Abstract
Drosophila Elav is the founding member of the conserved family of Hu RNA-binding proteins (RBPs), which play crucial and diverse roles in post-transcriptional regulation. Elav has long served as the canonical neuronal marker. Surprisingly, although Elav has a well-characterized neural cis-regulatory module, we find endogenous Elav is also ubiquitously transcribed and post-transcriptionally repressed in non-neural settings. Mutant clones of multiple miRNA pathway components derepress ubiquitous Elav protein. Our re-annotation of the elav transcription unit shows not only that it generates extended 3′ UTR isoforms, but also that its universal 3′ UTR isoform is much longer than previously believed. This longer common 3′ UTR includes multiple conserved, high-affinity sites for the miR-279/996 family. Of several miRNA mutants tested, endogenous Elav and a transgenic elav 3′ UTR sensor are derepressed in mutant clones of mir-279/996. We also observe cross-repression of Elav by Mei-P26, another RBP derepressed in non-neural miRNA pathway clones. Ubiquitous Elav has regulatory capacity, since derepressed Elav can stabilize an Elav-responsive sensor. Repression of Elav in non-neural territories is crucial as misexpression here has profoundly adverse consequences. Altogether, we define unexpected post-transcriptional mechanisms that direct appropriate cell type-specific expression of a conserved neural RBP.
KEY WORDS: Drosophila, Elav, Mei-P26, RNA-binding protein, MicroRNA, Neuron
Summary: Elav – the canonical Drosophila marker for post-mitotic neurons – has a previously unrecognized component of ubiquitous expression that is suppressed by the microRNA pathway, including by miR-279/996.
INTRODUCTION
microRNAs (miRNAs) are ∼22 nt RNAs that regulate broad target networks and play diverse biological roles (Bartel, 2009; Sun and Lai, 2013). Although it is difficult to identify processes that are not regulated by miRNAs, the general activity of the miRNA pathway and, by extension, that of the bulk of miRNAs, has often been considered to be important for differentiation. This concept is based on: (1) the broad diversity of miRNAs expressed in specific organs or terminally differentiated cells (Lagos-Quintana et al., 2002); (2) general downregulation of miRNAs in tumors compared with normal tissues (Lu et al., 2005); (3) that certain miRNA mutants, including the founding locus lin-4, reiterate early cell lineages (Chalfie et al., 1981; Lee et al., 1993); and (4) the fact that certain stem cell types, including embryonic stem cells (Wang et al., 2007) and neural stem cells (Andersson et al., 2010; Kawase-Koga et al., 2010), tolerate deletion of core miRNA biogenesis factors but are unable to differentiate. Nonetheless, it is clear that miRNAs affect the behavior of stem cells and other undifferentiated cells, and are otherwise embedded in a diverse array of biological settings (Flynt and Lai, 2008; Shenoy and Blelloch, 2014; Sun and Lai, 2013).
In this study, we report surprising observations on the role of post-transcriptional regulation in determining the spatial accumulation of Drosophila Elav. This was one of the first loci for which transcript and protein products were recognized as being restricted to neurons (Campos et al., 1987; Robinow et al., 1988). Antibodies against this nuclear RNA-binding protein (RBP) were the first reagent to label postmitotic Drosophila neurons (Robinow and White, 1991), and its status as the standard neuronal marker was solidified by the development of high-quality mouse and rat monoclonal Elav antibodies more than 20 years ago (O'Neill et al., 1994). Despite reports that Elav is transiently detected in embryonic neuroblasts and glial cells (Berger et al., 2007; Lai et al., 2012), its robust and specific accumulation in postmitotic neurons makes Elav the marker of choice for this terminally differentiated fate.
We unexpectedly find that endogenous Elav protein is ectopically expressed in non-neuronal mutant clones of miRNA pathway components due to loss of post-transcriptional repression via the elav 3′ UTR. Thus, this classic cell-specific differentiation marker is under spatially broad repression by the miRNA pathway. Moreover, we demonstrate that the seemingly background staining detected by Elav antibodies actually reflects native accumulation in wild-type non-neural cells. Out of many miRNAs bearing conserved target sites in the elav 3′ UTR, we identify a substantial role for mir-279/mir-996 in restricting Elav expression. We also provide evidence for an auxiliary repression mechanism for elav mediated by the RBP Mei-P26. Although basal levels of ubiquitous Elav are modest, its derepression in miRNA pathway clones has functional impact on a transgenic Elav sensor. Moreover, directed misexpression of Elav outside of the nervous system, but not within the nervous system, is profoundly deleterious. Altogether, we demonstrate unexpected post-transcriptional circuitry that restricts the expression and activity of the canonical postmitotic neural RBP Elav.
RESULTS
Loss of the miRNA pathway derepresses Elav in non-neuronal territories
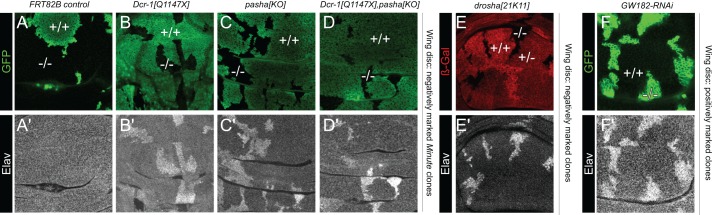
In the course of examining clonal phenotypes of miRNA pathway mutants (Smibert et al., 2011), we were surprised to observe that arbitrary imaginal disc clones expressed Elav, the canonical nuclear neuronal marker in Drosophila. This was notable because beyond the photoreceptors of the eye disc, few differentiated neurons are found in other larval imaginal discs. For example, whereas there are only a few Elav-positive neurons in the wing imaginal disc, homozygous mutant clones of core miRNA pathway factors such as drosha, pasha and Dicer-1 (Dcr-1) reliably exhibited cell-autonomous accumulation of Elav protein (Fig. S1A, Fig. 1E).
Fig. 1.
Loss of the miRNA pathway derepresses the canonical neural marker Elav in non-neural territories. Shown are regions of the wing imaginal disc pouch, co-stained for a clonal marker (A-F; GFP, green or β-Gal, red) and Elav (A′-F′; grayscale). Negatively marked clones were generated using the Minute technique in A-D or conventional technique in E. Positively marked clones using the MARCM technique are in F. Example wild-type territories are indicated by +/+ and example clones are noted by −/−; heterozygous (+/−) tissue was only generated in E. Elav protein is derepressed in mutant clones lacking diverse core miRNA pathway factors (Dcr-1, pasha or drosha), or that are depleted for the miRNA effector (GW182). At least five imaginal discs were assayed for each genotype, and each set of stainings was performed at least twice; representative clones are shown.
Although the clonal derepression we observed in multiple mutants was unequivocal, ectopic Elav accumulated to a lower level than in differentiated neurons, and was not restricted to the nucleus unlike in neurons. As miRNA pathway clones are growth disadvantaged (Herranz et al., 2010), we sought to improve their recovery using the Minute technique (Blair, 2003). Curiously, not only did we obtain larger clones, but these derepressed Elav protein more robustly than conventional clones (Fig. 1B-D). We previously showed substantial perdurance of miRNAs when depleting upstream miRNA biogenesis components (Smibert et al., 2013). Consequently, cells that are chromosomally null for miRNA biogenesis factors may retain variable amounts of miRNA functionality. We infer that the extent of Elav derepression is sensitive to the loss rate of cognate mRNA/protein products and/or existing miRNAs, which may be influenced by dilution upon cell division and/or potentially distinct turnover rates of individual miRNAs.
Post-transcriptional repression of elav might be due to miRNA-mediated silencing or, alternatively, might reflect direct mRNA cleavage by miRNA pathway nucleases (Han et al., 2009; Karginov et al., 2010; Smibert et al., 2011). We therefore examined Flp-out clones expressing a knockdown transgene against the AGO1 co-factor GW182 (Gawky – FlyBase) (Smibert et al., 2013), which specifically impairs miRNA regulatory activity. Clones expressing GW182-RNAi similarly accumulated Elav protein (Fig. 1F, Fig. S1A), indicating that miRNA activity per se restricts Elav in non-neuronal territories.
