Abstract
In the epithelial follicle stem cells (FSCs) of the Drosophila ovary, Epidermal Growth Factor Receptor (EGFR) signaling promotes self-renewal, whereas Notch signaling promotes differentiation of the prefollicle cell (pFC) daughters. We have identified two proteins, Six4 and Groucho (Gro), that link the activity of these two pathways to regulate the earliest cell fate decision in the FSC lineage. Our data indicate that Six4 and Gro promote differentiation towards the polar cell fate by promoting Notch pathway activity. This activity of Gro is antagonized by EGFR signaling, which inhibits Gro-dependent repression via p-ERK mediated phosphorylation. We have found that the phosphorylated form of Gro persists in newly formed pFCs, which may delay differentiation and provide these cells with a temporary memory of the EGFR signal. Collectively, these findings demonstrate that phosphorylated Gro labels a transition state in the FSC lineage and describe the interplay between Notch and EGFR signaling that governs the differentiation processes during this period.
KEY WORDS: Drosophila, EGFR, Six4, Epithelial stem cell, Groucho, Ovary
Summary: In the Drosophila ovary, the Six4 and Groucho proteins link the activities of the Notch and EGFR pathways to regulate the earliest cell fate decision in the follicle stem cell lineage.
INTRODUCTION
Adult stem cells are defined by their ability to divide with an asymmetric outcome: one cell retains the stem cell fate, while the other differentiates into a functionally distinct cell type in the tissue. Although many types of stem cells segregate differentiation determinants on the timescale of a single cell division, there is growing evidence that the differentiation programs of epithelial stem cell lineages proceed gradually, over the course of several cell divisions (Barker, 2014; Franz and Riechmann, 2010; Jones et al., 2007; Kronen et al., 2014). Indeed, many epithelial stem cell lineages contain a transit-amplifying phase downstream from the stem cell division that is defined by incomplete differentiation. Both stem cells and transit-amplifying cells share similar morphology, remain mitotically active, and often express an overlapping set of molecular markers (Chang et al., 2013; Itzkovitz et al., 2011; Mascré et al., 2012; Yan et al., 2012). Moreover, epithelial stem cells are regularly lost from the niche and replaced by the daughter of a neighboring stem cell during normal homeostasis (Clayton et al., 2007; de Navascués et al., 2012; Margolis and Spradling, 1995; Snippert et al., 2010), indicating that transit-amplifying cells retain the capacity to re-enter the niche and assume the stem cell fate. Thus, transit-amplifying cells must be able to respond to both self-renewal and differentiation factors present in their local environments.
In this study, we use the follicle stem cell (FSC) lineage in the Drosophila ovary (Losick et al., 2011; Sahai-Hernandez et al., 2012) to determine how the earliest differentiation decisions in the transit-amplifying population are controlled. The Drosophila ovary is composed of long strands of developing follicles, termed ovarioles, and a pair of FSCs resides in a structure at the anterior tip of each ovariole called the germarium (Fig. 1A-C). Within the germarium, a population of stromal inner germarial sheath cells (IGS cells, also known as escort cells) support germ cell development in regions 1 and 2a, and provide niche factors that anchor the FSCs at the region 2a/2b border to promote self-renewal. The niche that supports FSC self-renewal in this position has a very limited range, resulting in the activation of the Wingless (Wg) and EGFR pathways in the FSCs, but not in the immediately adjacent prefollicle cell (pFC) daughters (Castanieto et al., 2014; Sahai-Hernandez and Nystul, 2013). Wg and EGFR signaling are required for FSC self-renewal but do not appear to be required for pFC differentiation (Castanieto et al., 2014; Song and Xie, 2003). However, constitutive activation of either pathway inhibits pFC differentiation and, in the case of EGFR signaling, increases the propensity of mutant cells to occupy the FSC niche and self-renew (Castanieto et al., 2014; Song and Xie, 2003). These findings demonstrate that Wg and EGFR pathway activity are part of an FSC-specific program that is absent in the pFCs immediately downstream from the niche.
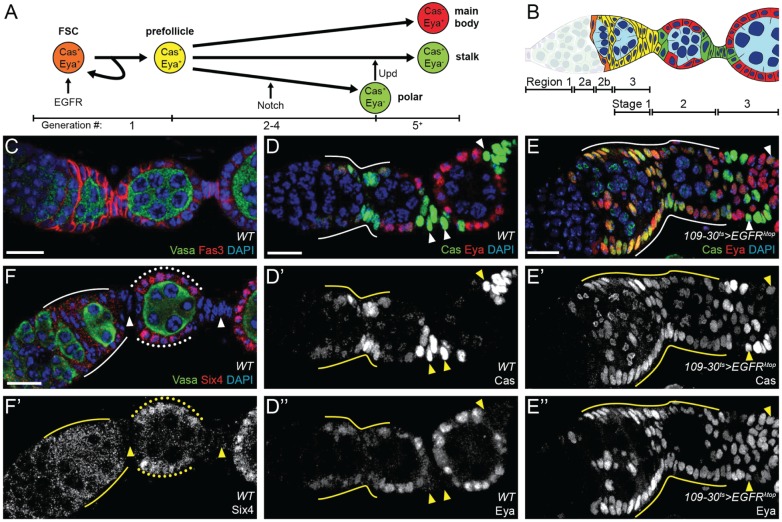
Fig. 1.
RNA-seq of follicle cells expressing constitutively active EGFR implicates the transcription factor Six4 in follicle cell differentiation. (A) Map of cell lineages in the follicle epithelium, including some of the known signaling inputs. Numbers at the bottom of the diagram indicate the approximate generation in the FSC lineage of each transition: the FSC division (generation 1) produces a pFC; pFCs divide 1–3 more times (generations 2–4) before committing to the polar cell fate; and differentiation towards the stalk and main body fates occurs over subsequent generations. EGFR promotes FSC self-renewal, Notch promotes the polar fate and Upd promotes the stalk fate. Expression of Cas or Eya is indicated on each cell type. (B) Schematic presentation of a germarium and the most anterior budded follicles, color coded to match the lineages of A. The regions of the germarium and stages of follicle development are indicated below. (C) Morphology of a wild-type germarium. Fas3 (red) staining outlines cell membranes in early follicle cells. Vasa (green) staining marks the germline cysts of the developing follicles. (D) Differentiation status of wild-type follicle cells, as monitored by staining for Cas (green) and Eya (red). Undifferentiated prefollicle cells express both Cas and Eya (solid line), whereas main body cells express only Eya and polar/stalk cells (arrowheads) express only Cas. (E) Follicle cells expressing EGFRλtop show an expansion of follicle cell staining for both Cas and Eya (solid line and arrowheads). D′,E′ and D″,E″ show the Cas and Eya channels, respectively. (F,F′) Six4 staining is uniform in the follicle cells of regions 2b and 3 of the germarium (solid lines), nuclear in the main body follicle cells of budded follicles (dotted lines) and absent from stalk cells (arrowheads). Scale bars: 10 μm. DAPI is in blue.
Newly produced pFCs either re-enter the niche to replace a resident FSC or, more commonly, move downstream from the FSC niche as they continue to divide, generating a pool of uncommitted transit-amplifying pFCs in region 2b. An orderly series of events directs the differentiation of pFCs into each of the three main follicle cell types: polar cells, stalk cells or main body follicle cells (Fig. 1A,B). First, approximately two or three divisions downstream from the FSC division, a subset of pFCs receive a Delta signal from the germline that activates Notch signaling and initiates differentiation towards the polar cell fate (Lopez-Schier and St Johnston, 2001; Nystul and Spradling, 2010). Next, these newly specified polar cells secrete the Jak-Stat ligand Unpaired (Upd) to initiate the differentiation of other pFCs into stalk cells (Assa-Kunik et al., 2007). It is unclear when the main body follicle cell fate is specified, but a single pFC division can produce one daughter cell that differentiates into a polar or stalk cell, and another daughter that differentiates into a main body follicle cell, suggesting that pFCs do not commit to an exclusively main body follicle cell fate prior to polar cell specification (Chang et al., 2013; Nystul and Spradling, 2010). Much less is understood about the signaling that occurs after newly produced pFCs have exited the niche and prior to the initiation of Notch signaling. pFCs that have exited the niche do not receive self-renewal signals and yet retain the ability to re-enter the niche and assume the stem cell fate. Additionally, these cells contact the germline almost immediately after they exit the niche, yet Notch activation and polar cell differentiation is delayed.
In this report, we demonstrate that two transcriptional regulators, Six4 and Groucho (Gro), promote differentiation in this early stage of the FSC lineage. We find that both are required for the activation of Notch signaling in pFCs, and that Gro is required for FSC self-renewal whereas Six4 is not. Conversely, loss of Six4 increases the rate at which pFCs replace wild-type FSCs, consistent with its role in promoting early pFC differentiation. These findings define a transition state of newly produced pFCs in which the cells are poised to differentiate but temporarily retain the capacity to re-enter the niche and re-acquire the stem cell fate.
RESULTS
Identification of new genes that regulate early prefollicle cell differentiation
Our previous finding that constitutively active EGFR signaling inhibits pFC differentiation (Castanieto et al., 2014) suggested that transcriptional targets of the EGFR pathway might be involved in early cell fate decisions. To investigate this possibility, we first expressed a constitutively active allele of EGFR, EGFRλtop (Queenan et al., 1997), using 109-30-Gal4 (Hartman et al., 2010), which drives expression in posterior IGS cells and throughout the early follicle cell lineage, combined with tub-Gal80ts (referred to herein as 109-30ts) to restrict expression to adulthood. We then assayed for differentiation defects by staining germaria for two transcription factors that control early follicle cell fate choices, Castor (Cas) and Eyes absent (Eya), as readouts for differentiation status (Bai and Montell, 2002; Chang et al., 2013). In wild-type tissue, FSCs and undifferentiated follicle cells in the germarium are Cas+ Eya+, whereas mature main body follicle cells are Cas– Eya+, and both stalk and polar cells are Cas+ Eya– (Fig. 1D). Consistent with our observation that expression of EGFRλtop inhibits differentiation, we found that most of the follicle cells remained Cas+ Eya+ and failed to acquire the morphological characteristics of main body follicle cells, polar cells or stalk cells (Fig. 1E).
