Abstract
Although much is known about the embryo during implantation, the architecture of the uterine environment in which the early embryo develops is not well understood. We employed confocal imaging in combination with 3D analysis to identify and quantify dynamic changes to the luminal structure of murine uterus in preparation for implantation. When applied to mouse mutants with known implantation defects, this method detected striking peri-implantation abnormalities in uterine morphology that cannot be visualized by histology. We revealed 3D organization of uterine glands and found that they undergo a stereotypical reorientation concurrent with implantation. Furthermore, we extended this technique to generate a 3D rendering of the cycling human endometrium. Analyzing the uterine and embryo structure in 3D for different genetic mutants and pathological conditions will help uncover novel molecular pathways and global structural changes that contribute to successful implantation of an embryo.
KEY WORDS: Implantation, Receptivity, Uterus, Blastocyst, Embryo, Confocal Imaging, Surface curvature, Wnt5a
Highlighted article: 3D imaging of the mouse uterine lumen and glands reveals dynamic changes in their architecture in preparation for embryo implantation.
INTRODUCTION
The uterus is the prerequisite for propagating mammalian species, as implantation in this tissue establishes pregnancy and protects the developing embryo throughout gestation. The window of uterine receptivity for embryo implantation is precisely timed (Ma et al., 2003). Rising levels of luteinizing hormone in concert with estradiol (E2) lead to ovulation and fertilization, termed gestational day (GD) 0 of murine pregnancy (reviewed by Cha et al., 2012; Wang and Dey, 2006). Formation of the corpus lutea at the site of ovulation releases progesterone (P4), which peaks at GD3 of pregnancy. Receptivity is achieved between GD3 and GD4 by the concurrence of high levels of P4 with a transient increase in E2 (Wang and Dey, 2006). Similarly, following human ovulation and corpus luteum formation, an increase in P4 during the secretory phase prepares the uterus for embryo implantation (Cha et al., 2012). In the mouse, blastocysts home and attach in the anti-mesometrial region of slit-like structures termed uterine crypts (Daikoku et al., 2011). The existence of crypts in the human uterus remains unknown. It stands to reason that ovarian hormones that coordinate uterine receptivity must also regulate uterine crypt formation; however, the underlying mechanisms are not defined.
In preparation for implantation, uterine epithelial cells undergo morphological changes, termed plasma membrane transformation (PMT) (Murphy, 2004). In the presence of E2, epithelial cells possess microvilli on their apical surfaces and short tight junctions on the lateral plasma membrane, whereas they lose microvilli, acquire flattened morphology and increase their number of tight junctions upon concurrent exposure to E2 and P4. PMT is observed at the time of implantation in eutherian mammals (Murphy, 2004), viviparous squamate reptiles (Hosie et al., 2003) and marsupials (Laird et al., 2014). These dramatic changes at the cellular level are accompanied by changes in the organization of the uterine lumen, as suggested by scanning electron microscopy studies (Winkelmann and Spornitz, 1997); however, the three-dimensional uterine architectural changes remain unknown.
Establishment of pregnancy also depends upon specialized uterine glands that provide nourishment to the embryo before the placenta forms (Filant and Spencer, 2014). A key factor secreted by the glands, leukemia inhibitory factor (LIF), is required for uterine receptivity (Daikoku et al., 2004; Stewart et al., 1992). The importance of uterine glands and their secretions is demonstrated by the drastic impact of their depletion on fertility (Filant et al., 2012). Studies involving uterine glands have been limited to histological analysis and currently there is no information on the 3D branching morphogenesis or structural organization of these glands during development, estrous or implantation.
In the past, localizing embryos and studying their interactions with the uterine lining required generating numerous tissue sections. This process is labor intensive and generates only two-dimensional images. We now combine the strengths of confocal imaging with quantitative image analysis to enable visualization of the intact uterus in three dimensions. We observe dynamic changes in the architecture of the uterine lumen surrounding implantation. We highlight the utility of this method by studying implantation defective Wnt5a mutants, as previously characterized by molecular marker expression, defective crypt formation and embryonic lethality (Cha et al., 2014). Our method reveals defects in the folding pattern of Wnt5a mutant uteri that cannot be discerned by traditional histology. Furthermore, we define the organization of the uterine glands in three dimensions and the large-scale changes that coincide with implantation. This methodology will help to resolve long-standing conundrums in the field, such as determinants of embryo spacing, closure of the uterine lumen, and the regulation and function of the poorly studied uterine glands.