Evidence for endogenous, ubiquitous accumulation of Elav
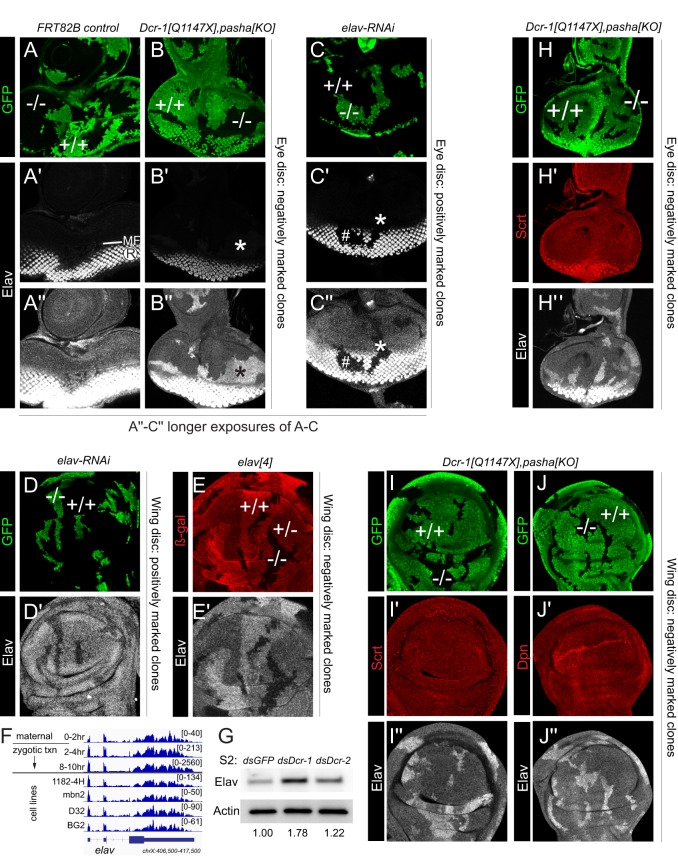
Since Elav is generally considered to accumulate in postmitotic neurons, the phenomenon described above might be interpreted at face value as reflecting ectopic Elav. However, since miRNAs operate post-transcriptionally, we considered whether Elav might be deployed more broadly than currently appreciated. In the larval eye imaginal disc, Elav is well known to accumulate in photoreceptor neurons located posterior to the morphogenetic furrow (MF). Eye discs bearing control clones showed the normal pattern of strong signal in differentiating photoreceptors posterior to the MF (Fig. 2A,A′), with longer exposure showing ubiquitous signals that are typically interpreted as background staining (Fig. 2A″). Essentially, all of the thousands of publications that utilize this standard eye marker employ imaging settings that minimize non-photoreceptor signals. Eye discs bearing Dcr-1, pasha clones showed substantial derepression of Elav anterior to the MF and in the antennal disc. This was evident even with ‘typical' exposure (Fig. 2B′) but became obvious upon increasing the gain (Fig. 2B″). We examined other larval tissues and detected Elav derepression in miRNA pathway clones in haltere and leg discs, as well as in non-imaginal tissues such as the gastric cecum (Fig. S1B), indicating spatially broad repression of Elav by the miRNA pathway.
Fig. 2.
Elav is expressed outside of the nervous system and is specifically derepressed in miRNA pathway clones. Shown are eye imaginal discs (A-C,H) and pouch regions of wing imaginal discs (D,E,I,J) stained for clonal markers (GFP, green or β-Gal, red), Elav (grayscale) and/or other pan-neural markers (red). (A-B″) Comparison of control wild-type clones (A) and Dcr-1, pasha double-mutant clones (B), all made using the Minute system. ‘Conventional' imaging technique for Elav shows typical photoreceptor (R) expression posterior to the morphogenetic furrow (MF), which is not substantially affected by miRNA pathway loss (A′,B′). Longer exposure (A″,B″) emphasizes ectopic Elav anterior to the MF and in the antennal region. (C-C″) Clonal expression of elav-RNAi eliminates Elav in R cells (hash mark), and longer exposure also shows loss of basal Elav in non-neural disc regions (asterisk). (D,D′) Clonal expression of elav-RNAi eliminates basal Elav in the wing disc. (E,E′) Mitotic clonal analysis of null allele elav[4] shows graded expression of basal, non-neural Elav in heterozygous and mutant regions. (F) RNA-seq data across an embryo timecourse show clear maternal expression and zygotic expression of elav long before the earliest presence of neurons (8-10 h). (G) Western blot of S2 cells shows that Elav is specifically derepressed upon Dcr-1 knockdown. Values beneath lanes indicate fold derepression compared with dsGFP control (H-J″) Analysis of canonical neural factors Scratch (Scrt; H,I) and Deadpan (Dpn; J) shows that they are not derepressed in miRNA pathway Minute clones in eye (H) or wing (I,J) discs. At least five imaginal discs were assayed for each genotype, and each set of stainings was performed at least twice; representative clones are shown.
We used an elav-RNAi transgene to assess whether non-neuronal Elav signals were genuine. Indeed, positively marked Flp-out Gal4 clones expressing elav-RNAi exhibited cell-autonomous depletion of Elav in both photoreceptors and non-neuronal territories of the eye-antennal disc (Fig. 2C). Similarly, we observed that wing disc clones expressing elav-RNAi eliminated endogenous Elav staining (Fig. 2D). These data suggest that the miRNA pathway clones do not reveal spatially ectopic Elav, but rather cause derepression of an unappreciated basal level of Elav present in most cells.
We confirmed these results by staining mitotic clones of the characterized null allele elav[4]. We used a negatively marked clone strategy in females (as elav resides on the X chromosome), in which homozygous mutant cells, their wild-type twinspots, and the heterozygous unrecombined tissue can all be distinguished. Remarkably, we observed distinct levels of Elav protein in territories bearing two, one or no copies of the elav locus (Fig. 2E). These tests firmly establish ubiquitous, non-neuronal accumulation of endogenous Elav protein in imaginal discs.
With this revised perspective in mind, we searched for evidence of non-neural expression of elav using modENCODE mRNA-seq (Graveley et al., 2011) and total RNA-seq (Brown et al., 2014) data. Surprisingly, we observe that elav is maternally deposited (in 0-2 h embryos), clearly elevated at the onset of zygotic expression (in 2-4 h embryos, Fig. 2F) and continues to be upregulated prior to neurogenesis (Fig. S2), which occurs at 9-10 h (Hartenstein, 1993). This provides definitive evidence for non-neuronal, even non-neuroblast (prior to 4 h), transcription of elav in the early embryo.
We also detected elav transcripts in all Drosophila cell lines profiled (Cherbas et al., 2011), most of which lack any documented neural character (Fig. 2F, Fig. S2). We detect Elav protein by western blotting in the commonly used S2 cell line (Fig. 2G), which has hemocyte character. To address whether the miRNA pathway restricts Elav in this setting, we treated S2 cells with dsRNA against GFP, Dcr-2 or Dcr-1 and observed specific upregulation of Elav protein in cells depleted of Dcr-1 (Fig. 2G).
In summary, even though elav bears a neural cis-regulatory enhancer (Yao and White, 1994) and has been used for two decades as the canonical neural antigen in Drosophila, elav is also transcribed in non-neuronal cells of diverse developmental stages and cell types. Moreover, Elav is detectably translated in non-neuronal cells and tissues, but its basal protein accumulation is restricted by the miRNA pathway.
miRNA pathway loss does not result in neural transformation or transcriptional activation of elav
Although the miRNA pathway mutant data support the scenario that one or more miRNAs repress elav, one could still hypothesize indirect mechanisms leading to elav derepression. For example, loss of the miRNA pathway might reveal a default neural program, which was reported as the ground differentiation state of the vertebrate ectoderm (Chang and Hemmati-Brivanlou, 1998; Ozair et al., 2013). Curiously, Mei-P26 is another factor with specific (although not exclusive) neural expression that is derepressed in non-neuronal miRNA pathway mutant clones (Herranz et al., 2010).
We addressed this by staining miRNA pathway clones for a panel of neural markers. For example, the transcription factors Scratch and Deadpan are long-established neural markers in CNS and PNS (Emery and Bier, 1995). We detected their endogenous expression in eye disc (Fig. 2H; data not shown) and wing disc (Fig. 2I,J), but did not observe elevated Scratch and Deadpan proteins in miRNA pathway mutant clones, as observed with parallel Elav stainings. We assayed other neural antigens (Futsch, Shep, FasII, FasIII and Orb), and none of these exhibited ectopic staining in Dcr-1 and/or pasha clones (Fig. S3A; data not shown). The specific effects on Elav argue against a general neural transformation in the loss of miRNA activity.
