Abstract
The derivation and maintenance of human pluripotent stem cells (hPSCs) in stable naïve pluripotent states has a wide impact in human developmental biology. However, hPSCs are unstable in classical naïve mouse embryonic stem cell (ESC) WNT and MEK/ERK signal inhibition (2i) culture. We show that a broad repertoire of conventional hESC and transgene-independent human induced pluripotent stem cell (hiPSC) lines could be reverted to stable human preimplantation inner cell mass (ICM)-like naïve states with only WNT, MEK/ERK, and tankyrase inhibition (LIF-3i). LIF-3i-reverted hPSCs retained normal karyotypes and genomic imprints, and attained defining mouse ESC-like functional features, including high clonal self-renewal, independence from MEK/ERK signaling, dependence on JAK/STAT3 and BMP4 signaling, and naïve-specific transcriptional and epigenetic configurations. Tankyrase inhibition promoted a stable acquisition of a human preimplantation ICM-like ground state via modulation of WNT signaling, and was most efficacious in efficiently reprogrammed conventional hiPSCs. Importantly, naïve reversion of a broad repertoire of conventional hiPSCs reduced lineage-primed gene expression and significantly improved their multilineage differentiation capacities. Stable naïve hPSCs with reduced genetic variability and improved functional pluripotency will have great utility in regenerative medicine and human disease modeling.
KEY WORDS: Differentiation, Ground state, Human embryonic stem cell, Induced pluripotent stem cell, Naïve pluripotency
Summary: A broad repertoire of conventional human ESCs and transgene-independent iPSC lines can be reverted to stable naive states using WNT, MEK/ERK and tankyrase inhibition.
INTRODUCTION
Although human induced pluripotent stem cells (hiPSCs) share highly similar transcriptional and epigenomic signatures with human embryonic stem cells (hESCs) (Chin et al., 2009; Bock et al., 2011), they demonstrate greater interline multilineage differentiation variability than hESCs (Osafune et al., 2008; Choi et al., 2009; Feng et al., 2009; Hu et al., 2010; Boulting et al., 2011). The discrepancy between highly variable ‘functional pluripotency’ among hiPSC lines, despite similar molecular and phenotypic pluripotency to hESCs, might be partially due to imperfect induction of the somatic donor cell epigenome to a bona fide ESC-like state (Lister et al., 2011; Nishino et al., 2011; Ruiz et al., 2012). Previous studies suggested that reprogramming-associated errors of retention of donor cell-specific epigenetic memory bias the differentiation potency of hiPSCs toward some lineages (Kim et al., 2010b, 2011; Polo et al., 2010). However, other studies did not confirm such correlations, or alternatively suggested that donor-specific genetic variability affecting lineage-primed gene expression might play a more dominant role (Ohi et al., 2011; Hu et al., 2011; Kajiwara et al., 2012; Kyttälä et al., 2016).
Complex determinants may collectively influence the functional pluripotency of both hiPSCs and hESCs. For example, one critical variable impacting the functional pluripotency of conventional hPSCs is their developmental, molecular and epigenetic commonality with ʻprimed' mouse post-implantation epiblast stem cells (mEpiSCs) (Tesar et al., 2007; Brons et al., 2007; Chou et al., 2008; Kojima et al., 2014; Weinberger et al., 2016), which possess a less primitive pluripotency than inner cell mass (ICM)-derived mouse ESC (mESCs). For example, mEpiSCs cannot fully contribute to a blastocyst chimera and are resistant to chemical reversion to ICM-like naïve ʻground state' pluripotency with ‘LIF-2i’ (MEK/ERK and GSK3β signal inhibition) (Bernemann et al., 2011; Ying et al., 2008; Marks et al., 2012). Conventional human pluripotent stem cells (hPSCs) rely on self-renewal signaling pathways more similar to those of mEpiSCs than ESCs, and these hPSCs might exist in developmentally primed states that display mEpiSC-like lineage skewing following directed differentiation. Although several hPSC naïve reversion approaches were recently described, none was maintained with classical MEK/ERK/WNT 2i signaling inhibition alone (Hanna et al., 2010; Chan et al., 2013; Gafni et al., 2013; Takashima et al., 2014; Theunissen et al., 2014; Ware et al., 2014). Thus, although various methods may achieve pluripotent states reminiscent of the human ICM, the determinants required for stable human rewiring to an mESC-like ground state remain undefined and might represent unknown species-specific differences.
The roles of the derivation method and of lineage priming of conventional hiPSCs in the amenability to naïve reversion have not been fully evaluated. For example, although human hematopoietic progenitors are more efficiently reprogrammed than fibroblast donors via standard methods (Eminli et al., 2009; Park et al., 2012; Guo et al., 2014), both donor types generated hiPSCs with diminished and lineage-skewed differentiation potencies that were attributed to the retention of donor epigenetic memory (Kim et al., 2011; Hu et al., 2011). By contrast, hiPSCs reprogrammed efficiently from cord blood-derived CD33+ CD45 (PTPRC)+ myeloid progenitors (MPs) (Park et al., 2012) displayed reduced interline variability or differentiation bias (Burridge et al., 2011; Park et al., 2014). These MP-iPSCs generated vascular progenitors (VPs) with less culture senescence, decreased sensitivity to DNA damage, and greater in vivo engraftment potential than VPs generated from standard fibroblast-derived hiPSCs (Park et al., 2014). MP-iPSCs also generated physiologically functional photoreceptors that elicited action potentials in a three-dimensional retinal differentiation system (Zhong et al., 2014).
Since murine and human MPs may represent a ‘privileged’ somatic donor type (Park et al., 2012; Guo et al., 2014), we tested the hypothesis that efficient myeloid reprogramming generates an improved primed functional pluripotency with reduced lineage priming and increased amenability to naïve ground state reversion. Here, we demonstrate that effective reprogramming of human CD33+ CD45+ MP donors generates hiPSCs with an improved multilineage differentiation potency that lacks the lineage-priming differentiation bias characteristic of hiPSCs derived via standard reprogramming methods. Moreover, supplementation of classical LIF-2i with only the tankyrase inhibitor XAV939 (LIF-3i) permitted a large repertoire of hiPSCs to efficiently revert to a stable mESC-like naïve state that possessed further improved multilineage functional pluripotency. Interestingly, MP-iPSCs reverted to this stable naïve state more efficiently than hiPSCs derived via less efficient methods.
RESULTS
STAT3-activated MP donors generate hiPSCs with decreased reprogramming-associated genetic variability and high functional pluripotency
Previous studies demonstrated that stromal-activated (sa) human MPs can be reprogrammed with four (4F-E) or seven (7F-E) episomal factors with extremely high efficiencies (Fig. S1A-C) (Park et al., 2012). Sa-MP-iPSCs arose directly from CD33+ CD34− CD14+ MP donor cells differentiated from CD34+ cord blood (CB), bone marrow (BM), fetal liver (FL) and GCSF (CSF3)-mobilized peripheral blood (PB) in these reprogramming systems. 4F-E-nucleofected CD33+ sa-MPs sustained high endogenous levels of phosphorylated STAT3 (P-STAT3) throughout critical phases of myeloid culture compared with fibroblasts or non-activated MPs (Fig. S1D,E), and upregulated their expression of targets as well as core pluripotency circuits known to potentiate both somatic cell reprogramming and naïve pluripotency reversion in mEpiSCs (Fig. S1F, Table S1) (Yang et al., 2010; van Oosten et al., 2012; Boyer et al., 2005).
To evaluate the quality of sa-MP reprogramming, we generated a library of over 40 unique MP-iPSC lines derived with and without sa from PB-, CB- and FL-derived CD33+ MPs (Table S2, supplementary Materials and Methods). To delineate the effects of reprogramming-associated donor-specific genetic variability (Kyttälä et al., 2016), independent MP-iPSC lines from unique as well as identical MP donors were generated. This repertoire of MP-iPSCs was complemented with hiPSCs generated via standard methods: 7F-E mononuclear CB cell-derived hiPSCs (Hu et al., 2011), 7F-E and 4F viral (4F-V) fetal (f)/adult (Ad) fibroblast-derived iPSCs (fibro-iPSCs: fF-iPSCs, AdF-iPSCs) and 7F-E adult skin keratinocyte-derived iPSCs (Ker-iPSCs) (Park et al., 2012; Byrne et al., 2009). We compared whole-genome transcriptomes of this MP-iPSC repertoire with comparable passage standard hiPSC and hESC lines (Fig. S2A). In contrast to standard fibro-iPSCs, which incompletely resemble hESCs in their gene signatures (Chin et al., 2009), CB-derived sa-MP-iPSCs attained global expression profiles that were indistinguishable [Pearson coefficient (R2)=0.99] from standard hESCs, and in a manner that was irrespective of donor genome origin (Fig. S2A). Whole-genome CpG DNA methylation analysis further revealed that sa-MP-iPSCs (from both unique and the same donors) clustered as a function of sa-MP reprogramming into an epigenetically distinct group relative to hESCs and standard fibro-iPSCs (Fig. S2B).
To evaluate the functional pluripotency of conventional (primed) hPSCs, we differentiated a repertoire of hiPSCs to mesodermal, endodermal and neural ectodermal lineages (Fig. 1, Figs S3 and S4). In contrast to previously reported lineage skewing preferences and diminished potencies of standard CB-iPSCs and fibro-iPSCs for osteogenic, neural and endothelial differentiation (Osafune et al., 2008; Choi et al., 2009; Feng et al., 2009; Hu et al., 2010), and regardless of whether they were derived from unique or identical MP donors, we found no evidence for lineage preference or interline donor-dependent differentiation bias of sa-MP-iPSC lines. For example, all sa-MP-iPSC lines tested generated comparable or greater numbers of hematopoietic progenitors (i.e. CD34+ CD45+), erythro-myeloid colony-forming unit (CFU) progenitor frequencies, and percentages and absolute numbers of total CD34+ and CD45+ cells relative to hESCs (Fig. 1A, Fig. S3A-C). Sa-MP-iPSCs differentiated just as robustly to CD31 (PECAM1)+ vascular cells (Fig. S3D), CXCR4+ SOX17+ FOXA2+ endodermal progenitors (Fig. 1C, Fig. S4H), nestin+ PAX6+ NCAM1+ neural progenitors and rhodopsin+ retinal cells (Fig. S3F-H), and Alizarin Red+ COL1A1+ osteopontin (SPP1)+ bone lineage cells (Fig. 1B,C, Fig. S4A-G). Overall, in all assays tested, sa-MP-iPSCs differentiated with reduced interline variance to all three germ layers, and regardless of individual donor origin.
