Abstract
Objective
The American College of Rheumatology guidelines for the treatment of lupus nephritis recommend change in induction therapy when response to therapy has not occurred within 6 months. Response is not defined, and renal fibrosis can occur while waiting for this end point. Therefore, a decision support tool to better define response is needed to guide clinicians when starting patients on therapy. This study was undertaken to identify biomarker models with sufficient predictive power to develop such a tool.
Methods
Urine samples from 140 patients with biopsy-proven lupus nephritis who had not yet started induction therapy were analyzed for a panel of urinary biomarkers. Univariate receiver operating characteristic (ROC) curves were generated for each individual biomarker and compared to the ROC area under the curve values from machine learning models developed using random forest algorithms. Biomarker models of outcome developed with novel markers in addition to clinical markers were compared to those developed with traditional clinical markers alone.
Results
Models developed with the combined traditional and novel biomarker panels demonstrated clinically meaningful predictive power. Markers most predictive of response were chemokines, cytokines, and markers of cellular damage.
Conclusion
This is the first study to demonstrate the power of low-abundance biomarker panels and machine learning algorithms for predicting lupus nephritis outcomes. This is a critical first step in research to develop clinically meaningful decision support tools.
Lupus nephritis is an immune complex–mediated glomerulonephritis that affects approximately half of all patients with systemic lupus erythematosus (SLE) (1). Currently, renal failure occurs in up to 50% of patients at 5 years (2). Clinicians use blood pressure, serum complement levels, anti–double-stranded DNA (anti-dsDNA) antibody levels, urinary sediment, urinary protein-to-creatinine ratio, and surrogates of renal function to monitor response to therapy in lupus nephritis. The American College of Rheumatology (ACR) guidelines for the treatment of lupus nephritis (3) recommend change in treatment if response to therapy has not been achieved after 6 months of induction therapy. However, response to therapy is not well defined. In addition, renal damage can occur within 6 months while waiting to define this response. Decision support tools could help define response at the start of induction therapy and have the potential to improve outcomes. Machine learning modeling can assist clinicians in defining such a response. However, even with the machine learning models, currently used clinical biomarkers provide only 69% accuracy for predicting lupus nephritis as a diagnosis (4). This lack of success may stem from the heterogeneity of the disease at presentation. Therefore, more reliable and less invasive methods of predicting outcomes are required for therapeutic decision-making and drug development in lupus nephritis. With such tools, induction therapy can be more appropriately tailored to disease severity in order to prevent renal damage or unnecessary drug toxicity.
Rational development of a lupus nephritis decision tool should take into account heterogeneity in the stages of onset and progression of lupus nephritis. Therefore, markers should indicate initial resident and inflammatory cell activation (cytokines), signals for homing to the kidney (chemokines), activation of inflammatory cell types (growth factors), and damage to resident cell types (5). Analysis of biomarkers in urine as a proximal fluid rather than serum/plasma as a systemic fluid could increase the sensitivity and specificity of signals for renal rather than systemic processes. We hypothesized that a targeted panel of urinary biomarkers reflecting the above processes, with machine learning modeling, could be used to develop and validate an early lupus nephritis decision support tool predictive of lupus nephritis outcomes that is superior to a tool created with standard clinical laboratory biomarkers. The long-range goal is to aid clinical decision-making in lupus nephritis to improve outcomes. Currently, no clinically useful tool exists to predict outcomes using baseline measures. This study demonstrates the feasibility of using machine learning models of baseline biomarker assessments to predict 1-year outcomes in lupus nephritis.