An alternative possibility is that loss of miRNA function leads to transcriptional derepression of elav. To address this possibility, we generated Dcr-1 clones in wing and eye-antennal imaginal discs bearing an elav-lacZ transcriptional reporter (Yao and White, 1994). We confirmed higher expression of β-Gal in neurons posterior to the MF of the eye-antennal disc, but did not observe upregulation of elav-lacZ in Dcr-1 clones (Fig. S3B). In fact, elav-lacZ activity appeared to be dampened, an effect that was more evident in wing disc clones that still maintained elevated Elav protein (Fig. S3B). We conclude that there is no transcriptional basis for the effects that we documented, and that transcriptional regulation at the locus might even antagonize the net elevation of Elav seen in cells lacking the miRNA pathway.
miRNA-mediated repression via the elav 3′ UTR can generate its spatial expression pattern
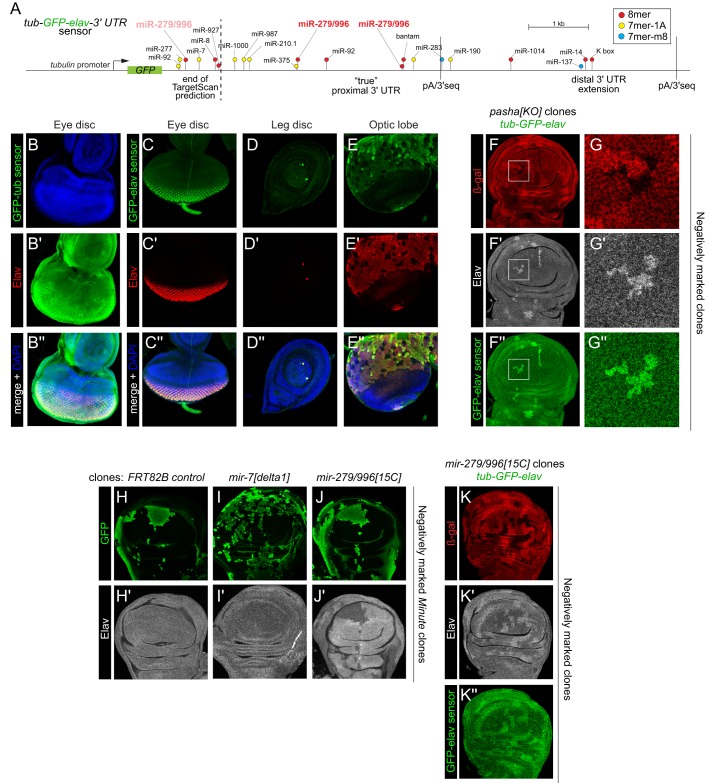
The above negative data implied that the miRNA pathway directly represses elav. To test this, we generated a transgenic tub-GFP-elav 3′ UTR (GFP-elav) sensor (Fig. 3A). We note that extensive transcriptome data (Brown et al., 2014) do not support the annotated elav 3′ UTR utilized in TargetScan predictions (www.targetscan.org) as a bona fide expressed isoform. Instead, we identified a longer proximal isoform as well as genuinely extended isoforms (Fig. 3A), consistent with previous northern blotting experiments (Smibert et al., 2012). Our GFP-elav sensor includes the full untranslated region. Interestingly, whereas control tub-GFP-tub 3′ UTR sensor lacked patterned expression (Fig. 3B), GFP-elav precisely recapitulated the endogenous Elav pattern (Fig. 3C-E). That is, GFP was low throughout imaginal discs but accumulated in Elav+ photoreceptors and neurons in the brain and ventral nerve cord. This was particularly striking in the case of specific cells that express high levels of GFP-elav in the vicinity of early arising sensory organs of leg discs. Double labeling with Elav confirmed that these were indeed neurons (Fig. 3D). Thus, post-transcriptional repression via the elav 3′ UTR is sufficient to generate the Elav spatial expression pattern.
Fig. 3.
Direct repression of the elav 3′ UTR via the miRNA pathway and miR-279/996. (A) The tub-GFP-elav 3′ UTR sensor transgene. We find that the 3′ terminus used in public miRNA annotations (e.g. TargetScan) is not detected in vivo. Rather, the genuine proximal 3′ UTR isoform is nearly 3 kb longer, and we identify an extended 3′ UTR isoform, both of which are supported by 3′-seq tags (as annotated). (B-K″) Images depict eye imaginal discs (B,C), leg imaginal disc (D), brain optic lobe (E) and pouch regions of wing imaginal discs (F-K) stained for reporter GFP (B-E,F,G,K), clonal markers (GFP, green or β-Gal, red) and Elav (grayscale). (B-E″) Comparison of tub-GFP-tubulin-3′ UTR and tub-GFP-elav-3′ UTR sensor transgenes. (B) The tub 3′ UTR sensor is broadly expressed in the eye disc, as well as other tissues (not shown). (C-E) The elav 3′ UTR restricts ubiquitously expressed GFP into the pattern of endogenous Elav protein, as seen by their colocalization in several tissues. (F-G″) Mitotic pasha[KO] clones (marked by absence of β-Gal staining) show coincident derepression of Elav and the elav 3′ UTR sensor. Boxed regions in F-F″ are magnified in G-G″. (H-J′) Tests of individual miRNA loci on post-transcriptional repression of Elav. (H) Control clones and (I) mir-7 null clones do not affect Elav, whereas clones of mir-279/996 derepress Elav protein (J). (K-K″) Clonal deletion of mir-279/996 causes concomitant elevation of Elav and the elav 3′ UTR sensor. At least five imaginal discs were assayed for each genotype, and each set of stainings was performed at least twice; representative clones are shown.
We then introduced the GFP-elav sensor into backgrounds suitable for generating negatively marked miRNA pathway clones. Cells lacking pasha (Fig. 3F,G) or drosha (not shown) exhibit concomitant, cell-autonomous derepression of both endogenous Elav and GFP-elav sensor. Thus, a major aspect of elav spatial control is mediated post-transcriptionally.
The mir-279/mir-996 cluster is an endogenous repressor of elav
As with many genes, the elav 3′ UTR contains conserved binding sites for multiple miRNAs (Fig. 3A, Fig. S4A). Among these, miR-7 and miR-8 are known to be active in imaginal discs. However, null clones of mir-7 and mir-8 did not derepress Elav (Fig. 3H,I, Fig. S5). In general, few miRNA targets have been shown to exhibit cell-autonomous derepression in miRNA mutant clones, so this is a very stringent test. These negative results do not distinguish whether potential regulation is non-existent or very mild, or requires the coordinated action of multiple miRNAs.
Cognizant of the longer elav 3′ UTR, we performed de novo assessment of potential conserved miRNA sites in elav 3′ extended regions. Notably, the ‘true' longer proximal 3′ UTR contains two deeply conserved 8mer sites for the miR-279/996/286 seed family (Fig. 3A). The 8mer site constitutes the highest affinity type of canonical site (Grimson et al., 2007), and both sites reside in locally conserved domains in the newly recognized common 3′ UTR (Fig. S4B). We also identified a less-conserved 7mer-1A site for the miR-279/996/286 family. Expression of miR-286 is restricted to the early embryo, whereas miR-279 and miR-996 are co-expressed from a genomic cluster and are detected throughout development (Mohammed et al., 2014; Sun et al., 2015). We recently characterized a double-deletion allele [15C] that specifically removes mir-279/mir-996 (Sun et al., 2015), and showed that both miRNAs contribute to a variety of neural phenotypes previously ascribed to the sole loss of miR-279 (Cayirlioglu et al., 2008; Luo and Sehgal, 2012).
We examined mir-279/996[15C] null clones and observed clearly elevated Elav in mutant cells (Fig. 3J), in contrast to the other miRNAs tested (Fig. S5). Therefore, this individual miRNA locus is a substantial mediator of non-neural repression of Elav. With this knowledge in hand, we assayed for direct regulation of the GFP-elav sensor in mir-279/996[15C] clones. Indeed, accumulation of this sensor was elevated in cells deleted for miR-279/996 (Fig. 3K), demonstrating that they are crucial effectors of the miRNA pathway for post-transcriptional suppression of ubiquitous elav.