Fig. 1.
Multilineage differentiation and genetic variability of conventional hiPSCs derived from unique donors via different reprogramming methods. (A) Hematopoietic differentiation [% day 14 human embryoid body (hEB) cells] of independent donor-derived hPSCs (see Table S2). hESC lines: H9, H7, H1, ES03 (gold). Fibro-iPSCs: IMR4, HUF3, HUF5, 7ta, WT2 (green). 7F-E and 4F-E hematopoietic-iPSCs: lines generated without stromal activation (sa) were mononuclear CB-7F-E (iCB 2.5, iCB8, iCB9; purple) and MP-CB-4F-E (E17C1, E20C2, E32C7; purple) and lines generated with sa were saMPCB-4FE (E5C3, E12C5, E17C6; red). Included are MP-PB-4FE lines from the same donor (ZPB) that were derived with sa (E29C1, E29C4, E29C6; red) and without sa (E29C10, E29C11, E29C12; purple). Four 7F-E CB-derived sa-MP-iPSC lines were all from the same donor (D003) generated with sa (6.2, 6.13, 19.11) (Fig. S2). *P<0.05, **P<0.005 (one-way ANOVA) for averaged groups relative to the hESC group. (B) Osteogenic differentiation (quantitative Alizarin Red dye), and (C) endodermal differentiations of individual lines in each hPSC class from the same unique donors as above. Error bars indicate s.e.m. (D) Number of genes differentially expressed between hiPSCs and hESCs (# hiPSC ≠ hESC) (ANOVA, P<0.05; fold change >1.5×). (E) Dendrogram of hPSCs from above clustered on lineage-primed gene targets of the PRC2 complex (Table S1). (F) PCA of ESC and core module genes (Table S1) showing variance in gene expression among donor cells and the hPSC lines shown in Fig. S2A. Red spheres indicate early passage sa-CB-derived MP-iPSCs (n=12, average passage=11.4); gold circles indicate hESCs (n=5, average passage=58.5); green circles indicate AdF-iPSCs (n=9, average passage=20.4); gray diamonds indicate donor fibroblasts; gray triangles indicate donor CB cells.
To determine the effects of sa-MP reprogramming on genetic variability, we evaluated MP-iPSCs from independent, unique MP donors generated with and without stromal STAT3 activation, and compared their whole-genome transcriptomes with standard hiPSCs and hESCs. These bioinformatics analyses revealed that sa-MP reprogramming significantly reduced hiPSC gene variability relative to hESC controls, and in a manner that was independent of individual donor source (Fig. 1D). Gene ontology (GO) and gene set enrichment analysis (GSEA) revealed that, relative to standard reprogrammed hiPSCs, sa-MP reprogramming generated hiPSCs with significantly reduced expression of genes associated with lineage priming (e.g. Polycomb complex targets), and distinct changes in the expression of genes involved in cell cycle regulation and metabolism (Fig. 1E, Fig. S5).
We next assessed the quality of sa-MP-iPSC pluripotency circuits by performing principal component analysis (PCA) of microarray data for the expression of pluripotency-associated ESC and core module circuits (Table S1) (Kim et al., 2010a; Boyer et al., 2005) for each hiPSC class. These analyses revealed that, in comparison to standard fibro-iPSC lines, low-passage sa-MP-iPSCs had already attained high-fidelity transcription of these pluripotency circuits that was indistinguishable from hESCs (hESC to CB-derived sa-MP-iPSC ESC module, R2=0.99; core module, R2=0.98; Fig. 1F). Collectively, these multilineage differentiation and bioinformatics studies revealed that sa-MP-iPSCs, as a class, possessed high molecular and functional pluripotency and lacked the lineage-specific differentiation skewing and increased lineage-primed gene expression variability typically observed in hiPSCs derived via standard methods.
Dual GSK3β and tankyrase inhibitions synergize with MEK/ERK blockade to rapidly revert conventional primed hPSCs to a stable mESC-like phenotype
mEpiSCs possess hierarchies of primed pluripotency with variable propensities for reversion to a naïve ground state, and reversion of EpiSCs via LIF-2i may be limited by residual lineage-primed gene expression (Bao et al., 2009; Bernemann et al., 2011). We tested the capacity of our broad repertoire (Table S2, Fig. 1) of conventional, bFGF (FGF2)-dependent, non-integrated hPSCs to revert to a naive ICM-like state following chemical 2i WNT-MEK/ERK modulation. Supplementation of hPSC cultures with the classical two small-molecule MEK/ERK (PD0325901) and GSK3β (CHIR99021) inhibitors (Ying et al., 2008) was insufficient to stably revert any hPSCs into clonogenic mESC-like lines. To identify novel conditions that stabilize human naïve pluripotency, we screened over 130 LIF-supplemented hPSC culture conditions comprising combinations of more than 15 small molecules known to modulate ESC self-renewal (see strategy for small molecule screening in the supplementary Materials and Methods; Table S3). We initially assayed for conditions that supported stable reversion (>5-10 passages) of conventional hPSC lines to mESC-like dome-shaped morphologies in WNT-MEK/ERK blockade conditions. This screen revealed that supplementing classical LIF-2i with the WNT pathway modulator XAV939 – a tankyrase inhibitor that potentiates axin-mediated cytoplasmic stabilization of activated β-catenin in primed PSCs (Huang et al., 2009; Kim et al., 2013) – uniquely permitted rapid reversion of conventional H9 hESC and multiple transgene-free 4F-E sa-MP-iPSC lines into uniform, compact, dome-shaped, clonogenic colonies with normal karyotypes (Fig. 2A, Table S3). qRT-PCR, FACS and immunofluorescence studies of LIF-3i-reverted sa-MP-iPSCs and hESCs revealed high TRA-1-81+ SSEA4+ (>95%) surface expression, and increased transcript and protein expression of naïve-specific epiblast factors [e.g. NANOG, E-cadherin (CDH1), NR5A2, STELLA (DPPA3), KLF2, KLF4, KLF5, KLF17, HERV-H and TFCP2L1] (Fig. 2B-D, Fig. S6A-D, Fig. S8C,D). The pluripotencies of multiple LIF-3i-reverted hPSCs were validated by robust tri-lineage teratoma formation in NOG-SCID mice (Fig. 2E, Fig. S6E, Table S3B). Furthermore, conventional female sa-MP-iPSCs with detectable XIST expression and an X-inactivated phenotype expressed significantly lower levels of XIST transcripts following LIF-3i reversion, which is consistent with the bi-allelic X-activation status observed in naïve mESCs (Ying et al., 2008) (Fig. 2B,F). LIF-3i-reverted hPSCs also exhibited decreased levels of HLA-A and HLA-B (Fig. 5E, Fig. S8D). LIF-3i-reverted H9 hESCs and 4F-E sa-MP-iPSCs maintained stable, robust clonal growth proliferation kinetics for at least 30 passages with normal karyotypes in standard mouse embryonic fibroblast (MEF)/hESC conditions (Fig. S7 and Fig. S8B). Phenotypically naive hPSCs reverted to flattened mEpiSC-like morphologies when transferred back to conventional bFGF hPSC culture.
Fig. 2.
Tankyrase inhibition promotes stable reversion of hPSCs to an mESC-like pluripotency in classical WNT-MEK/ERK 2i conditions. (A) Clonal passaging of conventional hPSCs in LIF-3i permitted stable reversion to dome-shaped colony morphology, with bi-directional reversion of phenotype when re-cultured in bFGF. (B) qRT-PCR analysis of pluripotency-associated transcripts of (female) sa-MP-iPSC line 6.2 cultured before (primed) and after four single-cell passages in LIF-3i. Transcript levels of NANOG, STELLA, NR5A2 and XIST are shown. **P<0.001 (paired t-test); NS, not significant. (C) Representative SSEA4+ TRA-1-81+ expression of stable LIF-3i-reverted hPSCs. (D) Confocal immunofluorescence microscopy of LIF-3i-cultured hPSCs. Shown are representative nuclear and cytoplasmic protein co-expressions of NANOG, OCT4, TRA-1-81 and SSEA4 with naïve-specific factors (E-cadherin, NR5A2 and STELLA). (E) Representative teratoma of LIF-3i-reverted E5C3 (Table S3B). Mesoderm (Meso; e.g. cartilage), ectoderm (Ecto; e.g. pigmented retinal epithelium) and endoderm (Endo; e.g. glandular epithelium) are indicated. (F) Immunofluorescence (right) and quantitation per field (left) of X-chromosome activation status of primed versus LIF-3i-reverted line 6.2 with XIST RNA-FISH probe; normal human dermal fibroblasts (NHDF) provided controls. Error bars indicate s.e.m. Scale bars: 500 µm in A; 50 µm in D; 100 µm in E; 20 µm in F.
Fig. 5.
