Abstract
Olfactory epithelium (OE) has a lifelong capacity for neurogenesis due to the presence of basal stem cells. Despite the ability to generate short-term cultures, the successful in vitro expansion of purified stem cells from adult OE has not been reported. We sought to establish expansion-competent OE stem cell cultures to facilitate further study of the mechanisms and cell populations important in OE renewal. Successful cultures were prepared using adult mouse basal cells selected for expression of c-KIT. We show that c-KIT signaling regulates self-renewal capacity and prevents neurodifferentiation in culture. Inhibition of TGFβ family signaling, a known negative regulator of embryonic basal cells, is also necessary for maintenance of the proliferative, undifferentiated state in vitro. Characterizing successful cultures, we identified expression of BMI1 and other Polycomb proteins not previously identified in olfactory basal cells but known to be essential for self-renewal in other stem cell populations. Inducible fate mapping demonstrates that BMI1 is expressed in vivo by multipotent OE progenitors, validating our culture model. These findings provide mechanistic insights into the renewal and potency of olfactory stem cells.
KEY WORDS: Olfaction, Neurogenesis, Stem cells, KIT, BMI1
Summary: Adult c-KIT+ olfactory basal stem cells may be propagated as expansion-competent BMI1+ progenitors in culture and provide insight into olfactory homeostasis.
INTRODUCTION
The olfactory epithelium (OE) has served as a model system in which to study adult neurogenesis and neuroepithelial renewal (Graziadei and Graziadei, 1979). Lining portions of the nasal cavity, the OE consists of olfactory receptor neurons, sustentacular and microvillar supporting cells, as well as basal cells, and replaces mature cell types throughout life. Stem and progenitor cells reside in the basal germinal zone of the OE and have been the focus of active investigation (Caggiano et al., 1994; Calof et al., 2002; Cau et al., 1997; Fletcher et al., 2011; Goldstein and Schwob, 1996; Joiner et al., 2015; Leung et al., 2007; Schultz, 1941). However, basic questions remain regarding the identification and regulation of basal cell subsets required for ongoing OE maintenance or homeostasis.
One major limitation in the study of olfactory basal stem cell regulatory mechanisms, or in manipulating OE stem cells for other potential uses, has been the inability to efficiently expand adult OE basal cells in culture (Jang et al., 2008; Krolewski et al., 2011; Nivet et al., 2011). Existing culture models have generally involved primary or short-term studies. Short-term embryonic or postnatal OE explants have provided key insights into the regulation of migratory neural precursor basal cells (Calof and Chikaraishi, 1989; Gokoffski et al., 2011; Jang et al., 2008). Other models have utilized mesenchymal-like cells capable of ongoing renewal in culture (Murrell et al., 2005, 2008). However, mesenchymal cells do not appear to be in the OE neuronal lineage and arise from the underlying lamina propria. Careful analysis of olfactory mucosa sphere-forming cultures indicates that a mixture of sphere growth may arise from OE basal cells or mesenchyme (Tomé et al., 2009). Indeed, successful growth of purified OE basal cells as expansion-competent stem cells has not been reported. Given the complexity of the OE basal populations in vivo, which include quiescent horizontal basal cells (HBCs) and heterogenous subsets of proliferative globose basal cells (GBCs) (Schwob, 2002), we reasoned that the study of purified expansion-competent cells in culture would provide a means to examine regulatory mechanisms essential for OE self-renewal.
Although the lineage progression from a stem cell to various intermediate progenitors and, finally, to differentiated progeny is often conceptualized as a linear ʻone-way' path, there is evidence that OE basal cells may transition from stem to progenitor cell in a reversible fashion, responsive to local cues (Gokoffski et al., 2011; Schnittke et al., 2015). As in skin and intestine, there is a rate of chronic replacement; injury or other conditions influence both the rate and the cells that source the renewing epithelium (Leung et al., 2007; Ritsma et al., 2014; Solanas and Benitah, 2013). For instance, normal OE turnover is accomplished by GBCs, but severe damage activates HBCs and GBCs (Fletcher et al., 2011; Leung et al., 2007; Packard et al., 2011). Previous investigations regarding OE basal cell regulation suggest overlap with other well-characterized stem cell niches. Mechanisms informing basal cells of epithelial status or controlling cell cycle or lineage decisions are likely to include Lgr5 and Wnt signaling, TGFβ superfamily signaling, c-Kit, p63, the Notch pathway and basic helix-loop-helix (bHLH) factors (Chen et al., 2014; Fletcher et al., 2011; Gokoffski et al., 2011; Goldstein et al., 2015; Guillemot et al., 1993; Guo et al., 2010; Schnittke et al., 2015; Wang et al., 2011). Nonetheless, regulation of OE homeostasis, and strategies for correction of failures in this process leading to olfactory dysfunction, remain incompletely understood.
Accordingly, we sought to purify adult OE basal cells for culture, and also considered the importance of repressive signals in stem cell niches (Bai et al., 2007; Méndez-Ferrer et al., 2010; Solanas and Benitah, 2013; Yan et al., 2012). We reasoned that culture conditions manipulating appropriate repressive signals might prevent the exhaustion or differentiation of renewal-competent OE stem cell pools. We find here that adult OE GBCs, purified on the basis of surface expression of the c-KIT receptor, may be cultured as expansion-competent stem cells. We utilized these cultures to investigate several interacting regulatory pathways and their impact on renewal and differentiation in vitro, and report novel corresponding findings in vivo. Of interest, our culture and in vivo studies point to the previously unrecognized importance of Polycomb proteins, including BMI1, in OE maintenance.
RESULTS
Basal cell isolation
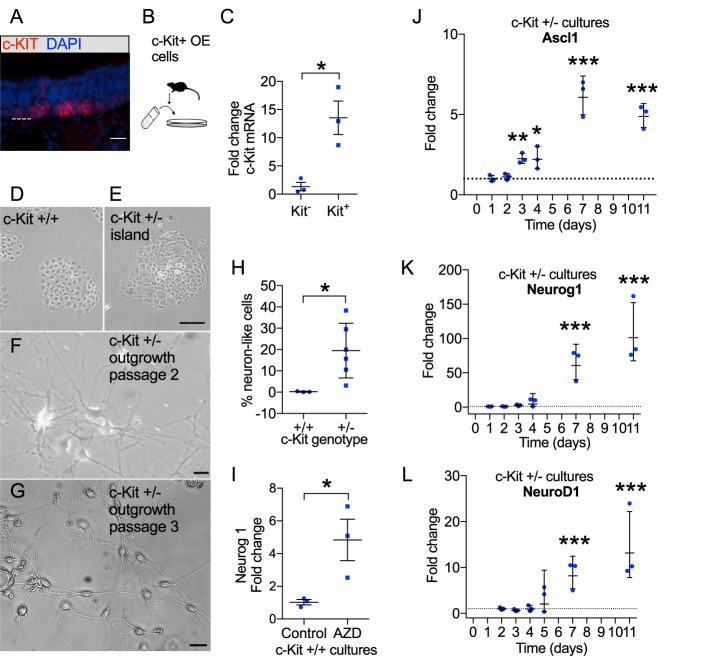
To obtain adult olfactory basal cells, we utilized the mouse methimazole lesion model (Bergman et al., 2002). Following a single intraperitoneal injection of methimazole, OE rapidly degenerates. Epithelial loss leads to proliferative expansion of the basal stem cell layers, which reconstitute the neuroepithelium over the next several weeks. We dissociated olfactory tissue from mice 8-10 days after lesion to obtain a cell suspension enriched in basal progenitor cells. We previously showed that GBCs expressing the cell surface receptor c-KIT are required for adult olfactory neurogenesis (Goldstein et al., 2015; Goss et al., 2016). In tissue sections from mice sacrificed 10 days following methimazole lesion, antibody to c-KIT labels clusters of GBCs in the basal regions of the regenerating OE (Fig. 1A). Thus, we immunomagnetically selected the GBC population from primary cell suspensions using antibodies against c-KIT (Fig. 1B). Note that c-KIT sorting-grade antibodies are validated and widely used for selection of hematopoietic stem cells based on their surface phenotype (Shizuru et al., 2005). In suspensions from regenerating OE, 5-10% of cells were recovered in the immunomagnetic selection. By contrast, the yield after selection was only ∼1% of cells in suspensions from non-lesioned adult OE preparations. As assessed by RT-qPCR, our c-KIT+ post-sort cell fraction included 13.53±2.97-fold more c-Kit mRNA than the c-KIT− fraction (±s.d.; n=3 separate preparations, three mice per preparation; P=0.017, t-test), confirming that the immunoselection technique enriches the GBC population and yields suitable starting material for culture (Fig. 1C).
Fig. 1.
Generation of olfactory basal stem cell cultures and effects of deficient c-KIT signaling. (A) Representative section of adult mouse OE 10 days following methimazole lesion, showing immunostaining of c-KIT+ basal cells. DAPI nuclear stain is blue. Dashed line indicates basal lamina. (B) The c-KIT+ basal cell populations were purified from dissociated tissue for establishing cultures. (C) RT-qPCR verification of the purification of the c-KIT+ cell fraction (n=3 preparations). *P=0.017, t-test. (D-G) Cultures were established from wild-type mice or c-Kit+/− mutants. Representative phase-contrast images 14 days after plating (D-F) or after further passage (G) show undifferentiated basal cell islands that emerge in both settings. Note that abundant neuronal-appearing outgrowth replaces many of these islands over time in c-Kit+/− cultures. (H) Quantification of neuron-like outgrowth from c-Kit+/− cultures (n=6 replicates from two culture preparations at passage 2), versus c-Kit+/+ cultures (n=3 replicates). *P=0.039, t-test. (I) Treatment of wild-type basal cell cultures with the c-KIT inhibitor AZD2932 resulted in increased Neurog1 expression within 48 h (n=3). *P=0.012, t-test. (J-L) Gene expression changes in c-Kit+/− cultures reflect the shift from undifferentiated cells toward the emergence of neurogenic cells over time (see Table 1 and Fig. 3C for additional characterization). *P<0.05, **P<0.01, ***P<0.001, t-tests. Error bars indicate geometric s.d. Scale bars: 20 μm in A; 50 μm in E; 10 μm in F,G.