Complementary expression of mir-279/996 and Elav in the peripheral nervous system
In some sense, our finding that miR-279/996 suppress non-neural Elav was as unexpected as our realization of ubiquitous Elav. In particular, our previous studies indicated that miR-279/996 are specifically deployed in sensory organs and regulate neural specification (Cayirlioglu et al., 2008; Sun et al., 2015), and thus did not hint at their ubiquitous expression.
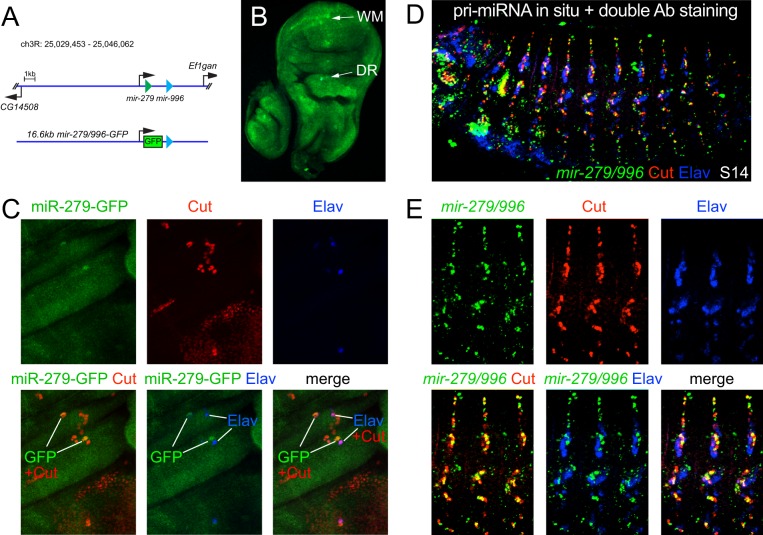
To address whether there was truly broader expression of the mir-279/996 locus outside of sensory organs, we analyzed the expression of a miR-279-GFP transcriptional reporter. We knocked GFP into a 16.6 kb mir-279/996 genomic backbone (Fig. 4A), which we showed fully rescues mir-279/996 null animals (Sun et al., 2015). This reporter generated ubiquitous GFP in imaginal discs (Fig. 4B), consistent with our finding that arbitrary clones of mir-279/996 derepress endogenous Elav. Interestingly, in very late third instars we noted elevated expression of the miR-279-GFP reporter in the vicinity of the wing margin and dorsal radius (Fig. 4B); these regions generate the first peripheral sensory organs in the wing disc during the transition to pupariation. We assessed cells expressing miR-279-GFP relative to Elav and the pan-sensory organ marker Cut. We observed that miR-279-GFP indeed accumulated in sensory organ cells that were Cut positive but Elav negative (Fig. 4C). Thus, transcription of the mir-279/996 locus generates ‘miRNA-lo' and ‘miRNA-hi' domains that are antagonistic and complementary to Elav accumulation.
Fig. 4.
Complementary expression of mir-279/996 and Elav. (A) The bicistronic mir-279/996 locus, and a 16.6 kb knock-in reporter that contains all regulatory elements for genetic rescue (Sun et al., 2015), in which GFP replaces the mir-279 hairpin. (B) Wing imaginal disc carrying the mir-279-GFP transcriptional reporter shows ubiquitous expression, as well as upregulation in presumptive neural territories: the wing margin (WM) and dorsal radius (DR). (C) Magnification of the dorsal radius region co-stained for miR-279-GFP, the pan-sensory organ marker Cut, and Elav. The cells that upregulate mir-279-GFP are Cut+ Elav− sensory organ cells. (D) Stage 14 embryo subjected to in situ hybridization for primary mir-279/996 transcripts and double labeling for Cut and Elav proteins. (E) Magnification of three lateral segments bearing PNS structures. In the embryo, Cut accumulates to a higher level in non-neural PNS cells. Endogenous mir-279/996 transcripts accumulate in Cut+ Elav− cells. At least five imaginal discs or embryos were assayed for each genotype; representative images are shown.
We extended these observations by analyzing endogenous transcription of the mir-279/996 locus. While imaginal discs were not amenable to miRNA in situ hybridization (not shown), we previously observed robust nascent transcription of mir-279/996 in the embryonic CNS and PNS (Aboobaker et al., 2005). It has been reported that mir-279 transcription does not occur in Elav+ cells in the CNS (Stark et al., 2005). However, the cell types expressing the miRNAs were not determined at that time. We studied the expression of mir-279/996 in detail in the embryonic PNS, using double in situ hybridization/antibody staining. Although this method necessitates a compromise for detection of primary miRNA transcripts, which can be challenging to detect, we were able to obtain clear PNS signals for mir-279/996 co-stained with Elav and Cut antibodies (Fig. 4D). In the embryonic PNS, Cut is expressed by all lineage cells but accumulates to a higher level in non-neuronal cell types such as the socket and shaft (Blochlinger et al., 1990). When examining segments at higher magnification to identify specific PNS clusters, we observed strong overlap of miRNA transcription in Cut+ PNS cells, but not in Elav+ neurons (Fig. 4E). We also observed some PNS accessory cells that were labeled only for mir-279/996, which, based on the known cell lineage, are likely to correspond to sheath cells.
Thus, the complementary expression of miR-279/996 and Elav applies in locations where they are both abundantly deployed (within diverse aspects of the nervous system) and basally deployed (ubiquitously throughout imaginal discs). In particular, within PNS sensory organs mir-279/996 are transcribed in non-neuronal cell types, complementary to Elav, which is upregulated in the differentiated mature neuron.
Cross-regulatory interactions of the RBPs Mei-P26 and Elav
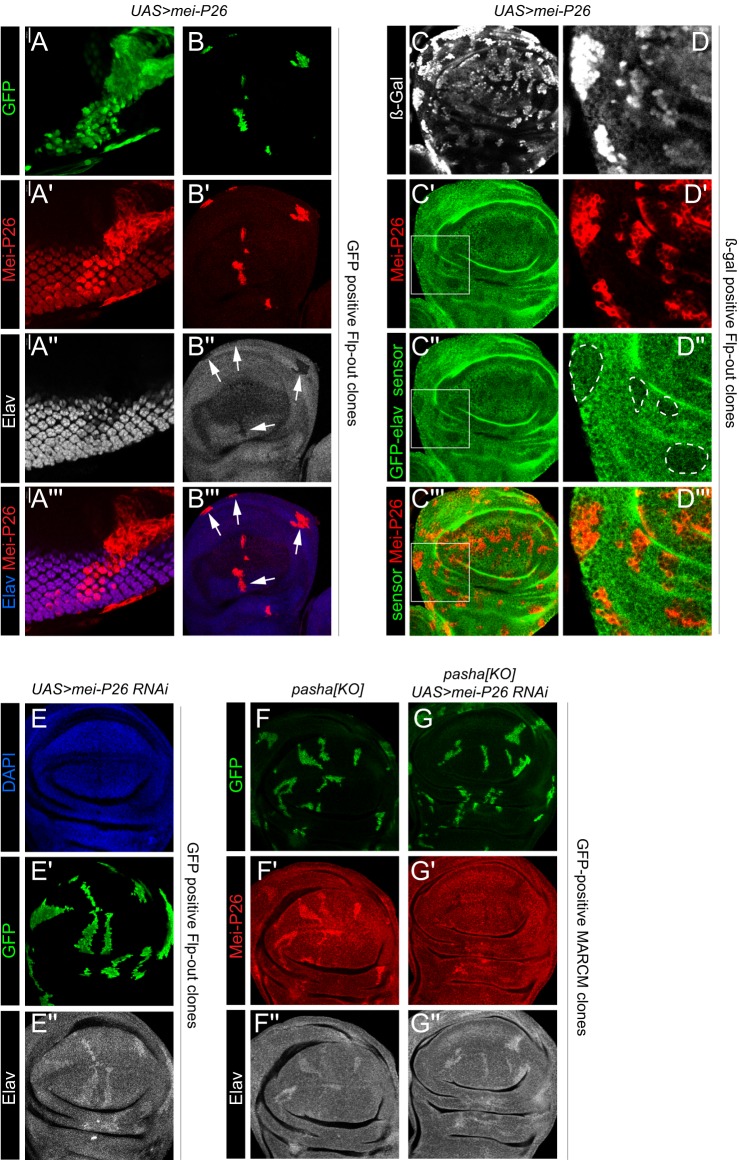
Mei-P26 is also broadly depressed in miRNA pathway disc clones (Herranz et al., 2010), and Mei-P26 was proposed to inhibit the miRNA pathway (Neumuller et al., 2008). Accordingly, ectopic Mei-P26 might be hypothesized to mimic miRNA pathway mutant clones, leading to derepression of Elav. Unexpectedly, we observed the opposite effect, in that Flp-out Gal4 clones expressing Mei-P26 showed strong loss of basal Elav. This was true in both an ‘Elav-hi' domain, such as the photoreceptor field (Fig. 5A), and ‘Elav-lo' domains, such as the wing pouch (Fig. 5B). Therefore, Mei-P26 appears to repress Elav, and this is not apparently due to an effect on the miRNA pathway in general.