PATIENTS AND METHODS
Study protocol
The goal of this study was to develop models of lupus nephritis outcome using a novel panel of urinary biomarkers and machine learning algorithms. Patients with active lupus nephritis but without high chronicity were enrolled if there was intent to advance to induction therapy based on active lupus nephritis. All patients had cortical renal biopsies performed as part of routine care. At baseline, patients underwent a physical examination, medical history was obtained, and blood and urine samples were obtained for evaluation of traditional markers of SLE disease activity and nephritis activity (Systemic Lupus International Collaborating Clinics renal activity index [6] data elements). Urine samples obtained from a subset of patients at the initiation of induction therapy were analyzed for a discovery panel of biomarkers of inflammation and damage. Predictive urinary biomarkers from that discovery panel were used to develop a smaller biomarker panel. Levels of markers from this smaller panel were then analyzed in urine samples obtained prior to induction therapy using univariate models and machine learning techniques with 1-year complete response as the end point. The machine learning models using traditional biomarkers and those including novel urinary biomarkers as input variables were compared to determine if the combined novel and traditional biomarker panel models were superior to models including only traditional biomarkers.
Urine collection
Urine was collected by clean catch, centrifuged to remove sediment, divided into aliquots, and frozen at −80°C for batch analysis. If urine was not immediately processed, it was stored at 0–4°C for less than 4 hours prior to centrifugation and aliquotting for freezing. No protease inhibitors were used because they are not necessary to maintain the stability of acute kidney injury markers (7).
Patient populations
All research was conducted in accordance with the Declaration of Helsinki and was approved by the Medical University of South Carolina (MUSC) Institutional Review Board. Patients were recruited from 4 different lupus nephritis populations. The MUSC Division of Rheumatology and Immunology Lupus Erythematosus clinical research group (MUSCLE) assisted in the recruitment of participants in Charleston, SC as part of a prospective cohort designed to study lupus nephritis outcomes. Participants were also recruited from the Hopkins Lupus Cohort, a well-established and rigorously described prospective outcome cohort begun in 1987 by Dr. Michelle Petri (8). A third patient population came from those in the Genentech lupus nephritis study of active class III or IV lupus nephritis entitled “A Study to Evaluate the Efficacy and Safety of Rituximab in Subjects With ISN/RPS Class III or IV lupus nephritis (Lupus Nephritis Assessment of Rituximab [LUNAR])” (ClinicalTrials.gov identifier: NCT00282347) (9,10). A fourth population came from patients participating in the Bristol-Myers Squibb trial of abatacept in lupus nephritis entitled “Efficacy and Safety Study of Abatacept to Treat Lupus Nephritis (Abatacept in Lupus Nephritis)” (ClinicalTrials.gov identifier: NCT004306 77) (10,11). The latter two cohorts were from randomized, controlled trials. Characteristics of these 4 cohorts are available from the corresponding author upon request.
Inclusion criteria
All patients met the 1997 update of the revised ACR criteria for SLE (12). All patients had International Society of Nephrology/Renal Pathology Society active class II, III, IV, or V nephritis determined by biopsy performed by the treating clinician or had newly active nephritis (a new increase in protein of 500 mg in a 24-hour urine test or urinary protein-to-creatinine ratio of 0.5 in a spot urine specimen). Only patients who met these criteria within 2 months of the baseline assessment were included. Only patients with complete input and output variable measures were included.
Exclusion criteria
Patients could not have an active infection or ongoing pregnancy or serum creatinine level of >2.5 (an exclusion in the clinical trials to show treatment effect). The LUNAR and Bristol-Myers Squibb trials had more rigorous exclusion criteria, as previously described (10,11).
Routine laboratory analysis of traditional biomarkers of lupus nephritis
Tests for serum C3, C4, creatinine, anti-dsDNA antibody (positive or negative per the individual laboratory), and 24-hour urine test for protein and urinary protein-to-creatinine ratio were performed by Clinical Laboratory Improvement Amendments–certified central laboratories at MUSC, Johns Hopkins, Covance, and Quintiles.
