescue condition, we co-electroporated the Nfix-CAS9 plasmid with INSC pCAGIG (Fig. S3). In this rescue condition there was an increased proportion of TBR2+ GFP+ cells compared with the knockout condition (P=0.023). Moreover, the number of TBR2+ GFP+ cells in the rescue condition was not significantly different from that of the control condition (P=0.62) (Fig. 7A-D). Thus, Insc overexpression in Nfix−/− radial glia was sufficient to restore IPC numbers to wild-type levels, confirming that NFIX regulation of INSC contributes to IPC production during hippocampal development.
Fig. 7.
Rescue of IPC number in Nfix−/− radial glia through INSC overexpression. (A-C) GFP (green) and TBR2 (magenta) in hippocampal sections of E15.5 CD1 wild-type mice electroporated at E12.5. The dashed line indicates the VZ border. (A) The control condition (glacZ-CAS9+pCAGIG), (B) the knockout condition (gNfix-CAS9+pCAGIG) and (C) the rescue condition (gNfix-CAS9+INSC pCAGIG). Arrowheads indicate TBR2+ GFP+ cells. IPCs were identified as cells expressing TBR2. (D) The proportion of total GFP+ cells that express TBR2. Mean±s.e.m. of three (control), seven (knockout) and nine (rescue) embryos *P<0.05, ANOVA. Scale bar: 30 µm.
A prolonged neurogenic window results in increased neuron number in the hippocampus and neocortex of P20 Nfix−/− mice
What effects do the increased self-expanding divisions of radial glia and the delayed IPC generation have on the late embryonic phenotype of Nfix−/− mice? We had previously observed that the reduction in TBR2+ cells in the Nfix−/− hippocampus was not permanent, so that at E18.5 there were greater numbers of these cells than in wild-type mice (Heng et al., 2014). This suggested that the expansion of the radial glial population in the presumptive hippocampus of Nfix−/− mice during early development would sustain the pool of radial glia and prolong the period of neurogenesis, thereby resulting in an overall greater production of neurons.
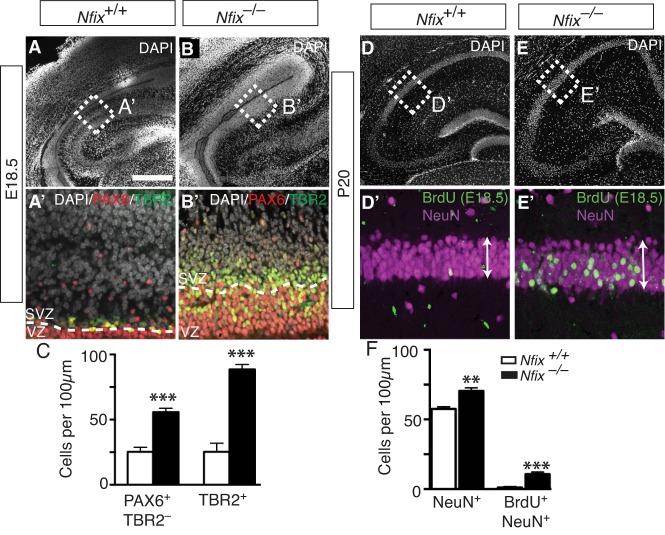
We investigated this by immunostaining wild-type mouse hippocampal sections for PAX6 and TBR2 at E18.5. This revealed very few radial glia or IPCs in the ammonic neuroepithelium, signaling the end of hippocampal neurogenesis. By contrast, in Nfix−/− mice at E18.5 the radial glial (P=2.9×10−5) and IPC (P=1×10−5) populations were ∼2.5-fold and ∼3.5-fold larger than in the control, respectively (Fig. 8A-C). We then asked whether this increase in radial glia and IPC number culminated in the increased production of CA pyramidal neurons in the postnatal hippocampus. Counts of the mature neuronal marker NeuN (Rbfox3) at postnatal day (P) 20 revealed significantly more neurons within both the CA1 and CA3 regions of the mutant compared with the controls (P=0.0011; Fig. 8D-F). Moreover, we tracked the fate of E18.5 progenitor cells by injecting pregnant dams with BrdU and immunostaining for BrdU and NeuN in Nfix−/− and wild-type mice at P20. In wild-type mice there were very few neurons in the P20 CA regions that were born at E18.5 (BrdU+ NeuN+). Remarkably, however, the number of late-born neurons in Nfix−/− mice was ∼10-fold higher (P=0.0002; Fig. 8D-F), which is likely to have contributed to the overall greater number of CA neurons. Additionally, we found a significantly greater number of S100β+ astrocytes in the stratum oriens of postnatal Nfix−/− mice (P=0.0107; Fig. S4). Despite the increases in neuron and glial number in these regions, the overall size of the hippocampus was not significantly different in Nfix−/− mice compared with controls, probably because the dentate gyrus, as previously described (Heng et al., 2014), is much smaller in these mutant mice.