Despite formation of primary spheres in standard neurosphere medium, attempts at expanding sphere-forming cultures were unsuccessful, consistent with prior reports (Jang et al., 2008). Thus, we abandoned floating cultures and allowed cultures derived from c-KIT+ cells to form adherent islands of polygonal cells (Fig. 1D), which routinely formed within 1-2 days of plating cells. Within 7-10 days, enlarging islands approached confluence. When maintained with appropriate medium supplements, cells were passaged into fresh wells successfully, as detailed below. Expansion-competent cultures, passaged approximately weekly, were characterized and used for subsequent experiments.
c-KIT signaling is necessary for ongoing self-renewal in basal cell cultures
Genetic fate mapping studies have indicated that adult c-KIT+ GBCs produce neurons in vivo during regeneration (Goldstein et al., 2015; Goss et al., 2016), although the functional role of c-KIT was not addressed. We hypothesized that c-KIT signaling might promote self-renewal of undifferentiated OE basal progenitors, analogous to its role in maintenance of the bone marrow hematopoietic niche (Ding et al., 2012) or in salivary gland morphogenesis (Matsumoto et al., 2016). Here, our culture model utilizing purified basal cells provided a means to examine c-KIT signaling in GBCs in isolation, i.e. separate from the effects of other populations such as HBCs, which can replenish the GBC population in vivo (Fletcher et al., 2011; Leung et al., 2007; Schnittke et al., 2015).
To test whether c-KIT plays an essential role in the expansion of basal cells, we established cultures from c-Kit+/+ and c-Kit+/− mice. Wild-type cultures consisted almost entirely of confluent islands of undifferentiated polygonal cells. By contrast, c-Kit+/− cultures included well-formed islands (similar to those in c-Kit+/+ cultures) as well as large, more diffuse clusters of cells with phase-bright somata and long processes, resembling neurons (Fig. 1D-G). The latter emerged 1-2 weeks after cultures were initially plated. In two independent cultures for each genotype, each prepared from separate mice, we found after 14 days of growth that 19.5±5.2% of cells in c-Kit+/− cultures (136 cells from 6 wells) possessed neuronal morphology, whereas only 0.2±0.1% of cells in wild-type cultures (35 cells from 3 wells) appeared neuronal.
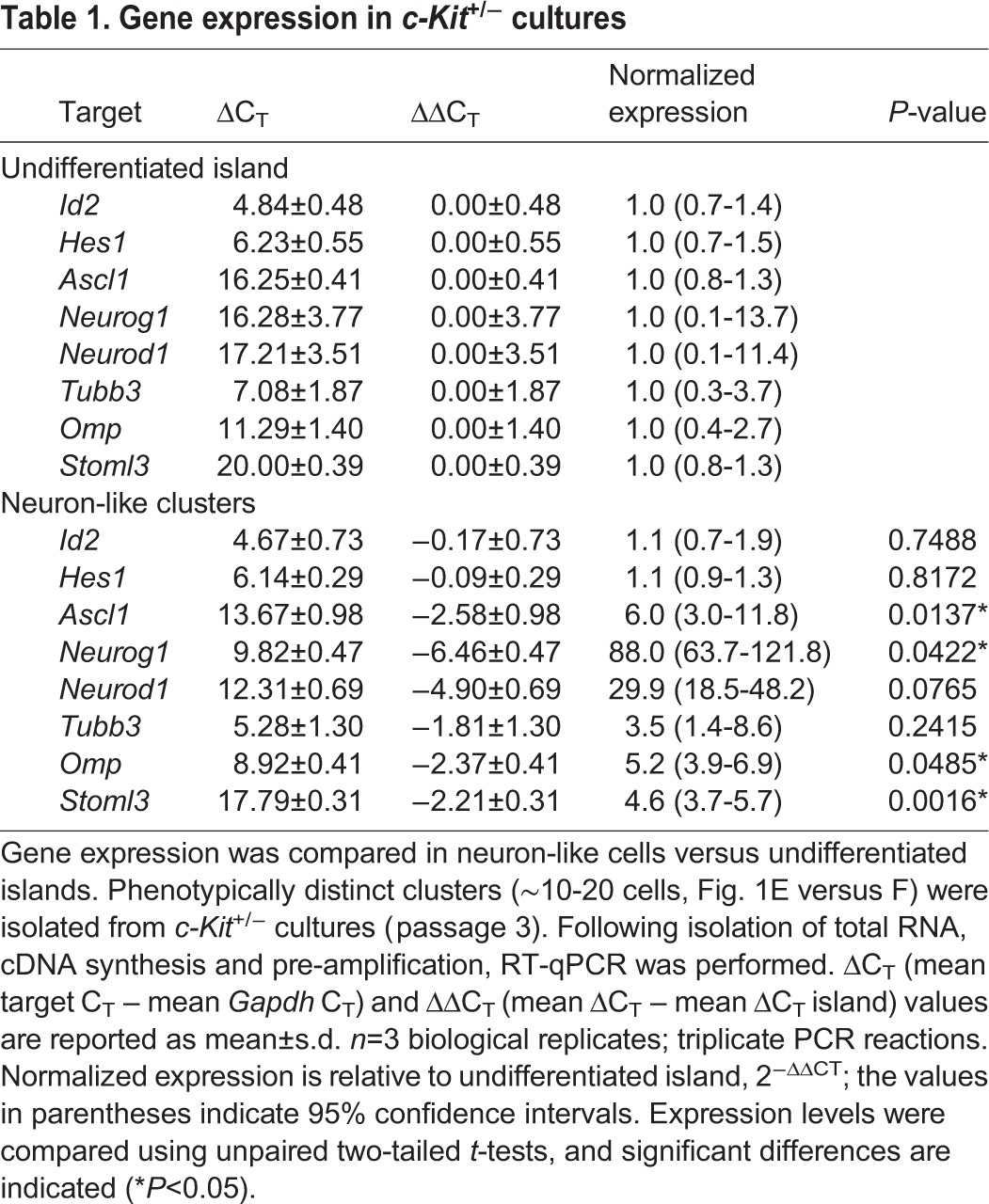
To test whether the outgrowth in c-Kit+/− cultures represented basal cells differentiating into neurons, we harvested individual islands using cloning cylinders and tested them by RT-qPCR for genes known to be expressed in olfactory sensory neurons or their precursors. Clusters that included neuron-like outgrowth expressed several genes associated with the neuronal lineage, strongly expressing Ascl1, neurogenin 1 (Neurog1) and Neurod1, as well as other olfactory neuronal markers such as Omp and Stoml3 (Goldstein et al., 2003), in contrast to undifferentiated basal cell islands (Table 1). We have found that cultures derived from c-Kit+/− mice stop producing islands after 3-4 passages and cannot be further expanded, in contrast to wild-type cultures, which we have routinely carried to >12 passages. We interpret this result to indicate that the c-Kit+/−-derived GBCs exhibit haploinsufficiency, consistent with the overall phenotype of the donor mice (Klein et al., 2013). Defects such as pigment loss, due to c-KIT expression in the melanocytic lineage, are comparable in the mice used here and in spontaneous Kit or stem cell factor [Scf; also known as kit ligand (Kitl)] mutants (Motro et al., 1991).
Table 1.
Gene expression in c-Kit+/− cultures
To further test this, we treated wild-type cultures with AZD2932 (10 nM), which inhibits c-KIT and other class III receptor tyrosine kinases. After 48 h in culture, we found that an early marker of neuronal differentiation, Neurog1 mRNA, was upregulated nearly 5-fold (Fig. 1I). Finally, we tracked gene expression changes over time in c-Kit+/− cultures by RT-qPCR, preparing RNA from whole wells after varying durations in vitro (Fig. 1J-L). With time, we found increased expression of genes marking the neuronal lineage, as compared with initial c-Kit+/− or passaged c-Kit+/+ cultures (see Fig. 3).
Fig. 3.
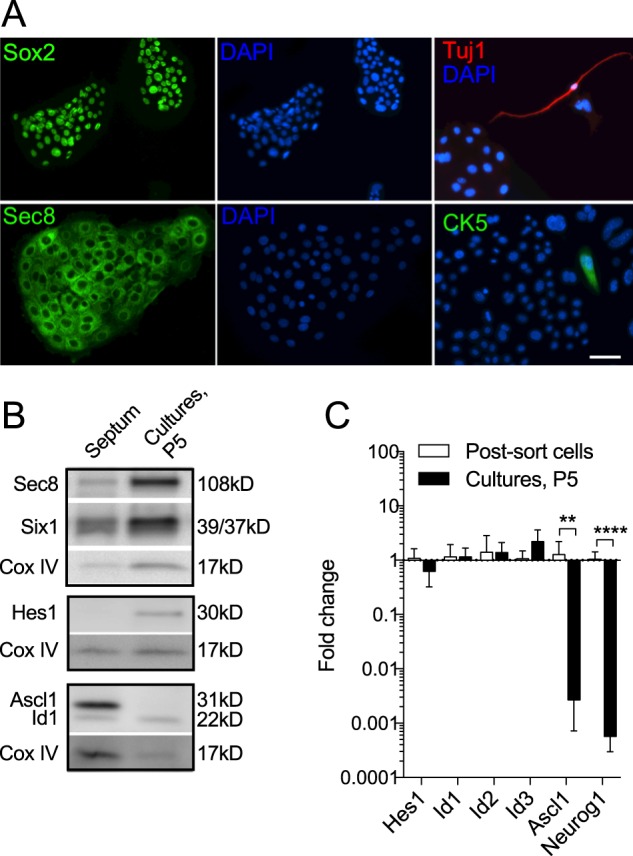
Expansion-competent culture islands express GBC markers. (A) Immunocytochemistry demonstrates that undifferentiated cell islands express SOX2 and SEC8 (green). Note the nuclear expression of SOX2, a transcription factor, and cytoplasmic localization of SEC8, a vesicle trafficking protein. DAPI nuclear counterstain (blue) of the same field is shown (middle). Adjacent to islands, rare cells with neuronal morphology and labeled by the neuronal marker Tuj1 (red) were detected. GBC islands were not labeled by Tuj1 or the HBC marker CK5; one CK5+ cell is seen here adjacent to an island. Scale bar: 25 μm. (B) By western blot, SEC8, SIX1, HES1 and ID1 proteins, as known basal cell markers, are detectable in passage 5 cultures. Tissue rich in GBCs, namely the septal mucosa prepared from mice 10 days post-methimazole lesion, was used as a positive control. ASCL1, a proneural protein repressed by HES1 and typically expressed in neural precursors, is not detected in adherent cultures, consistent with culture growth as undifferentiated basal stem cells rather than neural-committed progenitors. The mitochondrial protein COX IV was used as a loading control. (C) Gene expression changes in passage 5 (P5) cultures versus immediate c-KIT sorted OE basal cells. Expansion-competent GBCs maintain expression of Hes1 and the Id genes, whereas Ascl1 and Neurog1, which are typically expressed by neuronal-committed GBCs in vivo, are reduced. n=4 independent cultures. **P=0.004, ****P<0.0001, t-test. Error bars indicate s.d.