Determination of urinary biomarker panel
Most urinary biomarkers were determined by multiplex bead array using commercially available multiplex human cytokine assays and a Bioplex multiplex bead array reader from Bio-Rad that uses a Luminex 100 system. The system was used according to the manufacturer’s suggestion, with the exception that a diluent containing phosphate buffered saline (pH 7.4) and 0.5% bovine serum albumin was used to prepare the standards and dilute the urine samples 1:10 (to reduce urine matrix interference with the assay) (13,14). The discovery array was chosen based on the availability of analytes from the manufacturers and reports in the literature of biomarkers and pathogenic mediators of lupus nephritis and markers of renal damage. Urinary biomarkers from premixed kits (Bio-Rad) were included in an initial discovery array of the following analytes: eotaxin 1, granulocyte–macrophage colony-stimulating factor (GM-CSF), interferon-α2 (IFNα2), IFNγ, interleukin-1α (IL-1α), IL-1β, IL-2, IL-2 receptor α (IL-2Rα), IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-13, IL-15, IL-16, IL-17, IL-19, IFN-inducible protein 10 (IP-10), leukemia inhibitory factor, M-CSF, monocyte chemoattractant protein 1 (MCP-1; CCL2), MCP-3 (CCL7), macrophage migration inhibitory factor (MIF), monokine induced by IFNγ, macrophage inflammatory protein 1α (MIP-1α), MIP-1β, platelet-derived growth factor BB (PDGF-BB), RANTES (CCL5), stem cell factor, human stem cell growth factor β, vascular cell adhesion molecule 1, vascular endothelial growth factor, nerve growth factor β, stromal cell–derived factor 1α, tumor necrosis factor, and TRAIL (apolipoprotein 2 ligand). Lipocalin 2 (R&D Systems), TWEAK (eBioscience), osteoprotegerin (OPG), and cystatin C (BioVender) were analyzed by enzyme-linked immunosorbent assay using commercially available kits according to the manufacturers’ protocol at a 1:100 dilution (R&D Systems). An N-acetyl-β-d-glucosaminidase (NAG) assay (Roche Applied Science) was performed according to the manufacturer’s protocol.
Receiver operating characteristic (ROC) area under the curve (AUC) analysis was performed on a subset of patients, with biopsy class at onset of renal disease as an outcome. To select markers of more aggressive disease, those predictive of proliferative disease (class III or IV) with ROC AUC values of >0.6 or with literature supporting their use were combined to develop a panel of biomarkers that were analyzed using the same methods for the entire combined cohort of patients. These biomarkers were eotaxin 1, GM-CSF, IFNα2, IFNγ, IL-1α, IL1β, IL-6, IL-8, IP-10, MCP-1, MIP-1β, PDGF-BB, lipocalin 2, TWEAK, OPG, cystatin C, and NAG. Initial modeling in a subset of patients indicated that there was no increased discriminative power when biomarker levels were normalized to urine creatinine level.
Statistical analysis
All models/analyses used the novel and traditional renal biomarkers listed above, and induction therapy (mycophenolate mofetil, azathioprine, cyclophosphamide, abatacept, or rituximab) as separate input variables, and complete response as the outcome. An exploratory analysis included study cohort as a variable. The outcome variable was complete response, as defined by Wofsy et al and originally described in the LUNAR trial (15). This outcome was chosen among several comparative outcomes of 1-year complete response because it required fewer subjects to discriminate complete response among the lupus nephritis patients in this study who were treated with abatacept. Normal renal function was defined as an estimated glomerular filtration rate (GFR) of ≥90 based on the abbreviated Modification of Diet in Renal Disease equation (16).
Power analysis
A sample size of 140 including 37 complete responders and 103 nonresponders provides >80% power to detect an area under the ROC curve (AUC) of 0.65 compared to the null hypothesis of an AUC of 0.50 (equivalent to no diagnostic value) using a 2-sided z-test at a significance level of 0.05. This sample size also provides 80% power to detect a 0.14 unit difference between a statistical model with an AUC of 0.65 and another statistical model with an AUC of 0.79 using a 2-sided z-test at the significance level α = 0.05.