In order to identify genes that regulate early follicle cell differentiation, we isolated follicle cells with 109-30ts driving the expression of mCD8::GFP either alone or in combination with EGFRλtop, performed RNA-seq and compared the gene expression profiles of the two populations of cells. We identified 2286 genes with significant differences in expression (Table S1, P-adj<0.01, DESeq2 method). To identify genes that regulate pFC differentiation, we performed an RNAi screen using 109-30ts and RNAi lines from the TRiP collection (Ni et al., 2011) for 26 of the 40 transcription factors with the most statistically significant differences in gene expression (Fig. S1 and Table S2). Ovarioles were examined for gross disruption of the follicle epithelium, as monitored by staining for Fas3 to mark cell membranes of early follicle cells, and Vasa to mark germline cysts of the developing follicles. The most severe and highly penetrant follicle cell phenotype we observed was caused by knockdown of Six4.
Six4 is required for the specification of the polar and stalk cell lineages
Six4 is a well-conserved member of the SIX (sine oculus homeobox) family of transcription factors (Kumar, 2009), which have a DNA-binding domain that provides target specificity and a protein interaction domain that mediates binding to transcriptional co-regulators. There are three known members of the SIX family in Drosophila, and two of them, optix and sine oculis, are required for eye development (Pignoni et al., 1997). By contrast, Six4 does not seem to be important for eye development (Clark et al., 2006; Kirby et al., 2001) but is required for mesoderm development and in follicle cells at late stages of oogenesis (Borghese et al., 2006; Clark et al., 2006, 2007; Kirby et al., 2001).
To determine the expression pattern of Six4 in wild-type tissue, we performed immunofluorescence staining with an anti-Six4 antibody (Hwang and Rulifson, 2011). We detected uniform staining in all follicle cells of the germarium, nuclear staining in the main body follicle cells of later stages, and no signal in stalk cells (Fig. 1F). This signal was absent in follicle cell clones expressing Six4 RNAi, which confirms that the antibody is specific for Six4 and that RNAi knockdown is efficient (Fig. S2A). Our RNA-seq data indicated a 3.2-fold increase in Six4 transcript levels in the population of follicle cells expressing EGFRλtop (Fig. S1 and Table S1). However, Six4 may not be a direct target of EGFR signaling, as we do not observe elevated Six4 staining in FSCs where p-ERK is detected (Castanieto et al., 2014). Instead, because constitutive activation of EGFR signaling blocks pFC differentiation (Fig. 1E) and mature stalk cells downregulate Six4 (Fig. 1F), the relative increase in Six4 transcript levels may be due to the lack of mature stalk cells in the EGFRλtop-expressing population.
We found that RNAi knockdown of Six4 using 109-30ts prevents stalks from forming, resulting in partially fused egg chambers (Fig. 2A and Fig. S3). In addition, we consistently observed Cas+ Eya+ dual-positive cells located outside of the germarium, typically near the interface between adjacent follicles (Fig. 2C), which we never see in wild-type ovarioles. Notably, consistent with studies of Six4 in follicle cells at later stages of oogenesis (Borghese et al., 2006), we noticed that knockdown of Six4 increased the levels of Eya in the cytoplasm (compare Fig. 2C with Fig. 1D). We next used CRISPR (Bassett and Liu, 2014; Gratz et al., 2014) to induce a frameshift mutation after the first 36 codons (position 108) of Six4 in a stock containing FRT2A. This new allele, Six4108, is homozygous lethal and fails to complement Six4289 (Kirby et al., 2001). In addition, FSC clones that are homozygous mutant for Six4108 exhibit all of the phenotypes we observed by RNAi, including a lack of signal with the Six4 antibody and failure to form stalk-like structures (Fig. S2B). Thus, we conclude that Six4108 behaves as a functional null and the Six4 RNAi phenotypes we observed are due to loss of Six4.
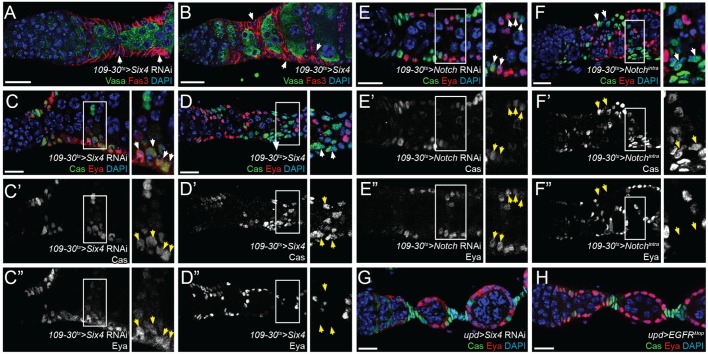
Fig. 2.
Six4 is required for polar and stalk cell formation. (A) Knockdown of Six4 prevents stalk formation between adjacent follicles (arrowheads). (B) Overexpression of Six4 causes the accumulation of extra cells between adjacent follicles (arrowheads). (C-C″,E-E″) Knockdown of Six4 (C-C″) or Notch (E-E″) by RNAi causes some follicle cells (arrowheads) to remain Cas+ Eya+ in follicles that have budded from the germarium. (D-D″,F-F″) Overexpression of Six4 (D-D″) or Nintra (F-F″) leads to ectopic Cas+ Eya– cells (arrowheads) between adjacent follicles. (G,H) Using the polar cell driver upd-Gal4, neither knockdown of Six4 nor overexpression of EGFRλtop cause follicle cell phenotypes, indicating that polar cell maintenance is unaffected by these inputs. Insets in C-F″ are enlarged to the right of each panel. Scale bars: 10 μm. DAPI is in blue.
Overexpression of Six4 in early follicle cells caused an excess of Cas+ Eya– cells to accumulate in the stalk regions and extend onto the surface of the adjacent follicles (Fig. 2B,D). The phenotypes caused by knockdown or overexpression of Six4 resemble the follicle cell phenotypes caused by downregulation (Keller Larkin et al., 1999; Lopez-Schier and St Johnston, 2001) or constitutive activation (Larkin et al., 1996; Lopez-Schier and St Johnston, 2001; Vied and Kalderon, 2009) of Notch signaling, respectively. Indeed, RNAi-mediated knockdown of Notch caused fused egg chambers and the perdurance of Cas+ Eya+ cells downstream from the germarium (Fig. 2E), whereas constitutive activation of Notch signaling by expression of the Notch intracellular domain (Nintra) in early follicle cells caused an accumulation of Cas+ Eya– cells in the stalk regions (Fig. 2F). These observations indicate that both Notch and Six4 are required for the establishment of the polar and stalk cell fates, and are sufficient to ectopically induce a polar/stalk-like cell fate in a subset of pFCs.
To determine whether Six4 is also required for the maintenance of the polar cell fate, we expressed Six4 RNAi using unpaired-Gal4 (upd-Gal4), which is expressed in mature polar cells but not stalk cells or pFCs within the germarium (Assa-Kunik et al., 2007). Interestingly, this did not cause any discernable phenotype (Fig. 2G). Polar and stalk cells were present in the proper positions with normal morphology and were Cas+ Eya–. Additionally, expression of EGFRλtop using upd-Gal4 also failed to produce a phenotype (Fig. 2H). This suggests that polar cells become refractory to these perturbations once they are established.
Six4 promotes the polar cell lineage via Notch signaling
Previous studies have shown that specification of the stalk cell fate requires the secretion of Upd from polar cells (Assa-Kunik et al., 2007; Torres et al., 2003). Accordingly, mutations that disrupt polar cell specification subsequently also disrupt stalk cell specification. To determine where Six4 functions in the progression toward the polar and stalk cell fates, we measured the expression of an early polar cell reporter, neuralized-lacZ (neur-lacZ) (Lopez-Schier and St Johnston, 2001), in the context of Six4 knockdown or overexpression using the 109-30ts driver. In wild-type tissue, neur-lacZ is weakly expressed in two to four cells between the cysts in regions 2b and 3 of the germarium, and is more strongly expressed in pairs of polar cells on budded follicles (Fig. 3A and Fig. S4). Knockdown of Six4 caused a loss of neur-lacZ+ cells throughout the germarium and in budded follicles (Fig. 3B and Fig. S4). Conversely, overexpression of Six4 caused neur-lacZ expression to expand beyond the developing polar cell region to include some stalk cells and cells in the main body of the follicle (Fig. 3C and Fig. S4). Thus, Six4 functions in undifferentiated pFCs to promote specification of the polar cell fate choice, which accounts for the loss of polar and stalk cells upon Six4 knockdown (Fig. 2A,C).