Taken together, we interpret these results as evidence that adequate levels of c-KIT signaling support the maintenance or renewal of undifferentiated olfactory GBCs, rather than neuropoiesis, analogous to the function of c-KIT in supporting the hematopoietic stem cell niche (Ding et al., 2012).
GBCs expand in culture when a TGFβ receptor is blocked
In our initial efforts to develop culture conditions supportive of purified GBC expansion, we postulated that negative-feedback signaling might prevent cells from remaining in an undifferentiated and self-renewing state, limiting their growth. One feedback signal previously identified in OE neurogenesis in vivo involves the TGFβ superfamily ligands GDF11 and activin B (Kawauchi et al., 2009; Wu et al., 2003), which activate the receptors Alk4 (Acvr1b) or Alk5 (Tgfβr1), signaling through Smad2/3 phosphorylation. We therefore tested an Alk5/4 inhibitor, SB431542, on our basal cell cultures. In initial screening using short-term GBC sphere culture conditions (Chen et al., 2014), treatment with SB431542 (10 μM) resulted in an increase in primary sphere generation from 28±4 to 52±9 spheres per well (Fig. 2A; ±s.e.m.; n=6 independent cultures, P=0.035, t-test).
Fig. 2.
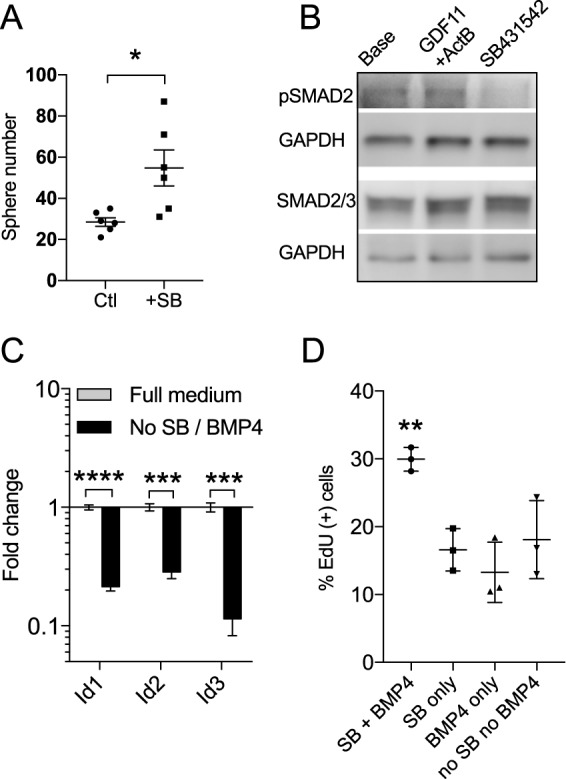
Blocking TGFβ signaling promotes culture expansion of purified GBCs. (A) Short-term primary cultures of purified c-KIT+ GBCs in sphere-forming conditions were treated with SB431542 (SB), an inhibitor of the Alk5 receptor. Alk5 mediates the TGFβ family ligands activin B and GDF11. SB431542 treatment resulted in increased primary sphere formation versus control (Ctl) medium (n=6 replicates). *P=0.035, t-test. (B) SB431542 blocks Alk5 receptor-mediated phosphorylation of Smad2/3 in GBC cultures. Western blot demonstrates a 43±2.3% (s.e.m.) decrease in Smad2/3 phosphorylation in cultures treated for 20-30 min with SB431542, as normalized to GAPDH (n=3; P<0.01, t-test). Note the presence of pSmad2/3 with GDF11/activin B treatment; total Smad2/3 remains present. (C) A 48 h withdrawal of SB431542 and BMP4, which signals via a different receptor, results in a marked decrease in Id gene expression as assayed by RT-qPCR (n=3). ***P<0.001, ****P<0.0001, t-test. A trend towards decreased Id expression was also measured upon independent withdrawal of each factor, although this was not statistically significant (not shown). (D) Proliferation is also reduced upon 48 h withdrawal of SB431542 or BMP4 (n=3). **P=0.005, ANOVA. Error bars indicate s.e.m. in A, s.d. in C,D.
As mentioned above, we found that suspension or sphere cultures grew poorly with attempted passaging, consistent with prior reports (Jang et al., 2008; Krolewski et al., 2011). Therefore, we transitioned to adherent substrates, such as vitronectin, but found cells remained difficult to expand with standard neurosphere medium (Reynolds and Weiss, 1992). However, when maintained in adherent conditions in medium with SB431542 (10 μM) to block Smad2/3 activation, along with appropriate growth factors including EGF (20 ng/ml), FGF2 (10 ng/ml) and BMP4 (10 ng/ml), purified GBC cultures grew consistently as islands that were undifferentiated in appearance (e.g. Fig. 1D). Western blot confirmed that SB431542 treatment of the GBC cultures indeed blocked Smad2/3 phosphorylation, demonstrating that this feedback pathway was associated with expansion of basal cell cultures (Fig. 2B).
Of note, BMP4 acts via a distinct BMP receptor (Alk3/6; BMPR1A/B) that is not blocked by SB431542. We included BMP4 based on its utility in other neural stem cell culture systems and its known expression in embryonic OE (Ikeda et al., 2007; Shi and Massagué, 2003), as well as its effects on promoting ʻupstream' GBCs (Shou et al., 2000). Following a 48 h removal of BMP4 and SB431542, we identified a rapid decrease in expression of the Id gene family (Fig. 2C), which is a known BMP4 target expressed in embryonic OE basal cells (Bai et al., 2007; Tietjen et al., 2003). Additionally, proliferation was reduced upon withdrawal of SB431542 or BMP4, as assessed by EdU assay (Fig. 2D). All subsequent culture work employed adherent conditions, with SB431542 and BMP4 present in standard expansion medium. To date, we have successfully expanded and carried such cultures through >12 passages, and cryopreserved and thawed cultures routinely.
Phenotype of expansion-competent GBC cultures: sustaining the expression of regulators that repress proneural transcription
The GBC population is heterogeneous in vivo (Goldstein and Schwob, 1996; Krolewski et al., 2013). Subsets of GBCs express differing levels of transcriptional regulators, likely reflecting lineage decisions or functional status as either a reserve stem cell, a transit amplifying cell, or an immediate neuronal precursor (Cau et al., 1997; Gokoffski et al., 2011; Jang et al., 2014). Our sorting technique, purifying OE c-KIT+ cells for culture starting material, enriches for a GBC population. But, how stem-like are the expanded cultures? To address this issue, we tested expanded cultures for the expression of known markers of stem and progenitor cells in OE or other systems.
We confirmed that expanded cultures of adherent islands indeed expressed GBC markers, including SOX2, a marker of multipotent GBCs (Krolewski et al., 2012), and SEC8 (EXOC4), a pan-GBC marker (Joiner et al., 2015) (Fig. 3A). Notably, the undifferentiated-appearing islands did not express neuronal markers. In wild-type cultures, the rare process-bearing cells identifiable outside of basal cell islands were immunoreactive for the neuron marker Tuj1 (Tubb3), whereas islands were not labeled (Fig. 3A). Also, the islands did not stain for cytokeratin 5 (CK5; KRT5), which is expressed by the relatively quiescent HBCs in the OE (Fig. 3A). Only rarely is a CK5+ cell identifiable in our cultures, such as the labeled cell shown in Fig. 3A adjacent to an island.
Expansion-competent cultures also expressed several other proteins typical of neural stem cells, including SIX1, Id gene products and HES1 (Fig. 3B). SIX1, a homolog of the Drosophila Sine oculis transcriptional regulator, is an early marker for cranial sensory placode progenitors during development and has been identified in embryonic OE progenitors (Moody and LaMantia, 2015; Tietjen et al., 2003) and adult OE (Rodriguez et al., 2008). We were unable to detect the proneural protein ASCL1 in culture islands (Fig. 3B). Protein from regenerating septal mucosa, which is enriched for ASCL1+ progenitors, was used for comparison. A lack of ASCL1 expression in the cultures suggests that expansion-competent GBCs remain as an ʻupstream' SOX2+/ASCL1− GBC subpopulation.
Id gene expression in expanded GBC cultures (Fig. 3B,C) is of particular interest, as Id is a dominant-negative regulator that prevents proneural gene transcriptional activity (Bai et al., 2007). Here, RT-qPCR detected Id1, Id2 and Id3 transcripts, and maintenance of strong ID1 protein expression was also identified. Consistent with our protein expression results, we found by RT-qPCR that basal cell islands express only low levels of Ascl1 or Neurog1 (Fig. 3C). Expanded GBCs retain responsiveness to Notch signaling, as evidenced by increased Neurog1 expression upon brief exposure to the gamma-secretase inhibitor DAPT (not shown). The culture phenotype identified here suggests a model in which expansion-competent GBCs express Id genes and the inhibitory bHLH factor HES1, preventing the proneural factors (ASCL1, NEUROG1) from driving cells towards neuropoiesis.
The Polycomb complex protein BMI1 is expressed in GBC cultures
Despite the known regulatory roles of Notch and the bHLH transcription factors, the master regulation of OE stem cells remains incompletely understood. The Polycomb family is an attractive candidate for OE regulation given its key roles in stem cell maintenance in other tissues, such as the intestinal crypt and bone marrow (Lopez-Arribillaga et al., 2015; Oguro et al., 2010). Accordingly, we probed our culture model for expression of the Polycomb complex protein BMI1 (Fig. 4A). By immunocytochemistry, nuclear-localized BMI1 signal was identified, of varying intensity, in 64.2±19.7% of cells (±s.d.; n=3 cultures). BMI1 protein was also readily detectable by western blot using protein extract from passaged cultures (Fig. 4B). BMI1 expression has not been reported previously in the adult OE.
Fig. 4.