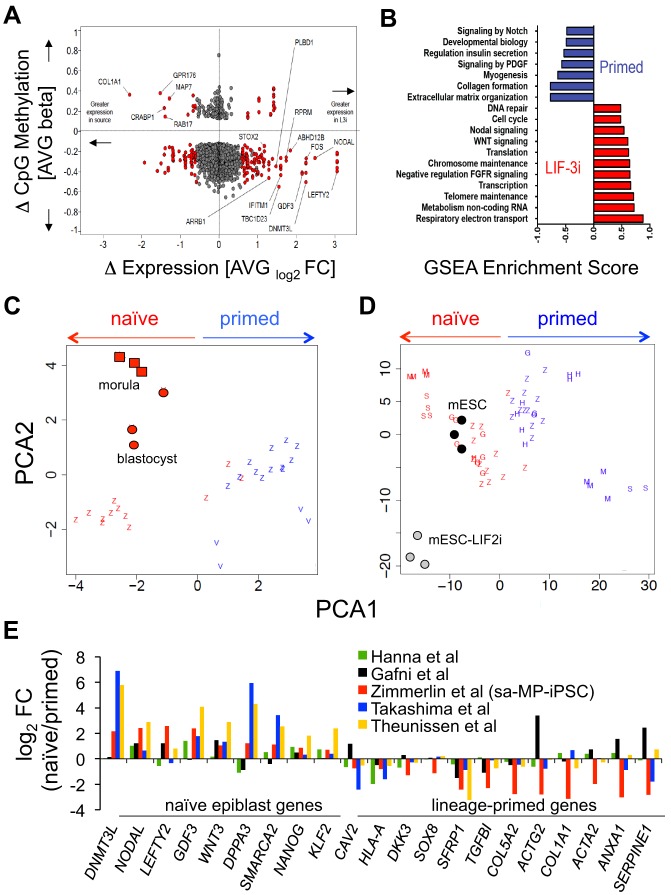
Comparative transcriptional/epigenetic meta-analyses of LIF-3i-reverted hPSCs. (A) Mean genome-wide gene expression versus CpG methylation DMRs crossplot of LIF-3i-reverted hPSCs versus their isogenic primed hPSC counterparts [same 12 samples as in Fig. 4, i.e. hESCs (n=3), MP-iPSCs (n=6), fibro-iPSCs (n=3)]. Shown are DMRs (CpG beta values) of LIF-3-reverted versus isogenic primed hPSC counterparts (y-axis, P≤0.05) versus their corresponding differential gene expressions [red, x-axis, log2 fold changes (FC); P≤0.05, FC ±≥1.5]. (B) Curated GSEA reactome pathways over-represented (FDR<0.01; P<0.01) in LIF-3i-reverted sa-MP-iPSCs versus their isogenic primed sa-MP-iPSC counterparts (n=6; same samples as Fig. 4A). (C,D) Comparative platform-normalized PCA of whole-genome expression of primed (blue) or naïve (red) PSC lines from different laboratories. (C) Human morula/blastocyst (red circles) or (D) mESC-serum/mESC-LIF-2i (black/gray circles) samples were used as benchmarks. Human morula/blastocyst PCA in C is clustered on a module of the most differentially expressed genes in E4-E5 human pluripotent epiblast (Petropoulos et al., 2016) (see Table S1). Z, this study and includes the n=12 independent hPSC lines in Fig. 4A; H, Hanna et al., 2010; G, Gafni et al., 2013; M, Takashima et al., 2014; S, Theunissen et al., 2014; V, Vassena et al., 2011. (E) Comparison of differentially expressed (P≤0.05, FC±≥1.5) naïve-specific and lineage-primed transcripts in naïve hPSCs derived in this work or other labs. FC: normalized ratios of naïve/primed expression microarray signal intensities. Ratios are of LIF-3i-reverted versus primed hPSC samples (n=12 hPSCs, as above) versus samples of those published as indicated.
LIF-3i reversion induces LIF-JAK/STAT3 signaling, BMP4 responsiveness and augmentation of activated β-catenin in both nuclear and cytoplasmic compartments
LIF-3i-reverted hPSCs did not require supplementation with primed PSC growth factors (e.g. bFGF, activin A, TGFβ) or apoptosis inhibitor cocktails to maintain long-term viability and robust proliferation. Furthermore, western blotting and chemical inhibition assays demonstrated that naïve-reverted hPSCs adopted authentic mESC-like signaling pathways that included increased active nuclear STAT3 phosphorylation, dependence on JAK/STAT3, LIF/gp130, CREB and PI3K signaling, and independence from FGF and MEK/ERK signaling (Fig. 3A,B). Notably, although LIF withdrawal reduced proliferation of naïve hiPSCs after three passages (12 days) by ∼50%, supplementation with bFGF (or TGFβ) did not exert further proliferative effects on viability of SSEA4+ TRA-1-81+ cells. Supplementation of LIF-2i with XAV939 resulted in elevated axin levels with an apparent stabilization and augmented expression of the activated isoform of non-phosphorylated β-catenin in both cytoplasmic and nuclear compartments of hPSCs (Fig. 3A,C-E). Interestingly, conventional sa-MP-iPSCs already possessed higher basal nuclear and cytoplasmic β-catenin activities than other hPSCs. One distinctive effect of LIF-3i reversion of sa-MP-iPSCs was a potent mESC-like BMP4 proliferative responsiveness (∼5-fold) of naïve SSEA4+ TRA-1-81+ cells, with concordant susceptibility to BMP4 inhibition (dorsomorphin) (Fig. 3B).
Fig. 3.
Acquisition of mESC-like signaling pathways in LIF-3i-reverted hPSCs. (A) Stabilization of nuclear P-STAT3 and cytoplasmic activated (activ) β-catenin in LIF-3-reverted hPSCs. Nuclear (Nu), cytoplasmic (Cy) and total (Tot) fractions are shown for H9 hESC, E5C3 sa-MP-iPSC and C1.2 fibro-iPSC lines. TBP and actin are protein loading controls. +, LIF-3i; −, primed culture. (B) Inhibition/proliferation assays of sa-MP-iPSC line E5C3. Shown are mESC signaling pathways [e.g. LIF/JAK/STAT, LIF-receptor/gp130, CREB, BMP4 (dorsomorphin) and PI3K]. Cumulative proliferations of SSEA4+ TRA-1-81+ E5C3 cells were measured after 12 days of culture in the presence of indicated inhibitors, normalized to LIF-3i-alone conditions (at 100%). (C) Western blots of key WNT components AXIN1 and activated β-catenin in both nuclear and cytoplasmic fractions in primed (−) versus LIF-3i reverted (+) hPSC cultures. (D,E) Confocal microscopy of WNT proteins in indicated primed (bFGF) versus LIF-3i-reverted hPSC lines. Activated β-catenin is shown to be distinctly sequestered outside of the nuclear compartment, whereas uniform expression of OCT4 was localized strictly within nuclei. Scale bars: 20 µm.
A broad repertoire of conventional hPSC lines stably reverts to naïve morphologies in LIF-3i
We evaluated 23 independent non-integrated, conventional, primed hPSC lines for their capacity to tolerate stable, clonogenic self-renewal of SSEA4+ TRA-1-81+ cells for at least ten passages in LIF-3i. (Fig. S8A,B, Table S3B). Long-term stability of colonies with undifferentiated dome-shaped morphologies for >10-20 passages via direct LIF-3i reversion alone was most reproducible for sa-MP-iPSCs and select hESC lines (e.g. H9). However, brief adaptation (one passage) in LIF-3i plus two additional molecules, namely forskolin and purmorphamine (LIF-5i), increased the initial survival of enzyme-digested hPSC single cells, and facilitated a broader repertoire of hPSCs to tolerate subsequent stable clonal self-renewal in LIF-3i alone (Fig. S7A-C and Fig. S8A,B, Table S3B). This initial LIF-5i modification permitted a wide repertoire of ∼16 conventional hPSC lines to revert with long-term stability in LIF-3i alone.
LIF-3i-reverted hPSCs increase expression of core pluripotency circuits and acquire mESC-like transcriptional and epigenetic features
mESCs possess molecular signatures, clearly distinct from those of EpiSCs and hPSCs, that are characterized by more open, derepressed chromatin configurations, decreased global CpG DNA methylation, and a transcriptome reflective of the naïve preimplantation epiblast (Marks et al., 2012; Leitch et al., 2013). To ascertain if preimplantation epiblast-like states were achieved in LIF-3i-reverted hPSCs, we evaluated whole-genome transcriptional and epigenomic signatures of 12 independent LIF-3i-reverted lines and their isogenic conventional counterparts [i.e. six sa-MP-iPSCs, three hESCs and three fibro-iPSCs before and after LIF-3i reversion at early post-reversion passages (p5)] (Figs 4 and 5, Fig. S8). These studies established that LIF3i-reverted hPSCs acquired signatures distinct from conventional primed states, with robust upregulation of genes associated with both the preimplantation human epiblast and mESC ground state. Cross-species whole-genome hierarchical clustering using mEpiSCs, mESCs and LIF-2i-reverted mESCs as benchmarks for measuring primed and naïve pluripotent states (Fig. 4A) revealed that conventional hPSCs clustered with mEpiSCs, whereas the same isogenic LIF3i-reverted hPSC lines alternatively clustered distinctly alongside serum-grown mESCs and LIF-2i-reverted naïve mESCs.
Fig. 4.