RESULTS AND DISCUSSION
Confocal imaging of the mouse and human endometrium
We used whole-mount immunofluorescence and confocal imaging (Faire et al., 2015) to visualize and characterize changes in the mouse endometrium from fertilization through implantation. Luminal and glandular epithelium in the uterus were identified using the epithelial marker E-cadherin (E-CAD) (Reardon et al., 2012), whereas FOXA2 (Besnard et al., 2004) was used to distinguish glandular epithelium throughout the length and depth of the uterine horn (Fig. 1A; Movie 1). Developing embryos at the blastocyst and epiblast stages were identified within the uterine horns using E-CAD staining along with the nuclear stain Hoechst (Fig. 1B-E). We applied similar methodology to image a full thickness biopsy of the proliferative phase human uterus obtained by hysterectomy (Fig. 1F; Movie 2). As expected, 3D renderings of the E-CAD+ endometrial lumen and glands revealed increased glandular complexity of the human uterus compared with mouse (Fig. 1G; Movie 3).
Fig. 1.
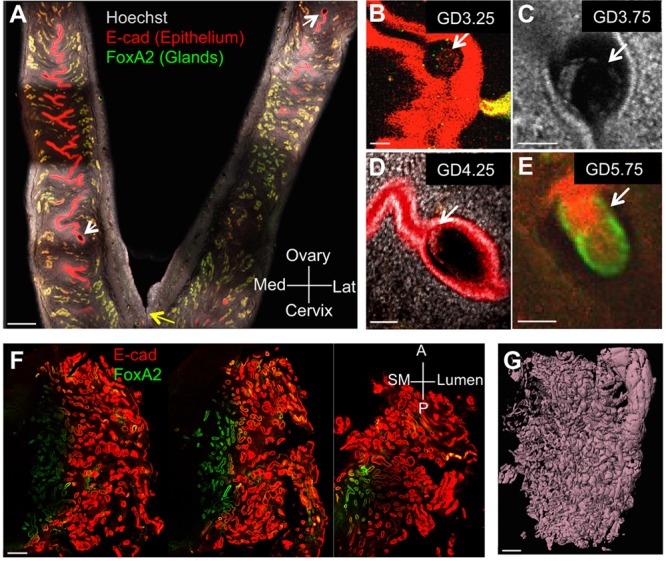
Confocal imaging of pregnant mouse uterus and cycling human endometrium. (A) Optical z slice showing two-thirds of both horns of a GD4.25 mouse uterus attached at the cervix (yellow arrow) stained with nuclear marker Hoechst (gray), epithelial marker E-CAD (red) and glandular marker FOXA2 (green). (B-D) Identification of blastocysts (white arrows) in optical slices of intact uteri at GD3.25 (B), GD3.75 (C) and GD4.25 (D). (E) Epiblast at GD5.75. (F,G) Three z slices (F) through a full-thickness segment of human endometrium stained for E-CAD (red) and FOXA2 (green), and corresponding surface rendering of the same specimen based on E-CAD staining (G). Scale bars: 500 μm in A,F,G; 50 μm in B-E. Lat, lateral; Med, medial; A, anterior; P, posterior; SM, smooth muscle.
Uterine crypts are folds in the uterine epithelium
To visualize the structure of the mouse uterine lumen, we computationally subtracted the FOXA2+ glandular signal from E-CAD+ total epithelium (Fig. 2A,B; Movies 4 and 5). After separating lumen and glands, generation of 3D surfaces through the length of the uterine horn permitted analysis of luminal structure and glandular organization (Movies 6 and 7). At GD 4.25, 2D optical sections revealed the overlap of 3D surface luminal folds with potential uterine crypts or chambers (Fig. 2C,D), which house the incoming embryo (Daikoku et al., 2011).
Fig. 2.