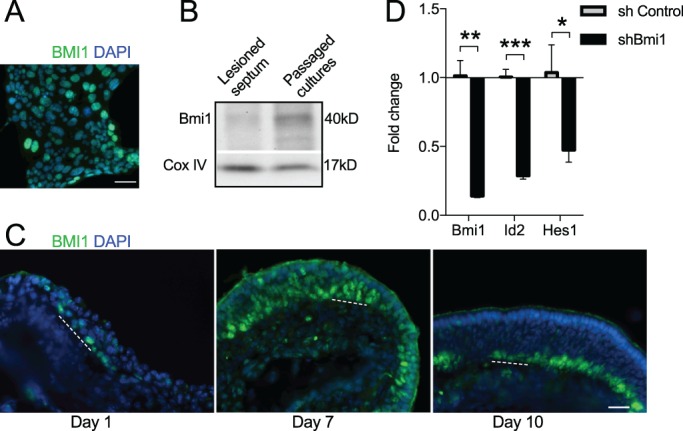
The Polycomb protein BMI1 is expressed by expansion-competent GBCs in culture and in regenerating OE in vivo. (A) Antibody to BMI1 (green) detects protein of varying intensity in GBC islands by immunocytochemistry; note the nuclear localization. (B) Western blot confirms robust BMI1 expression in cultures, with lower expression in total septal mucosal samples obtained 10 days post-methimazole lesion. (C) By immunohistochemistry, BMI1+ cells are present in vivo during lesion-induced OE reconstitution. Scattered cells are identifiable 1 day post-lesion, while much of the OE is labeled at day 7. By day 10, BMI1+ cells are present only in the basal GBC region. This BMI1 expression pattern is consistent with the derivation of our cultures from GBCs purified from 10 day post-methimazole samples. Dashed line marks the basal lamina. (D) shBmi1 lentivirus treatment of GBC cultures results in ∼80% knockdown of Bmi1 expression by 4 days versus shControl. By RT-qPCR, both Id2 and Hes1 expression are reduced, compared with shControl-treated wells, suggesting that BMI1-dependent mechanisms regulate Id/Hes levels in expansion-competent GBCs. n=3 cultures per condition. ***P=0.0003, **P=0.0015, *P=0.031, t-test. Error bars indicate s.d. Scale bars: 20 μm in A; 50 μm in C.
In vivo, BMI1+ cells replenish the adult regenerating OE
The identification of BMI1 expression in culture-expanded olfactory stem cells prompted us to assess BMI1 activity in vivo in olfactory tissue. Since our cultures were established from cells isolated from the regenerating OE of mice recovering from methimazole-induced lesion, we likewise examined olfactory mucosa from mice post-methimazole lesion (Fig. 4C). Two different anti-BMI1 antibodies, one raised against the N-terminus and the other raised against the C-terminus, produced an identical staining pattern in olfactory tissue sections. At 1 day post-methimazole, scattered BMI1+ cells were identifiable in the thin OE that remains immediately following chemical lesion. However, at 7 days, much of the regenerating epithelium is populated by BMI1+ progenitors, especially in the deeper basal cell layers. By 10 days post-methimazole, the BMI1+ cells appeared more confined to the basal layers, as the OE is reconstituted. By this stage, many newly produced differentiating neurons have emerged in the layers apical to the basal cells, and the OE pseudostratified laminar organization is re-established (Goldstein et al., 2015).
Treating GBC cultures with lentiviral vectors expressing short hairpin RNA against Bmi1 (shBmi1) resulted in a decrease in Bmi1 expression, as assessed by RT-qPCR after 4 days (Fig. 4D; n=3 independent cultures). Screening of shBmi1-treated cultures for other gene expression changes, we identified a decrease in both Id2 and Hes1, as compared with shControl-treated wells (Fig. 4D), suggesting that BMI1-dependent mechanisms might regulate Id/Hes expression levels in expansion-competent GBCs. This signaling pathway is of particular interest, given that the expression of Id genes and of Hes1 marks renewal-competent GBCs (Fig. 3) and Hes/Hey is a conserved Polycomb target (Ringrose, 2007).
Bmi1 lineage tracing in vivo
No data exist regarding Bmi1 in adult OE, so we performed inducible genetic fate mapping of Bmi1+ cells in olfactory tissue in vivo. Bmi1CreER+/+ mice were crossed to the R26RlacZ Cre-reporter strain (Sangiorgi and Capecchi, 2008). The mice were generated with a Bmi1-IRES-CreER construct and homozygotes are of normal phenotype (Sangiorgi and Capecchi, 2008). Newborn pups (n=6) were given a single dose of tamoxifen to induce reporter expression. In initial experiments, pups were euthanized after 2 days, when reporter label first becomes detectable (Fig. 5A,B). Positive control cryosections of small intestine displayed a strong signal for β-galactosidase, where Bmi1+ cells are known to underlie crypt tissue renewal (Sangiorgi and Capecchi, 2008; Yan et al., 2012). In the same mice, the OE displayed scattered basal cells stained for β-galactosidase, as well as occasional cells in the neuronal or sustentacular layers (Fig. 5B), confirming that Bmi1+ cells are active in the postnatal OE. We next examined adult methimazole-lesioned Bmi1CreER+/−;R26RlacZ mice (n=3) and identified robust reporter labeling during injury-induced OE reconstitution (Fig. 5C). In an effort to broadly capture Bmi1+ progenitor activity, mice were treated with intraperitoneal tamoxifen every other day for 2 days before methimazole lesion through day 5 post-lesion and sacrificed at day 12 post-lesion. Quantification revealed a mean of 44.7±12.4 (±s.e.m.) reporter-labeled cells per 0.5 mm of OE, examining medial turbinates II and III in sections from the middle anteroposterior portions of the nasal cavity, for consistency. β-galactosidase+ cells were found in a patchy distribution among the neuronal layers of the OE (Fig. 5C), as well as in cells of Bowman's glands and ducts (Fig. 5C, inset), consistent with Bmi1 expression by multipotent basal stem cells.
Fig. 5.
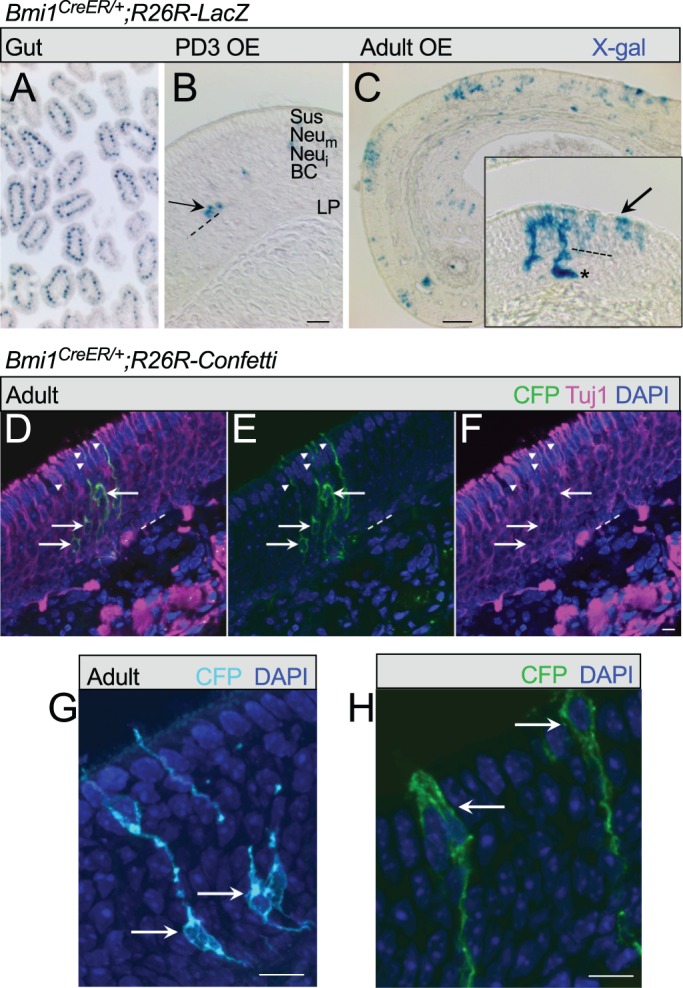
Genetic fate mapping indicates that Bmi1 is expressed by multipotent basal cells in vivo. (A) Bmi1CreER+/−;R26RlacZ mice were initially used for inducible fate mapping. As a positive control, tamoxifen was administered by intraperitoneal injection of newborn pups and tissue was harvested at day 3 (PD3). X-gal-stained sections of intestine were positive for β-galactosidase in crypts, which are known to possess Bmi1+ stem cells. (B) In nasal tissue sections from the same mice, scattered reporter-labeled cells are present in the OE at PD3. Arrow marks a pair of GBCs. OE cell layers are indicated: sustentacular/microvillar cells (Sus) are situated apically; mature neurons (Neum) are below the Sus cells; immature neurons (Neui) are deeper; and basal cells (BC) are deepest, near the basal lamina (dashed line). LP, lamina propria. (C) Adult mice were methimazole lesioned and tamoxifen treated, and sacrificed at day 12. Reporter-labeled cells are scattered along the newly reconstituted OE, confirming that Bmi1+ basal cells are active in the adult OE. Multiple cell types arise from Bmi1-expressing precursors (inset); representative field showing X-gal label in Bowman's gland (asterisk), cells in neuronal layers, and cells in the apical Sus layer (arrow). (D-F) Bmi1CreER+/−;R26R-Confetti methimazole-lesioned mice treated with a single tamoxifen dose 2 days post-lesion and sacrificed at day 13 post-lesion provided sparse reporter labeling. Membrane-tethered CFP+ cells are visualized with anti-GFP, outlining cellular morphology. Tuj1 immunolabeling (magenta) of neurons was combined with anti-GFP (green) to confirm the reporter-labeled cell phenotype. Examples of double-labeled neuronal somata are marked (arrows); dendrites are indicated with arrowheads. (G) Membrane-tethered CFP+ neurons are visualized at higher magnification. Arrows mark differentiating neurons near basal layers. (H) CFP+ cells in the sustentacular/microvillar layer (arrows) are also identifiable. Scale bars: 25 μm in A,B; 50 μm in C; 10 μm in D-H.
To better define reporter labeling, we crossed Bmi1CreER mice with the Confetti Cre-reporter strain containing the Brainbow 2.1 construct (Livet et al., 2007), and treated mice with a single tamoxifen dose 2 days post-methimazole lesion with sacrifice at day 13 (Fig. 5D-H). Sparse reporter labeling permitted identification of individual Bmi1+-derived cell clusters, and visualization of the membrane-tethered CFP reporter provided detailed cell morphology, as we have reported previously for c-Kit fate mapping (Goss et al., 2016). Reporter-labeled olfactory neurons were evident and colabeled with Tuj1 (Fig. 5D-G), confirming the neurogenic potential of Bmi1+ cells. Finally, high-magnification images of CFP+ clusters in regenerating OE identified immature neurons deep in the epithelium, just above the basal layers (Fig. 5G), and other cells situated apically in the sustentacular/microvillar layer with a morphology typical of these non-neuronal cells (Fig. 5H), including an elongated cell body and a thin basal process.