Transcriptional and epigenetic profiling of LIF-3i-reverted hPSCs. (A) Genome-wide cross-species hierarchical clustering. Shown is a dendrogram of expression microarrays of mESC [serum/LIF; naïve (LIF-2i)], primed mEpiSC, and isogenic hPSC samples from this study before (hPSC primed) and after 5 passages in LIF-3i (hPSC naïve). Human PSC lines (n=12) included: three hESC lines H9, H7 and ES03 (gold); six sa-MP-iPSC lines E5C3, E5C1, E17C6, LZ6+2, LZ6+10 and 6.2 (red); and three fibro-iPSC lines 7ta, C1.2 and C2 (green). (B) (Top) CpG methylation. Box plot shows beta values of genome-wide autosomal differentially methylated region (DMR) CpG probes from Infinium methylation arrays [16,282 of 473,864 autosomal probes significantly (P<0.05) differentially methylated (SD>0.15); see supplementary Materials and Methods for further details] in the same isogenic primed (−) versus LIF-3i-reverted (+) hPSC samples used for the microarrays above. Gold, hESCs (n=3); red, sa-MP-iPSCs (n=6); green, fibro-iPSCs (n=3). ***P<0.001 (paired two-way t-test). (Bottom) Global 5MC and 5hMC levels from dot blot immunoassays (relative to primed) for representative LIF-3i-reverted hPSCs. Genomic DNA samples were collected before (−) and after (+) LIF-3i reversion from H9 (gold), E5C3 (red) and C1.2 (green). (C) Activities of proximal enhancer (PE) and distal enhancer (DE) elements of the human OCT4 promoter in primed (bFGF) versus LIF-3i-reverted E5C3. Shown are relative firefly luciferase activities following normalization with Renilla luciferase and negative control basal activities ±s.d. (n=3). *P<0.05 (paired t-test). (D) Stable BAC reporter transgenic OCT4 PE/DE mutant lines. (Top) Cytometry plots of representative LIF-3i-reverted C2 hiPSC subclones (n=3) stably transfected with full-length OCT4-GFP-2A-PURO PE/DE sequences (control), mutant ΔPE-OCT4-GFP-PURO constructs, or non-transfected (no construct) controls. (Bottom) Percentage GFP+ cells among naïve cultures of individual hiPSC subclones (n=3) expressing control or mutant ΔPE sequences. (E) Pluripotency circuits in LIF-3i-reverted hPSCs. (Top) Mean beta values of core module-specific CpG DMRs in primed (−) versus LIF-3i-reverted (+) hPSC; (bottom) corresponding log2 mean subtracted normalized expression of core module genes (Table S1) of the same independent hPSC samples (identical to those used above for expression microarrays). Gold, hESCs (n=3); red, sa-MP-iPSCs (n=6); green, fibro-iPSCs (n=3). (F) Pluripotency gene-specific promoter CpG methylation. Heatmap-dendrogram clustering and box plots of mean beta values of ESC module gene-specific CpG DMRs [P<0.001 (paired two-way t-test)] of LIF-3i (+) versus primed (−) hPSCs. Samples are the same 12 hPSC lines in each category, as described above. Percentages represent reduction of median beta value following LIF-3i reversions.
To characterize the epigenetic status of naïve-reverted hPSCs, we performed Infinium CpG DNA 450K methylation array analysis of these 12 LIF-3i-reverted lines and their isogenic conventional counterparts. This methodology interrogates more than 485,000 methylation sites at single nucleotide resolution, and covers 96% of CpG islands of 99% of RefSeq genes, with an average of 17 CpG sites per gene region distributed across the promoter, 5′UTR, first exon, gene body, and 3′UTR. These studies revealed that all LIF-3i-reverted hPSCs possess similar and significantly decreased (P<0.001) epigenome-wide CpG DNA methylation at differentially methylated regions (DMRs) compared with their primed hPSC sources (Fig. 4B, top panel). Additionally, quantification of dot blot immunoassays of global CpG 5-methylcytosine isoforms (5hMC versus 5MC) revealed reduced global 5MC activities and increased ratios of 5hMC/5MC CpG DNA methylation, which was most evident for sa-MP-iPSC E5C3 (Fig. 4B, bottom panel). Taken together, these data were consistent with a more epigenetically open configuration, and a potential role for TET-mediated CpG DNA demethylation activities in sustaining naïve pluripotency in LIF-3i-reverted hPSCs (Leitch et al., 2013). Interestingly, in contrast to previous reports that naïve reversion results in loss of CpG methylation at known imprinted genomic sites (Pastor et al., 2016), allele-specific analysis of over 1400 known imprinted CpG sites in these 12 independent isogenic hPSC lines (before and after LIF-3i reversion) revealed stability of methylation imprints established in conventional hPSCs, with no systematic loss of imprinted methylation patterns resulting from LIF-3i culture (Table S4A).
To query for naïve-specific epigenetic functionality, we assayed for the activation of the proximal (PE) and distal (DE) enhancers of the OCT4 (POU5F1) promoter in LIF-3i-reverted versus primed hPSCs. Using both transient luciferase reporter assays and stable transgenic genomic DE/PE sequence mutant reporter hPSC lines (Gafni et al., 2013), we demonstrated that LIF-3i reversion potentiated naïve ESC-like activation of the DE of the OCT4 promoter, whereas primed hPSCs displayed preferential mEpiSC-like PE OCT4 activity (Fig. 4C,D).
Finally, to probe the status of pluripotency circuits in naïve-reverted hPSCs, we conducted modular GSEA and bioinformatics analysis of expression and methylation arrays for key pluripotency-associated stem cell circuits (e.g. the ESC module and SOX2-NANOG-OCT4-regulated core module; Table S1) in these 12 lines, before and after isogenic LIF-3i-reversion. These studies revealed that LIF-3i reversion significantly rewired both core and ESC module genes in naïve hPSCs, with (∼50%) decreases in gene promoter CpG DNA methylation and corresponding increases in gene expression of these pluripotency circuits (Fig. 4E,F).
LIF-3i induces human preimplantation epiblast-specific genes and increases expression of naïve-specific STAT3 and WNT transcriptional targets
We next aimed to determine the transcriptional correlation of LIF-3i-reverted hPSCs to human preimplantation epiblasts and to naïve hPSCs derived by other methods. We conducted comparative meta-analyses with published expression data from human embryonic day (E)3-E7 ICM epiblast cells (Petropoulos et al., 2016), as well as naïve hPSCs from several laboratories.
We found that the most differentially expressed (P<0.05) transcripts in LIF-3i-reverted hPSCs were also among the highest-ranked overexpressed genes in E3-E6 human morula and epiblast ICM cells (e.g. DNMT3L, NODAL, GDF3, IFITM1, LEFTY2, WNT3) (Petropoulos et al., 2016) or, alternatively, were known core regulators of naïve mESC pluripotency (e.g. NANOG, STELLA, KLF2, NR5A2) (Fig. 5, Fig. S8C-E). Many of these human epiblast-specific genes overexpressed in LIF-3i-reverted hPSCs are direct downstream targets of activated STAT3 signaling that are known to enhance mESC self-renewal and inhibit meso-endoderm differentiation (e.g. STAT3, FOS, SALL3, KLF2, KLF4, MYCN, IFITM1, EOMES) (Table S4B) (Bourillot et al., 2009). By contrast, the expression of lineage-primed developmental pathways (Fig. 5B) and genes (e.g. COL1A1, DKK3, TGFB1, SOX3, SOX8, SOX9) (Fig. 5A,E, Fig. S8D) were significantly underexpressed in LIF-3i-reverted hPSCs, relative to their primed states. To further determine over-represented pathways acquired in LIF-3i-reverted hiPSCs, we conducted genome-wide GSEA of the microarray data from LIF-3i-reverted hiPSCs versus their isogenic conventional counterparts using curated pathway databases. This analysis revealed that, relative to conventional hiPSCs, LIF-3i-reverted hiPSCs possess significantly higher transcriptional activities, increased chromatin remodeling, increased telomere function, increased expression of targets of WNT activation, and decreased expression of pathways associated with lineage-specific differentiation (FDR<0.01, P<0.01; Fig. 5B).
We next compared the whole-genome transcriptional signatures of LIF-3i-reverted hPSCs with both human ICM-derived epiblasts and mESCs by PCA normalized to published human blastocyst/morula, and with mESC/mESC-LIF-2i data sets as benchmark controls (Fig. 5C,D) (Bao et al., 2009; Vassena et al., 2011). We employed modular bioinformatics using the most differentially expressed genes in E4-E5 human epiblast cells (Petropoulos et al., 2016), and revealed that the patterns of gene expression in LIF-3i-reverted hPSCs cluster closely with human morula/blastocyst epiblast cells. Additional PCA demonstrated that LIF-3i-reverted hPSCs were not only transcriptionally similar to mESCs, but also to naïve-reverted hPSCs derived by others with alternate methods.
LIF-3i reversion reduces lineage-primed gene expression and improves multilineage differentiation potency for a broad repertoire of conventional hiPSCs
Since core pluripotency circuits were expressed at higher levels in LIF-3i-reverted hPSCs (Fig. 4E,F) with concomitant dramatic reduction in lineage-primed gene expression, including the broad network of lineage-specifying targets of the Polycomb PRC2 circuitry (Fig. 6A), we tested the hypothesis that naïve reversion improves multilineage functional pluripotency. We differentiated seven representative isogenic 4F-E sa-MP-iPSC and 7F-E fibro-iPSC lines (before and after LIF-3i reversion) to multiple representative derivatives of all three germ layers (Fig. 6B-F, Fig. S9). These studies revealed that LIF-3i reversion of multiple independently derived hiPSCs significantly improved their differentiation efficiency to endodermal (e.g. FOXA2+ and CXCR4+ SOX17+), ectodermal neural progenitor (e.g. SOX1+ nestin+; PAX6+ nestin+) and mesodermal vascular-pericytic [e.g. CD31+ CD146 (MCAM)+; KDR+ CD73 (NT5E)+ progenitor] populations. In multiple differentiation protocols, LIF-3i-reverted hPSC lines differentiated more efficiently, with less interline variability, and in some cases with more rapid kinetics (e.g. to neural ectoderm) than their conventional states. Altogether, these data suggested that LIF-3i reversion produced a more homogenous PSC population, with reduced lineage-primed gene variability and increased functional pluripotency (Fig. 7A).
Fig. 6.