Fig. 8.
A prolonged neurogenic window increases neuron number in the hippocampus of Nfix−/− mice. (A,B) DAPI (white) staining of wild-type and Nfix−/− hippocampi at E18.5. (A′,B′) Boxed regions at higher magnification showing DAPI (white), PAX6 (red) and TBR2 (green) staining. Dashed lines demarcate VZ/SVZ. (C) Cell counts of radial glia and IPCs in the E18.5 wild-type and Nfix−/− hippocampi. Mean±s.e.m. of five embryos. ***P<0.001, t-test. (D,E) DAPI staining in wild-type and Nfix−/− hippocampi at P20, with arrows spanning the width of the CA neuronal layer. (D′,E′) Boxed regions at higher magnification showing BrdU-labeled cells (green) from a BrdU injection at E18.5 and NeuN (magenta). (F) Cell counts within the cornu ammonis (CA) neuronal layer of wild-type and Nfix−/− mice showing of the number of NeuN+ cells and BrdU+ NeuN+ cells following a BrdU chase at E18.5. Mean±s.e.m. of five pups. **P<0.01, ***P<0.001, t-test. Scale bar in A: A,B,D,E, 350 µm; A′,B′, 50 µm; D′,E′, 40 µm.
Given that postnatal Nfix−/− mice have a dorsoventral expansion of the cingulate cortex and neocortex (Campbell et al., 2008), we asked whether the increased neuronal generation observed in the postnatal hippocampus was evident within the broader dorsal telencephalon, and so may contribute towards this phenotype. At E18.5 we saw similar phenotypes within the neocortex as in the hippocampal ammonic neuroepithelium, with increased radial glial (P=0.031) and IPC (P=0.0002) numbers compared with controls, although the phenotype was substantially less severe than that observed in the hippocampus (Fig. S5A-C). BrdU labeling at E18.5 also revealed significantly more late-born neurons (BrdU+ NeuN+) in the upper layers of the neocortex (P=0.0167; Fig. S5D-F). Thus, the prolonged neurogenic period of Nfix−/− mice is likely to contribute to the increased dorsal-ventral size of these regions.
Collectively, these findings highlight the crucial role that NFIX plays in promoting indirect neurogenesis within the developing dorsal telencephalon, significantly enhancing our understanding of the transcriptional mechanism underpinning the development of this brain region.
DISCUSSION
Studies have begun to reveal the factors that drive the progression of radial glia into IPCs during development (Farkas et al., 2008; Sessa et al., 2008; Saffary and Xie, 2011). For example, loss of the transcription factor PAX6 from radial glia downregulates the expression of the microtubule-associated protein SPAG5 and, in turn, radial glia undergo more divisions with oblique cleavage planes and generate more basal mitoses (Götz et al., 1998; Asami et al., 2011). However, these basal mitoses retain the hallmarks of radial glial cells, indicating that PAX6 regulates the delamination of radial glia progenitors from adherens junction complexes, but does not regulate other key features of IPC differentiation. Likewise, HES transcription factors inhibit the neurogenic divisions of radial glia by repressing proneural genes (Mizutani et al., 2007; Pierfelice et al., 2011), but it is unclear how this repression directly affects the maintenance of apical-basal polarity, cell cycle progression, or other aspects of radial glial cell biology. In this study, we have uncovered a novel role for members of the NFI transcription factor family in orchestrating the transition of radial glia to IPCs. Although NFIs have previously been shown to be important for astrogliogenesis and stem cell maintenance in the neocortex and hippocampus, here we reveal that NFIX-mediated activation of Insc expression promotes the timely generation of IPCs.