In summary, inducible fate mapping results demonstrate that Bmi1-expressing progenitors are active in vivo in the adult OE, producing neurons and non-neuronal cells during lesion-induced epithelial reconstitution.
BMI1 in the intact, unlesioned OE
We further examined BMI1 protein expression with antibodies to BMI1 and other cell type-specific markers in unlesioned olfactory tissue (Fig. 6). As expected from fate mapping experiments, a subset of GBCs was found to be BMI1+. Interestingly, although BMI1 is not detectable in the immature differentiating neuronal layers, it appears to be strongly expressed again in the fully differentiated OMP+ neuron layers. Several lines of evidence support this conclusion: we stained tissue from multiple animals, used antibodies recognizing different regions of BMI1 protein, and verified the expression pattern on tissue prepared from Bmi1GFP+/− reporter mice, in which GFP was inserted as a knock-in to replace the coding region of Bmi1 (Hosen et al., 2007). Cre recombination in existing fully mature OMP+ neurons is unlikely to explain neuronal β-galactosidase labeling in our inducible Bmi1CreER fate mapping results, since we examined lesioned tissue in which the reporter-labeled neurons were newly generated during reconstitution of the epithelium after lesion, and OMP+/BMI1+ neurons are only sparsely present by 10-12 days (Goldstein et al., 2015). Importantly, reporter-labeled neurons were evident in the deeper, immature OMP−/BMI1− layers following a single tamoxifen dose at day 2 post-methimazole in Bmi1CreER;R26R-Confetti mice, consistent with emergence from Bmi1+ GBCs.
Fig. 6.
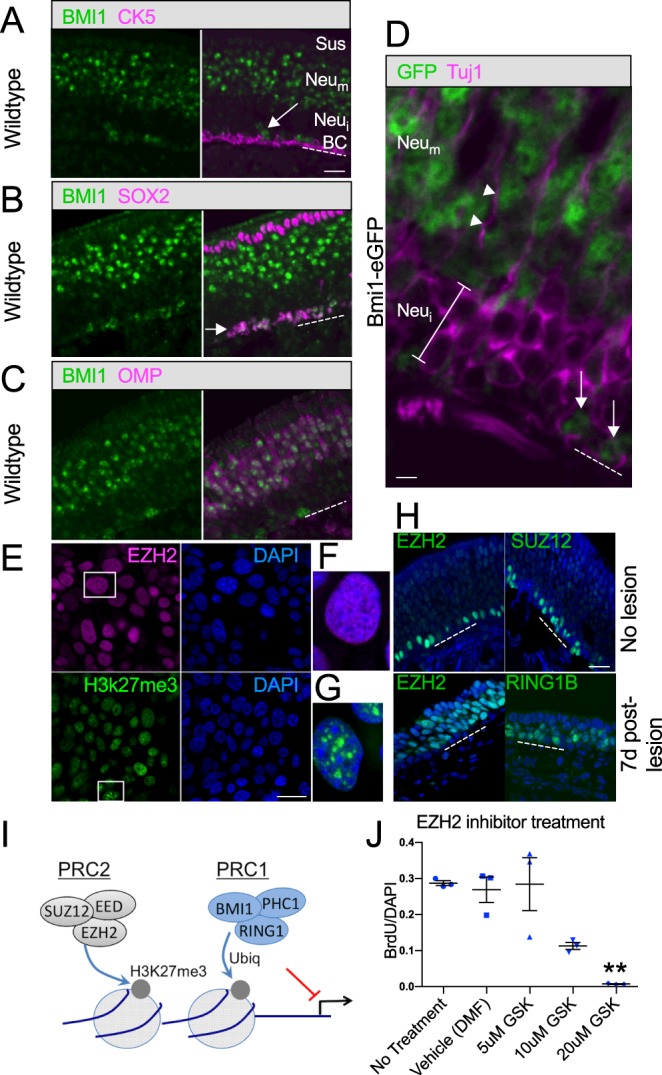
BMI1 and associated PRC proteins in vivo and in GBC cultures. An immunohistochemical panel applied to OE sections from 3-week-old wild-type mice confirms colocalization of BMI1 and markers for GBCs or mature neurons, whereas HBCs, immature neurons and sustentacular cells are BMI1−. (A) Mouse anti-BMI1-labeled basal cells (arrow) are not labeled by antibody to cytokeratin 5 (CK5) and are situated just above the CK5+ horizontal cell layer. OE cell layers are indicated as in Fig. 5. (B) SOX2 and rabbit anti-BMI1 colocalize extensively in basal cell layers (arrow); the apical SOX2+ sustentacular cell layer is not BMI1 labeled. (C) OMP+ neurons are BMI1+, whereas the immature neuron cell layers below the OMP+ neurons and above the GBCs do not express BMI1. (D) Tissue sections from Bmi1GFP knock-in mice confirm the BMI1 expression pattern. High-magnification view of the basal region of the OE. Tuj1 antibody labels immature differentiating neuron somata below the OMP+ layers (bracket). Note that the Tuj1-labeled cells do not express GFP. Arrows mark GFP+ basal cells; GFP is also present in the mature neuronal layers apical to the Tuj1+ immature cells; arrowheads mark a Tuj1+ dendrite. Dashed line marks basal lamina. (E-J) Additional PRC1/PRC2 proteins were localized to nuclei in GBC cultures (E-G) and in vivo (H). The H3K27me3 mark labels cultured GBCs (E), consistent with PRC2 transcriptional repression (see schematic I). Perturbation of Bmi1 in GBC cultures resulted in gene expression changes but no obvious phenotype at 48 h (see Fig. 4D). However, treatment with GSK343, an inhibitor of the PRC2 component EZH2, resulted in a rapid decrease in proliferation (J). **P=0.0037, ANOVA, versus vehicle (dimethyl formamide). Error bars indicate s.d. Scale bars: 25 μm A-C,E,H; 5 μm in D.
Co-staining anti-BMI1-labeled sections for other cell type-specific markers was used to confirm that the BMI1+ basal cells are GBCs (Fig. 6). We found that BMI1+ cells are generally located immediately above the CK5+ HBC layer (Fig. 6A) (Holbrook et al., 1995). However, most BMI1+ cells near the base of the epithelium were colabeled by anti-SOX2 (Fig. 6B). SOX2 is a known marker for multipotent GBCs that are thought to act upstream of the ASCL1+ transit-amplifying progenitors (Krolewski et al., 2012). Finally, nearly all OMP+ neurons are also BMI1+, whereas there is little detectable BMI1 signal in the apical SOX2+ sustentacular layer, or in the OMP− immature neuronal layers between the GBCs and mature neurons (Fig. 6C). To confirm that immature neurons are BMI1−, we stained tissue sections from Bmi1GFP+/− knock-in reporter mice (Hosen et al., 2007) with Tuj1 antibody to label immature neuronal cell bodies (Fig. 6D). Consistent with the anti-BMI1 results, the Tuj1+ neurons are in the GFP− layer, deep beneath the GFP+ mature neurons.
BMI1-associated proteins: members of the Polycomb repressive complex (PRC) in OE GBC regulation
Because the function of BMI1 in GBCs remained unclear, we examined GBC cultures and in vivo tissue for the expression of BMI1 partners, in particular other PRC proteins (Boyer et al., 2006) (Fig. 6E-H). BMI1 can function as part of PRC heteromeric complexes to repress transcription (Fig. 6I). In stem cells, PRC2 proteins, including EZH2 and SUZ12, function upstream of BMI1 to trimethylate Lys27 of histone H3 (H3K27me3); this serves to recruit PRC1, including BMI1 and RING proteins, to stabilize transcriptional repression via ubiquitylation activity (Rajasekhar and Begemann, 2007).
Here, we identified expression of the PRC2 components EZH2 and SUZ12 in our expanded GBC cultures and in OE tissue sections (Fig. 6E-H), along with the PRC1 protein RING1B (RNF2). In addition, the H3K27me3 mark was identified in culture. Of interest, EZH2 and SUZ12 expression in vivo appears confined to GBCs (Fig. 6H), in contrast to BMI1, which also localized to mature neurons. Given the expression of other PRC components, we reasoned that BMI1 is likely to act in GBCs as part of the complex. Knockdown of Bmi1 expression in cultures did produce gene expression changes (Fig. 4D), but culture phenotype was not rapidly altered. Therefore, we tested whether perturbation of the upstream partners of BMI1, specifically PRC2 components, might interfere with GBC self-renewal. Indeed, treatment of cultures with an EZH2 inhibitor, GSK343, resulted in a rapid decrease in proliferation (Fig. 6J). These results suggest that, in OE basal stem cells, Polycomb proteins are likely to function as part of PRC1 and PRC2 repressive complexes, and one role is to regulate self-renewal.
DISCUSSION
In this study, we established a novel adult OE GBC culture system, demonstrated that it is highly enriched for GBCs at an ʻearly' or undifferentiated stage, and demonstrated that pathways that regulate progression to neuronal differentiation are active in culture. Among the signaling mechanisms that appear to keep these cells in an early stage, capable of self-renewal and expansion in culture, are the c-KIT receptor and the TGFβ family (Fig. 7). Expression of repressive factors, Id genes and Hes1 mark renewing cultures. In addition, we identified Polycomb proteins, including BMI1, in expansion-competent adult GBC cultures and in the OE in vivo. In terms of understanding the regulation of olfactory neurogenesis, Polycomb proteins are of interest given their function in vertebrates in the maintenance of stem cell identity and in epigenetic regulation, as well as the multitude of BMI1 targets that have been identified in other systems (Oguro et al., 2010; Ringrose, 2007).
Fig. 7.