Multilineage differentiation of isogenic primed versus LIF-3i-reverted hiPSC lines. (A) Differential expression of lineage-primed genes in the Polycomb (PRC2) circuit (ANOVA, P<0.001; Table S1) in seven hiPSC lines before (−) and after (+) LIF-3i reversion. Shown are heatmaps and associated log2 mean subtracted expression of PRC2 module genes of the LIF-3i-reverted versus isogenic-primed hiPSCs used in the differentiation studies below. Red, 4F-E sa-MP-iPSCs (n=4): circle, E5C3; square, E5C1; and triangle, E17C6 (or LZ6+10 for neural differentiations). Green, fibro-iPSCs (n=3): circle, C1.2; square, C2; and triangle, 7ta. *P<0.05 (paired t-tests). (B) Definitive endoderm differentiations (FOXA2+, CXCR4+ SOX17+) of isogenic LIF-3i-reverted versus primed hPSCs. Neural differentiations. (C,D) Kinetics of SOX1+ nestin+ and PAX6+ nestin+ neural progenitors in the same primed versus LIF-3i-reverted isogenic sa-MP-PSC (n=3) and fibro-hiPSC (n=3) lines described above. *P<0.05, **P<0.01 (paired t-tests). (E) Confocal microscopy of CDr3+ dye-binding neural progenitor rosettes (Yun et al., 2012). Neural rosettes were evaluated following passage of day 7 neural-induced LIF-3i-reverted (+) versus isogenic primed (−) E5C1 hiPSCs. Scale bars: 100 µm. (F) Isogenic vascular-endothelial hEB differentiations. Flow cytometry kinetics of CD31+ CD146+ (left) and KDR+ CD73+ (right) VP populations of the same isogenic sa-MP-iPSC lines as above (n=3). Error bars indicate s.e.m.
Fig. 7.

Summary schemas. (A) Relationships between primed pluripotency, WNT signaling, and amenability to stable naïve LIF-3i reversion. (B) Proposed molecular mechanisms underlying naïve-promoting effects of tankyrase inhibition in the presence of LIF-2i.
DISCUSSION
Stable reversion to a naïve epiblast-like ground pluripotent state may improve the functional utility of conventional hPSCs. Here, we comprehensively evaluated how the variables of derivation method, lineage priming, and baseline-primed functional pluripotency influence the stability of subsequent reversion to a human naïve epiblast-like pluripotent state. Our studies revealed that stable, long-term reversion to an mESC-like state could be achieved from a wide spectrum of EpiSC-like lineage-primed hPSC states via supplementation of classical LIF-2i with only a tankyrase inhibitor (LIF-3i). LIF-3i-reverted hiPSCs were highly proliferative, generated well-differentiated tri-lineage teratomas, possessed normal karyotypes and stable genomic CpG methylation imprints within a globally more hypomethylated genome that was highly transcriptional, and could be stably passaged as undifferentiated, clonal SSEA4+ TRA-1-81+ dome-shaped colonies for at least 30 passages.
Although human chimera generation and germ line contribution is the most stringent measure of naïve pluripotency and could not be tested here, LIF-3i-reverted hPSCs possessed most of the accepted characteristics of mESCs that we tested. These characteristics included high clonal proliferation rates, MEK/ERK independence, bFGF signaling unresponsiveness, STAT3 phosphorylation and signaling, JAK/STAT3 and BMP4 signal dependence, increased naïve-specific transcript expression (e.g. STELLA, NR5A2), upregulation of core pluripotency networks with concomitant decrease in lineage-primed gene circuits, whole-genome transcriptomic clustering with both human preimplantation epiblasts and mESCs, dominant distal OCT4 enhancer usage, global DNA CpG hypomethylation with increased 5hMC/5MC ratios, X-chromosome activation, decreased class I MHC, increased E-cadherin expression, and augmented expression of cytoplasmic and nuclear activated β-catenin. Importantly, LIF-3i-reverted hPSCs had significantly reduced lineage-primed gene expression and improved multilineage differentiation potency relative to their primed states. The derivation of naïve hPSC lines with improved functional pluripotency has broad impact for optimizing future hiPSC-based cellular therapies.
Although efficiently reprogrammed sa-MP-iPSC and select hESC lines demonstrated increased stability to LIF-3i reversion, transient supplementation of LIF-3i with forskolin and purmorphamine (LIF-5i) allowed the reversion of a broader repertoire (>16 independent hPSC lines) of variably lineage-primed hPSC lines. Interestingly, sa-MP-iPSCs already possess an improved functional pluripotency, with reduced lineage-primed differentiation skewing at baseline. Previous studies noted that STAT3 signaling was rate limiting for donor cell reprogramming completion (Yang et al., 2010; van Oosten et al., 2012). Thus, we hypothesize that the sustained STAT3 activation of ‘privileged’ CD33+ CD45+ sa-MP donors might play a crucial role not only in their efficient reprogramming, but also in facilitating the acquisition of a high-quality primed pluripotency with reduced lineage priming (Park et al., 2012). Similarly, mEpiSCs with increased lineage-primed gene expression possess reduced capacities for reverting to ICM-like pluripotency following exposure to LIF-STAT3 signaling, and ‘non-permissive’ mouse strains require augmented WNT pathway stimulation (Bernemann et al., 2011; Ten Berge et al., 2011). In those studies, only mEpiSCs with developmentally immature primed ground states reverted to naïve pluripotency with LIF-2i [likely by processes orchestrated by Krüppel-like factors KLF2/KLF4 (Greber et al., 2010; Guo et al., 2009; Gillich et al., 2012), NANOG (Silva et al., 2009) or co-regulators of transcriptional activities (e.g. MBD3, NR5A2) (Rais et al., 2013; Guo and Smith, 2010)]. Although hPSCs share molecular overlap with mEpiSCs, a similar stable reversion with 2i alone was not attainable, and GSK3β inhibition may not be sufficient to tune human cell β-catenin levels to a mouse-like subcellular distribution. We hypothesize that the expression of primed hPSC pluripotency circuits with decreased lineage-primed gene expression might similarly facilitate naïve reversion. We also propose that additional reinforcing signals provided by tankyrase inhibition might allow β-catenin to maintain pluripotency core signals in the context of MEK/ERK inhibition. Thus, we propose that more effective reprogramming, reduced lineage priming, and optimized WNT signaling in hPSCs are important general determinants of facile reversion to a stable human naïve state under MEK/ERK inhibition conditions (Fig. 7A).
Recently, hPSC naïve reversion approaches have variably required the imposition of transgenic core factor overexpression, complex anti-apoptosis cocktails (e.g. ROCK, BRAF, SRC or JNK inhibition) to sustain survival/proliferation, HDAC inhibition to reset global epigenetic barriers, and/or EpiSC-specific growth factor reinforcement (e.g. bFGF, activin, TGFβ or BMP inhibition) (Table S5). These studies suggest that hPSCs might be generally ‘non-permissive’ to classical mESC 2i WNT and MEK/ERK pathway reversion, or that human and murine naïve states might be fundamentally non-equivalent. Although our comparative bioinformatics meta-analyses suggested a common pathway between other reversion methods and ours, we demonstrated that a stable human naive epiblast-like state could be maintained in conventional hPSCs via LIF-2i and only a tankyrase inhibitor.
The mechanism of stabilizing human naïve pluripotency by tankyrase inhibition currently remains unclear and potentially complex, but is likely to involve a synergy with GSK3β inhibition (CHIR99021) to further augment WNT signaling (Fig. 7B). Canonical WNT signaling is determined by a post-translational balance between activated non-phosphorylated β-catenin and its phosphorylation by its destruction complex (axin/APC/GSK3β). Inhibition of the protein kinase GSK3β impedes β-catenin destruction, and allows its non-phosphorylated form to reach the nucleus where both pluripotency- and differentiation-associated factors are targeted (Ye et al., 2012; Buehr et al., 2008; Sato et al., 2004). Distinct mechanisms of reinforcement of pluripotency by GSK3β inhibition include TCF repression (Wray et al., 2011) and direct targeting of NANOG, OCT4, KLF4, ESRRB and CBP (CREBBP) (Takao et al., 2007; Tam et al., 2008; Kelly et al., 2011; Evans et al., 2010; Martello et al., 2012). WNT activation was previously shown to be not only rate limiting for naïve reversion of mEpiSCs, but also facilitated mESC derivation from ‘non-permissive’ mouse strains (Faunes et al., 2013). Additionally, both nuclear (transcriptional) and cytoplasmic (non-transcriptional) activities of β-catenin have been linked to stabilizing mouse naïve pluripotency via interactions with E-cadherin and cytoplasmic OCT4 and NANOG (Marucci et al., 2014). Furthermore, although 2i sufficiently stabilized naïve reversion of mEpiSCs, some mouse strains required reinforcement of KLF4, c-MYC or β-catenin activities (Hanna et al., 2010; Ye et al., 2012).
XAV939, an inhibitor of the poly-ADP-ribosylating enzymes tankyrase 1 and 2 (TNKS and TNKS2; also known as PARP5A/B, ARTDF5/6), was originally identified for its capacity to stimulate β-catenin degradation via stabilizing axin. The sole use of XAV939 in cancer cells inhibited WNT signaling (Huang et al., 2009) by reinforcing the β-catenin destruction complex. However, dual combination of tankyrase inhibition (XAV939) and GSK3β inhibition (CHIR99021) in primed rodent mEpiSCs and primed conventional hESCs paradoxically increased WNT signaling by increasing axin expression, stabilizing the axin-catenin complex, and increasing cytoplasmic retention of β-catenin (Kim et al., 2013; Schmitz et al., 2013). Our hPSC studies herein demonstrated that XAV939 similarly synergizes with CHIR99021 in naïve conditions (i.e. in the absence of MEK/ERK signal; PD0325901) to paradoxically stabilize and augment the expression of activated β-catenin in both nuclear and cytoplasmic compartments.