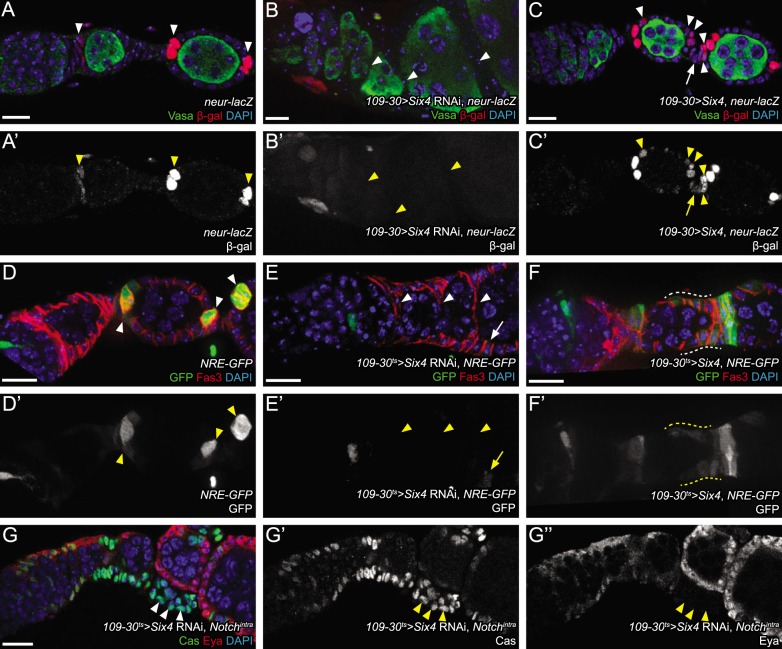
Fig. 3.
Six4 promotes the polar cell lineage via Notch signaling. (A,A′) Expression of neur-lacZ (red, arrowheads) in wild type. Expression is low in two to four pFCs on the anterior face of a region 3 follicle and high in pairs of polar cells at subsequent stages. (B,B′) Knockdown of Six4 by RNAi eliminates neur-lacZ reporter expression in cells positioned to become polar cells (arrowheads). (C,C′) Overexpression of Six4 causes ectopic activation of neur-lacZ (red) in main body (arrowheads) and stalk cells (arrow) near the polar cells, but not in all follicle cells. (D,D′) Expression of NRE-GFP (green) in wild type. GFP is detected in four to six cells at the anterior face of a follicle budding from the germarium, but is restricted to pairs of polar cells in later stages (arrowheads). (E,E′) Knockdown of Six4 by RNAi causes loss of NRE-GFP activity in cells positioned to become polar cells (arrowheads). Occasional follicle cells can still activate NRE-GFP (arrow), but fail to induce stalk formation. (F,F′) Overexpression of Six4 causes ectopic activation of NRE-GFP (green) in many cells near the border between adjacent follicles (e.g. yellow dashed lines) but not in all follicle cells. (G-G″) Expression of Nintra is epistatic to Six4 RNAi, indicating that Six4 is likely upstream of Notch cleavage. Many ectopic Cas+ Eya– cells are observed (arrowheads). Scale bars: 10 μm. DAPI is in blue.
The expression of neur-lacZ in pFCs requires Notch signaling (Lopez-Schier and St Johnston, 2001). Therefore, we next investigated whether Six4 is required for the activation of Notch target genes using a Notch pathway activity reporter, NRE-GFP (Saj et al., 2010). Expression of NRE-GFP in wild-type ovarioles is detectable in pFCs that are in contact with the anterior face of the germline in regions 2b and 3 of the germarium, and is then restricted to pairs of mature polar cells of early follicles in stages 2-5 (Fig. 3D). At stage 6, a separate wave of Notch signaling activates NRE-GFP expression in all follicle cells (Lopez-Schier and St Johnston, 2001). We found that the effect of Six4 knockdown or overexpression on NRE-GFP expression closely paralleled the effects we observed on neur-LacZ expression. Specifically, upon RNAi knockdown of Six4, the NRE-GFP signal was undetectable in follicle cells throughout the germarium, and in the polar cell regions of most stage 2-5 follicles (Fig. 3E), whereas overexpression of Six4 caused an expansion in the region of Notch-responsive cells at the poles of each follicle (Fig. 3F). These results indicate that Six4 is necessary and sufficient to promote Notch signaling in the pFCs of the germarium and in the subset of follicle cells at the poles of early follicles. To determine where in the Notch signaling pathway Six4 exerts its influence, we established an epistatic relationship by co-expressing Six4 RNAi and Nintra using 109-30ts. This caused an accumulation of Cas+ Eya– cells between follicles (Fig. 3G), which resembles the phenotype caused by overexpression of Nintra alone (Fig. 2F), rather than a loss of Cas+ Eya– cells, which is caused by knockdown of Six4 (Fig. 2C), indicating that Six4 is not required downstream of Notch cleavage in Notch signaling and polar cell specification.
The global co-repressor Groucho promotes prefollicle cell differentiation
Our finding that Six4 promotes Notch signaling and polar cell differentiation prompted us to search for other genes that may participate in the process. Although little is known about Six4, the founding SIX family member sine oculis has been extensively studied (Kumar, 2009). During retinal development Sine oculis forms a complex with Eya to promote transcription or with Gro to inhibit transcription (Silver et al., 2003). Gro was of interest in our study because it is a highly conserved and broadly expressed transcriptional repressor that is an effector of many signaling pathways, including EGFR and Notch signaling (Hasson et al., 2005). In wing and notal bristle patterning, Gro functions as a positive effector of Notch signaling by cooperating with Enhancer-of-split proteins, and is antagonised by ERK-mediated phosphorylation of Gro (Hasson et al., 2005).
To examine potential roles for Gro in the germarium, we first stained wild-type ovarioles using an anti-Gro antibody (Delidakis et al., 1991). We found that Gro is expressed strongly in all follicle cells of the germarium and more weakly in germ cells and IGS cells (Fig. 4A). Knockdown of gro in follicle cells produced a phenotype that strongly resembles the phenotype caused by overexpression of EGFRλtop. Specifically, upon expression of gro RNAi with 109-30ts, we observed an accumulation of follicle cells in the germarium and an absence of stalks between follicles (Fig. 4B) in 100% (n=64) of ovarioles. In addition, as with overexpression of EGFRλtop, follicle cells remained Cas+ Eya+ beyond the germarium (Fig. 4D). In combination with previous studies (Hasson et al., 2005; Helman et al., 2011), these observations suggest that the inhibition of follicle cell differentiation caused by ectopic activation of EGFR signaling in pFCs may be due to ERK-mediated phosphorylation and inhibition of Gro.
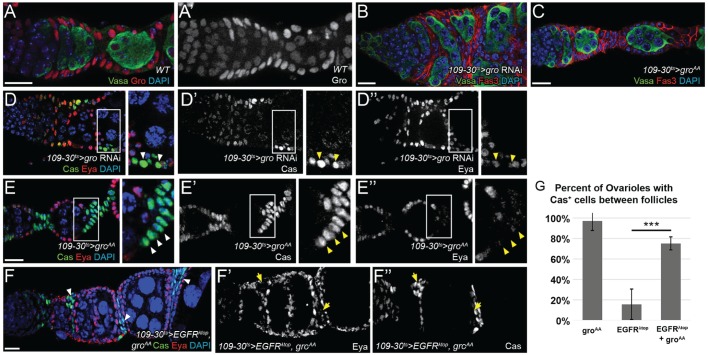
Fig. 4.
Gro is required for the specification of the polar and stalk cell lineages. (A,A′) Gro is expressed most strongly in follicle cells, but also in IGS and germ cells. (B,D-D″) Knockdown of gro causes severe multilayering, fusion of adjacent follicles and ectopic Cas+ Eya+ cells (arrowheads) beyond the germarium, indicative of excess undifferentiated follicle cells and a lack of stalk cells. (C-E″) Overexpression of groAA causes elongated, multilayered stalks and elicits the accumulation of excess Cas+ Eya– cells between adjacent follicles (arrowheads). (F-F″) Stalk-like Cas+ Eya− cells (arrowheads) are present between follicles in ovarioles expressing groAA and EGFRλtop. (G) Percentage of ovarioles with Cas+ cells between follicles overexpressing groAA or EGFRλtop, or both.***P<0.001 using a two-tailed t-test. Scale bars: 10 μm. DAPI is in blue.
To investigate the role of Gro phosphorylation in FSCs and early pFC differentiation, we examined the phenotypes caused by overexpression of either wild-type gro or alleles of gro with point mutations at the ERK target sites. We found that overexpression of an allele of gro (groAA) that is refractory to ERK inhibition (Helman et al., 2011) caused excessive differentiation towards the polar/stalk lineage, resulting in elongated and multilayered stalks with extra Cas+ Eya– cells in 78±11% of germaria (Fig. 4C,E and Fig. S5). By contrast, overexpression of wild-type gro produced less severe phenotypes with lower penetrance (15±2% of ovarioles had at least one elongated or multilayered stalk, Fig. S5). Overexpression of phosphomimetic gro (groDD), which behaves as if it is constitutively repressed by ERK-mediated phosphorylation, produced these phenotypes with a similar low penetrance (10±8%, Fig. S5). These results indicate that the ERK target sites of Gro are important for its function in promoting polar and stalk cell differentiation, and suggest that overexpression of EGFRλtop inhibits polar and stalk cell formation by repressing Gro. To test this possibility, we investigated whether expression of groAA could restore polar/stalk cell differentiation in ovarioles expressing EGFRλtop. Indeed, we found that 75±6% of ovarioles expressing both groAA and EGFRλtop had polar/stalk-like Cas+ Eya– cells between follicles, compared with just 16±6% of ovarioles expressing EGFRλtop alone (Figs 1E and 4F,G).