Univariate and random forest analysis
Univariate associations between demographic factors, therapeutic agent usage, and all biomarkers were initially evaluated using simple logistic regression models. The univariate area under the ROC curve (AUC) with 95% Wald confidence intervals were calculated for all traditional and novel biomarkers from the logistic regression models. Hypothesis tests comparing the AUCs for each biomarker to a null value of 0.5 were conducted by comparing an intercept-only model to each univariate model. For multivariable classification analysis, random forest models with 1) clinical variables only and 2) clinical variables and novel biomarkers (combined) were considered. Parameters in the random forest models, including the number of variables considered at each step of the algorithm and maximum tree size, were selected using the train function in the caret package in R. In order to evaluate the prediction performance of each model, in a separate study (17), the predictive power of random forest modeling was determined in separate training and test sets from these data. The test (validation) set performed equally as well as the training set, which is consistent with the findings of Breiman (18) that random forest models produce an unbiased estimate of model error. Because the random forest modeling did not overtrain as other modeling techniques did, the entire data set was used as a training set for this study. The predictive performance of all univariate and multivariable models was assessed using the AUC, accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV). Comparisons between univariate AUCs and random forest model AUCs were conducted using the DeLong test (19). All analyses were conducted in R version 3.1.1.
RESULTS
Patient characteristics by outcome
For this analysis, patients were classified as complete responders or nonresponders. The overall percentage of complete responders was 26.4%, and the response rate was not different between cohorts. General characteristics of patients with different outcomes are represented in Table 1. All patients were receiving mycophenolate background oral therapy unless otherwise specified as taking cyclophosphamide or azathioprine. Therapy with a biologic agent was added to oral or intravenous cyclophosphamide background therapy. A majority of the patients were female, and both African Americans and whites were well represented. Given that the responder definition relies on renal function and urinary protein-to-creatinine ratio, it is not surprising that urinary protein-to-creatinine ratio and estimated GFR were significantly different between groups.
Table 1.
Characteristics of the patients with lupus nephritis*
| Variable | Nonresponders (n = 103) |
Responders (n = 37) |
|---|---|---|
| White race | 66 (64.1) | 24 (64.9) |
| Azathioprine treatment | 5 (4.85) | 0 (0.00) |
| Mycophenolate treatment | 95 (92.2) | 37 (100.0) |
| Cyclophosphamide treatment | 1 (0.97) | 0 (0.00) |
| Rituximab treatment | 14 (13.6) | 7 (18.9) |
| Abatacept treatment | 24 (23.3) | 10 (27) |
| dsDNA positive | 77 (74.8) | 29 (78.4) |
| Age, mean ± SEM years | 31.7 ± 1.05 | 29.2 ± 1.66 |
| Class III or IV nephritis | 94 (91.3) | 33 (89.2) |
| Urinary protein-to-creatinine ratio, mean ± SEM gm/gm | 3.64 ± 0.30 | 2.34 ± 0.40† |
| C3, mean ± SEM mg/dl | 71.1 ± 2.82 | 66.5 ± 4.39 |
| C4, mean ± SEM mg/dl | 14.7 ± 0.73 | 13.2 ± 1.13 |
| Estimated GFR, mean ± SEM ml/minute | 92.7 ± 4.32 | 108.1 ± 5.19 |
Responders were patients in whom complete response had been achieved at 1 year. Except where indicated otherwise, values are the number (%).
dsDNA = double-stranded DNA; GFR = glomerular filtration rate.
P = 0.020 versus nonresponders.
Single biomarkers are poor to fair markers of complete response
To determine the extent to which individual biomarkers predict treatment response in lupus nephritis, AUCs were generated for all biomarkers. The following is a general guide for the discriminative power of a test based on ROC AUC: 0.9–1.0 is excellent, 0.8–0.9 is good, 0.7–0.8 is fair, and 0.6–0.7 is poor (20). The test is considered a failure if the ROC AUC is 0.5–0.6. The ROC AUC for the top 9 most discriminative clinical and novel biomarker variables are shown in Table 2. The AUCs for the individual clinical and urinary biomarkers ranged from 0.42 to 0.67 (for eotaxin and OPG, respectively). Among the traditional clinical markers, only estimated GFR and urinary protein-to-creatinine ratio had AUC values significantly greater than 0.5 (P = 0.019 and P = 0.005, respectively). Among the 18 novel biomarkers, IL-2Rα, OPG, and IL-8 were the only markers that had AUC values significantly greater than 0.5 (P = 0.034, P = 0.002, and P = 0.008, respectively).