The function of INSC has been extensively studied during D. melanogaster neurogenesis. INSC acts in D. melanogaster neuroblasts as an adaptor protein, linking two protein complexes that assemble at the apical cell cortex to control spindle orientation (Wodarz et al., 1999; Schaefer et al., 2000). The control of mitotic spindle orientation by INSC during neuroblast division leads to unequal inheritance of the cell fate determinants Prospero, Numb and Brat to the basal daughter cell and results in neuronal differentiation (Knoblich, 2008). There is a single insc homolog in mice (Katoh and Katoh, 2003). INSC is expressed at very low levels within the developing cerebral cortex, which is perhaps why changes in INSC expression result in only modest fluctuations in spindle orientation in mice compared with flies. These modest fluctuations in spindle orientation are, however, correlated with large increases in IPC number with increased INSC expression, and vice versa (Postiglione et al., 2011). Despite the clear relationship between the level of Insc expression and the rate of IPC production in the mouse cortex, the mechanism through which Insc expression is regulated within the cortex has, until now, remained unclear.
Although we found that the impaired IPC development and spindle defects of Nfix-deficient radial glia closely mirror those of Insc conditional knockout mice during early development (Postiglione et al., 2011), there are some important differences between these mutants. One of these differences is that the number of radial glial cells in Nfix-deficient mice is substantially increased during early development compared with controls. By contrast, although the radial glia in Insc conditional knockout mice were quantified using only PAX6 as a marker (thereby including newborn IPCs that retain PAX6 expression), there were no gross changes in the number of radial glia in these mice. As a corollary to this, we found that the expansion of radial glia in Nfix−/− mice sustained the pool of radial glia progenitors into late development, thereby ensuring an overall increase in the number of neurons in the hippocampal CA1/CA3 neuronal layers and neocortex, whereas the neocortical neuron number in Insc conditional knockout mice was reduced (Postiglione et al., 2011). These differences are likely to be explained by the diverse cohort of genes that are likely to be under NFI transcriptional control, in addition to Insc. For example, we have previously shown that NFIs can repress genes involved in stem cell maintenance during cortical development, such as Sox9, Ezh2 and Hes1 (Piper et al., 2010, 2014; Heng et al., 2014). Therefore, although the downregulation of Insc contributes to the impaired IPC generation in Nfix null mice, the misregulation of these additional factors probably explains the expansion of the radial glial pool beyond that observed following loss of Insc alone. NFIX is likely to have functions in cell types other than radial glia that could also account for differences between these mouse lines. For example, NFIX is expressed by IPCs. Does NFIX regulate the differentiation of IPCs to mature neurons, and if so, does this contribute to the accumulation of IPCs at E18.5? Future studies using a Tbr2- or Dcx-driven cre could address this question.
Our data further highlight the pleiotropic roles of NFIs during forebrain development. How NFI proteins promote the generation of IPCs by activating Insc promoter-driven transcription, while repressing genes involved in stem cell maintenance, and also being crucial in astrocytic lineage progression, is a fascinating and open question. Future studies aimed at identifying the genome-wide chromatin binding profile of NFIs in purified populations of neural stem cells, neurons and glia across developmental time will lead to a broader understanding of how these transcription factors fulfil these functions during development.
MATERIALS AND METHODS
Animal ethics
The work performed in this study conformed to The University of Queensland's Animal Welfare Unit Guidelines for Animal Use in Research (AEC approval numbers QBI/353/13/NHMRC and QBI/355/13/NHMRC/BREED) and those of the IACUC at Roswell Park Cancer Institute. All experiments were performed in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes, and were carried out in accordance with The University of Queensland Institutional Biosafety Committee.
Animals
Nfix−/− and Nfix+/+ littermates used in this study have been described previously (Campbell et al., 2008). Conditional Nfix and Nfib alleles, each of which contains loxP sites flanking exon 2 of the respective gene were generated as described previously (Messina et al., 2010; Hsu et al., 2011). The conditional lines were then bred together to produce the double conditional strain (Nfixfl/fl; Nfibfl/fl), which was subsequently crossed to Rosa26-CreERT2 mice (#008463, The Jackson Laboratory). The Nfixfl/fl; Nfibfl/fl; cre nomenclature is used to reflect the fact that the Nfib allele did not recombine as efficiently. Animals of either sex were used.