Next, we examined NRE-GFP expression to test whether gro regulates Notch pathway activity. Indeed, we found that RNAi knockdown of gro eliminated NRE-GFP activity in the germarium and early-stage egg chambers (Fig. 5A), whereas overexpression of groAA ectopically activated NRE-GFP activity throughout the FSC lineage in the germarium, including the FSCs (Fig. 5B). Follicle cells expressing groAA also exhibit ectopic expression of the polar cell reporter neur-lacZ in cells typically positioned to become stalk cells (Fig. 5C and Fig. S4). Since Six4 and Gro both promote Notch signaling in pFCs, we investigated whether either gene is required for the expression of fringe (fng), which encodes a secreted protein that potentiates Notch signaling and has been shown to be regulated by EGFR signaling in late-stage egg chambers (Zhao et al., 2000). We found that fng is strongly expressed in region 1 of wild-type germaria and then substantially downregulated at subsequent stages (Fig. S6A,B). However, we did not detect any decrease in fng expression in FSCs and pFCs upon RNAi knockdown of either Six4 or Gro (Fig. S6C-F), suggesting that fng expression in the FSC lineage does not depend on either Six4 or Gro. Collectively, these results indicate that Gro promotes Notch signaling and polar cell differentiation, and that, as in other tissues, Gro function is antagonized by ERK-mediated phosphorylation of Gro at ERK target sites.
Fig. 5.
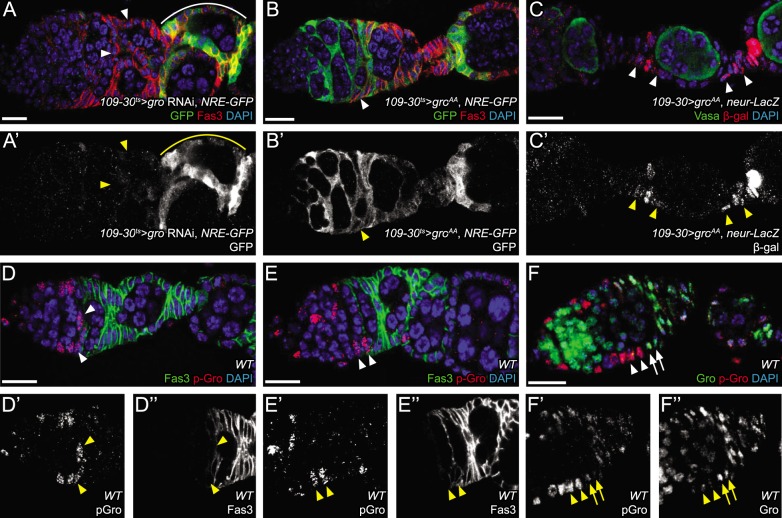
Phosphorylation of Gro prevents Notch signaling and polar cell specification in early pFCs. (A,A′) Expression of gro RNAi causes loss of NRE-GFP activity in early stage follicles (arrowheads). A separate wave of Notch activation occurs surrounding stage 6 follicles (solid line), which is beyond the range of 109-30 expression. (B,B′) Expression of groAA, which is refractory to ERK-mediated phosphorylation, causes ectopic Notch activity throughout the early follicle cell lineage, including FSCs (arrowhead). (C,C′) Expression of groAA causes ectopic activation of neur-lacZ in stalk cells (arrowheads). (D-E″) Phosphorylated Gro (p-Gro) is observed only in FSCs and pFCs within three cell diameters of the niche (arrowheads). (F-F″) Co-staining for Gro (green) and p-Gro (red) indicates that FSCs and early pFCs are p-Gro+, Gro− (arrowheads), whereas later pFCs are p-Gro−, Gro+ (arrows). Scale bars: 10 μm. DAPI is in blue.
Groucho phosphorylation is enriched in the FSC niche
We hypothesized that, if Gro undergoes phosphorylation in response to EGFR signaling as the above results suggest, then it should be detected in its phosphorylated state in cells with active EGFR signaling. To visualize the pattern of Gro phosphorylation in the FSC lineage, we performed immunofluorescence with an antibody that specifically recognizes Gro protein that has been phosphorylated at the ERK target sites (Helman et al., 2011). Interestingly, we found that phosphorylated Gro (p-Gro) is found at high levels not only in FSCs and IGS cells, both of which have active EGFR signaling, but also in newly produced pFCs, located within approximately three cell diameters from the FSCs (Fig. 5D,E). Beyond this stage, p-Gro staining is still detectable in follicle cells but at substantially lower levels in the germarium, with a return to higher levels outside the germarium, where EGFR signaling becomes active again. We confirmed this result using a staining protocol that allows for simultaneous and mutually exclusive detection of the phosphorylated and non-phosphorylated forms of Gro (Cinnamon et al., 2008). This co-staining revealed very low Gro signal in the p-Gro+ FSCs and pFCs (Fig. 5F), suggesting that the majority of Gro protein is phosphorylated in these cells. To confirm that Gro phosphorylation depends on EGFR signaling, we generated MARCM clones homozygous for EGFRF2, a null allele, and stained for p-Gro. These clones were rapidly lost from the niche, as we have reported previously (Castanieto et al., 2014), and thus clones that included cells at the region 2a/2b border were very rare. However, the p-Gro signal was clearly reduced in large EGFRF2 clones outside the germarium (Fig. S7), demonstrating that EGFR is required for Gro phosphorylation in follicle cells.
Gro is required for FSC maintenance whereas Six4 loss induces hypercompetition
Our findings thus far demonstrate a clear role for Six4 and gro in promoting differentiation of pFCs toward the polar cell fate. To test whether either of these transcriptional regulators are also part of the programs that promote FSC self-renewal and occupancy of the niche, we performed an FSC competition assay (Kronen et al., 2014). This assay compares the fitness of a mutant FSC lineage to a wild-type FSC lineage in the same germarium. Mutations that disrupt a function required for FSC self-renewal or niche occupancy cause ‘hypocompetition’ in which the mutant FSCs are lost at an increased rate and replaced by daughters of the wild-type FSC lineage. Conversely, other mutations cause ‘hypercompetition’ in which the mutant FSC lineage expands at the expense of the wild-type lineage. The causes of hypercompetition are not fully understood but, in the FSC lineage, the phenotype is associated with mutations that delay pFC differentiation (Kronen et al., 2014).
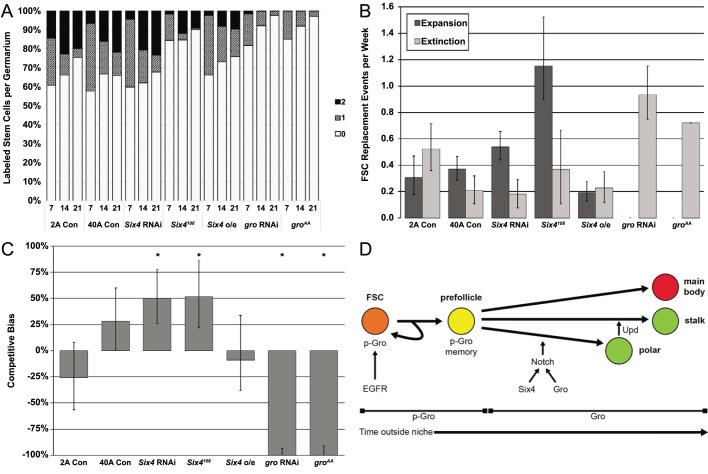
We measured the proportion of germaria containing 0 (unlabeled), 1 (single-labeled) or 2 (double-labeled) GFP+ FSC clones at 7, 14, and 21 days post-clone induction (Fig. 6A-C and Tables S3,S4). In germaria that are single labeled at the time of clone induction, the replacement of one FSC by a daughter cell of the other FSC results in a decrease in the proportion of single-labeled germaria with a concomitant increase in either the unlabeled or double-labeled population. Thus, the changes in the proportion of germaria with 0, 1, or 2 labeled FSCs can be used to calculate rates of FSC turnover, as well as a competitive bias value (b) which describes the fitness of a mutant stem cell relative to a wild-type stem cell (Kronen et al., 2014). Bias values range from +100% to −100%, with positive values indicating that the mutant is hypercompetitive for the niche relative to wild type, negative values indicating that the mutant is hypocompetitive relative to wild type, and 0% indicating that mutant and wild type are equally competitive (neutral competition).
Fig. 6.
Six4 and Gro are required to maintain neutral FSC competition. (A) FSC competition assay using MARCM clones. The proportion of germaria that were double labeled, single labeled or unlabeled are indicated by the black, hatched or white bars, respectively. Timepoints observed were 7, 14 and 21 days after clone induction. At least 250 ovarioles scored for each genotype and time point. (B) Rates of FSC clonal extinction or expansion. Error bars indicate the 95% confidence intervals. (C) Competitive bias for the indicated genotypes. FRT 2A and FRT 40A controls exhibit neutral competition. Six4 loss, either by Six4 RNAi or homozygosity for the Six4108 allele, causes hypercompetition. Six4 overexpression is neutral. Expression of either gro RNAi or groAA causes strong FSC hypocompetiton. Error bars indicate the 95% confidence intervals. *P<0.05 for the null hypothesis that b=0. (D) Interactions of Six4 and Gro in the FSC niche. Within the niche, EGFR maintains Gro phosphorylation to inhibit Notch signaling, and thus differentiation. Cells that have recently left the niche temporarily maintain p-Gro, thus resisting differentiation to allow for replacement events and transit amplifying divisions. As the prevalence of p-Gro decreases, non-phosphorylated Gro becomes available to engage in Notch signaling. Six4 and Gro both promote Notch signaling to initiate differentiation of pFCs specifically towards the polar cell fate.
We found that loss of Six4, mediated either by RNAi knockdown or homozygosity for Six4108, caused a significant hypercompetition phenotype (b=50±23% and 52±30%, respectively; P<0.05 for the null hypothesis that b=0). This indicates that Six4 is not necessary for FSC self-renewal and is consistent with a role for Six4 in pFC differentiation, as mutant pFCs that fail to differentiate may be more competitive for the niche. Despite the essential role for Six4 in pFC differentiation, overexpression of Six4 did not result in a significant niche competition phenotype (b=−9±5%; P=0.73), indicating that it is not sufficient to cause FSCs to differentiate prematurely. By contrast, overexpression of groAA caused a severe hypocompetition phenotype (b=−100%±0; P<0.001), indicating that phosphorylation of Gro at ERK target sites is essential for FSC self-renewal. Surprisingly, RNAi knockdown of gro also caused a severe hypocompetition phenotype (b=−100%±0; P<0.001), suggesting that either a small amount of Gro remains unphosphorylated in FSCs and is required for self-renewal, or the phosphorylated form of Gro has an unexpected function that promotes self-renewal.