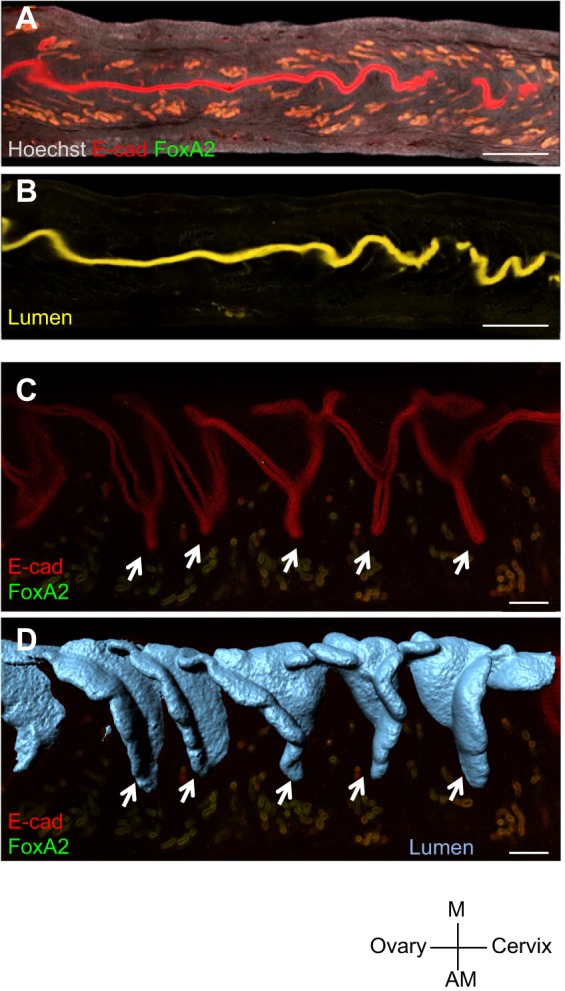
Uterine crypts are generated by folding the luminal epithelium. (A,B) Optical slices through a segment of the uterus at GD4.25 immunolabeled for FOXA2 signal (green) and ECAD signal (red) (A), and the resulting subtraction of FOXA2 from ECAD signal to obtain the uterine lumen (yellow) (B). (C) Optical slice showing uterine crypts (arrows) revealed by luminal epithelial staining at GD4.25. (D) Luminal folds (arrows) in the 3D surface model of the uterus coincide with crypts in the optical slice. Scale bars: 500 μm in A,B; 200 μm in C,D. M, mesometrial; AM, anti-mesometrial.
Uterine architecture changes dynamically from fertilization to implantation
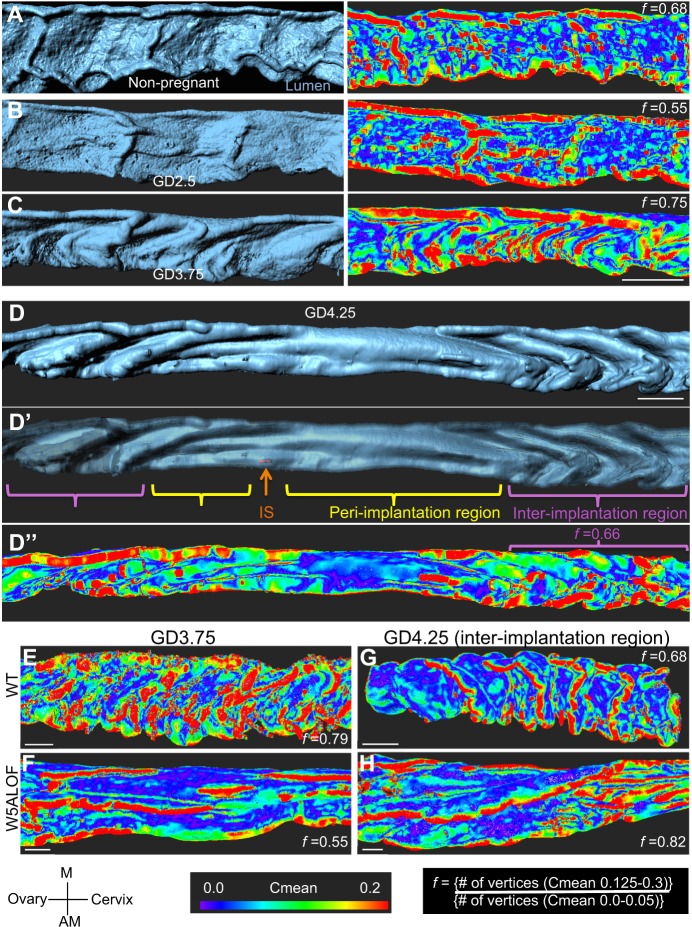
We observed that the murine uterine lumen undergoes dynamic changes in topology on the day preceding implantation (Fig. 3A-D). Three-dimensional imaging revealed an increase in the number of folds at gestational day (GD) 3.5-3.75, specifically along the mesometrial–anti-mesometrial (M-AM) axis, which lies perpendicular to both the oviductal-cervical axis and the dorsalventral axis of the uterine horn (Fig. 3C). Luminal closure ensues at GD4.25 to lock the embryo into position (Chen et al., 2013; Yoshinaga, 2013). Concomitant with closure, we observed that the lumen flanking the embryo, the peri-implantation region, becomes entirely devoid of folds in the M-AM axis; folds also grow more shallow along the oviductal-cervical axis, while folding along the M-AM axis is retained in the inter-implantation region (Fig. 3D,D′; Movie 6). This is consistent with uterine folding changes first described by scanning electron microscopy of epithelial cells (Winkelmann and Spornitz, 1997). We developed an algorithm to quantify topological changes in the E-CAD+ FOXA2– luminal epithelium by measuring curvature. The Surface Curvature algorithm splits the 3D surface into vertices and estimates the radius of the curve formed by connecting neighbor vertices. Radii calculated with multiple neighbor vertices are averaged to obtain curvature mean (Cmean). Topology of uterine luminal segments is expressed as the folding factor (f), which is the ratio of the Cmean for the most highly curved luminal epithelium (0.125<Cmean>0.3), to the least curved luminal epithelium regions (0<Cmean>0.05); (Fig. S1A-C). An increase in the depth of folds was observed from GD2.5 (Fig. 3B, f=0.55) to GD3.5-GD3.75 (Fig. 3C, f=0.75). At GD4.25, when embryo attachment has occurred, folds in the uterine lumen resolved specifically at implantation sites but were retained in intervening inter-implantation regions (Fig. 3D″, f=0.66; Movie 6). These quantitative descriptions of endometrial topology will advance understanding of mutants in uterine epithelial organization and in luminal closure.