Fig. 5.
Mei-P26 represses elav via its 3′ UTR. Shown are (A) eye imaginal disc and (B-G) pouch regions of wing imaginal discs. (A-B‴) Clonal activation of Mei-P26 leads to cell-autonomous decrease in Elav protein in both a high-expression domain (photoreceptors, A) and a low-expression domain (wing pouch, B, arrows). (C-D‴) Ectopic Mei-P26 represses the tub-GFP-elav-3′ UTR sensor. Although the effect is quantitatively mild, it is more clearly observed in higher magnification clones (D, outlined regions). (E) Clonal knockdown of mei-P26 causes cell-autonomous increase in basal Elav. (F-G″) MARCM analysis of pasha[KO] clones. (F) Control pasha clones derepress both Mei-P26 and Elav proteins. (G) Knockdown of mei-P26 reverses the accumulation of Mei-P26 protein in pasha clones, but does not reliably superactivate Elav. At least five imaginal discs were assayed for each genotype, and each set of stainings was performed at least twice; representative clones are shown.
Given that Mei-P26 was shown to bind RNA via its NHL domain (Loedige et al., 2014), we tested whether Mei-P26 directly regulates elav via its 3′ UTR. Indeed, clonal misexpression of Mei-P26 resulted in cell-autonomous reduction of the GFP-elav sensor (Fig. 5C). This effect was milder than observed with endogenous Elav protein, but close examination of well-positioned clones clearly showed that the GFP-elav sensor was lower in clones that misexpress Mei-P26 (Fig. 5D). Thus, Mei-P26 may contribute to post-transcriptional repression of Elav outside of the nervous system. Consistent with this, clonal knockdown of Mei-P26 results in mild upregulation of endogenous Elav (Fig. 5E).
Since ectopic Mei-P26 acts oppositely to the miRNA pathway with respect to basal Elav, and both RBPs are derepressed in miRNA pathway clones, we assessed the consequences of simultaneously removing Mei-P26 and miRNAs. We compared control pasha-KO MARCM clones with those that expressed UAS-mei-P26-RNAi. The former exhibited cell-autonomous derepression of Mei-P26 protein (Fig. 5F), whereas the latter did not (Fig. 5G), confirming efficacy of the mei-P26-RNAi transgene. The accumulation of Elav protein was sometimes higher in pasha-KO+UAS-mei-P26-RNAi than in pasha-KO clones, but they were not consistently different.
These data suggest that activity of the miRNA pathway predominates over Mei-P26 with respect to repression of basal Elav. Nevertheless, the existence of multiple strategies for post-transcriptional repression of Elav supports the notion that it is a biologically significant imperative to restrict Elav outside of neurons.
Regulatory and phenotypic impact of non-neural Elav
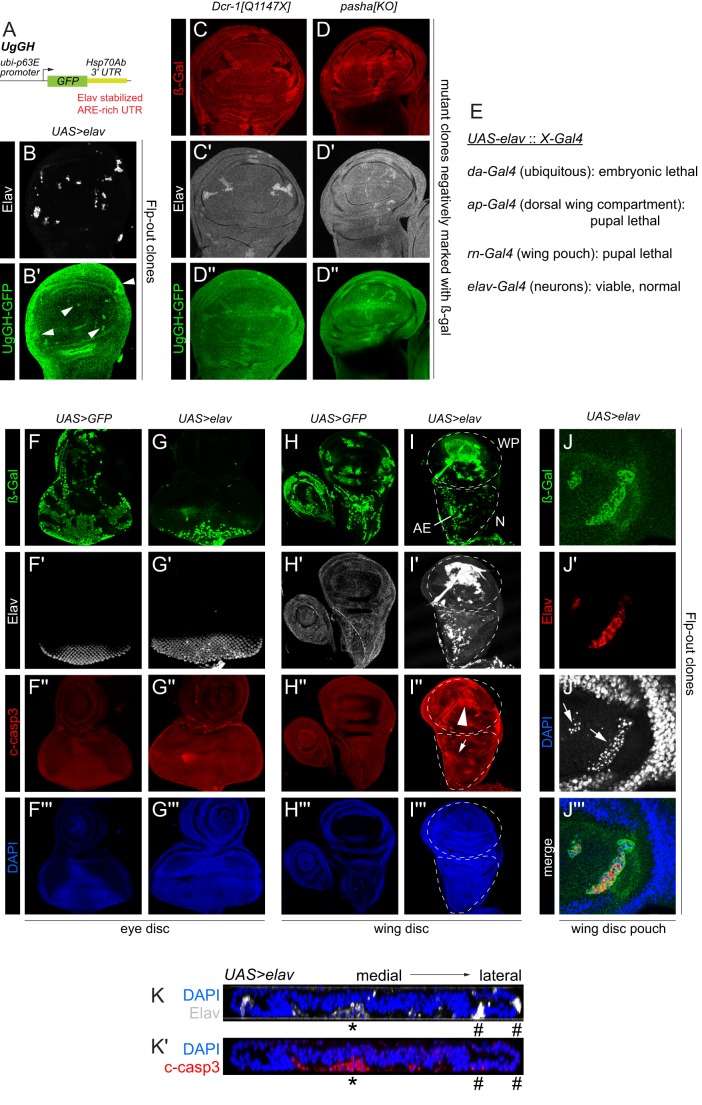
Elav is mostly considered to influence neuronal gene expression. Do the lower levels of ubiquitous Elav have detectable regulatory impact? To address this, we took advantage of a transgenic Elav activity sensor (ub-GFP-Hsp70Ab 3′ UTR or UgGH, Fig. 6A) to assess the in vivo function of Elav (Toba et al., 2002). We confirmed that Flp-out clones expressing Elav can upregulate the UgGH reporter (Fig. 6B). We then introduced UgGH into a background bearing either Dcr-1 or Pasha null mutant clones. Both types of mutant clones elevated both Elav and UgGH (Fig. 6C,D), demonstrating palpable regulatory activity of derepressed, basal Elav outside of the nervous system.
Fig. 6.
Functional consequences of elevating Elav in wing imaginal discs. (A) The UGgH Elav sensor consists of ubiquitously expressed GFP followed by the AU-rich element (ARE)-rich 3′ UTR of Hsp70Ab, a sensor previously shown to be stabilized by Elav. (B,B′) Clonal expression of UAS-elav increases UGgH expression (arrowheads). (C-D″) Dcr-1 (C) or pasha (D) mutant clones that derepress Elav also stabilize the UGgH Elav activity sensor. (E) Summary of elav misexpression tests shows that it induces lethality when activated with multiple non-neuronal drivers, but not when activated neuronally. (F-G‴) Flp-out expression clones in eye discs, marked by activation of UAS-lacZ (β-Gal, green). (F) Control GFP clones are recovered throughout the eye disc. (G) Elav-expressing clones are preferentially recovered in photoreceptors, and do not induce cell death as marked by cleaved caspase 3 (c-casp3; G″). (H-K′) Flp-out expression clones in wing discs, marked by activation of UAS-lacZ (β-Gal, green). (H) Control GFP clones are recovered throughout the wing disc and do not induce c-casp3 activity. (I) Elav-expressing clones are recovered in the wing pouch (WP) and prospective notum (N), but cells in the latter region are not in the disc epithelium but rather reside in the adepithelial layer (AE). Elav-expressing cells in the wing pouch accumulate high levels of c-casp3 (I″, arrowhead), whereas Elav-expressing adepithelial cells do not (I″, arrow). (J) High magnification of wing pouch region shows that Elav-expressing clones are fragmented. Lack of continuous DAPI signal is due to visualization of a narrow z-section, and the pyknotic nuclei (J″, arrows) delaminate from the epithelium. (K) Transverse section through the wing pouch illustrates how dying, Elav+ c-casp-3+ clones (asterisks) in the center of the wing pouch are removed from the epithelium, whereas laterally located clones remain integrated and express little or no c-casp3+ (hash marks). At least five imaginal discs were assayed for each genotype, and each set of stainings was performed at least twice; representative clones are shown.