DISCUSSION
In this study, we have demonstrated that two transcriptional regulators, Six4 and gro, initiate differentiation of pFCs toward the polar cell fate by promoting Notch signaling. Overall, our findings support a model (Fig. 6D) in which EGFR signaling inhibits FSC differentiation within the niche by inhibiting the repressive function of Gro via p-ERK-mediated phosphorylation. Outside the niche, newly produced pFCs enter a transition state that is defined by the lack of exposure to self-renewal cues, such as active Wg and EGFR signaling (Castanieto et al., 2014; Sahai-Hernandez and Nystul, 2013), and by a dependence on continued Gro inhibition to resist differentiation. During embryonic development, the perdurance of p-Gro in the neuroectoderm allows the effects of transiently active signaling from receptor tyrosine kinases such as EGFR to persist beyond the window of pathway activation (Helman et al., 2011). Likewise, the perdurance of p-Gro may provide early pFCs with a molecular memory of the niche signaling, thus delaying differentiation and allowing these cells to participate in stem cell replacement or to increase in number before committing to a cell fate choice. Following a prolonged absence of EGFR signaling, p-Gro is replaced by an active unphosphorylated form of Gro that is able to promote Notch signaling in some pFCs, causing them to differentiate into polar cells.
The possibility that p-ERK represses Gro activity in FSCs by directly phosphorylating it is consistent with previous findings that p-ERK is detectable in FSCs (Castanieto et al., 2014), that Gro is a direct substrate of p-ERK, and that p-ERK phosphorylation inhibits the activity of Gro (Cinnamon et al., 2008; Hasson et al., 2005; Helman et al., 2011). This possibility is also supported by data from this study. First, we found that the phosphorylated form of Gro predominates in FSCs (Fig. 5D-F) where p-ERK levels are high and that the p-Gro signal in follicle cells is largely eliminated by loss of EGFR (Fig. S7). Notably, we could not directly assay p-Gro signal in FSCs lacking EGFR, and p-Gro was still detectable in some EGFRF2 follicle cells, so we cannot exclude the possibility that other serine-threonine kinases also phosphorylate Gro and regulate its activity. Second, we found that overexpression of groAA partially suppressed the polar/stalk cell differentiation phenotype caused by overexpression of EGFRλtop (Fig. 4F). Last, the hypocompetition phenotype (Fig. 6A-C) and differentiation phenotype (Fig. 4E) caused by groAA overexpression demonstrates the importance of phosphorylation at these sites for promoting FSC self-renewal and repressing early pFC differentiation, which are two functions that are associated with EGFR signaling in the early FSC lineage.
Previous studies have found that a subset of pFCs in region 2b activate Notch signaling in response to a spatially and temporally restricted Delta signal from the germline (Lopez-Schier and St Johnston, 2001; Nystul and Spradling, 2010). Our observations that loss of gro or Six4 attenuated Notch pathway activation and polar cell specification while overexpression of groAA or Six4 had the opposite effect, strongly suggest that both genes promote Notch-mediated differentiation of pFCs towards the polar cell fate. In addition, groDD had a much weaker effect on pFC differentiation than overexpression of groAA, indicating that loss of phosphorylation at these sites is important for the pro-differentiation function of Gro. Our observation that overexpression of groWT produced a mild phenotype similar to the effect of groDD overexpression suggests that, as with endogenously expressed Gro, the overexpressed wild-type Gro protein is also predominantly phosphorylated in FSCs and early pFCs, and thus is functionally equivalent to groDD at these stages. Interestingly, our finding that NRE-GFP is active in FSCs overexpressing groAA (Fig. 5B) but not in wild-type FSCs (Fig. 3D) suggests that FSCs are also exposed to Notch ligand and capable of activating Notch signaling, but the pathway activity is suppressed by the phosphorylation of Gro. The activation of Notch signaling in FSCs expressing groAA may cause premature differentiation, which is consistent with the finding that Nintra increases the rate of FSC loss (Vied and Kalderon, 2009), and would account for the hypocompetition phenotype caused by groAA overexpression.
In many cell types, Gro functions downstream of Notch target gene expression by interacting with primary Notch targets such as Enhancer of Split [E(spl)] to carry out the effects of Notch pathway activation (Bailey and Posakony, 1995; Fisher et al., 1996; Lecourtois and Schweisguth, 1995; Paroush et al., 1994). It is unclear whether this occurs in the FSC lineage because E(spl) is not required for follicle cell differentiation (Lopez-Schier and St Johnston, 2001). Instead, our NRE-GFP data suggest that both Gro and Six4 function upstream of primary Notch target gene expression in pFCs.
Niche signals are commonly thought to promote stem cell self-renewal in part by antagonizing the signals that promote differentiation. In support of this idea, studies of the epithelial stem cell lineages in the mammalian hair follicle bulge and the intestine found that the stem cell self-renewal and differentiation cues are activated in opposing gradients (Barker, 2014; Rompolas and Greco, 2014; Sato et al., 2011; Tian et al., 2015), but the mechanisms of interaction between the opposing cues in these tissues are unclear. Our findings suggest that Gro may balance the decision between self-renewal and differentiation in the FSC lineage in a manner that is similar to the mechanisms that operate in the Drosophila male and female germline stem cell niches (de Cuevas and Matunis, 2011; Losick et al., 2011; Xie, 2013). Specifically, just as EGFR signaling inhibits Gro in the FSC niche, BMP signaling in the germline stem cell niches inhibits two repressors that promote germ cell differentiation, bag of marbles (bam) and benign gonial cell neoplasia (bgcn) (Chen et al., 2014; Gönczy et al., 1997; McKearin and Ohlstein, 1995; Shen et al., 2009). The structure and function of epithelial stem cell niches are similar in many fly and mammalian tissues, and the structure of both Gro and Six4 are evolutionarily conserved. Moreover, as in the FSC lineage, EGFR and Notch signaling also promote stem cell self-renewal and differentiation in mammalian epithelia such as the intestinal lining, the hair follicle and the interfollicular epidermis (Aubin-Houzelstein, 2012; Doma et al., 2013; Sato et al., 2011; Tian et al., 2015; Watt et al., 2008). Thus, it will be interesting to determine whether a similar Gro-mediated transition state exists in other epithelial tissues.
MATERIALS AND METHODS
Fly stocks
Fly stocks were maintained on standard molasses food.
Genotypes used in this work were obtained from the Bloomington Stock Center or as indicated : (1) UAS-EGFRλtop (active EGFR) (Trudi Schupbach, Princeton University, NJ, USA); (2) UAS-Six4.HA (FlyORF; Line ID F000049) (Bischof et al., 2013); (3) upd-Gal4 (Denise Montell, UC Santa Barbara, Santa Barbara, CA, USA); (4) y,w, hsFLP, tub-Gal4, UAS-GFP/FM7 ; tub-Gal80 FRT40A/CyO (Yuh-Nung Jan, UC San Francisco, San Francisco, CA, USA); and (5) y, w, hsFLP, tub-Gal4, UAS-GFP/FM7 ; ; tub-Gal80, FRT2A/TM3 (Yuh-Nung Jan).
TRiP RNAi lines (Ni et al., 2011) from the Bloomington Stock Center used in the main figures are as follows: (1) UAS-Six4 RNAi (TRiP; BL# 30510); (2) UAS-gro RNAi (TRiP; BL# 35759); and (3) UAS-Notch RNAi (TRiP; BL# 31383).
The following genotypes were used for the isolation of pFCs for RNA-seq: (1) wild type: +; 109-30-Gal4/UAS-mCD8::GFP; tub-Gal80ts/+; and (2) EGFRλtop: +; 109-30-Gal4/UAS-mCD8::GFP; tub-Gal80ts/UAS-EGFRλtop.
The following genotypes were used in Gal4 experiments: (1) wild type: y, w;+; +; (2) 109-30ts>EGFRλtop: +; 109-30-Gal4/+; tub-Gal80ts/UAS-EGFRλtop; (3) 109-30ts>Six4 RNAi: +; 109-30-Gal4/+; tub-Gal80ts/UAS-Six4 RNAi; (4) 109-30ts>Six4.HA: +; 109-30-Gal4/+; tub-Gal80ts/UAS-Six4.HA; (5) 109-30ts>Notch RNAi: +; 109-30-Gal4/+; tub-Gal80ts/UAS-Notch RNAi; (6) 109-30ts>Notchintra: +; 109-30-Gal4/+; tub-Gal80ts/UAS-Notchintra; (7) upd>Six4 RNAi: upd-Gal4/+; +; UAS-Six4 RNAi/+; (8) upd>EGFRλtop: upd-Gal4/+; +; UAS-EGFRλtop/+; (9) neur-lacZ: +; +; neur-lacZ/+; (10) 109-30>Six4 RNAi, neur-lacZ: +; 109-30-Gal4/+; neur-lacZ/UAS-Six4 RNAi; (11) 109-30>Six4.HA, neur-lacZ: +; 109-30-Gal4/+; neur-lacZ/UAS-Six4.HA; (12) NRE-GFP: w ; NRE-GFP; +; (13) 109-30ts>Six4 RNAi, NRE-GFP: +; 109-30-Gal4/NRE-GFP; tub-Gal80ts/UAS-Six4 RNAi; (14) 109-30ts>Six4.HA, NRE-GFP: +; 109-30-Gal4/NRE-GFP; tub-Gal80ts/UAS-Six4.HA; (15) 109-30ts>gro RNAi: +; 109-30-Gal4/+; tub-Gal80ts/UAS-gro RNAi; (16) 109-30ts>groAA: +; 109-30-Gal4/+; tub-Gal80ts/UAS-groAA; (17) 109-30ts>gro RNAi, NRE-GFP: +; 109-30-Gal4/NRE-GFP; tub-Gal80ts/UAS-gro RNAi; (18) 109-30ts>groAA, NRE-GFP: +; 109-30-Gal4/NRE-GFP; tub-Gal80ts/UAS-groAA; and (19) 109-30>groAA, neur-lacZ: +; 109-30-Gal4/+; neur-lacZ/UAS-groAA.