Fig. 3.
Dynamic folding of the mouse uterine lumen along the mesometrial–anti-mesometrial axis and aberrant folding in Wnt5acKO uteri. (A-C) Uterine luminal surfaces are on the left and folding heat maps are on the right. (A) Luminal structure of non-pregnant mouse uterus. (B,C) Luminal folds appear at GD2.5 (B) and become predominant at GD3.75 (C). (D) Surface model of a GD4.25 luminal segment containing an embryo. (D′) Increased translucence reveals the site where the embryo is present (orange arrow, embryo surface). (D″) Curvature analysis in the same uterine horn measured using Cmean shows flatness in the peri-implantation region (Cmean values are primarily less than 0.05, blue/purple), whereas higher values, indicated by green-yellow and red, are found distant from the implantation site in the inter-implantation region. (E-H) Whereas wild-type uteri show folding along the M-AM axis (E,G), Wnt5acKO uterine lumens fold along the oviductal-cervical axis throughout the lumen at GD 3.75 (F) and at inter-implantation sites at GD4.25 (H). Scale bars: 500 μm in A-C; 300 μm in D-D″; 200 μm in E-H. M, mesometrial; AM, anti-mesometrial; IS, implantation site.
Wnt5a-deficient uteri have altered fold morphology
Conditional deletion of Wnt5a (Wnt5acKO) in the uterine luminal epithelium and mesenchyme, using the postnatally expressed Pgrcre, leads to defective crypt formation, aberrant gene expression at implantation sites and mid-gestation lethality (Cha et al., 2014). When the Surface Curvature algorithm was applied to the Wnt5acKO uteri at GD3.75, we observed multiple folding defects (Fig. 3E,F). Lumens appeared flatter and the orientation of folds was altered, as ridges increasingly ran along the oviductal-cervical axis instead of the M-AM axis. Owing to resolution of folds at the implantation site at GD4.25 in wild type (Fig. 3D), we asked whether aberrant folding was retained in inter-implantation regions in the Wnt5acKO uteri. At GD4.25, folds were still misoriented along the oviductal-cervical axis in the inter-implantation regions (Fig. 3G,H) of the Wnt5acKO uteri. Orientation of folding along the incorrect axis is reminiscent of planar cell polarity phenotypes, such as incorrect orientation of cochlear cilia in Wnt5a-deficient mutants (Qian et al., 2007). Defects in folding pattern such as these are very difficult to visualize using 2D histology, thus demonstrating the application of the 3D imaging technique.