Table 2.
Most discriminating variables for the prediction of treatment response in univariate analysis*
| Variable | ROC AUC (95% CI) | P |
|---|---|---|
| Age | 0.58 (0.47–0.69) | 0.156 |
| Urinary protein-to-creatinine ratio | 0.65 (0.55–0.76) | 0.005 |
| Serum C4 | 0.57 (0.46–0.66) | 0.202 |
| Estimated GFR | 0.62 (0.52–0.72) | 0.019 |
| OPG | 0.67 (0.57–0.78) | 0.002 |
| IFNα2 | 0.58 (0.47–0.69) | 0.150 |
| IL-2Rα | 0.63 (0.51–0.75) | 0.034 |
| IL-8 | 0.64 (0.54–0.75) | 0.008 |
| IP-10 | 0.58 (0.47–0.69) | 0.149 |
ROC AUC = receiver operating characteristic area under the curve; 95% CI = 95% confidence interval; GFR = glomerular filtration rate; OPG = osteoprotegerin; IFNα2 = interferon α2; IL-2Rα = interleukin-2 receptor α; IP-10 = interferon-inducible protein
Increased discriminative power of novel biomarkers compared to traditional biomarkers in random forest models
We hypothesized that random forest models using a biomarker panel would outperform traditional or low-abundance biomarkers in univariate models. We also hypothesized that a random forest model including both traditional and novel biomarkers (combined) would predict outcomes better than one with traditional biomarkers alone. Because random forest models do not overtrain in test sets as other machine learning models tend to do (18), we did not split the data into test and training data. Random forest models of response to treatment were developed using either traditional markers only or novel biomarkers in addition to clinical markers (combined models). The full model included 500 decision trees (results are available from the corresponding author upon request). In the random forest model, each of the 500 different decision trees makes a prediction about whether or not a subject is a responder.
The combined models had a greater AUC and significance than the models created with traditional clinical markers alone (AUC 0.79 [P < 0.001] versus AUC 0.61 [P = 0.05], respectively). The random forest combined model provided better discrimination of outcomes than the random forest model that included only traditional biomarkers (P = 0.002). The random forest combined model had a significantly better AUC than a majority of the individual biomarkers. This increased the percentage of correctly predicted complete responders from 27% in the traditional model to 76% in the novel model (threshold 0.3) (data are available from the corresponding author upon request). OPG was the only biomarker that did not have an AUC that was significantly lower than the random forest model, although there was a trend toward significance (P = 0.055). However, the OPG model AUC was not superior to those with traditional markers.
Predictions from a random forest model are based on the proportion of trees (threshold) in the model that predict a person to be a responder versus the proportion that predict a person to be a nonresponder. The default is to assume that if ≥50% of the trees in the forest predict that a person is a responder, then the person is a responder; otherwise, they are predicted to be a nonresponder. The AUC was chosen as the primary measure to evaluate a model’s discriminative power because it does not depend on the prediction threshold chosen for a model. Sensitivity and specificity can also provide information about a model’s discriminative power but can change dramatically based on the threshold probability chosen. Therefore, we estimated sensitivity and specificity for a range of threshold probabilities. Results for selected threshold probabilities for the random forest model with traditional markers only and for the combined model are presented in Table 3. Additionally, we evaluated the PPV, the NPV, and the accuracy (data are available from the corresponding author upon request).
Table 3.
Sensitivity and specificity at different thresholds for random forest models with traditional clinical markers only and with both traditional and novel markers*
| Threshold | |||
|---|---|---|---|
| Model | t = 0.3 | t = 0.4 | t = 0.5 |
| Traditional markers only | |||
| Sensitivity (95% CI) | 0.32 (0.24–0.40) | 0.14 (0.08–0.20) | 0.05 (0.01–0.09) |
| Specificity (95% CI) | 0.82 (0.76–0.88) | 0.90 (0.85–0.95) | 0.94 (0.90–0.98) |
| Traditional and novel markers | |||
| Sensitivity (95% CI) | 0.76 (0.69–0.83) | 0.49 (0.41–0.57) | 0.24 (0.17–0.31) |
| Specificity (95% CI) | 0.73 (0.66–0.80) | 0.86 (0.80–0.92) | 0.98 (0.96–1.00) |
The threshold is the proportion of trees in the random forest model that classify response to therapy required to predict that a person is a responder.