The following genotypes were used to generate MARCM clones: (1) wild type 40A: hsFlp, tub-Gal4, UAS-GFP/+; tub-Gal80, FRT 40A/FRT 40A; +; (2) wild type 2A: hsFlp, tub-Gal4, UAS-GFP/+; +; tub-Gal80, FRT 2A/FRT 2A; (3) Six4 RNAi: hsFlp, tub-Gal4, UAS-GFP/+; tub-Gal80, FRT 40A/FRT 40A; UAS-Six4 RNAi/+; +; (4) Six4108: hsFlp, tub-Gal4, UAS-GFP/+; +; tub-Gal80, FRT 2A/Six4108, FRT 2A; (5) Six4.HA: hsFlp, tub-Gal4, UAS-GFP/+; tub-Gal80, FRT 40A/FRT 40A; UAS-Six4.HA/+; (6) gro RNAi: hsFlp, tub-Gal4, UAS-GFP/+; tub-Gal80, FRT 40A/FRT 40A; UAS-gro RNAi/+; and (7) groAA: hsFlp, tub-Gal4, UAS-GFP/+; tub-Gal80, FRT 40A/FRT 40A; UAS-groAA/+.
Immunofluorescence
Adult flies were fed wet yeast for at least 2 days prior to dissection to ensure plump ovaries. Ovaries were dissected in Schneider's Insect Medium, fixed in phosphate-buffered saline (PBS)+4% paraformaldehyde for 15 min. Tissue was rinsed in PBS+0.1% Triton X-100 (0.1% PBT) twice for 1 min per rinse prior to incubation with blocking solution (0.1% PBT+0.5% bovine serum albumin) for 15 min. Primary antibodies were diluted in blocking solution and incubated with tissue overnight at 4°C, while rocking on a nutator. Tissue was rinsed twice for 1 min per rinse then washed for 1 h in 0.1% PBT. Secondary antibodies were diluted in blocking solution and incubated with tissue for 2 h at room temperature, while rocking on a nutator. Tissue was rinsed twice for 1 min per rinse and washed for 1 h in PBS, then mounted on a glass slide using Hard Set Vectashield plus DAPI mounting medium (Vector Labs).
All images were acquired using a Zeiss M2 Axioimager with Apotome unit or Nikon C1si Spectral Confocal microscope. For multicolor fluorescence images, each channel was acquired separately. Post-acquisition processing, such as image rotation, cropping, brightness or contrast adjustment, stitching of two overlapping fields (in Fig. 3F) (Preibisch et al., 2009) and z-projections, were performed using FIJI (Schindelin et al., 2012). Comparable staging of follicles between samples was accomplished by counterstaining for Fas3 or Vasa to identify the region 2a/b border in the germarium. The size and shape of each cyst were recorded, using (as guides) the number of germline cysts posterior to region 2a/b border and, in the case of budded follicles, their position relative to the germarium.
The following primary antibodies were used: ms α-β-gal (1:1000, Promega Z3781), rb α-Cas (1:5000, a gift from Ward Odenwald, National Institutes of Health, NINDS, Bethesda, MD, USA) (Kambadur et al., 1998), ms α-Eya (1:100, DSHB 10H6) (Bonini et al., 1993), ms α-Fas3 (1:100, DSHB 7G10) (Patel et al., 1987), gp α-GFP (1:1000, Synaptic Systems 132005), ms α-Gro (1:1000, DSHB anti-Gro) (Delidakis et al., 1991), rb α-p-Gro (1:1000) (Cinnamon et al., 2008), rt α-Six4 (1:1000, a gift from Eric Rulifson, UC San Francisco, San Francisco, CA, USA) (Hwang and Rulifson, 2011), goat α-fringe (1:500, Santa Cruz sc-15782) and rb α-Vasa (1:1000, Santa Cruz sc-30210). The following secondary antibodies were purchased from Thermo Fisher Scientific and used at 1:1000: gt α-gp 488 (A-11073), gt α-rb 488 (A-11008), gt α-rb 555 (A-21428), gt α-ms 488 (A-11029), gt α-ms 555 (A-21424) and gt α-rt 555 (A-21434).
For the co-staining of Gro and p-Gro in Fig. 4H, ovaries were dissected in PBS and fixed in PBS+5% formaldehyde for 20 min. Ovaries were rinsed twice in PBS containing 1% Triton X-100 (1% PBT), washed in 1% PBT for 10 min, then washed again with 1% PBT for 1 h. Ovaries were blocked in PBS+0.3% Triton X-100+1% BSA (PBTB) for 1 h, then incubated with mouse anti-Gro (1:1000) and rabbit anti-p-Gro (1:1000) antibodies in PBTB overnight at 4°C. Ovaries were washed in PBTB twice for 30 min per wash, then blocked with PBTB+5% normal goat serum (NGS, Sigma) for 1 h. Secondary antibodies were donkey α-ms 488 and donkey α-rb Rhodamine Red-X (Jackson Laboratories) diluted 1:500 in PBTB+5% NGS. Following 2 h incubation with the secondary antibodies, ovaries were washed in 0.3% PBT three times for 30 min per wash, rinsed twice in PBS, incubated in PBS+DAPI (1:1000) for 5 min, then washed twice in PBS. Finally, ovaries were mounted using Vectashield mounting medium. Images were acquired using a LSM710 confocal microscope.
Clone induction
Flies of the appropriate genotype were cultured and fed wet yeast for 2 days prior to clone induction. Heat shock was performed by transferring flies to empty plastic vials and immersing these vials in a 37°C water bath for 1 h. Flies were then allowed to recover at 25°C in vials containing supplemental wet yeast. This process was repeated twice daily for 2 days for a total of four 1 h heat shocks. Control flies were always simultaneously subjected to the same heat-shock regimen as experimental flies. Flies were then maintained at 25°C and fed wet yeast daily until dissection.
FSC competition assay
In this assay, MARCM clones of various genotypes were induced and the frequency of germaria containing zero, one or two labeled stem cells was measured at 7, 14 or 21 days after clone induction. Stem cell labeling counts were analyzed as described previously (Kronen et al., 2014). In brief, replacement events can be measured within the subset of germaria that have one stem cell labeled at the initial time point. Over time, single-labeled germaria can become unlabeled or double-labeled, indicating the replacement has occurred. (Germaria containing zero or two labeled stem cells at the earliest timepoint are already homogenous and replacement cannot be detected.) An increase in the proportion of double-labeled germaria is related to the rate of clone expansion, whereas an increase in the proportion of unlabeled germaria is related to the rate of clone extinction. Under neutral competition, the rates of extinction and expansion should be equal. Unless otherwise stated, all confidence intervals in this paper are 95%.
RNA-seq of pFCs expressing EGFRλtop
109-30ts was used to express UAS-EGFRλtop+UAS-mCD8::GFP or UAS-mCD8::GFP alone. Flies were bred and reared to adulthood at 18°C, then transferred to 29°C for 7-10 days prior to dissection. During the incubation at 29°C, flies were provided with wet yeast daily until 24 h prior to dissection; removal of wet yeast on the final day allows mature egg chambers to clear from the ovary and reduces clogging of cell filters.
For each replicate, over 200 flies were dissected and collected in Schneider's Insect Medium+10% FBS. Two replicates were collected for each genotype for a total of four samples. To maintain tissue health, the total dissection time was limited to 45 min, and the dissection dish and tissue collection tubes were maintained on ice. Pipette tips used for transferring ovaries to collection tubes were coated in 35% BSA immediately prior to use to prevent tissue from adhering to the inside of the tips.
Ovaries were rinsed three times in Cell Dissociation Buffer (CDB; Thermo Fisher Scientific 13151014), then incubated in CDB+4 mg/ml elastase (Worthington Biochemical LS002292)+2.5 mg/ml collagenase (Thermo Fisher Scientific 17018029) for 15 min at room temperature to disrupt the tissue. During this 15 min incubation, tissue was agitated by inverting the tubes and by passing the tissue through a P200 pipette tip. Dissociated cells were passed through a 50 µm filter (Partec 04-0042-2317), the dissociation enzymes were quenched by adding 0.5 volumes Schneider's Insect Medium+10% FBS, and the cells were maintained on ice. Dissociated cells were pelleted by centrifugation at 1000 g for 7 min at 4°C, then resuspended in 90 μl Schneider's Insect Medium+10 μl α-CD8a MicroBeads (Miltenyi Biotec 130-049-401) per 15 flies dissected. Dissociated cells were allowed to incubate with the α-CD8 MicroBeads for 15 min at 4°C. The CD8+ cells were then isolated by passing the cells over a magnetic column in an OctoMACS separator (Miltenyi Biotec 130-042-108). Cells were eluted from the column, pelleted by centrifugation at 1000 g for 7 min at 4°C, resuspended in ∼150 μl Lysis Solution (Thermo Fisher Scientific AM1931), then stored at −80°C until RNA isolation.