Uterine glands reorient towards site of implantation
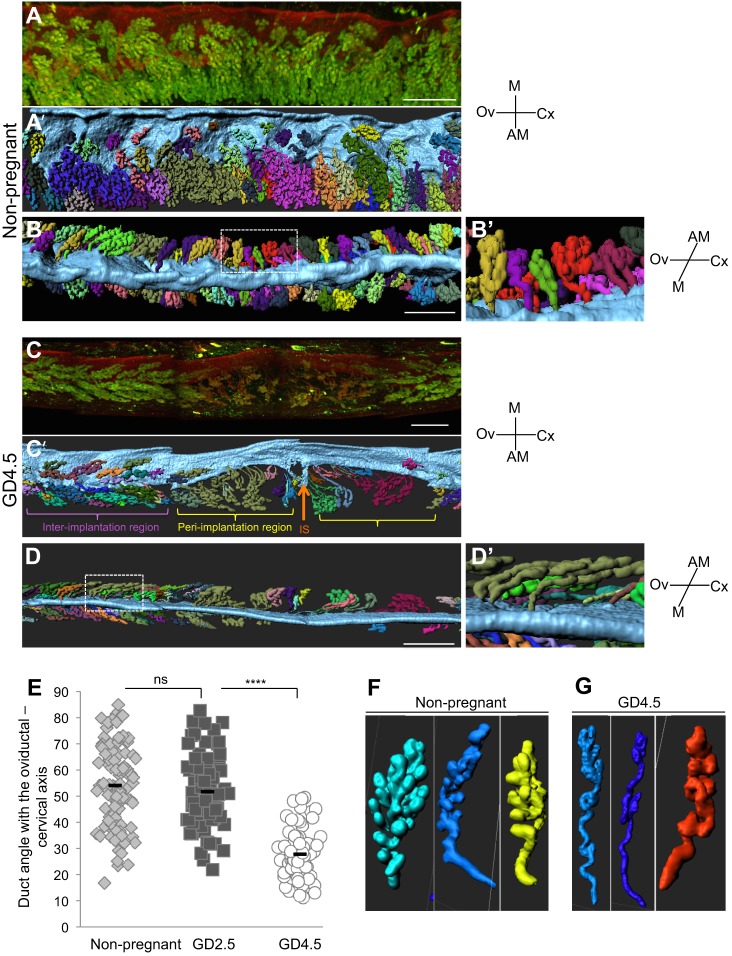
While visualizing the uterine glands using FOXA2 immunofluorescence, we observed that the glandular ducts bend, elongate and coil with increasing gestational age (Fig. 4). Generating separate surfaces of each glandular tree revealed large-scale changes in their morphology during early pregnancy, which culminates in the bending of the flanking glands towards the site of implantation (Fig. 4A-D, Fig. S2A; Movies 6 and 7). We measured the angle between the ducts of the glands and luminal epithelial plane perpendicular to the M-AM axis (Fig. S2B). This revealed a drastic decrease in the ductal angle at the time of implantation (GD 4.5, ∼28°) when compared with the ∼54° inclination at GD2.5 or in a non-pregnant mouse (Fig. 4E). We also observed a qualitative increase in coiling and ductal length when comparing non-pregnant and GD4.25 glandular trees (Fig. 4F,G; Figs S3 and S4). Spatially, the elongation of glandular ducts and the bending of the uterine glands positions the glandular epithelium in close proximity to stroma that surround the implantation site. This reorganization can help explain the mechanisms behind emerging evidence that implicates uterine glands in decidualization and placentation post-implantation (Filant and Spencer, 2014).
Fig. 4.
Glandular ducts reorient towards the site of implantation. (A-D′) 3D images and surface renderings of luminal segments and uterine glands, with separate glandular structures randomly pseudocolored for easy visualization. (A-B′) View from the ventral side (A,A′) and mesometrial side (B,B′) of non-pregnant uterine segment. (C-D′) View from the ventral side (C,C′) and mesometrial side (D,D′) of GD4.5 uterine segment. Boxed areas in B,D represent magnified regions in B′,D′, respectively. (E) The glandular ducts in non-pregnant uteri (B,B′) are at a mean angle of ∼54° to the uterine oviductal-cervical axis. (D,D′) With the introduction of the embryo (orange arrow) at the anti-mesometrial pole at GD4.5, the glandular ducts bend drastically until they are at a mean angle of ∼28° to the oviductal-cervical axis towards the site of implantation. The duct angle was measured in a total of 70-80 ducts from two different mice and around three embryos at GD4.5 (E). A t-test was used for statistical analysis and significance was defined as P<0.001. ****P<10−12; ns, not significant. (F,G) Representative examples of glandular branching in non-pregnant uteri (F), and glandular branching, coiling and duct elongation in GD4.5 uteri (G). Scale bars: 500 μm in A-D′. M, mesometrial; AM, anti-mesometrial; Ov, ovary; Cx, cervix; IS, implantation site.