In the course of Elav activity sensor tests (Fig. 6B), we noticed that clones of cells that overexpress Elav were much smaller than control clones or even miRNA pathway clones. This suggested that elevation of Elav in non-neural settings might not be tolerated. To investigate this further, we activated Elav with a panel of Gal4 drivers. We found that activation of Elav using da-Gal4 (ubiquitous), ap-Gal4 (dorsal compartment of wing disc) and rn-Gal4 (wing pouch) were all fully lethal (Fig. 6E). Therefore, elevation of Elav in non-neural settings is highly deleterious. By contrast, misexpression of Elav in neurons using elav-Gal4 was compatible with viability, consistent with its normally high levels of expression in this cell type.
Immunostaining of Flp-out expression clones provided cellular insight into region-specific effects of Elav. Control GFP-expressing clones were easily induced throughout the eye disc (Fig. 6F). However, inspection of eye discs bearing Elav Flp-out clones showed that labeled cells persisted robustly only within the normal ‘Elav-hi' domain, namely in the photoreceptor field (Fig. 6G). Elav-expressing clones were poorly recovered elsewhere in undifferentiated portions of the retina or antennal domain. This is consistent with the viability of elav-Gal4>UAS-elav animals, and the overall notion that Elav exerts its normal function in neurons but is poorly tolerated at high levels elsewhere.
In the wing disc, control GFP clones were again recovered throughout (Fig. 6H). By contrast, Elav-expressing clones exhibited distinct behaviors in different wing disc regions. They were poorly recovered in the prospective notum epithelium (Fig. 6I,N), although the large adepithelial cells that cover the notum could express ectopic Elav (Fig. 6I, AE). We also observed clones in the wing pouch disc proper (Fig. 6I, WP), but these exhibited poor morphology suggestive of apoptosis. Indeed, pouch clones that overexpressed Elav reacted strongly with the apoptosis marker cleaved caspase 3 (c-casp3), whereas adepithelial cells with high Elav did not react similarly (Fig. 6I″). Close examination of wing pouch clones showed fragmented, pyknotic nuclei that were in the process of being removed from the disc epithelium (Fig. 6J). This was more evident in transverse sections through the wing pouch, which showed that medially located Elav+ cells accumulated high levels of c-casp3 and delaminated (Fig. 6K, asterisk), whereas lateral Elav+ cells in the wing hinge area did not robustly activate c-casp3 and largely remained in the epithelium (Fig. 6K, hash marks). Overall, these results highlight a biological imperative to restrict accumulation of Elav outside of the nervous system, particularly within distinct disc compartments.
DISCUSSION
Unexpected expression of cell-specific or compartment-specific markers
Qualitative techniques for assessing mRNA and protein accumulation in tissues can sometimes provide impressions that are unsettlingly distinct from those of quantitative techniques. A classic example is that wholemount in situ hybridizations of Drosophila egg chambers and oocytes provide striking visual evidence for highly localized transcripts of key anterior-posterior patterning determinants. Nevertheless, quantitative analysis reveals that such signals reflect a small minority of total cellular transcripts. For example, the sharp posterior localization of nanos and oskar in situ signals represent only 4% and 18% of total oocyte transcripts, respectively (Bergsten and Gavis, 1999). Thus, although it originally appeared that mRNA localization determines protein localization, later observations demonstrated that translational control is crucial for the appropriate spatial restriction of cognate proteins.
As another example, antibodies to the Notch transcription factor Su(H) have long served to mark socket cells of peripheral sense organs (Gho et al., 1996), and its characteristic expression there is driven by an autoregulatory socket enhancer (Barolo et al., 2000). Nevertheless, as most cells can execute Notch signaling, as evidenced by profound cell-autonomous effects of activated Notch, it is implicit that Su(H) must be ubiquitously expressed. In these, as in all staining experiments, the investigator chooses when to stop a colorimetric reaction or how much to expose a fluorescent image. However, when utilizing cell-specific or subcellular-specific markers, one typically tries to minimize apparent background signals.
The case we present for Elav is particularly surprising given its broad usage as a postmitotic, neural-specific Drosophila antigen. In fact, Elav was reported to accumulate transiently in embryonic glia (Berger et al., 2007) and that it can be detected in a small fraction (∼10%) of larval neuroblasts (Lai et al., 2012). Nevertheless, these findings have not detracted from its broad use and reliable utility to mark mature neurons.
Here, we show that Elav protein is modestly but ubiquitously expressed, and is substantially derepressed in miRNA pathway mutant clones. We acknowledge that the endogenous regulatory impact of basal Elav remains to be demonstrated. For example, tissue-specific knockdown of Elav in the wing pouch did not overtly affect wing development (data not shown). Nevertheless, Elav is a powerful and multifaceted post-transcriptional regulator that orchestrates alternative splicing, 3′ end formation and alternative polyadenylation (Hilgers et al., 2012; Koushika et al., 2000; Lisbin et al., 2001; Soller and White, 2003), and it would not be surprising for basal Elav to have demonstrable effects. Indeed, we visualized that ectopic Elav generated in miRNA pathway mutant clones upregulates a transgenic Elav sensor.
Elav was previously shown to be transcriptionally repressed in neuroblasts by the intrinsic factor Worniu, and that this is required to prevent their premature neural differentiation (Lai et al., 2012). Our data indicate another unanticipated tier of Elav repression acting via miRNAs, and further suggest that the RBP Mei-P26 also contributes to post-transcriptional repression of the elav 3′ UTR. It is conceivable that other genetic situations might activate Elav in unexpected ways in non-neuronal settings. Thus, the reality of broadly transcribed and translated Elav should be taken into consideration in Drosophila studies.
miR-279/996 repress Elav within sensory organs and outside the nervous system
Amongst the many miRNAs that have captured elav within their target cohorts, the mir-279/996 locus is particularly notable. Current knowledge points to important roles of this miRNA operon in sensory organ development. This locus was one of the first to be characterized by primary transcript in situ hybridization, revealing expression in embryonic CNS and PNS (Aboobaker et al., 2005) but not in differentiated neurons (Stark et al., 2005). This was coupled to bioinformatic evidence that the miR-279/996 seed is enriched for conserved targets that are neurally expressed (Stark et al., 2005). Finally, deletion mutants of mir-279/996 reveal defects in olfactory sensory organs, causing inappropriate specification of ectopic CO2-sensing, Elav+ neurons within the maxillary palp (Cayirlioglu et al., 2008; Sun et al., 2015). The developmental basis of this phenotype remains unknown, but a plausible hypothesis based on their expression and computational patterns as ‘anti-neuronal' determinants might be that they stem from the transformation of a non-neuronal sensory lineage cell into an ectopic neuron.
These observations stem from the locations of overt expression of mir-279/996 and Elav within the nervous system, comprising ‘miRNA-hi' and ‘Elav-hi' territories, which we extend using in situ and transcriptional reporter studies. However, we unexpectedly show that their antagonistic relationships extend more broadly to ‘miRNA-lo' and ‘Elav-lo' territories, which comprise all imaginal cells and possibly include other settings. In particular, we use stringent knockout analyses to show derepression of Elav protein and GFP-elav sensor in mir-279/996 mutant clones. We do not rule out a potential contribution of other miRNAs to elav control, but miR-279/996 exert substantial regulation and constitute a notable example of a potent single miRNA locus-target interaction. Strikingly, post-transcriptional regulation is sufficient to generate the appropriate Elav spatial pattern. Indeed, a ubiquitous reporter linked to the elav 3′ UTR actually mimics Elav expression more closely than the elav transcriptional reporter, since GFP-elav is neural restricted but is responsive to the miRNA pathway and to miR-279/996 in non-neural territories.