RNA was isolated from tissue using the RNAqueous-Micro Total RNA Isolation Kit (Thermo Fisher Scientific AM1931), including DNase I treatment. From a preparation of ∼200 ovaries, ∼100 ng of total RNA was isolated. PolyA selection was performed on the total RNA using oligo-dT Dynabeads (Thermo Fisher Scientific 61006). The ScriptSeqv2 kit (Illumina SSV21106) with Index Primers Set 1 (Illumina RSBC10948) was used to generate indexed paired-end RNA-seq libraries.
Libraries were sequenced at the UCSF Institute for Human Genetics on an Illumina HiSeq 2500 with a High Output PE 2×100 bp flow cell. Sequenced reads were trimmed using Scythe (v 0.991, https://github.com/vsbuffalo/scythe) and Sickle (v 1.29, https://github.com/najoshi/sickle). Quality check was performed using FastQC (v 0.11.2, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmed reads were aligned to the Drosophila melanogaster genome annotation from ENSEMBL (BDGP 5.74; downloaded from Illumina iGenomes, https://support.illumina.com/sequencing/sequencing_software/igenome.html) using TopHat2 (v 2.0.10) (Kim et al., 2013). Differential expression of aligned reads was tested by two methods: (1) Cufflinks Cuffdiff (v 2.1.1) (Trapnell et al., 2012) or (2) HTSeq-count (v 0.6.1) (Anders et al., 2015) and DESeq2 (v 1.2.10) (Love et al., 2014). To limit subsequent analysis to transcription factors, only genes referenced in Trusted TFs (v1) on FlyTF.org (http://www.flytf.org/flytfmine/bag.do) were considered. Plotting of FPKM values for transcription factors was performed using matplotlib (Hunter, 2007).
CRISPR generation of the Six4108 allele
The following primers were used to generate a chimeric RNA for the CRISPR Cas9 system: sense, CTTCGCGAGCTGAGATTGTCCTGA; and anti AAACTCAGGACAATCTCAGCTCGC. These primers were annealed and inserted into pU6-BbsI-gRNA according to the U6-gRNA (chiRNA) protocol available at flyCRISPR (Bassett and Liu, 2014; Gratz et al., 2014). This construct was co-injected into flies containing FRT2A with Cas9 RNA by Rainbow Transgenic Flies. Injected flies (P0) were crossed to (w; Sp/CyO; TM2/TM6) to introduce balancers. Independent lines were established by backcrossing individual F1 males to w; Sp/CyO; TM2/TM6. As previously described, Six4 alleles are known to be recessive lethal (Clark et al., 2007), so lines were screened for lethality. Lines containing a lethal mutation were sequenced by first PCR amplifying the region of Six4 targeted for mutation, then sequencing this PCR product using the following primers: forward, GACAAGTGAATGCAGTTTAGTG; reverse, AAATGTGTACTCCTCAGCAG; and sequencing, CTCTGGACTATTTGCACCGA.
Of 100+ lines screened, only one line with a Six4 disruption was isolated. Based on the mixed sequencing peaks present, this line likely has a deletion of G108 relative to the ATG start. We have designated this allele Six4108.
Acknowledgements
We are grateful to Pat O'Farrell, Marco Conti, Katja Brückner and Lucy O'Brien for critical comments on the manuscript and to our colleagues in the fly community (cited in the Materials and Methods section) for fly stocks and antibodies. We also thank the Bloomington Stock Center and The Developmental Studies Hybridoma Bank for curation of many of the stocks and reagents used in this study.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.J.J.,T.G.N.; Methodology: M.J.J., S.B.C., Z.P.,T.G.N.; Software: M.J.J., T.G.N.; Validation: M.J.J., S.B.C., Z.P., T.G.N.; Formal Analysis: M.J.J., T.G.N.; Investigation; M.J.J., S.B.C.; Resources: Z.P.,T.G.N.; Data Curation: M.J.J.; Writing – Original Draft: M.J.J., T.G.N.; Writing – Review and Editing: M.J.J., S.B.C., Z.P., T.G.N.; Visualization: M.J.J.,T.G.N.; Supervision: Z.P., T.G.N.; Project Administration: T.G.N.; Funding Acquisition: Z.P.,T.G.N.
Funding
This work was supported by the National Institutes of Health (R01 GM097158 to T.G.N.), by a predoctoral Genentech Foundation Fellowship to M.J.J., and by funds from the Israel Science Foundation (Center of Excellence 1772/13) and the Jan M. and Eugenia Król Charitable Foundation to Z.P., who is an incumbent of the Lady Davis Chair in Experimental Medicine and Cancer Research. S.B.C. was supported by a Bester PhD Scholarship. Deposited in PMC for release after 12 months.
Data availability
The RNAseq data has been deposited in Dryad Digital Repository (Johnson et al., 2016): http://dx.doi.org/10.5061/dryad.5bf77.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.143263.supplemental
References
- Anders S., Pyl P. T. and Huber W. (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assa-Kunik E., Torres I. L., Schejter E. D., Johnston D. S. and Shilo B.-Z. (2007). Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development 134, 1161-1169. 10.1242/dev.02800 [DOI] [PubMed] [Google Scholar]
- Aubin-Houzelstein G. (2012). Notch signaling and the developing hair follicle. Adv. Exp. Med. Biol. 727, 142-160. 10.1007/978-1-4614-0899-4_11 [DOI] [PubMed] [Google Scholar]
- Bai J. and Montell D. (2002). Eyes absent, a key repressor of polar cell fate during Drosophila oogenesis. Development 129, 5377-5388. 10.1242/dev.00115 [DOI] [PubMed] [Google Scholar]
- Bailey A. M. and Posakony J. W. (1995). Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609-2622. 10.1101/gad.9.21.2609 [DOI] [PubMed] [Google Scholar]
- Barker N. (2014). Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 15, 19-33. 10.1038/nrm3721 [DOI] [PubMed] [Google Scholar]
- Bassett A. R. and Liu J.-L. (2014). CRISPR/Cas9 and genome editing in Drosophila. J. Genet. Genomics 41, 7-19. 10.1016/j.jgg.2013.12.004 [DOI] [PubMed] [Google Scholar]
- Bischof J., Björklund M., Furger E., Schertel C., Taipale J. and Basler K. (2013). A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434-2442. 10.1242/dev.088757 [DOI] [PubMed] [Google Scholar]
- Bonini N. M., Leiserson W. M. and Benzer S. (1993). The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell 72, 379-395. 10.1016/0092-8674(93)90115-7 [DOI] [PubMed] [Google Scholar]
- Borghese L., Fletcher G., Mathieu J., Atzberger A., Eades W. C., Cagan R. L. and Rørth P. (2006). Systematic analysis of the transcriptional switch inducing migration of border cells. Dev. Cell 10, 497-508. 10.1016/j.devcel.2006.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanieto A., Johnston M. J. and Nystul T. G. (2014). EGFR signaling promotes the identity of Drosophila follicle stem cells via maintenance of partial cell polarity. eLife 10.7554/eLife.04437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-C., Jang A. C.-C., Lin C.-H. and Montell D. J. (2013). Castor is required for Hedgehog-dependent cell-fate specification and follicle stem cell maintenance in Drosophila oogenesis. Proc. Natl. Acad. Sci. USA 110, E1734-E1742. 10.1073/pnas.1300725110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Wu C., Zhao S., Geng Q., Gao Y., Li X., Zhang Y. and Wang Z. (2014). Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila. PLoS Genet. 10, e1004797 10.1371/journal.pgen.1004797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnamon E., Helman A., Schyr R. B.-H., Orian A., Jiménez G. and Paroush Z. (2008). Multiple RTK pathways downregulate Groucho-mediated repression in Drosophila embryogenesis. Development 135, 829-837. 10.1242/dev.015206 [DOI] [PubMed] [Google Scholar]
- Clark I. B. N., Boyd J., Hamilton G., Finnegan D. J. and Jarman A. P. (2006). D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev. Biol. 294, 220-231. 10.1016/j.ydbio.2006.02.044 [DOI] [PubMed] [Google Scholar]
- Clark I. B. N., Jarman A. P. and Finnegan D. J. (2007). Live imaging of Drosophila gonad formation reveals roles for Six4 in regulating germline and somatic cell migration. BMC Dev. Biol. 7, 52 10.1186/1471-213X-7-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton E., Doupé D. P., Klein A. M., Winton D. J., Simons B. D. and Jones P. H. (2007). A single type of progenitor cell maintains normal epidermis. Nature 446, 185-189. 10.1038/nature05574 [DOI] [PubMed] [Google Scholar]
- de Cuevas M. and Matunis E. L. (2011). The stem cell niche: lessons from the Drosophila testis. Development 138, 2861-2869. 10.1242/dev.056242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Navascués J., Perdigoto C. N., Bian Y., Schneider M. H., Bardin A. J., Martínez-Arias A. and Simons B. D. (2012). Drosophila midgut homeostasis involves neutral competition between symmetrically dividing intestinal stem cells. EMBO J. 31, 2473-2485. 10.1038/emboj.2012.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C., Preiss A., Hartley D. A. and Artavanis-Tsakonas S. (1991). Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer of split locus of Drosophila melanogaster. Genetics 129, 803-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma E., Rupp C. and Baccarini M. (2013). EGFR-ras-raf signaling in epidermal stem cells: roles in hair follicle development, regeneration, tissue remodeling and epidermal cancers. Int. J. Mol. Sci. 14, 19361-19384. 10.3390/ijms141019361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A. L., Ohsako S. and Caudy M. (1996). The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16, 2670-2677. 10.1128/MCB.16.6.2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz A. and Riechmann V. (2010). Stepwise polarisation of the Drosophila follicular epithelium. Dev. Biol. 338, 136-147. 10.1016/j.ydbio.2009.11.027 [DOI] [PubMed] [Google Scholar]
- Gönczy P., Matunis E. and DiNardo S. (1997). bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124, 4361-4371. [DOI] [PubMed] [Google Scholar]
- Grammont M. and Irvine K. D. (2001). fringe and Notch specify polar cell fate during Drosophila oogenesis. Development 128, 2243-2253. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M. and O'Connor-Giles K. M. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961-971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman T. R., Zinshteyn D., Schofield H. K., Nicolas E., Okada A. and O'Reilly A. M. (2010). Drosophila Boi limits Hedgehog levels to suppress follicle stem cell proliferation. J. Cell Biol. 191, 943-952. 10.1083/jcb.201007142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P., Egoz N., Winkler C., Volohonsky G., Jia S., Dinur T., Volk T., Courey A. J. and Paroush Z. (2005). EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat. Genet. 37, 101-105. [DOI] [PubMed] [Google Scholar]