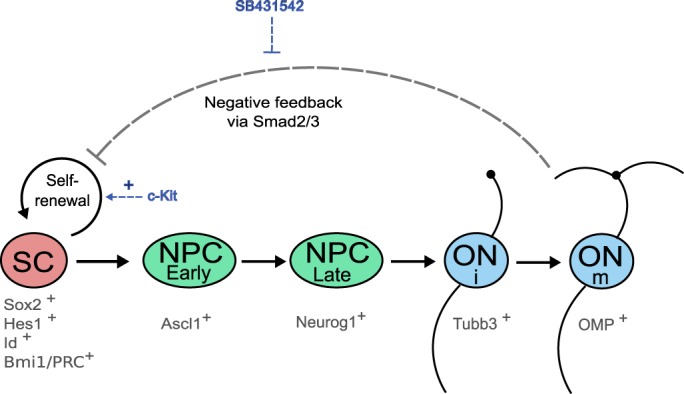
Schematic depiction of aspects of adult olfactory GBC regulation. Expansion-competent stem cells (SC) are defined by expression of SOX2, HES1, Id genes and BMI1/PRC proteins. c-KIT signaling promotes self-renewal rather than progression to progenitor cells that will commit to differentiation. PRC proteins regulate proliferation. Neural progenitor cell GBCs (NPC) express the bHLH factors ASCL1 (early) or NEUROG1 (late), while immature olfactory neurons (ONi) express neurotubulin (Tubb3) and mature neurons (ONm) express OMP. TGFβ ligands signal via Alk5 receptor-mediated Smad2/3 phosphorylation to inhibit GBC activity, and blockage of this pathway using SB431542 facilitates GBC culture expansion.
We focused on the c-KIT+ OE GBC population as starting material for cultures. Although there are multiple subpopulations of GBCs, we had shown previously, using inducible diphtheria toxin (DTA) ablation, that the c-KIT+ population is required for adult olfactory neurogenesis (Goldstein et al., 2015). c-KIT is a receptor tyrosine kinase, and its ligand SCF has been found to support the self-renewal of hematopoietic stem cells (Ding et al., 2012). However, a role for SCF-KIT signaling in OE basal cells had not been defined.
Indeed, in the OE of Ascl1 mutant mice, in which neurogenesis is severely perturbed, SCF expression was found to be upregulated, strongly suggesting that the c-KIT signaling pathway may be involved in olfactory neuron production (Guillemot et al., 1993). However, spontaneous c-Kit mutants, such as the W or Wv mouse lines, do not have an obvious olfactory phenotype, despite abnormalities in other tissues that include pigment defects and anemia (Motro et al., 1991; Orr-Urtreger et al., 1990). A possible explanation might be that c-KIT can signal via multiple intracellular pathways, and mutations such as Wv cause only loss of a specific kinase domain, with function remaining at other protein regions (Lennartsson and Ronnstrand, 2012). Alternatively, the substantial redundancy and complexity of the basal populations in the OE might provide an explanation. For instance, it is clear that reserve cells in the HBC population, which do not express c-KIT, can give rise to GBCs when necessary in order to replenish the pools of GBC stem and progenitor cells (Fletcher et al., 2011; Leung et al., 2007; Schnittke et al., 2015). Thus, in vivo, a defect in c-KIT-mediated GBC self-renewal might be masked by the replenishment of GBCs from HBCs via KIT-independent mechanisms.
Our culture model provided a means to test this idea by examining purified expansion-competent GBCs in isolation. Importantly, the GBC islands in these cultures were characterized by the expression of upstream GBC markers such as SOX2, as well as SEC8, but did not express the HBC-specific CK5. Indeed, when cultures were prepared from c-Kit mutant mice (a Cre-expressing line that is heterozygous for c-Kit expression), typical GBC islands initially arose normally, but efforts to expand and passage the cultures uniformly failed by passage 3-4, and robust basal cell islands were instead replaced by clusters of neurons. That mice with c-Kit mutation at only one kinase domain (Wv) or with c-Kit haploinsufficiency display a typical c-Kit mutant phenotype (pigment defects) indicates that even partial alteration of c-KIT signaling perturbs normal function in certain cells. We interpret these results as evidence that c-KIT signaling in GBCs promotes self-renewal rather than differentiation.
Successful growth of purified GBCs in culture provided an opportunity to identify other relevant regulatory mechanisms in expansion-competent OE stem cells. A role for the Polycomb group (PcG) genes had not been investigated in adult OE basal cells. Polycomb genes are highly conserved between Drosophila and human, and are particularly attractive candidates to regulate aspects of OE stem cell function given their regulation of gene targets involved in maintaining pluripotency, or regulating proliferation, differentiation or cell fate specification (Lopez-Arribillaga et al., 2015; Mich et al., 2014; Oguro et al., 2010; Ringrose, 2007). Here, we identified the PcG protein BMI1 in expansion-competent GBC cultures. The in vivo expression pattern of BMI1 during OE reconstitution following chemical lesion with methimazole, as well as the results from BMI1 inducible genetic fate mapping, validate our culture model, in that BMI1 is expressed in multipotent GBCs in vivo.
To further assess the role of BMI1 in GBCs, we looked at functional partners of BMI1 in PRC1 and PRC2. We showed that many of the requisite subunits, including EZH2, SUZ12 and RING1B, are expressed. Importantly, inhibiting the action of the upstream complex (PRC2) dramatically decreased cell proliferation. That is, the Polycomb protein complexes in GBCs are likely to regulate self-renewal, as in certain other stem cells. Elucidating further details of this regulation is the subject of ongoing work in our lab.
Aside from the utility of culturing purified adult GBCs for mechanistic study, the model reported here might provide a basis for broader applications. For instance, it is intriguing that a cell suspension of GBCs delivered intranasally can engraft into the damaged OE of rats (Goldstein et al., 1998) or mice (Chen et al., 2004) following experimental lesion, suggesting the potential to utilize cultured stem cells to help restore the OE for disease states marked by neuronal loss. One such condition, with the histopathological hallmark of replacement of neurons by non-neural respiratory epithelium, is presbyosmia, an age-related loss of olfactory function. The importance of presbyosmia has been detailed repeatedly in human studies (Doty et al., 1984; Murphy et al., 2002; Pinto et al., 2014). In animal models, aging-related declines in OE basal proliferation and corresponding neuronal loss are well documented but not well understood (Jia and Hegg, 2015; Loo et al., 1996; Weiler and Farbman, 1997). Nonetheless, it is clear that the presence of intact OE stem cell populations is necessary for the maintenance of olfactory function throughout adulthood (Mobley et al., 2014).
In summary, in the present work we sought to define how specific basal cell populations contribute to cellular renewal in the adult OE, a model system for understanding adult neurogenesis. Our establishment of purified expansion-competent GBC cultures from adult mice provides a novel model system that will be of use in the investigation and manipulation of olfactory basal stem cells.
MATERIALS AND METHODS
Animal strains and genetic fate mapping
All experimental procedures were approved by the University of Miami Institutional Animal Care and Use Committee, and were performed in full compliance with NIH Guidelines for the Care and Use of Laboratory Animals. Mice were obtained from Jackson Laboratory and included: C57BL/6J wild type, Bmi1CreER (stock 010531; in which an IRES followed by CreER is knocked-in downstream of the Bmi1 coding sequence), R26RlacZ Cre reporter (stock 003474; in which β-galactosidase is expressed from the ROSA26 locus after Cre-mediated recombination) and Gt(ROSA)26Sortm1(CAG-Brainbow2.1)Cle/J (stock 013731, abbreviated here as R26R-Confetti; in which a Cre-dependent multicolor fluorescent reporter is expressed from the ROSA26 locus). The Cre and reporter strains are all on a C57BL/6 background and were maintained in-house as homozygotes; mating produced Bmi1CreER+/−;R26RlacZ or Bmi1CreER+/−;R26R-Confetti reporters used for conditional fate mapping experiments. Bmi1GFP knock-in mice, expressing eGFP from the Bmi1 locus (replacing exon 2) were obtained from Jackson Laboratory (stock 017351) and maintained as heterozygotes. The c-KitCreERT2 strain (with knocked-in CreERT2 at the c-Kit locus) was obtained from Dr Dieter Saur (Klein et al., 2013); expression in olfactory progenitors was described previously (Goldstein et al., 2015).
Methimazole lesion and tamoxifen treatment are described in the supplementary Materials and Methods.
Cell culture and reagents
Olfactory tissue was obtained from adult wild-type mice or from c-KitCreERT2+/− mice. For culture, 2-4 mice were euthanized by exsanguination by perfusion with saline under ketamine-xylazine anesthesia, decapitated, and nasal septal and turbinate mucosa free of bone were harvested and pooled. Tissue was dissociated in collagenase, Dispase and DNase I in Hank's buffered salt solution (HBSS) for 15 min, followed by 3 min with 0.125% trypsin with gentle trituration. Cells were pelleted at 500 g for 5 min, washed in HBSS with 10% FBS and then passed through a 70 μm cell strainer, pelleted and resuspended in sort medium. GBCs were purified using APC-conjugated antibody to c-KIT (1:20; eBioscience 17-1171; RRID:AB_469430) for 15 min, followed by APC magnetic selection using the EasySep kit (Stem Cell Tech) as per instructions. Cells were then plated on vitronectin-coated dishes at ∼105 cells per well of 6-well plates and incubated in full growth medium: NeuroCult NSC medium, 20 ng/ml EGF, 10 ng/ml FGF2, 2 µg/ml heparin, 10 μM Y27632 (all from Stem Cell Tech), 10 μM SB431542 (Selleck Chemicals), penicillin-streptomycin (Invitrogen) and 10 ng/ml BMP4 (Peprotech). Medium was changed every other day, and cells were passaged splitting 1:3 with trypsinization/cell scraper when ∼80% confluent. Y27632 was only included when splitting cells. Cryopreservation in mFreSR medium (Stem Cell Tech) provided good viability upon thawing. Additional treatments with GSK343 or AZD2932 are detailed in the supplementary Materials and Methods.
shRNA treatment
Lentiviral shRNA plasmids targeting Bmi1 and containing a puromycin resistance gene were obtained (Sigma), along with shControl plasmid with no gene target, and packaging constructs to produce replication-deficient particles. Virus was produced in HEK293 cells following calcium phosphate transfection with the above plasmids. Three shBmi1 constructs were tested in GBC cultures and produced 50-80% knockdown of Bmi1 as assessed by RT-qPCR. We used construct #3, which produced the greatest knockdown, in subsequent experiments. Lentiviral suspension was added to triplicate GBC cultures in 6-well plates along with 4 μg/ml polybrene (Qiagen). Medium was changed after 18-20 h. Puromycin was added the following day, and total RNA was prepared from cultures at day 5.