This new methodology allows observations in an intact organ, and the quantification of structure and morphological changes, yet still permits optical sectioning in any desired plane comparable with traditional histological views. Our analysis integrates surface curvature measurements with confocal 3D imaging. The algorithm developed for Surface Curvature can easily be modified to account for object size and complexity for broader applications in other organ systems.
We expect that this technique will reveal novel aspects of uterine architecture during development, estrus and pregnancy. The understudied uterine glands that are key to the development of the embryo until the formation of the placenta are now accessible for studies of branching morphogenesis. This technique will allow for detailed investigation of the roles of different signaling pathways in development of the uterine lining and implantation. Elucidating such contributions to the spatial uterine environment will aid in: (1) better understanding of pregnancy failures due to implantation defects; (2) improving implantation rates when using assisted reproductive technologies such as in vitro fertilization; (3) opening doors for novel contraceptive targets; and (4) better understanding diseases such as uterine hyperplasia and endometrial cancer.
MATERIALS AND METHODS
Mice
CD1 mice were purchased from Charles River. Wnt5acKO (PgrCre;Wnt5aflox/flox, mixed 129Sv × C57BL/6 background) pregnant uteri were obtained from S. K. Dey (Cincinnati Children's Hospital Medical Center, Cincinnati, USA) (Cha et al., 2014). Uteri were dissected from non-pregnant adult females or from timed matings at: GD2.5, GD3.5 and GD4.5 (between 12:00 and 15:00 hours); GD4.25 at 09:00 hours; and GD3.75 between 19:00 and 21:00 hours on the day of dissection. The dark period was 19.00 to 05.00 h and the day of mating plug was identified as gestational day (GD) 0.5. All mouse work was carried out under University of California San Francisco Institutional Animal Care and Use Committee guidelines, in an AAALAC approved facility.
Human sample collection
Human adult uterine samples were obtained from the NIH UCSF Human Endometrial Tissue and DNA Bank, which contains samples of women undergoing endometrial biopsy or hysterectomy for nonmalignant indications. The samples were collected under approved Institutional Review Board protocols and informed consent to participate in the study was obtained from the patients.
Whole-mount immunofluorescence
Uteri were fixed in DMSO:methanol (1:4) and blocked (using PBS+1%Triton+2% powdered milk) for 2 h at room temperature. Primary antibodies for mouse E-CAD (M108, Clontech), human E-CAD (ab1416, Abcam) and FOXA2 (NBP1-95426, Novus Biologicals; or ab108422, Abcam) were diluted (1:200) in block, and uteri were incubated for 5 nights at 4°C. Uteri were washed (PBS+1% Triton) six times for 30 min each and incubated with secondary antibodies [fluorescently conjugated Alexa Fluor IgGs (Invitrogen)] for 2 nights at 4°C. Uteri were washed (in PBS+1% Triton) six times for 30 min each, dehydrated in methanol and incubated overnight in 3% H2O2 diluted in methanol. Uteri were washed in 100% methanol twice for 15 min each and cleared overnight using BABB (benzyl alcohol:benzyl benzoate, 1:2). Uteri were imaged using a Leica SP5 TCS confocal microscope with white-light laser, using a 10× air objective with z stacks that were 7 µm apart. Full uterine horns were imaged using 18×2 tile scans and tiles were merged using the mosaic merge function of the Leica software. For further details on the immunofluorescence, see supplementary Materials and Methods.
Image analysis
LIF files (Leica software) were analyzed using Imaris v8.1 (Bitplane). Using the channel arithmetic function, the glandular FOXA2+ signal was removed from the E-CAD+ signal to create lumen-only signal. Surfaces were created in surpass 3D mode for the luminal signal and the FOXA2+ glandular signal. Images and videos were captured using the snapshot function and the animation function, respectively. For further details of image analysis, see supplementary Materials and Methods.
Description of Matlab script and calculation of Cmean
The Matlab script was adapted from http://www.mathworks.com/matlabcentral/fileexchange/32573-patch-curvature. For further details, including equations used to calculate Cmean, see supplementary Materials and Methods.