- Helman A., Cinnamon E., Mezuman S., Hayouka Z., Von Ohlen T., Orian A., Jiménez G. and Paroush Z. (2011). Phosphorylation of Groucho mediates RTK feedback inhibition and prolonged pathway target gene expression. Curr. Biol. 21, 1102-1110. 10.1016/j.cub.2011.05.043 [DOI] [PubMed] [Google Scholar]
- Hunter J. D. (2007). Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90-95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Hwang H. J. and Rulifson E. (2011). Serial specification of diverse neuroblast identities from a neurogenic placode by Notch and Egfr signaling. Development 138, 2883-2893. 10.1242/dev.055681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzkovitz S., Lyubimova A., Blat I. C., Maynard M., van Es J., Lees J., Jacks T., Clevers H. and van Oudenaarden A. (2011). Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat. Cell Biol. 14, 1-10. 10.1038/ncb2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Bar-Cohen S., Paroush Z. and Nystul T. (2016). Data from: Phosphorylated Groucho delays differentiation in the follicle stem cell lineage by providing a molecular memory of EGFR signaling in the niche Dryad Digital Repository. doi: 10.5061/dryad.5bf77 10.5061/dryad.5bf77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. H., Simons B. D. and Watt F. M. (2007). Sic transit gloria: farewell to the epidermal transit amplifying cell? Cell Stem Cell 1, 371-381. 10.1016/j.stem.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Kambadur R., Koizumi K., Stivers C., Nagle J., Poole S. J. and Odenwald W. F. (1998). Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 12, 246-260. 10.1101/gad.12.2.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller Larkin M., Deng W.-M., Holder K., Tworoger M., Clegg N. and Ruohola-Baker H. (1999). Role of Notch pathway in terminal follicle cell differentiation during Drosophila oogenesis. Dev. Genes Evol. 209, 301-311. 10.1007/s004270050256 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. and Salzberg S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby R. J., Hamilton G. M., Finnegan D. J., Johnson K. J. and Jarman A. P. (2001). Drosophila homolog of the myotonic dystrophy-associated gene, SIX5, is required for muscle and gonad development. Curr. Biol. 11, 1044-1049. 10.1016/S0960-9822(01)00319-0 [DOI] [PubMed] [Google Scholar]
- Kronen M. R., Schoenfelder K. P., Klein A. M. and Nystul T. G. (2014). Basolateral junction proteins regulate competition for the follicle stem cell niche in the Drosophila ovary. PLoS ONE 9, e101085 10.1371/journal.pone.0101085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J. P. (2009). The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell. Mol. Life Sci. 66, 565-583. 10.1007/s00018-008-8335-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M. K., Holder K., Yost C., Giniger E. and Ruohola-Baker H. (1996). Expression of constitutively active Notch arrests follicle cells at a precursor stage during Drosophila oogenesis and disrupts the anterior-posterior axis of the oocyte. Development 122, 3639-3650. [DOI] [PubMed] [Google Scholar]
- Lecourtois M. and Schweisguth F. (1995). The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 9, 2598-2608. 10.1101/gad.9.21.2598 [DOI] [PubMed] [Google Scholar]
- Lopez-Schier H. and St Johnston D. (2001). Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes Dev. 15, 1393-1405. 10.1101/gad.200901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick V. P., Morris L. X., Fox D. T. and Spradling A. (2011). Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev. Cell 21, 159-171. 10.1016/j.devcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J. and Spradling A. (1995). Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121, 3797-3807. [DOI] [PubMed] [Google Scholar]
- Mascré G., Dekoninck S., Drogat B., Youssef K. K., Brohée S., Sotiropoulou P. A., Simons B. D. and Blanpain C. (2012). Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489, 257-262. 10.1038/nature11393 [DOI] [PubMed] [Google Scholar]
- McKearin D. and Ohlstein B. (1995). A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121, 2937-2947. [DOI] [PubMed] [Google Scholar]
- Ni J.-Q., Zhou R., Czech B., Liu L.-P., Holderbaum L., Yang-Zhou D., Shim H.-S., Tao R., Handler D., Karpowicz P. et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405-407. 10.1038/nmeth.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystul T. and Spradling A. (2010). Regulation of epithelial stem cell replacement and follicle formation in the Drosophila ovary. Genetics 184, 503-515. 10.1534/genetics.109.109538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z., Finley R. L. Jr, Kidd T., Wainwright S. M., Ingham P. W., Brent R. and Ish-Horowicz D. (1994). Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79, 805-815. 10.1016/0092-8674(94)90070-1 [DOI] [PubMed] [Google Scholar]
- Patel N. H., Snow P. M. and Goodman C. S. (1987). Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell 48, 975-988. 10.1016/0092-8674(87)90706-9 [DOI] [PubMed] [Google Scholar]
- Pignoni F., Hu B., Zavitz K. H., Xiao J., Garrity P. A. and Zipursky S. L. (1997). The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881-891. 10.1016/S0092-8674(00)80480-8 [DOI] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S. and Tomancak P. (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463-1465. 10.1093/bioinformatics/btp184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queenan A. M., Ghabrial A. and Schupbach T. (1997). Ectopic activation of torpedo/Egfr, a Drosophila receptor tyrosine kinase, dorsalizes both the eggshell and the embryo. Development 124, 3871-3880. [DOI] [PubMed] [Google Scholar]
- Rompolas P. and Greco V. (2014). Stem cell dynamics in the hair follicle niche. Semin. Cell Dev. Biol. 25-26, 34-42. 10.1016/j.semcdb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai-Hernandez P. and Nystul T. G. (2013). A dynamic population of stromal cells contributes to the follicle stem cell niche in the Drosophila ovary. Development 140, 4490-4498. 10.1242/dev.098558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai-Hernandez P., Castanieto A. and Nystul T. G. (2012). Drosophila models of epithelial stem cells and their niches. Wiley Interdiscip. Rev. Dev. Biol. 1, 447-457. 10.1002/wdev.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A., Arziman Z., Stempfle D., van Belle W., Sauder U., Horn T., Dürrenberger M., Paro R., Boutros M. and Merdes G. (2010). A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev. Cell 18, 862-876. 10.1016/j.devcel.2010.03.013 [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J. H., Snippert H. J., Stange D. E., Vries R. G., van den Born M., Barker N., Shroyer N. F., van de Wetering M. and Clevers H. (2011). Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 469, 415-418. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Weng C., Yu J. and Xie T. (2009). eIF4A controls germline stem cell self-renewal by directly inhibiting BAM function in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 106, 11623-11628. 10.1073/pnas.0903325106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver S. J., Davies E. L., Doyon L. and Rebay I. (2003). Functional dissection of eyes absent reveals new modes of regulation within the retinal determination gene network. Mol. Cell. Biol. 23, 5989-5999. 10.1128/MCB.23.17.5989-5999.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert H. J., van der Flier L. G., Sato T., van Es J. H., van den Born M., Kroon-Veenboer C., Barker N., Klein A. M., van Rheenen J., Simons B. D. et al. (2010). Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134-144. 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Song X. and Xie T. (2003). Wingless signaling regulates the maintenance of ovarian somatic stem cells in Drosophila. Development 130, 3259-3268. 10.1242/dev.00524 [DOI] [PubMed] [Google Scholar]
- Tian H., Biehs B., Chiu C., Siebel C. W., Wu Y., Costa M., de Sauvage F. J. and Klein O. D. (2015). Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 11, 33-42. 10.1016/j.celrep.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres I. L., López-Schier H. and St Johnston D. (2003). A Notch/Delta-dependent relay mechanism establishes anterior-posterior polarity in Drosophila. Dev. Cell 5, 547-558. 10.1016/S1534-5807(03)00272-7 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L. and Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562-578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vied C. and Kalderon D. (2009). Hedgehog-stimulated stem cells depend on non-canonical activity of the Notch co-activator Mastermind. Development 136, 2177-2186. 10.1242/dev.035329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt F. M., Estrach S. and Ambler C. A. (2008). Epidermal Notch signalling: differentiation, cancer and adhesion. Curr. Opin. Cell Biol. 20, 171-179. 10.1016/j.ceb.2008.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T. (2013). Control of germline stem cell self-renewal and differentiation in the Drosophila ovary: concerted actions of niche signals and intrinsic factors. Wiley Interdiscip. Rev. Dev. Biol. 2, 261-273. 10.1002/wdev.60 [DOI] [PubMed] [Google Scholar]
- Yan K. S., Chia L. A., Li X., Ootani A., Su J., Lee J. Y., Su N., Luo Y., Heilshorn S. C., Amieva M. R. et al. (2012). The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466-471. 10.1073/pnas.1118857109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Clyde D. and Bownes M. (2000). Expression of fringe is down regulated by Gurken/Epidermal Growth Factor Receptor signalling and is required for the morphogenesis of ovarian follicle cells. J. Cell Sci. 113, 3781-3794. [DOI] [PubMed] [Google Scholar]