95% CI = 95% confidence interval.
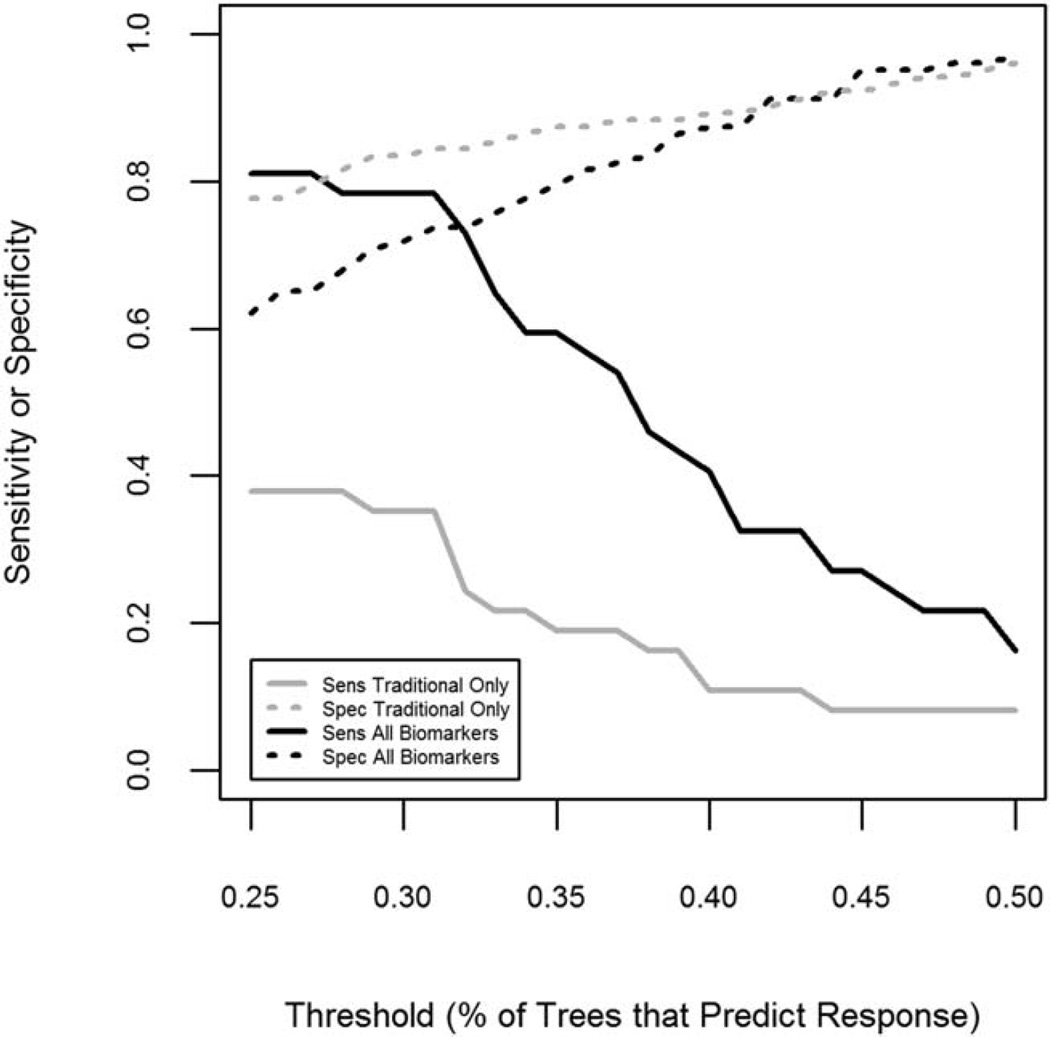
Figure 1 shows the change in sensitivity and specificity for each model as the threshold probability increases from 0.25 to 0.5. These results indicate that decreasing the threshold from 50% to 25% results in large increases in sensitivity (to >80%) and a smaller decrease in specificity in both models. However, the random forest combined model is able to achieve both sensitivity and specificity of >70% when the threshold is near 30%. In comparison, the traditional biomarker model is less sensitive at all thresholds.