We speculate that additional, potentially complex activities of tankyrase beyond WNT signaling may further support stabilization of a human naïve ground state (Fig. 7B). These mechanisms might include the promotion of genomic integrity via telomere recombination/elongation and stability of the non-homologous end-joining (NHEJ) protein DNA-PKC (Dregalla et al., 2010; Smith, 1998), pericentric heterochromatin regulation (Karantzali et al., 2011), centrosome and mitotic spindle regulation (Kim et al., 2012; Chang et al., 2009) and promotion of homologous recombination (Vidi et al., 2014). Thus, future studies will focus not only on whether differentiated derivatives of naïve hPSCs provide functional advantages over conventional primed hPSCs for cellular transplantation, but also on elucidating the mechanisms of naïve promotion by tankyrase inhibition.
MATERIALS AND METHODS
Naïve reversion screens with small molecules and cytokines
We tested >130 combinations of 23 small molecules/cytokines known to regulate ESC self-renewal. Permutations were tested using standard bFGF-supplemented KOSR-based hESC medium on irradiated MEF feeders without bFGF (Table S3). Modifications included N2B27 supplement (Life Technologies) and incubation in 5% O2. Molecules included bFGF (10 ng/ml; Peprotech, 100-18B), human LIF (hLIF) (20 ng/ml; Sigma, L5283; Cell Signaling, 8911LC; or Peprotech, 300-05), PD0325901 (1 µM; Stemgent, 040006; Sigma, PZ0162), CHIR99021 (3 µM; Stemgent, 04-0004; Tocris Bioscience, 4423), XAV939 (4 µM; Sigma, X3004), SB431542 (2 µM; Stemgent, 04-0010), forskolin (10 µM; Stemgent, 04-0025), ACTH peptide 1-24 (10 µM; American Peptide, 10-1-21), 2′,5′-dideoxyadenosine (500 µM; Sigma, D7408), AICA-riboside (100 µM; EMD Millipore, 123040), 8-(4-chlorophenylthio)-2′-O-methyladenosine 3′,5′-cyclic monophosphate monosodium hydrate (100 µM; Sigma, C8988), 3-isobutyl-1-methylxanthine (100 µM; Sigma, I5879), BayK8644 (1 µM; Stemgent, 04-0013), DLPC (100 µM; Tocris Bioscience, 4378), purmorphamine (2 µM; Stemgent, 04-0009), AM580 (10 nM; Sigma, AM580), CCL2 (200 ng/ml; 571406, Biolegend), SCF (10 ng/ml; Peprotech), IGF1 (10 ng/ml; Peprotech), IL6 (10 ng/ml; Peprotech), TPO (10 ng/ml; Peprotech), BMP4 (10 ng/ml; Peprotech) and thiazovivin (2 µM; Stemgent, 04-0017).
Initial testing was performed using conventional hPSCs (hESC-H9, sa-MP-iPSCs 6.2, E5C3); for ethics relating to the use of hESC lines and details of conventional hPSC culture see the supplementary Materials and Methods. Conventional hPSCs were expanded 5-6 days on MEFs. Single-cell passaging was performed using Accutase (Sigma), with initial cell plating densities of 50,000-100,000 cells/cm2 followed by 5000-20,000 cells/cm2 for subsequent passages. Exposure (24-48 h) to small molecules prior to initial passage greatly enhanced survival of conventional hPSCs. Naive-reverted hPSC cultures were passaged every 3-4 days on fresh irradiated MEFs. Morphological changes were photomicrographed using a Nikon Eclipse TE-2000 inverted microscope, DS-Fi1 camera and NIS-Elements software.
Cultures were assayed by flow cytometry using anti-SSEA1 (BD Biosciences), SSEA4 (R&D Systems) and TRA-1-60/TRA-1-81 (BD Biosciences) antibodies. For details of flow cytometry and associated western blotting see the supplementary Materials and Methods. Conditions that supported stable expansion of hPSC lines for >3-5 passages were further evaluated by qRT-PCR using TaqMan assays on a ViAA7 Real-Time PCR System (Life Technologies) for expression of OCT4, NANOG, ZFP42, KLF2, NR5A2 and STELLA. For details of qRT-PCR and primers see the supplementary Materials and Methods. Karyotype analysis was performed by the JHU Cytogenetics Core Facility. RNA-FISH for XIST is described in the supplementary Materials and Methods.
Extensive small-molecule screening (supplementary Materials and Methods, Table S3) identified XAV939 for permitting long-term survival of hPSCs in classical 2i (CHIR99021 and PD0325901). A combination of CHIR99021, PD0325901 and XAV939 was sufficient for directly reverting a series of hPSC lines to stable, clonal cultures. Other hPSC lines required additional pre-treatment for one passage with forskolin and purmorphamine along with LIF-3i (LIF-5i) to enhance initial clonal viability. Cultures were subsequently passaged with LIF-3i alone, and routinely evaluated for expression of SSEA1/SSEA4, TRA-1-60/TRA-1-81 by flow cytometry every two to five passages thereafter.
Characterization of naïve hPSCs
A description of the generation of a repertoire of episomal hiPSC lines for the functional and molecular characterization of the reprogrammed state is provided in Table S2 and in the supplementary Materials and Methods, along with a description of how hESCs and hiPSCs were differentiated into the various mesodermal, ectodermal and endodermal lineages.
Immunofluorescent staining
Cell cultures were passaged onto MEFs in 8-well Lab-Tek II chamber slides (Nunc, Thermo-Fisher) at 30,000 cells per cm2. Primed and naïve cultures were expanded for 4-6 days in respective culture media before fixation with 2% formaldehyde in PBS (Affymetrix) for 15-20 min at room temperature. Cultures were washed in sterile PBS, permeabilized for 10 min in Wash Buffer (DAKO) at room temperature, and incubated for 1 h in blocking solution [PBS, 5% goat serum (Sigma) and 0.05% Tween 20 (Sigma)]. Antibodies were diluted in blocking solution. Cells were incubated overnight at 4°C in humid chambers with: primary rabbit anti-human unconjugated antibodies for AXIN1 (1:200; 06-1049, Millipore), KLF2 (1:50; HPA055964, Sigma), KLF4 (1:50; HPA002926, Sigma), KLF5 (1:50; HPA040398, Sigma), KLF17 (1:200; HPA024629, Sigma), Nanog (1:100; 3369-1, Epitomics), NR5A2 (1:50; HPA005455, Sigma), OCT4 (POU5F1) (1:50; sc9081, Santa Cruz), STELLA (DPPA3) (1:200; HPA045695, Sigma) and TFCP2L1 (1:50; HPA029708, Sigma) or alternatively with mouse anti-human STELLA (1:50; MAB4388, Millipore) or AXIN1 (clone C76H11, 1:500; 2087, Cell Signaling). Washed slides were sequentially incubated for 1 h with biotinylated secondary goat anti-rabbit or anti-mouse antibodies (1:500; E0432 and E0433, DAKO) for 30 min with streptavidin-Cy3 (1:500; S6402, Sigma) at room temperature. Cells were washed twice, incubated for 2 h with a second primary mouse anti-human antibody (for initial rabbit anti-human primary antibodies) including active β-catenin (ABC clone 8E7, 1:100; 05-665, Millipore), β-catenin (total) (1:50; M3539, DAKO) and E-cadherin (1:25; M3612, DAKO). Slides were washed twice in Wash Buffer (DAKO), incubated for 1 h with either Alexa 488-conjugated highly cross-adsorbed goat anti-mouse secondary antibody (1:200; A11029, Life Technologies) or Alexa 488-conjugated highly cross-adsorbed goat anti-rabbit secondary antibody (1:200; A11034, Life Technologies). Alternatively, second immunostains were performed using directly conjugated anti-human SSEA4 NL493 (1:50; SC023 Kit, R&D Systems), TRA-1-81-Alexa 488 (1:10; 560174, BD Biosciences) or anti-TRA-1-60-Dylight 488 (1:100; 09-0068, Stemgent). Nuclear staining was performed with DAPI (10 µg/ml; Life Technologies). Labtek chambers were separated and mounted with Prolong Gold Anti-Fade Reagent (Life Technologies), and observed and photographed with a Zeiss LSM 510 Meta confocal microscope. Universal negative control for mouse and rabbit primary antibodies (DAKO) were used.
Signaling inhibitor assays
Stable LIF-3i/MEF co-cultures (hESC H9, MP-iPSC E5C3 and Fibro-iPSC C1.2) were supplemented with small-molecule inhibitors of BMP4 (dorsomorphin, 2 µM; P5499 Sigma), CBP-CREB (217505, 10 µM; Millipore), FGF (PD173074, 0.1 µM; S1264, Selleck Chemicals), JAK/STAT (420097, 2 µM; Millipore), Pi3K (GDC0941, 1 µM; 04-0047, Stemgent), TGFβ (SB431542, 2 µM; Stemgent) or hLIF/gp130 blocking antibody (MAB628, 0.05 µg/ml; R&D Systems). Addition of bFGF (10 ng/ml; Peprotech), BMP4 (1-10 ng/ml; Peprotech), TGFβ (1 ng/ml; Peprotech) or withdrawal of hLIF were also tested. All culture conditions were maintained for at least three passages (12 days) prior to flow cytometry for SSEA4/TRA-1 antigen expression. Cell numbers were estimated every passage using the Countess system.
Genomic CpG methylation
Assays of global 5-methylcytosine versus 5-hydroxymethylcytosine were carried out on genomic DNA of hPSC lines by dot blot as described in the supplementary Materials and Methods.
OCT4 enhancer usage
The use of the proximal versus distal enhancer of OCT4 was assessed by luciferase assays and using enhancer mutant reporter hPSC lines as described in the supplementary Materials and Methods.
Bioinformatics analysis
Details of bioinformatics analyses, including gene expression and CpG DNA methylation microarrays and transcriptome analysis, are provided in the supplementary Materials and Methods.