In utero electroporation
In utero electroporation was performed as previously described (Suarez et al., 2014), with minor modifications whereby 0.5-1 µl of plasmid DNA was injected into one lateral ventricle and electroporated caudomedially into the presumptive hippocampus using 35 V. Plasmid expression constructs were pCAG IRES GFP (pCAGIG) or NFIX pCAG IRES GFP (NFIX pCAGIG). For the rescue experiment, gNfix-CAS9, glacZ-CAS9, INSC pCAGIG and pCAGIG constructs were used. For details of plasmid construction, see the supplementary Materials and Methods.
Measurement of cleavage plane orientation in Nfix−/− radial glia
To analyze cleavage plane orientation within radial glia, we stained sections for phospho-histone H3 (PHH3), α-tubulin and with DAPI. Sections were imaged using a 63× objective on a spinning disk confocal microscope through a depth of 10 µm (consecutive 1 µm z-steps). Hippocampal radial glia undergoing mitosis were identified as PHH3-positive cells located at the ventricular surface of the ammonic neuroepithelium. For each cell three angle measurements were taken from adjacent z-stacks and averaged. Because the metaphase plate of radial glia rocks extensively until anaphase (Haydar et al., 2003; Sanada and Tsai, 2005), only cells that were in anaphase or telophase were analyzed, as revealed by chromosomal (DAPI and PHH3 staining) and mitotic spindle arrangement (α-tubulin staining). We measured cleavage plane orientation by the angle created by the vector that runs parallel to the ventricular surface and the vector that runs through the cleavage plane of the dividing cell.
Immunofluorescence and immunohistochemistry
Embryos were immersion fixed at E14.5 or younger in 4% paraformaldehyde (PFA) or perfused transcardially (E15.5 and older) and sectioned for immunohistochemistry using the primary and fluorescently conjugated secondary antibodies described in detail in the supplementary Materials and Methods.
Nfix−/− mouse hippocampal cell counts
For each section (two sections per animal) cell counts were performed from two 100 µm sampling fields, spanning the width of the hippocampal primordium (E13.5-E18.5) or neocortex (E18.5) as described in the supplementary Materials and Methods.
Nfix−/− mouse birth-dating experiments
BrdU birth-dating experiments were performed using a single low dose (50 mg/kg) or high dose (200 mg/kg) injection. For the low dose, dams were injected at E13.5 or E18.5 and examined 24 h or 20 days later, respectively. For the high dose, dams were injected at E13.5 and examined 48 h later. Cell counts were then performed as described in the supplementary Materials and Methods.
Measurement of cell cycle kinetics in Nfix−/− radial glia
Mean cell cycle (Tc) and S-phase (Ts) durations were determined by a dual-pulse labeling method whereby EdU (50 mg/kg) and 1 h later BrdU (50 mg/kg) were injected into pregnant dams, before the animals were sacrificed 1.5 h post EdU injection. Counts of EdU+ and BrdU+ cells were then performed to determine Tc and Ts as described in the supplementary Materials and Methods and Fig. S6.
Nfixfl/fl; Nfibfl/fl; Rosa26-CreERT2 tamoxifen treatment and cell analysis
Animals were injected with 2 mg tamoxifen dissolved in corn oil at E10.5 and E11.5 before being immersion fixed in PFA at E15.5. Cell counts were performed as in Nfix−/− mice, except for the clonal analysis, where a minimum of two clones per animal were assessed regardless of their rostral-caudal or lateral position within the hippocampus. Cellular analysis of these clones is described in the supplementary Materials and Methods.
Quantitative real-time PCR (qPCR)
The medial cortex (hippocampal primordium and medial neocortex) of E13.5 embryos or the entire hippocampus of E16.5 embryos was dissected and snap frozen. After RNA extraction (RNeasy Micro Kit, Qiagen) and reverse transcription (Superscript III, Invitrogen), qPCR was performed using SYBR Green (Qiagen) using 500 nM of forward and reverse primers. Primer sequences and further technical details are provided in the supplementary Materials and Methods.
Reporter gene assays
DNA was transfected into Neuro2A cells (1×104 cells) in a 96-well plate using Lipofectamine 2000 (Invitrogen), and Cypridina luciferase was added to each transfection as an internal control. After 24 h, luciferase activity was measured using a dual-luciferase system (Switchgear Genomics), as described in the supplementary Materials and Methods.