Immunochemistry and western blotting
Antibodies, tissue preparation and microscopy details are provided in the supplemental Materials and Methods. For staining, slides were rinsed in PBS, and blocking was performed with PBS containing 5% normal donkey serum (Jackson ImmunoResearch), 4% BSA (Sigma), 5% nonfat dried milk and 0.3% Triton X-100 (Sigma) for 30-60 min, followed by primary antibody diluted in the same solution overnight at 4°C. Heat-mediated antigen retrieval was utilized if necessary, using Tris buffer pH 8.0. Slides were rinsed in PBS and incubated with either fluorescent or biotinylated secondary antibody (Jackson ImmunoResearch) for 30-45 min in the same blocking solution. For visualization of biotinylated secondary, a fluorescein tyramide signal amplification kit was used (Perkin-Elmer). Slides were then rinsed and coverslipped with Vectashield containing DAPI (Vector Labs).
For western blotting, cell cultures or nasal septal and turbinate mucosal samples were collected in cold RIPA buffer with protease inhibitor cocktail (Sigma) and homogenized briefly. Lysate was centrifuged at 14,000 g for 15 min and the supernatant stored at −20°C. Protein concentration was determined by Bradford assay, and 25 μg was loaded per lane. Samples in Laemmli buffer were heated at 100°C for 5 min, separated by 10% bis-Tris PAGE and transferred to PVDF membranes (Bio-Rad). For staining, blots were blocked in 5% nonfat dried milk in TBS with 0.1% Tween 20 and then incubated overnight with primary antibodies. Rabbit anti-GAPDH (Cell Signaling Technology, 5174; RRID:AB_10828810) or anti-COX IV (COX4; Cell Signaling Technology, 4850; RRID:AB_2085424) at 1:1000 was used as a normalization control. After incubation with HRP-conjugated secondary antibody diluted in Tris-buffered saline (TBS) with 0.1% Tween 20 and 5% BSA, binding was visualized via chemiluminescence. For pSmad2/3 assays, cells were incubated for 30 min with GDF11 and activin B (10 ng/ml each, Peprotech) or SB431542 (10 μM) prior to collection.
Quantitative PCR
TaqMan qPCR assays were performed using 20 ng cDNA per assay on a Bio-Rad cycler. For details, see the supplementary Materials and Methods and Table S1.
Statistical analysis
Samples were compared using either ANOVA or Student's t-test using GraphPad Prism 7 or Microsoft Excel. P<0.05 was considered significant. All experiments were performed at least in triplicate.
Acknowledgements
We thank Lluis Morey from the Sylvester Comprehensive Cancer Center, University of Miami, for advice and for providing plasmids. We are grateful to members of the Interdisciplinary Stem Cell Institute at the University of Miami for their input and critical comments.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
B.J.G., J.M.H. and N.C. designed research; B.J.G., R.C. and G.M.G. performed research; B.J.G., G.M.G., R.C., N.C., D.S. and B.S. analyzed data; D.S. and B.S. provided reagents; B.J.G. and N.C. wrote the manuscript.
Funding
This work was funded by a National Institutes of Health grant [K08-DC013556] to B.J.G. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.142653.supplemental
References
- Bai G., Sheng N., Xie Z., Bian W., Yokota Y., Benezra R., Kageyama R., Guillemot F. and Jing N. (2007). Id sustains Hes1 expression to inhibit precocious neurogenesis by releasing negative autoregulation of Hes1. Dev. Cell 13, 283-297. 10.1016/j.devcel.2007.05.014 [DOI] [PubMed] [Google Scholar]
- Bergman U., Ostergren A., Gustafson A. L. and Brittebo B. (2002). Differential effects of olfactory toxicants on olfactory regeneration. Arch. Toxicol. 76, 104-112. 10.1007/s00204-002-0321-2 [DOI] [PubMed] [Google Scholar]
- Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K. et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349-353. 10.1038/nature04733 [DOI] [PubMed] [Google Scholar]
- Caggiano M., Kauer J. S. and Hunter D. D. (1994). Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron 13, 339-352. 10.1016/0896-6273(94)90351-4 [DOI] [PubMed] [Google Scholar]
- Calof A. L. and Chikaraishi D. M. (1989). Analysis of neurogenesis in a mammalian neuroepithelium: proliferation and differentiation of an olfactory neuron precursor in vitro. Neuron 3, 115-127. 10.1016/0896-6273(89)90120-7 [DOI] [PubMed] [Google Scholar]
- Calof A. L., Bonnin A., Crocker C., Kawauchi S., Murray R. C., Shou J. and Wu H.-H. (2002). Progenitor cells of the olfactory receptor neuron lineage. Microsc. Res. Tech. 58, 176-188. 10.1002/jemt.10147 [DOI] [PubMed] [Google Scholar]
- Cau E., Gradwohl G., Fode C. and Guillemot F. (1997). Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development 124, 1611-1621. [DOI] [PubMed] [Google Scholar]
- Chen X., Fang H. and Schwob J. E. (2004). Multipotency of purified, transplanted globose basal cells in olfactory epithelium. J. Comp. Neurol. 469, 457-474. 10.1002/cne.11031 [DOI] [PubMed] [Google Scholar]
- Chen M., Tian S., Yang X., Lane A. P., Reed R. R. and Liu H. (2014). Wnt-responsive Lgr5(+) globose basal cells function as multipotent olfactory epithelium progenitor cells. J. Neurosci. 34, 8268-8276. 10.1523/JNEUROSCI.0240-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Saunders T. L., Enikolopov G. and Morrison S. J. (2012). Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457-462. 10.1038/nature10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doty R. L., Shaman P., Applebaum S. L., Giberson R., Siksorski L. and Rosenberg L. (1984). Smell identification ability: changes with age. Science 226, 1441-1443. 10.1126/science.6505700 [DOI] [PubMed] [Google Scholar]
- Fletcher R. B., Prasol M. S., Estrada J., Baudhuin A., Vranizan K., Choi Y. G. and Ngai J. (2011). p63 regulates olfactory stem cell self-renewal and differentiation. Neuron 72, 748-759. 10.1016/j.neuron.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokoffski K. K., Wu H.-H., Beites C. L., Kim J., Kim E. J., Matzuk M. M., Johnson J. E., Lander A. D. and Calof A. L. (2011). Activin and GDF11 collaborate in feedback control of neuroepithelial stem cell proliferation and fate. Development 138, 4131-4142. 10.1242/dev.065870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. J. and Schwob J. E. (1996). Analysis of the globose basal cell compartment in rat olfactory epithelium using GBC-1, a new monoclonal antibody against globose basal cells. J. Neurosci. 16, 4005-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. J., Fang H., Youngentob S. L. and Schwob J. E. (1998). Transplantation of multipotent progenitors from the adult olfactory epithelium. Neuroreport 9, 1611-1617. 10.1097/00001756-199805110-00065 [DOI] [PubMed] [Google Scholar]
- Goldstein B. J., Kulaga H. M. and Reed R. R. (2003). Cloning and characterization of SLP3: a novel member of the stomatin family expressed by olfactory receptor neurons. J. Assoc. Res. Otolaryngol. 4, 74-82. 10.1007/s10162-002-2039-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B. J., Goss G. M., Hatzistergos K. E., Rangel E. B., Seidler B., Saur D. and Hare J. M. (2015). Adult c-Kit(+) progenitor cells are necessary for maintenance and regeneration of olfactory neurons. J. Comp. Neurol. 523, 15-31. 10.1002/cne.23653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss G. M., Chaudhari N., Hare J. M., Nwojo R., Seidler B., Saur D. and Goldstein B. J. (2016). Differentiation potential of individual olfactory c-Kit+ progenitors determined via multicolor lineage tracing. Dev. Neurobiol. 76, 241-251. 10.1002/dneu.22310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziadei G. A. and Graziadei P. P. (1979). Neurogenesis and neuron regeneration in the olfactory system of mammals. II. Degeneration and reconstitution of the olfactory sensory neurons after axotomy. J. Neurocytol. 8, 197-213. 10.1007/BF01175561 [DOI] [PubMed] [Google Scholar]
- Guillemot F., Lo L.-C., Johnson J. E., Auerbach A., Anderson D. J. and Joyner A. L. (1993). Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell 75, 463-476. 10.1016/0092-8674(93)90381-Y [DOI] [PubMed] [Google Scholar]
- Guo Z., Packard A., Krolewski R. C., Harris M. T., Manglapus G. L. and Schwob J. E. (2010). Expression of pax6 and sox2 in adult olfactory epithelium. J. Comp. Neurol. 518, 4395-4418. 10.1002/cne.22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook E. H., Szumowski K. E. M. and Schwob J. E. (1995). An immunochemical, ultrastructural, and developmental characterization of the horizontal basal cells of rat olfactory epithelium. J. Comp. Neurol. 363, 129-146. 10.1002/cne.903630111 [DOI] [PubMed] [Google Scholar]
- Hosen N., Yamane T., Muijtjens M., Pham K., Clarke M. F. and Weissman I. L. (2007). Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells 25, 1635-1644. 10.1634/stemcells.2006-0229 [DOI] [PubMed] [Google Scholar]
- Ikeda K., Ookawara S., Sato S., Ando Z., Kageyama R. and Kawakami K. (2007). Six1 is essential for early neurogenesis in the development of olfactory epithelium. Dev. Biol. 311, 53-68. 10.1016/j.ydbio.2007.08.020 [DOI] [PubMed] [Google Scholar]
- Jang W., Lambropoulos J., Woo J. K., Peluso C. E. and Schwob J. E. (2008). Maintaining epitheliopoietic potency when culturing olfactory progenitors. Exp. Neurol. 214, 25-36. 10.1016/j.expneurol.2008.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W., Chen X., Flis D., Harris M. and Schwob J. E. (2014). Label-retaining, quiescent globose basal cells are found in the olfactory epithelium. J. Comp. Neurol. 522, 731-749. 10.1002/cne.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia C. and Hegg C. C. (2015). Effect of IP3R3 and NPY on age-related declines in olfactory stem cell proliferation. Neurobiol. Aging 36, 1045-1056. 10.1016/j.neurobiolaging.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner A. M., Green W. W., McIntyre J. C., Allen B. L., Schwob J. E. and Martens J. R. (2015). Primary cilia on horizontal basal cells regulate regeneration of the olfactory epithelium. J. Neurosci. 35, 13761-13772. 10.1523/JNEUROSCI.1708-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawauchi S., Kim J., Santos R., Wu H.-H., Lander A. D. and Calof A. L. (2009). Foxg1 promotes olfactory neurogenesis by antagonizing Gdf11. Development 136, 1453-1464. 10.1242/dev.034967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Seidler B., Kettenberger A., Sibaev A., Rohn M., Feil R., Allescher H.-D., Vanderwinden J.-M., Hofmann F., Schemann M. et al. (2013). Interstitial cells of Cajal integrate excitatory and inhibitory neurotransmission with intestinal slow-wave activity. Nat. Commun. 4, 1630 10.1038/ncomms2626 [DOI] [PubMed] [Google Scholar]