Acknowledgements
We thank Igor Slukvin and Renee Reija-Pera for hiPSCs; Jiwon Ryu, Lynette Naler, Wayne Yu, Ada Tam, and Lee Blosser for technical support.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
All authors designed, performed and interpreted experiments. In addition, J.S.H., T.S.P., L.Z., C.C.T., S.B.B., M.C., L.C. and E.T.Z. designed, analyzed and interpreted bioinformatics analyses. E.T.Z., L.Z., T.S.P. and J.S.H. wrote and edited the manuscript. E.T.Z. provided administrative and financial support, and edited/approved the final manuscript.
Funding
This work was supported by grants from the National Institutes of Health (NIH)/NHLBI [U01HL099775 to E.T.Z., A.D.F., S.B.B.; PCBC2012Pilot_01 to E.T.Z.], NIH/NEI [R01EY023962 to E.T.Z.], NIH/NICHD [R01HD082098 to E.T.Z.] and NIH/NCI [CA60441 to J.S.H. and D.S.]; Research to Prevent Blindness Stein Innovation Award (E.T.Z.); Maryland Stem Cell Research Fund [2011-MSCRF-II-0008-00 to E.T.Z., 2014-MSCRF-II-118153 to T.S.P., 2012-MSCRF-III-033 to J.S.H., 2013-MSCRF-III-114936 to L.Z., 2012-MSCRFE-0207-00 to M.V.C.-S.]; Alex's Lemonade Stand Foundation for Childhood Cancer (J.S.H.); and core grants from the NIH [P30 CA006973 and EY01765]. Deposited in PMC for release after 12 months.
Data availability
Microarray data are available at Gene Expression Omnibus under accession number GSE44430: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE44430.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.138982.supplemental
References
- Bao S., Tang F., Li X., Hayashi K., Gillich A., Lao K. and Surani M. A. (2009). Epigenetic reversion of post-implantation epiblast to pluripotent embryonic stem cells. Nature 461, 1292-1295. 10.1038/nature08534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernemann C., Greber B., Ko K., Sterneckert J., Han D. W., Araúzo-Bravo M. J. and Schöler H. R. (2011). Distinct developmental ground states of epiblast stem cell lines determine different pluripotency features. Stem Cells 29, 1496-1503. 10.1002/stem.709 [DOI] [PubMed] [Google Scholar]
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z. D., Ziller M., Croft G. F., Amoroso M. W., Oakley D. H. et al. (2011). Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439-452. 10.1016/j.cell.2010.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting G. L., Kiskinis E., Croft G. F., Amoroso M. W., Oakley D. H., Wainger B. J., Williams D. J., Kahler D. J., Yamaki M., Davidow L. et al. (2011). A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 29, 279-286. 10.1038/nbt.1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourillot P.-Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N. and Savatier, P. (2009). Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells 27, 1760-1771. 10.1002/stem.110 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G. et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947-956. 10.1016/j.cell.2005.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons I. G., Smithers L. E., Trotter M. W. B., Rugg-Gunn P., Sun B., Chuva De Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A. et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191-195. 10.1038/nature05950 [DOI] [PubMed] [Google Scholar]
- Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., Mclay R., Hall J., Ying Q.-L. and Smith A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287-1298. 10.1016/j.cell.2008.12.007 [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Thompson S., Millrod M. A., Weinberg S., Yuan X., Peters A., Mahairaki V., Koliatsos V. E., Tung L. and Zambidis E. T. (2011). A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS ONE 6, e18293 10.1371/journal.pone.0018293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne J. A., Nguyen H. N. and Reijo Pera R. A. (2009). Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. PLoS ONE 4, e7118 10.1371/journal.pone.0007118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.-S., GöKe J., Ng J.-H., Lu X., Gonzales K. A. U., Tan C.-P., Tng W.-Q., Hong Z.-Z., Lim Y.-S. and Ng H.-H. (2013). Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 13, 663-675. 10.1016/j.stem.2013.11.015 [DOI] [PubMed] [Google Scholar]
- Chang P., Coughlin M. and Mitchison T. J. (2009). Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol. Biol. Cell 20, 4575-4585. 10.1091/mbc.E09-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin M. H., Mason M. J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J. et al. (2009). Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111-123. 10.1016/j.stem.2009.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. D., Yu J., Smuga-Otto K., Salvagiotto G., Rehrauer W., Vodyanik M., Thomson J. and Slukvin I. (2009). Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells 27, 559-567. 10.1002/stem.20080922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.-F., Chen H.-H., Eijpe M., Yabuuchi A., Chenoweth J. G., Tesar P., Lu J., McKay R. D. G. and Geijsen N. (2008). The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell 135, 449-461. 10.1016/j.cell.2008.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dregalla R. C., Zhou J., Idate R. R., Battaglia C. L. R., Liber H. L. and Bailey S. M. (2010). Regulatory roles of tankyrase 1 at telomeres and in DNA repair: suppression of T-SCE and stabilization of DNA-PKcs. Aging 2, 691-708. 10.18632/aging.100210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eminli S., Foudi A., Stadtfeld M., Maherali N., Ahfeldt T., Mostoslavsky G., Hock H. and Hochedlinger K. (2009). Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat. Genet. 41, 968-976. 10.1038/ng.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. M., Chen X., Zhang W. and Liu C. (2010). KLF4 interacts with beta-catenin/TCF4 and blocks p300/CBP recruitment by beta-catenin. Mol. Cell. Biol. 30, 372-381. 10.1128/MCB.00063-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F., Hayward P., Descalzo S. M., Chatterjee S. S., Balayo T., Trott J., Christoforou A., Ferrer-Vaquer A., Hadjantonakis A.-K., Dasgupta R. et al. (2013). A membrane-associated beta-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 140, 1171-1183. 10.1242/dev.085654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Lu S.-J., Klimanskaya I., Gomes I., Kim D., Chung Y., Honig G. R., Kim K.-S. and Lanza R. (2009). Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28, 704-712. 10.1002/stem.321 [DOI] [PubMed] [Google Scholar]
- Gafni O., Weinberger L., Mansour A. A. F., Manor Y. S., Chomsky E., Ben-Yosef D., Kalma Y., Viukov S., Maza I., Zviran A. et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282-286. 10.1038/nature12745 [DOI] [PubMed] [Google Scholar]
- Greber B., Wu G., Bernemann C., Joo J. Y., Han D. W., Ko K., Tapia N., Sabour D., Sterneckert J., Tesar P. et al. (2010). Conserved and divergent roles of FGF signaling in mouse epiblast stem cells and human embryonic stem cells. Cell Stem Cell 6, 215-226. 10.1016/j.stem.2010.01.003 [DOI] [PubMed] [Google Scholar]
- Gillich A., Bao S., Grabole N., Hayashi K., Trotter M. W. B., Pasque V., Magnúsdóttir E. and Surani M. A. (2012). Epiblast stem cell-based system reveals reprogramming synergy of germline factors. Cell Stem Cell 10, 425-439. 10.1016/j.stem.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G. and Smith A. (2010). A genome-wide screen in EpiSCs identifies Nr5a nuclear receptors as potent inducers of ground state pluripotency. Development 137, 3185-3192. 10.1242/dev.052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W. and Smith A. (2009). Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063-1069. 10.1242/dev.030957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Zi X., Schulz V. P., Cheng J., Zhong M., Koochaki S. H. J., Megyola C. M., Pan X., Heydari K., Weissman S. M. et al. (2014). Nonstochastic reprogramming from a privileged somatic cell state. Cell 156, 649-662. 10.1016/j.cell.2014.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., Cassady J. P., Muffat J., Carey B. W. and Jaenisch R. (2010). Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc. Natl. Acad. Sci. USA 107, 9222-9227. 10.1073/pnas.1004584107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.-Y., Weick J. P., Yu J., Ma L.-X., Zhang X.-Q., Thomson J. A. and Zhang S.-C. (2010). Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 107, 4335-4340. 10.1073/pnas.0910012107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Yu J., Suknuntha K., Tian S., Montgomery K., Choi K.-D., Stewart R., Thomson J. A. and Slukvin I. I. (2011). Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood 117, e109-e119. 10.1182/blood-2010-07-298331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.-M., Mishina Y. M., Liu S., Cheung A., Stegmeier F., Michaud G. A., Charlat O., Wiellette E., Zhang Y., Wiessner S. et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signaling. Nature 461, 614-620. 10.1038/nature08356 [DOI] [PubMed] [Google Scholar]