Determination of folding using Surface Curvature
Folding measurements were generated by modifying an existing Matlab script made available by Imaris Open (Bitplane), originally intended to calculate localized, small area, curvature. The script was modified to calculate the curvature in larger objects, such as the uterus (Matlab Script for Surface Curvature). The analysis was carried out using the vertices generated in the Surface mode in Imaris. Surface complexity is reduced by evenly reducing the number of vertices to one-tenth of the original number. Using the normals in each axis at each vertex, the radius of the curve formed by connecting neighbor vertices was calculated. Radii calculated with six neighbor vertices are averaged to obtain curvature mean and multiplied by 10 to calculate Cmean. Cmean is displayed as a heat map where the value of 0-0.05 is blue/purple and curvature values above 0.200 are displayed in red. Morphology of uterine luminal segments is expressed as the folding factor (f), which is the ratio of the Cmean for the highly curved luminal epithelium (0.125<Cmean>0.3), to the least curved luminal epithelium regions (0<Cmean>0.05) (Fig. S1A-C):
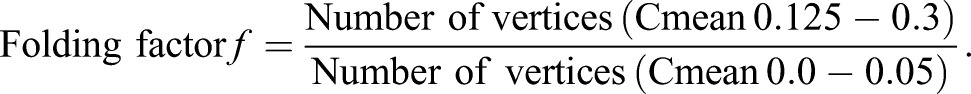 |
Cmean values above 0.3 were attributed to remnants of glandular ducts after subtraction and accordingly excluded from lumen curvature analysis.
Determination of bending angle for glandular ducts
The angle of glandular duct bending towards the implantation site was measured using the Imaris function measurement points. Three planes along the xy/yz and zx axes can be displayed using the orthogonal slicer. The three planes separate along the dorsal/ventral, anterior/posterior and mesometrial/anti-mesometrial axis, respectively. Using both the confocal z slices of the uterus and glandular surfaces, the three planes can be positioned to define four standardized measurement points. The first point is placed at the duct of the gland (where it branches from the luminal epithelium) and the intersection of the anterior/posterior axis and the dorsal/ventral axis. The second measurement point is placed at the farthest branched tip of the gland along the intersection of the dorsal/ventral axis and the mesometrial/anti-mesometrial axis. The third measurement point is placed at the intersection of the mesometrial/anti-mesometrial plane, the anterior/posterior (luminal) plane and the dorsal/ventral axis. The fourth measurement point is placed at the starting position that creates a right-angled triangle that dictates the degree to which the glandular duct is bending, with the luminal epithelium always being the standard comparison point.
Acknowledgements
We thank S. K. Dey and Jeeyeon Cha for the Wnt5acKO uteri, David Rowitch of the UCSF IVF core, Matthew Krummel of the Biological Imaging Development Center, and Zev Gartner for feedback on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
R.A. and D.J.L. conceived the project; R.A., K.S. and K.O. performed the experiments; R.A., A.F. and K.M. performed the image analysis; R.A., L.C.G. and D.J.L. analyzed the data and wrote the manuscript; K.S., A.F. and K.M. provided manuscript feedback.
Funding
We acknowledge support from the California Institute for Regenerative Medicine (TG2-01153), the UCSF Program for Breakthrough Biomedical Research and the National Institutes of Health (5T32HD007263-32 to R.A., P50HD055764 to L.C.G., and DP2OD007420 and 1R21ES023297 to D.J.L.). Deposited in PMC for release after 12 months.