- Krolewski R. C., Jang W. and Schwob J. E. (2011). The generation of olfactory epithelial neurospheres in vitro predicts engraftment capacity following transplantation in vivo. Exp. Neurol. 229, 308-323. 10.1016/j.expneurol.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski R. C., Packard A., Jang W., Wildner H. and Schwob J. E. (2012). Ascl1 (Mash1) knockout perturbs differentiation of nonneuronal cells in olfactory epithelium. PLoS ONE 7, e51737 10.1371/journal.pone.0051737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krolewski R. C., Packard A. and Schwob J. E. (2013). Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. J. Comp. Neurol. 521, 833-859. 10.1002/cne.23204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennartsson J. and Ronnstrand L. (2012). Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol. Rev. 92, 1619-1649. 10.1152/physrev.00046.2011 [DOI] [PubMed] [Google Scholar]
- Leung C. T., Coulombe P. A. and Reed R. R. (2007). Contribution of olfactory neural stem cells to tissue maintenance and regeneration. Nat. Neurosci. 10, 720-726. 10.1038/nn1882 [DOI] [PubMed] [Google Scholar]
- Livet J., Weissman T. A., Kang H., Draft R. W., Lu J., Bennis R. A., Sanes J. R. and Lichtman J. W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56-62. 10.1038/nature06293 [DOI] [PubMed] [Google Scholar]
- Loo A. T., Youngentob S. L., Kent P. F. and Schwob J. E. (1996). The aging olfactory epithelium: neurogenesis, response to damage, and odorant-induced activity. Int. J. Dev. Neurosci. 14, 881-900. 10.1016/S0736-5748(96)00046-9 [DOI] [PubMed] [Google Scholar]
- Lopez-Arribillaga E., Rodilla V., Pellegrinet L., Guiu J., Iglesias M., Roman A. C., Gutarra S., Gonzalez S., Munoz-Canoves P., Fernandez-Salguero P. et al. (2015). Bmi1 regulates murine intestinal stem cell proliferation and self-renewal downstream of Notch. Development 142, 41-50. 10.1242/dev.107714 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Kurimoto T., Taketo M. M., Fujii S. and Kikuchi A. (2016). The Wnt-Myb pathway suppresses KIT expression to control the timing of salivary proacinar differentiation and duct formation. Development 143, 2311-2324. 10.1242/dev.134486 [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T. V., Ferraro F., Mazloom A. R., MacArthur B. D., Lira S. A., Scadden D. T., Ma'ayan A., Enikolopov G. N. and Frenette P. S. (2010). Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829-834. 10.1038/nature09262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mich J. K., Signer R. A., Nakada D., Pineda A., Burgess R. J., Vue T. Y., Johnson J. E. and Morrison S. J. (2014). Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife 3, e02669 10.7554/elife.02669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley A. S., Rodriguez-Gil D. J., Imamura F. and Greer C. A. (2014). Aging in the olfactory system. Trends Neurosci. 37, 77-84. 10.1016/j.tins.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody S. A. and LaMantia A.-S. (2015). Transcriptional regulation of cranial sensory placode development. Curr. Top. Dev. Biol. 111, 301-350. 10.1016/bs.ctdb.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motro B., van der Kooy D., Rossant J., Reith A. and Bernstein A. (1991). Contiguous patterns of c-kit and steel expression: analysis of mutations at the W and Sl loci. Development 113, 1207-1221. [DOI] [PubMed] [Google Scholar]
- Murphy C., Schubert C. R., Cruickshanks K. J., Klein B. K., Klein R. and Nondahl D. M. (2002). Prevalence of olfactory impairment in older adults. JAMA 288, 2307-2312. 10.1001/jama.288.18.2307 [DOI] [PubMed] [Google Scholar]
- Murrell W., Féron F., Wetzig A., Cameron N., Splatt K., Bellette B., Bianco J., Perry C., Lee G. and Mackay-Sim A. (2005). Multipotent stem cells from adult olfactory mucosa. Dev. Dyn. 233, 496-515. 10.1002/dvdy.20360 [DOI] [PubMed] [Google Scholar]
- Murrell W., Wetzig A., Donnellan M., Féron F., Burne T., Meedeniya A., Kesby J., Bianco J., Perry C., Silburn P. et al. (2008). Olfactory mucosa is a potential source for autologous stem cell therapy for Parkinson's disease. Stem Cells 26, 2183-2192. 10.1634/stemcells.2008-0074 [DOI] [PubMed] [Google Scholar]
- Nivet E., Vignes M., Girard S. D., Pierrisnard C., Baril N., Devèze A., Magnan J., Lanté F., Khrestchatisky M., Féron F. et al. (2011). Engraftment of human nasal olfactory stem cells restores neuroplasticity in mice with hippocampal lesions. J. Clin. Invest. 121, 2808-2820. 10.1172/JCI44489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H., Yuan J., Ichikawa H., Ikawa T., Yamazaki S., Kawamoto H., Nakauchi H. and Iwama A. (2010). Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 6, 279-286. 10.1016/j.stem.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A., Avivi A., Zimmer Y., Givol D., Yarden Y. and Lonai P. (1990). Developmental expression of c-kit, a proto-oncogene encoded by the W locus. Development 109, 911-923. [DOI] [PubMed] [Google Scholar]
- Packard A., Schnittke N., Romano R.-A., Sinha S. and Schwob J. E. (2011). DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J. Neurosci. 31, 8748-8759. 10.1523/JNEUROSCI.0681-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto J. M., Wroblewski K. E., Kern D. W., Schumm L. P. and McClintock M. K. (2014). Olfactory dysfunction predicts 5-year mortality in older adults. PLoS ONE 9, e107541 10.1371/journal.pone.0107541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar V. K. and Begemann M. (2007). Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells 25, 2498-2510. 10.1634/stemcells.2006-0608 [DOI] [PubMed] [Google Scholar]
- Reynolds B. A. and Weiss S. (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707-1710. 10.1126/science.1553558 [DOI] [PubMed] [Google Scholar]
- Ringrose L. (2007). Polycomb comes of age: genome-wide profiling of target sites. Curr. Opin. Cell Biol. 19, 290-297. 10.1016/j.ceb.2007.04.010 [DOI] [PubMed] [Google Scholar]
- Ritsma L., Ellenbroek S. I. J., Zomer A., Snippert H. J., de Sauvage F. J., Simons B. D., Clevers H. and van Rheenen J. (2014). Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507, 362-365. 10.1038/nature12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez S., Sickles H. M., DeLeonardis C., Alcaraz A., Gridley T. and Lin D. M. (2008). Notch2 is required for maintaining sustentacular cell function in the adult mouse main olfactory epithelium. Dev. Biol. 314, 40-58. 10.1016/j.ydbio.2007.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E. and Capecchi M. R. (2008). Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40, 915-920. 10.1038/ng.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittke N., Herrick D. B., Lin B., Peterson J., Coleman J. H., Packard A. I., Jang W. and Schwob J. E. (2015). Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc. Natl. Acad. Sci. USA 112, E5068-E5077. 10.1073/pnas.1512272112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E. W. (1941). Regeneration of olfactory cells. Proc. Soc. Exp. Biol. Med. 46, 41-43. 10.3181/00379727-46-11882P [DOI] [Google Scholar]
- Schwob J. E. (2002). Neural regeneration and the peripheral olfactory system. Anat. Rec. 269, 33-49. 10.1002/ar.10047 [DOI] [PubMed] [Google Scholar]
- Shi Y. and Massagué J. (2003). Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685-700. 10.1016/S0092-8674(03)00432-X [DOI] [PubMed] [Google Scholar]
- Shizuru J. A., Negrin R. S. and Weissman I. L. (2005). Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annu. Rev. Med. 56, 509-538. 10.1146/annurev.med.54.101601.152334 [DOI] [PubMed] [Google Scholar]
- Shou J., Murray R. C., Rim P. C. and Calof A. L. (2000). Opposing effects of bone morphogenetic proteins on neuron production and survival in the olfactory receptor neuron lineage. Development 127, 5403-5413. [DOI] [PubMed] [Google Scholar]
- Solanas G. and Benitah S. A. (2013). Regenerating the skin: a task for the heterogeneous stem cell pool and surrounding niche. Nat. Rev. Mol. Cell Biol. 14, 737-748. 10.1038/nrm3675 [DOI] [PubMed] [Google Scholar]
- Tietjen I., Rihel J. M., Cao Y., Koentges G., Zakhary L. and Dulac C. (2003). Single-cell transcriptional analysis of neuronal progenitors. Neuron 38, 161-175. 10.1016/S0896-6273(03)00229-0 [DOI] [PubMed] [Google Scholar]
- Tomé M., Lindsay S. L., Riddell J. S. and Barnett S. C. (2009). Identification of nonepithelial multipotent cells in the embryonic olfactory mucosa. Stem Cells 27, 2196-2208. 10.1002/stem.130 [DOI] [PubMed] [Google Scholar]
- Wang Y.-Z., Yamagami T., Gan Q., Wang Y., Zhao T., Hamad S., Lott P., Schnittke N., Schwob J. E. and Zhou C. J. (2011). Canonical Wnt signaling promotes the proliferation and neurogenesis of peripheral olfactory stem cells during postnatal development and adult regeneration. J. Cell Sci. 124, 1553-1563. 10.1242/jcs.080580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler E. and Farbman A. I. (1997). Proliferation in the rat olfactory epithelium: age-dependent changes. J. Neurosci. 17, 3610-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-H., Ivkovic S., Murray R. C., Jaramillo S., Lyons K. M., Johnson J. E. and Calof A. L. (2003). Autoregulation of neurogenesis by GDF11. Neuron 37, 197-207. 10.1016/S0896-6273(02)01172-8 [DOI] [PubMed] [Google Scholar]
- Yan K. S., Chia L. A., Li X., Ootani A., Su J., Lee J. Y., Su N., Luo Y., Heilshorn S. C., Amieva M. R. et al. (2012). The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA 109, 466-471. 10.1073/pnas.1118857109 [DOI] [PMC free article] [PubMed] [Google Scholar]