ChIP-qPCR
A putative ChIP peak within the Insc promoter was identified by analysis of a published ChIP-seq dataset (supplementary table S3, NFI tab, in Mateo et al., 2015). E14.5 forebrains were dissociated and fixed in 1% formaldehyde for 8 minutes. Nuclei were lysed and chromatin sonicated using the Bioruptor Pico (Diagenode) to 100-500 bp. Immunoprecipitation was performed with 8 µg goat anti-NFI (sc-30918, Santa Cruz) or 8 µg goat IgG control (AB-108-C, R&D Systems) antibody coupled to 40 μl Protein G Dynabeads (10003D, Thermo Fisher Scientific). Enrichment of NFI at the Insc promoter was calculated relative to the IgG control using the delta CT method, and normalized to a negative control primer set. For primer sequences and further details see the supplementary Materials and Methods.
Statistical analyses
Student's t-tests were performed when comparing two groups (n>4) or a Mann-Whitney U-test (n=3). For comparisons of more than two groups, ANOVA was performed followed by multiple comparisons analysis (corrected using the Holm-Šídák method) using a pooled estimate of variance if appropriate (Prism 6.0, GraphPad). See the supplementary Materials and Methods for further details.
Acknowledgements
Microscopy was performed in the Queensland Brain Institute's Advanced Microscopy Facility. We thank Rowan Tweedale and Dominic Ng for critical analysis of the manuscript. For provision of reagents, we thank Juergen Knoblich, Nathan Godde and Patrick Humbert.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
L.H. designed, carried out, and interpreted the experiments and wrote the manuscript. O.Z., I.G., H.M., J.O., T.J.H., A.E., I.G.-N., J.I.H. carried out experiments, interpreted data and edited the manuscript. D.V., T.H.B., L.J.R. interpreted data and edited the manuscript. R.M.G. generated mouse lines, interpreted data and edited the manuscript. M.P. provided reagents and mouse lines, designed experiments, interpreted data and wrote the manuscript.
Funding
This work was supported by National Health and Medical Research Council project grants [1003462, 1057751 and 1022308 to M.P.]; an Australian Research Council Discovery Project grant [DP160100368]; and by National Institutes of Health and NYSTEM grants [HL080624, C026714 and C026429 to R.M.G.]. M.P. was supported by an Australian Research Council Future Fellowship [FT120100170] and a Research Award from the Hydrocephalus Association. L.H., A.E. and D.V. were supported by Australian Postgraduate Awards. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.140681.supplemental
References
- Altman J. and Bayer S. A. (1990). Prolonged sojourn of developing pyramidal cells in the intermediate zone of the hippocampus and their settling in the stratum pyramidale. J. Comp. Neurol. 301, 343-364. 10.1002/cne.903010303 [DOI] [PubMed] [Google Scholar]
- Anthony T. E., Klein C., Fishell G. and Heintz N. (2004). Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron 41, 881-890. 10.1016/S0896-6273(04)00140-0 [DOI] [PubMed] [Google Scholar]
- Arai Y., Pulvers J. N., Haffner C., Schilling B., Nüsslein I., Calegari F. and Huttner W. B. (2011). Neural stem and progenitor cells shorten S-phase on commitment to neuron production. Nat. Commun. 2, 154 10.1038/ncomms1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami M., Pilz G. A., Ninkovic J., Godinho L., Schroeder T., Huttner W. B. and Gotz M. (2011). The role of Pax6 in regulating the orientation and mode of cell division of progenitors in the mouse cerebral cortex. Development 138, 5067-5078. 10.1242/dev.074591 [DOI] [PubMed] [Google Scholar]
- Campbell C. E., Piper M., Plachez C., Yeh Y.-T., Baizer J. S., Osinski J. M., Litwack E. D., Richards L. J. and Gronostajski R. M. (2008). The transcription factor Nfix is essential for normal brain development. BMC Dev. Biol. 8, 52 10.1186/1471-213X-8-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A. and Walsh C. A. (2002). Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297, 365-369. 10.1126/science.1074192 [DOI] [PubMed] [Google Scholar]