Data availability
The Matlab script used to determine surface curvature, which was was modified from a script made available by Imaris Open (Bitplane), is provided in the supplementary data.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.144386.supplemental
References
- Besnard V., Wert S. E., Hull W. M. and Whitsett J. A. (2004). Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr. Patterns 5, 193-208. 10.1016/j.modgep.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Cha J., Sun X. and Dey S. K. (2012). Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 18, 1754-1767. 10.1038/nm.3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J., Bartos A., Park C., Sun X., Li Y., Cha S.-W., Ajima R., Ho H.-Y. H., Yamaguchi T. P. and Dey S. K. (2014). Appropriate crypt formation in the uterus for embryo homing and implantation requires Wnt5a-ROR signaling. Cell Rep. 8, 382-392. 10.1016/j.celrep.2014.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang Y., Elad D., Jaffa A. J., Cao Y., Ye X. and Duan E. (2013). Navigating the site for embryo implantation: biomechanical and molecular regulation of intrauterine embryo distribution. Mol. Aspects Med. 34, 1024-1042. 10.1016/j.mam.2012.07.017 [DOI] [PubMed] [Google Scholar]
- Daikoku T., Song H., Guo Y., Riesewijk A., Mosselman S., Das S. K. and Dey S. K. (2004). Uterine Msx-1 and Wnt4 signaling becomes aberrant in mice with the loss of leukemia inhibitory factor or Hoxa-10: evidence for a novel cytokine-homeobox-Wnt signaling in implantation. Mol. Endocrinol. 18, 1238-1250. 10.1210/me.2003-0403 [DOI] [PubMed] [Google Scholar]
- Daikoku T., Cha J., Sun X., Tranguch S., Xie H., Fujita T., Hirota Y., Lydon J., DeMayo F., Maxson R. et al. (2011). Conditional deletion of Msx homeobox genes in the uterus inhibits blastocyst implantation by altering uterine receptivity. Dev. Cell 21, 1014-1025. 10.1016/j.devcel.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faire M., Skillern A., Arora R., Nguyen D. H., Wang J., Chamberlain C., German M. S., Fung J. C. and Laird D. J. (2015). Follicle dynamics and global organization in the intact mouse ovary. Dev. Biol. 403, 69-79. 10.1016/j.ydbio.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filant J. and Spencer T. E. (2014). Uterine glands: biological roles in conceptus implantation, uterine receptivity and decidualization. Int. J. Dev. Biol. 58, 107-116. 10.1387/ijdb.130344ts [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filant J., Zhou H. and Spencer T. E. (2012). Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol. Reprod. 86, 141-149. 10.1095/biolreprod.111.097089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosie M. J., Adams S. M., Thompson M. B. and Murphy C. R. (2003). Viviparous lizard, Eulamprus tympanum, shows changes in the uterine surface epithelium during early pregnancy that are similar to the plasma membrane transformation of mammals. J. Morphol. 258, 346-357. 10.1002/jmor.10163 [DOI] [PubMed] [Google Scholar]
- Laird M. K., Thompson M. B., Murphy C. R. and McAllan B. M. (2014). Uterine epithelial cell changes during pregnancy in a marsupial (Sminthopsis crassicaudata; Dasyuridae). J. Morphol. 275, 1081-1092. 10.1002/jmor.20282 [DOI] [PubMed] [Google Scholar]
- Ma W.-G., Song H., Das S. K., Paria B. C. and Dey S. K. (2003). Estrogen is a critical determinant that specifies the duration of the window of uterine receptivity for implantation. Proc. Natl. Acad. Sci. USA 100, 2963-2968. 10.1073/pnas.0530162100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. R. (2004). Uterine receptivity and the plasma membrane transformation. Cell Res. 14, 259-267. 10.1038/sj.cr.7290227 [DOI] [PubMed] [Google Scholar]
- Qian D., Jones C., Rzadzinska A., Mark S., Zhang X., Steel K. P., Dai X. and Chen P. (2007). Wnt5a functions in planar cell polarity regulation in mice. Dev. Biol. 306, 121-133. 10.1016/j.ydbio.2007.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon S. N., King M. L., MacLean J. A. II, Mann J. L., DeMayo F. J., Lydon J. P. and Hayashi K. (2012). Cdh1 Is essential for endometrial differentiation, gland development, and adult function in the mouse uterus. Biol. Reprod. 86, 1-10. 10.1095/biolreprod.112.098871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. L., Kaspar P., Brunet L. J., Bhatt H., Gadi I., Köntgen F. and Abbondanzo S. J. (1992). Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359, 76-79. 10.1038/359076a0 [DOI] [PubMed] [Google Scholar]
- Wang H. and Dey S. K. (2006). Roadmap to embryo implantation: clues from mouse models. Nat. Rev. Genet. 7, 185-199. 10.1038/nrg1808 [DOI] [PubMed] [Google Scholar]
- Winkelmann A. and Spornitz U. M. (1997). Alkaline phosphatase distribution in rat endometrial epithelium during early pregnancy: a scanning electron-microscopic study. Cells Tissues Organs 158, 237-246. 10.1159/000147936 [DOI] [PubMed] [Google Scholar]
- Yoshinaga K. (2013). A sequence of events in the uterus prior to implantation in the mouse. J. Assist. Reprod. Genet. 30, 1017-1022. 10.1007/s10815-013-0093-z [DOI] [PMC free article] [PubMed] [Google Scholar]