- Kajiwara M., Aoi T., Okita K., Takahashi R., Inoue H., Takayama N., Endo H., Eto K., Toguchida J., Uemoto S. et al. (2012). Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 12538-12543. 10.1073/pnas.1209979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzali E., Lekakis V., Ioannou M., Hadjimichael C., Papamatheakis J. and Kretsovali A. (2011). Sall1 regulates embryonic stem cell differentiation in association with Nanog. J. Biol. Chem. 286, 1037-1045. 10.1074/jbc.M110.170050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. F., Ng D. Y., Jayakumaran G., Wood G. A., Koide H. and Doble B. W. (2011). beta-catenin enhances Oct-4 activity and reinforces pluripotency through a TCF-independent meism. Cell Stem Cell 8, 214-227. 10.1016/j.stem.2010.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Woo A. J., Chu J., Snow J. W., Fujiwara Y., Kim C. G., Cantor A. B. and Orkin S. H. (2010a). A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313-324. 10.1016/j.cell.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I. R. et al. (2010b). Epigenetic memory in induced pluripotent stem cells. Nature 467, 285-290. 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Zhao R., Doi A., Ng K., Unternaehrer J., Cahan P., Hongguang H., Loh Y.-H., Aryee M. J., Lensch M. W. et al. (2011). Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol 29, 1117-1119. 10.1038/nbt.2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. K., Dudognon C. and Smith S. (2012). Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 13, 724-732. 10.1038/embor.2012.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Wu J., Ye S., Tai C. I., Zhou X., Yan H., Li P., Pera M. and Ying Q. L. (2013). Modulation of beta-catenin function maintains mouse epiblast stem cell and human embryonic stem cell self-renewal. Nat. Commun. 4, 2403 10.1038/ncomms3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y., Kaufman-Francis K., Studdert J. B., Steiner K. A., Power M. D., Loebel D. A. F., Jones V., Hor A., De Alencastro G., Logan G. J. et al. (2014). The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 14, 107-120. 10.1016/j.stem.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Kyttälä A., Moraghebi R., Valensisi C., Kettunen J., Andrus C., Pasumarthy K. K., Nakanishi M., Nishimura K., Ohtaka M. and Weltner J. et al. (2016). Genetic variability overrides the impact of parental cell type and determines the iPSC differentiation potential. Stem Cell Rep. 6, 200-212. 10.1016/j.stemcr.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch H. G., McEwen K. R., Turp A., Encheva V., Carroll T., Grabole N., Mansfield W., Nashun B., Knezovich J. G., Smith A. et al. (2013). Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 20, 311-316. 10.1038/nsmb.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Kida Y. S., Hawkins R. D., Nery J. R., Hon G., Antosiewicz-Bourget J., O'Malley R., Castanon R., Klugman S. et al. (2011). Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68-73. 10.1038/nature09798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Francis Stewart A., Smith A. et al. (2012). The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149, 590-604. 10.1016/j.cell.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Sugimoto T., Diamanti E., Joshi A., Hannah R., Ohtsuka S., Göttgens B., Niwa H. and Smith A. (2012). Esrrb is a pivotal target of the Gsk3/Tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell 11, 491-504. 10.1016/j.stem.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marucci L., Pedone E., Di Vicino U., Sanuy-Escribano B., Isalan M. and Cosma M. P. (2014). beta-catenin fluctuates in mouse ESCs and is essential for Nanog-mediated reprogramming of somatic cells to pluripotency. Cell Rep. 8, 1686-1696. 10.1016/j.celrep.2014.08.011 [DOI] [PubMed] [Google Scholar]
- Nishino K., Toyoda M., Yamazaki-Inoue M., Fukawatase Y., Chikazawa E., Sakaguchi H., Akutsu H. and Umezawa A. (2011). DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 7, e1002085 10.1371/journal.pgen.1002085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y., Qin H., Hong C., Blouin L., Polo J. M., Guo T., Qi Z., Downey S. L., Manos P. D., Rossi D. J. et al. (2011). Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541-549. 10.1038/ncb2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K., Caron L., Borowiak M., Martinez R. J., Fitz-Gerald C. S., Sato Y., Cowan C. A., Chien K. R. and Melton D. A. (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 26, 313-315. 10.1038/nbt1383 [DOI] [PubMed] [Google Scholar]
- Park T. S., Huo J. S., Peters A., Talbot C. C., Verma K., Zimmerlin L., Kaplan I. M. and Zambidis E. T. (2012). Growth factor-activated stem cell circuits and stromal signals cooperatively accelerate non-integrated iPSC reprogramming of human myeloid progenitors. PLoS ONE 7, e42838 10.1371/journal.pone.0042838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park T. S., Bhutto I., Zimmerlin L., Huo J. S., Nagaria P., Miller D., Rufaihah A. J., Talbot C., Aguilar J., Grebe R. et al. (2014). Vascular progenitors from cord blood-derived iPSC possess augmented capacity for regenerating ischemic retinal vasculature. Circulation 129, 359-372. 10.1161/CIRCULATIONAHA.113.003000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A., Chen D., Liu W., Kim R., Sahakyan A., Lukianchikov A., Plath K., Jacobsen S. E. and Clark A. T. (2016). Naïve human pluripotent stem cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 18, 323-329. 10.1016/j.stem.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petropoulos S., Edsgärd D., Reinius B., Deng Q., Panula S. P., Codeluppi S., Plaza Reyes A., Linnarsson S., Sandberg R. and Lanner F. (2016). Single-cell RNA-Seq reveals lineage and X-chromosome dynamics in human preimplantation embryos. Cell 165, 1012-1026. 10.1016/j.cell.2016.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J. M., Liu S., Figueroa M. E., Kulalert W., Eminli S., Tan K. Y., Apostolou E., Stadtfeld M., Li Y., Shioda T. et al. (2010). Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 28, 848-855. 10.1038/nbt.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rais Y., Zviran A., Geula S., Gafni O., Chomsky E., Viukov S., Mansour A. A. F., Caspi I., Krupalnik V., Zerbib M. et al. (2013). Deterministic direct reprogramming of somatic cells to pluripotency. Nature 502, 65-70. 10.1038/nature12587 [DOI] [PubMed] [Google Scholar]
- Ruiz S., Diep D., Gore A., Panopoulos A. D., Montserrat N., Plongthongkum N., Kumar S., Fung H.-L., Giorgetti A., Bilic J. et al. (2012). Identification of a specific reprogramming-associated epigenetic signature in human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 109, 16196-16201. 10.1073/pnas.1202352109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Meijer L., Skaltsounis L., Greengard P. and Brivanlou A. H. (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55-63. 10.1038/nm979 [DOI] [PubMed] [Google Scholar]
- Schmitz Y., Rateitschak K. and Wolkenhauer O. (2013). Analysing the impact of nucleo-cytoplasmic shuttling of beta-catenin and its antagonists APC, Axin and GSK3 on Wnt/beta-catenin signalling. Cell. Signal. 25, 2210-2221. 10.1016/j.cellsig.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Silva J., Nichols J., Theunissen T. W., Guo G., Van Oosten A. L., Barrandon O., Wray J., Yamanaka S., Chambers I. and Smith A. (2009). Nanog is the gateway to the pluripotent ground state. Cell 138, 722-737. 10.1016/j.cell.2009.07.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. (1998). Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484-1487. 10.1126/science.282.5393.1484 [DOI] [PubMed] [Google Scholar]
- Takao Y., Yokota T. and Koide H. (2007). Beta-catenin up-regulates Nanog expression through interaction with Oct-3/4 in embryonic stem cells. Biochem. Biophys. Res. Commun. 353, 699-705. 10.1016/j.bbrc.2006.12.072 [DOI] [PubMed] [Google Scholar]
- Takashima Y., Guo G., Loos R., Nichols J., Ficz G., Krueger F., Oxley D., Santos F., Clarke J., Mansfield W. et al. (2014). Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 158, 1254-1269. 10.1016/j.cell.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W. L., Lim C. Y., Han J., Zhang J., Ang Y.-S., Ng H.-H., Yang H. and Lim B. (2008). T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells 26, 2019-2031. 10.1634/stemcells.2007-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Berge D., Kurek D., Blauwkamp T., Koole W., Maas A., Eroglu E., Siu R. K. and Nusse R. (2011). Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat. Cell Biol. 13, 1070-1075. 10.1038/ncb2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L. and Mckay R. D. G. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196-199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Theunissen T. W., Powell B. E., Wang H., Mitalipova M., Faddah D. A., Reddy J., Fan Z. P., Maetzel D., Ganz K., Shi L. et al. (2014). Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15, 471-487. 10.1016/j.stem.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosten A. L., Costa Y., Smith A. and Silva J. C. (2012). JAK/STAT3 signalling is sufficient and dominant over antagonistic cues for the establishment of naive pluripotency. Nat. Commun. 3, 817 10.1038/ncomms1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R., Boue S., Gonzalez-Roca E., Aran B., Auer H., Veiga A. and Belmonte J. C. I. (2011). Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 138, 3699-3709. 10.1242/dev.064741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidi P. A., Liu J., Salles D., Jayaraman S., Dorfman G., Gray M., Abad P., Moghe P. V., Irudayaraj J. M., Wiesmuller L. et al. (2014). NuMA promotes homologous recombination repair by regulating the accumulation of the ISWI ATPase SNF2h at DNA breaks. Nucleic Acids Res. 42, 6365-6379. 10.1093/nar/gku296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware C. B., Nelson A. M., Mecham B., Hesson J., Zhou W., Jonlin E. C., Jimenez-Caliani A. J., Deng X., Cavanaugh C., Cook S. et al. (2014). Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 111, 4484-4489. 10.1073/pnas.1319738111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger L., Ayyash M., Novershtern N. and Hanna J. H. (2016). Dynamic stem cell states: naïve to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 17, 155-169. 10.1038/nrm.2015.28 [DOI] [PubMed] [Google Scholar]
- Wray J., Kalkan T., Gomez-Lopez S., Eckardt D., Cook A., Kemler R. and Smith A. (2011). Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 13, 838-845. 10.1038/ncb2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Van Oosten A. L., Theunissen T. W., Guo G., Silva J. C. R. and Smith A. (2010). Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 7, 319-328. 10.1016/j.stem.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Tan L., Yang R., Fang B., Qu S., Schulze E. N., Song H., Ying Q. and Li P. (2012). Pleiotropy of glycogen synthase kinase-3 inhibition by CHIR99021 promotes self-renewal of embryonic stem cells from refractory mouse strains. PLoS ONE 7, e35892 10.1371/journal.pone.0035892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P. and Smith A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519-523. 10.1038/nature06968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S. W., Leong C., Zhai D., Tan Y. L., Lim L., Bi X., Lee J.-J., Kim H. J., Kang N.-Y., Ng S. H. et al. (2012). Neural stem cell specific fluorescent chemical probe binding to FAB7. Proc. Natl. Acad. Sci. USA 109, 10214-10217. 10.1073/pnas.1200817109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M. N., Cao L.-H., Peters A., Park T. S., Zambidis E. T., Meyer J. S. et al. (2014). Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5, 4047 10.1038/ncomms5047 [DOI] [PMC free article] [PubMed] [Google Scholar]