- Englund C., Fink A., Lau C., Pham D., Daza R. A. M., Bulfone A., Kowalczyk T. and Hevner R. F. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247-251. 10.1523/JNEUROSCI.2899-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas L. M., Haffner C., Giger T., Khaitovich P., Nowick K., Birchmeier C., Pääbo S. and Huttner W. B. (2008). Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron 60, 40-55. 10.1016/j.neuron.2008.09.020 [DOI] [PubMed] [Google Scholar]
- Götz M. and Huttner W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777-788. 10.1038/nrm1739 [DOI] [PubMed] [Google Scholar]
- Götz M., Stoykova A. and Gruss P. (1998). Pax6 controls radial glia differentiation in the cerebral cortex. Neuron 21, 1031-1044. 10.1016/S0896-6273(00)80621-2 [DOI] [PubMed] [Google Scholar]
- Haubensak W., Attardo A., Denk W. and Huttner W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. USA 101, 3196-3201. 10.1073/pnas.0308600100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar T. F., Ang E. Jr and Rakic P. (2003). Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc. Natl. Acad. Sci. USA 100, 2890-2895. 10.1073/pnas.0437969100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y. H. E., McLeay R. C., Harvey T. J., Smith A. G., Barry G., Cato K., Plachez C., Little E., Mason S., Dixon C. et al. (2014). NFIX regulates neural progenitor cell differentiation during hippocampal morphogenesis. Cereb. Cortex 24, 261-279. 10.1093/cercor/bhs307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y.-C., Osinski J., Campbell C. E., Litwack E. D., Wang D., Liu S., Bachurski C. J. and Gronostajski R. M. (2011). Mesenchymal nuclear factor I B regulates cell proliferation and epithelial differentiation during lung maturation. Dev. Biol. 354, 242-252. 10.1016/j.ydbio.2011.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner W. B. and Kosodo Y. (2005). Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr. Opin. Cell Biol. 17, 648-657. 10.1016/j.ceb.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Insolera R., Bazzi H., Shao W., Anderson K. V. and Shi S.-H. (2014). Cortical neurogenesis in the absence of centrioles. Nat. Neurosci. 17, 1528-1535. 10.1038/nn.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. and Katoh M. (2003). Identification and characterization of human Inscuteable gene in silico. Int. J. Mol. Med. 11, 111-116. 10.3892/ijmm.11.1.111 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583-597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Konno D., Shioi G., Shitamukai A., Mori A., Kiyonari H., Miyata T. and Matsuzaki F. (2008). Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 10, 93-101. 10.1038/ncb1673 [DOI] [PubMed] [Google Scholar]
- Laguesse S., Creppe C., Nedialkova D. D., Prévot P.-P., Borgs L., Huysseune S., Franco B., Duysens G., Krusy N., Lee G. et al. (2015). A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell 35, 553-567. 10.1016/j.devcel.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Martynoga B., Morrison H., Price D. J. and Mason J. O. (2005). Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113-127. 10.1016/j.ydbio.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Mateo J. L., van den Berg D. L. C., Haeussler M., Drechsel D., Gaber Z. B., Castro D. S., Robson P., Lu Q. R., Crawford G. E., Flicek P. et al. (2015). Characterization of the neural stem cell gene regulatory network identifies OLIG2 as a multifunctional regulator of self-renewal. Genome Res. 25, 41-56. 10.1101/gr.173435.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F. and Shitamukai A. (2015). Cell division modes and cleavage planes of neural progenitors during mammalian cortical development. Cold Spring Harb. Perspect. Biol. 7, a015719 10.1101/cshperspect.a015719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina G., Biressi S., Monteverde S., Magli A., Cassano M., Perani L., Roncaglia E., Tagliafico E., Starnes L., Campbell C. E. et al. (2010). Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell 140, 554-566. 10.1016/j.cell.2010.01.027 [DOI] [PubMed] [Google Scholar]
- Mizutani K., Yoon K., Dang L., Tokunaga A. and Gaiano N. (2007). Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351-355. 10.1038/nature06090 [DOI] [PubMed] [Google Scholar]
- Mori T., Buffo A. and Götz M. (2005). The novel roles of glial cells revisited: the contribution of radial glia and astrocytes to neurogenesis. Curr. Top. Dev. Biol. 69, 67-99. 10.1016/S0070-2153(05)69004-7 [DOI] [PubMed] [Google Scholar]