Non-neural expression of neural Elav family members is observed in a subset of human malignancies associated with co-occurring paraneoplastic syndromes. Remarkably, characterization of abundant immunoglobulins in these patients led to the discovery of the human Elav ortholog HuD (ELAVL4). Subsequent studies revealed that HuD, like Elav in flies, is expressed specifically in mature neurons but is aberrantly expressed in cancers such as small cell lung cancer. The ectopic expression outside of the immune-privileged nervous system mounts a strong immune response that crosses the brain-blood barrier. Here, HuD+ neurons are destroyed leading to neurological paraneoplastic syndromes that are often lethal (Albert and Darnell, 2004; Darnell, 1996). The mechanism leading to ectopic HuD outside of the nervous system has long been elusive, but HuD notably bears one of the most conserved mammalian 3′ UTRs (Siepel et al., 2005). Thus, it is plausible that a post-transcriptional mechanism similar to that which we identified in flies might help restrict HuD/Elav to the nervous system.
MATERIALS AND METHODS
Drosophila genetics
To generate 3′ UTR sensor transgenes, we used recombineering to insert the entire elav or tubulin 3′ UTRs, including ∼1 kb downstream of the most distal cleavage site, downstream of a tubulin-GFP cassette. Detailed cloning procedures are provided in the supplementary Materials and Methods. The resulting attB-p[acman]GFP-3′ UTR sensors were integrated into the attP2 site (BDSC#8622) by BestGene.
We used published alleles of miRNA pathway factors on FRT backgrounds: FRT42D drosha[21K11] (Smibert et al., 2011), FRT82B Dcr-1[Q1147X] (Lee et al., 2004), FRT82B pasha[KO] (Martin et al., 2009); a recombinant FRT82B, Dcr-1, pasha chromosome was constructed herein. RNAi lines were from the Vienna Drosophila RNAi Center: UAS-GW182-RNAi (VDRC 103581), UAS-elav-RNAi (VDRC 37915) and UAS-mei-P26-RNAi (VDRC 101060). Other published mutants and transgenes included elav[4] (Campos et al., 1985), elav-lacZ (Yao and White, 1994), mir-7[delta1] (Li and Carthew, 2005), mir-8[delta1] (Shcherbata et al., 2007), mir-279/996[15C] (Sun et al., 2015), 16.6 kb mir-279/996-GFP (Sun et al., 2015), UAS-elav[2e2], UAS-elav[3e3], ubi-GFP-UgGH (Toba et al., 2002) and UAS-mei-P26 (Page et al., 2000). A detailed summary of the genotypes analyzed is provided in the supplementary Materials and Methods.
Cell culture methods
RNAi knockdowns and western blots were performed in S2 cells (DGRC) as outlined in the supplementary Materials and Methods.
Immunohistochemistry and in situ hybridization
Imaginal disc clones were induced 72 h after egg-laying (AEL) with either 60-min (mitotic or MARCM) or 15-min (Flp-out) 37°C heat shock and fixed 72 h later. Primary antibodies were chicken anti-GFP (1:1000; Abcam, ab13970), mouse anti-β-galactosidase (1:50; 40-1a, DSHB), rabbit anti-Mei-P26 (1:1000; gift of P. Lasko, McGill University, Montreal, Quebec, Canada), rat anti-Elav (1:50; 7E8A10, DSHB), mouse anti-Orb (1:50; 4H8, DSHB), rabbit anti-Dpn (1:1000; gift of Y. Jan, University of California, San Francisco, USA), rabbit anti-Scrt (1:1000; our laboratory), rabbit anti-Shep (1:50; gift of E. Lei, NIH National Institute of Diabetes and Digestive and Kidney Diseases), mouse anti-Fas2 (1:50; 1D4, DSHB), mouse anti-Fas3 (1:50; 7G10, DSHB), mouse anti-Futsch (1:100; 22C10, DSHB), mouse-anti-Cut (1:100; 5G12-D3, DSHB) and rabbit anti-cleaved caspase-3 (1:250; 9661, Cell Signaling). Secondary antibodies were made in donkey and conjugated to Alexa 488, 568 or 647 (Jackson ImmunoResearch).
For miRNA in situ/antibody double labeling, we first cloned a genomic fragment covering the mir-279 hairpin (using mir279F, GCTCTAGAagacgccgcttatcaacgct; and mir279R, CCGCTCGAGgttccggtagaaaccggagc) into XbaI/XhoI sites of pBS-SK (Stratagene). The plasmid was cut with XbaI and digoxigenin-labeled RNA probes were generated using T7 polymerase. In situ hybridization was carried out using tyramide signal amplification (TSA).
Statement of reproducibility and consistency
We utilized clonal techniques wherein wild-type tissue serves as the internal control to marked clones of interest, which are either deleted for one or more genes and/or misexpress gene products or knockdown triggers. We typically examined five or more discs for each genotype, and analyzed tissues from independent batches of crosses to ensure reproducible and consistent data.
Acknowledgements
We thank the Bloomington Stock Center, Vienna Drosophila RNAi Center (VDRC), Drosophila Genomics Resource Center (DGRC) and the Developmental Studies Hybridoma Bank (DSHB) for fly stocks, cells and antibodies.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
P.Sa., P.Sm. and E.C.L. conceived and designed this work; P.Sa., P.Sm. and H.D. collected and analyzed data; P.Sa. and E.C.L. wrote the manuscript.
Funding
Work in the E.C.L. group was supported by the National Institutes of Health [R01-NS074037, R01-NS083833 and R01-GM083300]; and a Memorial Sloan-Kettering Cancer Center Core Grant [P30-CA008748]. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.141978.supplemental
References
- Aboobaker A. A., Tomancak P., Patel N., Rubin G. M. and Lai E. C. (2005). Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. USA 102, 18017-18022. 10.1073/pnas.0508823102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert M. L. and Darnell R. B. (2004). Paraneoplastic neurological degenerations: keys to tumour immunity. Nat. Rev. Cancer 4, 36-44. 10.1038/nrc1255 [DOI] [PubMed] [Google Scholar]
- Andersson T., Rahman S., Sansom S. N., Alsiö J. M., Kaneda M., Smith J., O'Carroll D., Tarakhovsky A. and Livesey F. J. (2010). Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE 5, e13453 10.1371/journal.pone.0013453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S., Walker R., Polyanovsky A., Freschi G., Keil T. and Posakony J. W. (2000). A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell 103, 957-970. 10.1016/S0092-8674(00)00198-7 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215-233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Renner S., Lüer K. and Technau G. M. (2007). The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev. Dyn. 236, 3562-3568. 10.1002/dvdy.21372 [DOI] [PubMed] [Google Scholar]
- Bergsten S. E. and Gavis E. R. (1999). Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development 126, 659-669. [DOI] [PubMed] [Google Scholar]
- Blair S. S. (2003). Genetic mosaic techniques for studying Drosophila development. Development 130, 5065-5072. 10.1242/dev.00774 [DOI] [PubMed] [Google Scholar]
- Blochlinger K., Bodmer R., Jan L. Y. and Jan Y. N. (1990). Patterns of expression of Cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes Dev. 4, 1322-1331. 10.1101/gad.4.8.1322 [DOI] [PubMed] [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G., Stoiber M., Duff M., Booth B., Wen K., Park S., Suzuki A. et al. (2014). Diversity and dynamics of the Drosophila transcriptome. Nature 512, 393-399. 10.1038/nature12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos A. R., Grossman D. and White K. (1985). Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J. Neurogenet. 2, 197-218. 10.3109/01677068509100150 [DOI] [PubMed] [Google Scholar]
- Campos A. R., Rosen D. R., Robinow S. N. and White K. (1987). Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J. 6, 425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P., Kadow I. G., Zhan X., Okamura K., Suh G. S. B., Gunning D., Lai E. C. and Zipursky S. L. (2008). Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319, 1256-1260. 10.1126/science.1149483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M., Horvitz H. R. and Sulston J. E. (1981). Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24, 59-69. 10.1016/0092-8674(81)90501-8 [DOI] [PubMed] [Google Scholar]
- Chang C. and Hemmati-Brivanlou A. (1998). Cell fate determination in embryonic ectoderm. J. Neurobiol. 36, 128-151. [DOI] [PubMed] [Google Scholar]
- Cherbas L., Willingham A., Zhang D., Yang L., Zou Y., Eads B. D., Carlson J. W., Landolin J. M., Kapranov P., Dumais J. et al. (2011). The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 21, 301-314. 10.1101/gr.112961.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R. B. (1996). Onconeural antigens and the paraneoplastic neurologic disorders: at the intersection of cancer, immunity, and the brain. Proc. Natl. Acad. Sci. USA 93, 4529-4536. 10.1073/pnas.93.10.4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery J. F. and Bier E. (1995). Specificity of CNS and PNS regulatory subelements comprising pan-neural enhancers of the deadpan and scratch genes is achieved by repression. Development 121, 3549-3560. [DOI] [PubMed] [Google Scholar]