- Namihira M., Kohyama J., Semi K., Sanosaka T., Deneen B., Taga T. and Nakashima K. (2009). Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev. Cell 16, 245-255. 10.1016/j.devcel.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Noctor S. C., Martínez-Cerdeño V., Ivic L. and Kriegstein A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136-144. 10.1038/nn1172 [DOI] [PubMed] [Google Scholar]
- Petros T. J., Bultje R. S., Ross M. E., Fishell G. and Anderson S. A. (2015). Apical versus basal neurogenesis directs cortical interneuron subclass fate. Cell Rep. 13, 1090-1095. 10.1016/j.celrep.2015.09.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierfelice T., Alberi L. and Gaiano N. (2011). Notch in the vertebrate nervous system: an old dog with new tricks. Neuron 69, 840-855. 10.1016/j.neuron.2011.02.031 [DOI] [PubMed] [Google Scholar]
- Piper M., Barry G., Hawkins J., Mason S., Lindwall C., Little E., Sarkar A., Smith A. G., Moldrich R. X., Boyle G. M. et al. (2010). NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J. Neurosci. 30, 9127-9139. 10.1523/JNEUROSCI.6167-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M., Harris L., Barry G., Heng Y. H. E., Plachez C., Gronostajski R. M. and Richards L. J. (2011). Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J. Comp. Neurol. 519, 3532-3548. 10.1002/cne.22721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M., Barry G., Harvey T. J., McLeay R., Smith A. G., Harris L., Mason S., Stringer B. W., Day B. W., Wray N. R. et al. (2014). NFIB-mediated repression of the epigenetic factor Ezh2 regulates cortical development. J. Neurosci. 34, 2921-2930. 10.1523/JNEUROSCI.2319-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postiglione M. P., Jüschke C., Xie Y., Haas G. A., Charalambous C. and Knoblich J. A. (2011). Mouse inscuteable induces apical-basal spindle orientation to facilitate intermediate progenitor generation in the developing neocortex. Neuron 72, 269-284. 10.1016/j.neuron.2011.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffary R. and Xie Z. (2011). FMRP regulates the transition from radial glial cells to intermediate progenitor cells during neocortical development. J. Neurosci. 31, 1427-1439. 10.1523/JNEUROSCI.4854-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanada K. and Tsai L.-H. (2005). G protein betagamma subunits and AGS3 control spindle orientation and asymmetric cell fate of cerebral cortical progenitors. Cell 122, 119-131. 10.1016/j.cell.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Schaefer M., Shevchenko A., Shevchenko A. and Knoblich J. A. (2000). A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr. Biol. 10, 353-362. 10.1016/S0960-9822(00)00401-2 [DOI] [PubMed] [Google Scholar]
- Sessa A., Mao C.-A., Hadjantonakis A.-K., Klein W. H. and Broccoli V. (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron 60, 56-69. 10.1016/j.neuron.2008.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitamukai A., Konno D. and Matsuzaki F. (2011). Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci. 31, 3683-3695. 10.1523/JNEUROSCI.4773-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez R., Fenlon L. R., Marek R., Avitan L., Sah P., Goodhill G. J. and Richards L. J. (2014). Balanced interhemispheric cortical activity is required for correct targeting of the corpus callosum. Neuron 82, 1289-1298. 10.1016/j.neuron.2014.04.040 [DOI] [PubMed] [Google Scholar]
- Vidovic D., Harris L., Harvey T. J., Evelyn Heng Y. H., Smith A. G., Osinski J., Hughes J., Thomas P., Gronostajski R. M., Bailey T. L. et al. (2015). Expansion of the lateral ventricles and ependymal deficits underlie the hydrocephalus evident in mice lacking the transcription factor NFIX. Brain Res. 1616, 71-87. 10.1016/j.brainres.2015.04.057 [DOI] [PubMed] [Google Scholar]
- Wang X., Tsai J.-W., Imai J. H., Lian W.-N., Vallee R. B. and Shi S.-H. (2009). Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461, 947-955. 10.1038/nature08435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A., Ramrath A., Kuchinke U. and Knust E. (1999). Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402, 544-547. 10.1038/990128 [DOI] [PubMed] [Google Scholar]