- Flynt A. S. and Lai E. C. (2008). Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 9, 831-842. 10.1038/nrg2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gho M., Lecourtois M., Geraud G., Posakony J. W. and Schweisguth F. (1996). Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 122, 1673-1682. [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., Yang L., Artieri C. G., van Baren M. J., Boley N., Booth B. W. et al. (2011). The developmental transcriptome of Drosophila melanogaster. Nature 471, 473-479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A., Farh K. K.-H., Johnston W. K., Garrett-Engele P., Lim L. P. and Bartel D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91-105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Pedersen J. S., Kwon S. C., Belair C. D., Kim Y.-K., Yeom K.-H., Yang W.-Y., Haussler D., Blelloch R. and Kim V. N. (2009). Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136, 75-84. 10.1016/j.cell.2008.10.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. (1993). Atlas of Drosophila Development. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Herranz H., Hong X., Pérez L., Ferreira A., Olivieri D., Cohen S. M. and Milan M. (2010). The miRNA machinery targets Mei-P26 and regulates Myc protein levels in the Drosophila wing. EMBO J. 29, 1688-1698. 10.1038/emboj.2010.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V., Lemke S. B. and Levine M. (2012). ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 26, 2259-2264. 10.1101/gad.199653.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov F. V., Cheloufi S., Chong M. M. W., Stark A., Smith A. D. and Hannon G. J. (2010). Diverse endonucleolytic cleavage sites in the mammalian transcriptome depend upon microRNAs, Drosha, and additional nucleases. Mol. Cell 38, 781-788. 10.1016/j.molcel.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y., Low R., Otaegi G., Pollock A., Deng H., Eisenhaber F., Maurer-Stroh S. and Sun T. (2010). RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell Sci. 123, 586-594. 10.1242/jcs.059659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika S. P., Soller M. and White K. (2000). The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol. Cell. Biol. 20, 1836-1845. 10.1128/MCB.20.5.1836-1845.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W. and Tuschl T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735-739. 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- Lai S.-L., Miller M. R., Robinson K. J. and Doe C. Q. (2012). The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Dev. Cell 23, 849-857. 10.1016/j.devcel.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Feinbaum R. L. and Ambros V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843-854. 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Lee Y. S., Nakahara K., Pham J. W., Kim K., He Z., Sontheimer E. J. and Carthew R. W. (2004). Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69-81. 10.1016/S0092-8674(04)00261-2 [DOI] [PubMed] [Google Scholar]
- Li X. and Carthew R. W. (2005). A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123, 1267-1277. 10.1016/j.cell.2005.10.040 [DOI] [PubMed] [Google Scholar]
- Lisbin M. J., Qiu J. and White K. (2001). The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 15, 2546-2561. 10.1101/gad.903101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loedige I., Stotz M., Qamar S., Kramer K., Hennig J., Schubert T., Loffler P., Langst G., Merkl R., Urlaub H. et al. (2014). The NHL domain of BRAT is an RNA-binding domain that directly contacts the hunchback mRNA for regulation. Genes Dev. 28, 749-764. 10.1101/gad.236513.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A. et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435, 834-838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Luo W. and Sehgal A. (2012). Regulation of circadian behavioral output via a microRNA-JAK/STAT Circuit. Cell 148, 765-779. 10.1016/j.cell.2011.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Smibert P., Yalcin A., Tyler D. M., Schaefer U., Tuschl T. and Lai E. C. (2009). A Drosophila pasha mutant distinguishes the canonical microRNA and mirtron pathways. Mol. Cell. Biol. 29, 861-870. 10.1128/MCB.01524-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed J., Siepel A. and Lai E. C. (2014). Diverse modes of evolutionary emergence and flux of conserved microRNA clusters. RNA 20, 1850-1863. 10.1261/rna.046805.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller R. A., Betschinger J., Fischer A., Bushati N., Poernbacher I., Mechtler K., Cohen S. M. and Knoblich J. A. (2008). Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454, 241-245. 10.1038/nature07014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E. M., Rebay I., Tjian R. and Rubin G. M. (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137-147. 10.1016/0092-8674(94)90580-0 [DOI] [PubMed] [Google Scholar]
- Ozair M. Z., Kintner C. and Brivanlou A. H. (2013). Neural induction and early patterning in vertebrates. Wiley Interdiscip. Rev. Dev. Biol. 2, 479-498. 10.1002/wdev.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S. L., McKim K. S., Deneen B., Van Hook T. L. and Hawley R. S. (2000). Genetic studies of mei-P26 reveal a link between the processes that control germ cell proliferation in both sexes and those that control meiotic exchange in Drosophila. Genetics 155, 1757-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S. and White K. (1991). Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443-461. 10.1002/neu.480220503 [DOI] [PubMed] [Google Scholar]
- Robinow S., Campos A. R., Yao K. M. and White K. (1988). The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science 242, 1570-1572. 10.1126/science.3144044 [DOI] [PubMed] [Google Scholar]
- Shcherbata H. R., Ward E. J., Fischer K. A., Yu J.-Y., Reynolds S. H., Chen C.-H., Xu P., Hay B. A. and Ruohola-Baker H. (2007). Stage-specific differences in the requirements for germline stem cell maintenance in the Drosophila ovary. Cell Stem Cell 1, 698-709. 10.1016/j.stem.2007.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy A. and Blelloch R. H. (2014). Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat. Rev. Mol. Cell Biol. 15, 565-576. 10.1038/nrm3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L. W., Richards S. et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034-1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Bejarano F., Wang D., Garaulet D. L., Yang J.-S., Martin R., Bortolamiol-Becet D., Robine N., Hiesinger P. R. and Lai E. C. (2011). A Drosophila genetic screen yields allelic series of core microRNA biogenesis factors and reveals post-developmental roles for microRNAs. RNA 17, 1997-2010. 10.1261/rna.2983511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Miura P., Westholm J. O., Shenker S., May G., Duff M. O., Zhang D., Eads B., Carlson J., Brown J. B. et al. (2012). Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 1, 277-289. 10.1016/j.celrep.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P., Yang J. S., Azzam G., Liu J. L. and Lai E. C. (2013). Homeostatic control of Argonaute stability by microRNA availability. Nat. Struct. Mol. Biol. 20, 789-795. 10.1038/nsmb.2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M. and White K. (2003). ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 17, 2526-2538. 10.1101/gad.1106703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R. B. and Cohen S. M. (2005). Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123, 1133-1146. 10.1016/j.cell.2005.11.023 [DOI] [PubMed] [Google Scholar]
- Sun K. and Lai E. C. (2013). Adult-specific functions of animal microRNAs. Nat. Rev. Genet. 14, 535-548. 10.1038/nrg3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Jee D., de Navas L. F., Duan H. and Lai E. C. (2015). Multiple in vivo biological processes are mediated by functionally redundant activities of Drosophila mir-279 and mir-996. PLoS Genet. 11, e1005245 10.1371/journal.pgen.1005245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toba G., Qui J., Koushika S. P. and White K. (2002). Ectopic expression of Drosophila ELAV and human HuD in Drosophila wing disc cells reveals functional distinctions and similarities. J. Cell Sci. 115, 2413-2421. [DOI] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R. and Blelloch R. (2007). DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 39, 380-385. 10.1038/ng1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K.-M. and White K. (1994). Neural specificity of elav expression: defining a Drosophila promoter for directing expression to the nervous system. J. Neurochem. 63, 41-51. 10.1046/j.1471-4159.1994.63010041.x [DOI] [PubMed] [Google Scholar]