Figure 1.
Effect of varying threshold on the sensitivity (Sens) and specificity (Spec) of random forest models of response to therapy that included traditional clinical markers alone or clinical markers plus novel biomarkers. Random forest models were created for the prediction of complete response to therapy for lupus nephritis at 1 year.
Analytes that contribute to the models in sensitivity analysis
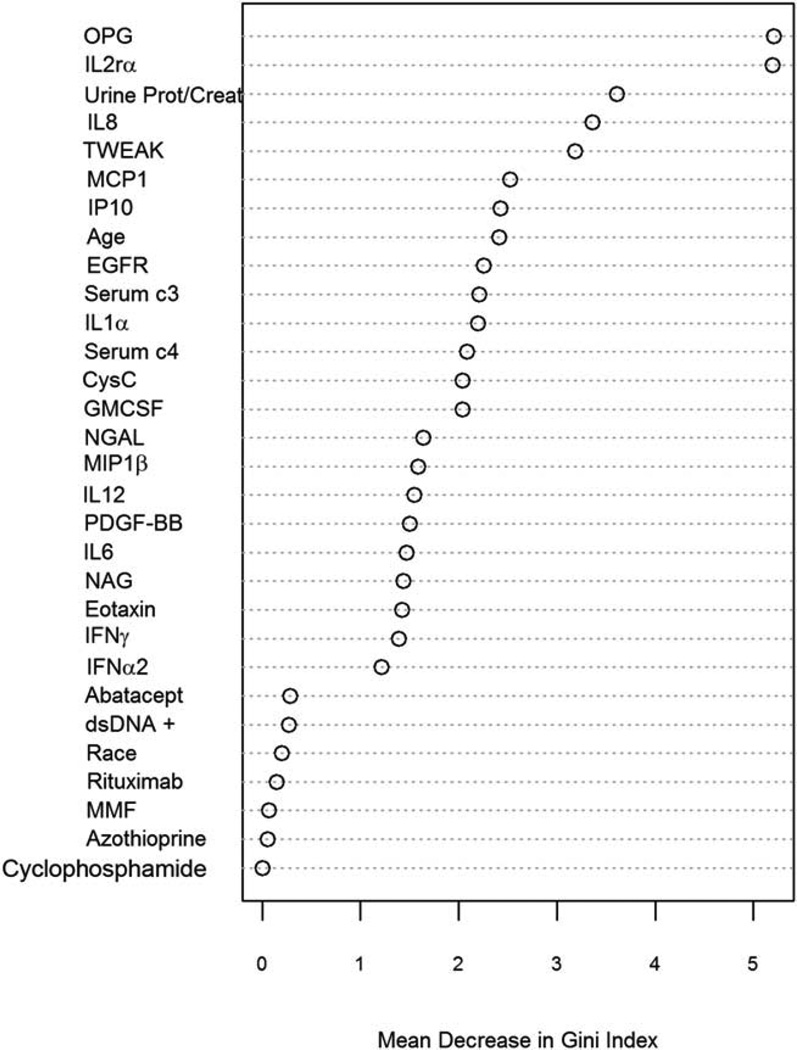
Identifying analytes that are important in modeling outcome can be useful in generating hypotheses about nephritis pathogenesis. The random forest method also provides a measure of variable importance for all variables evaluated in a model based on the mean decrease in Gini Index (a measure of prediction purity) when a variable is permuted. These importance measures allow us to examine the relative importance of each biomarker in the prediction of treatment response. The variable importance plot for the random forest model fit to all of the data is shown in Figure 2. Five biomarkers were distinct from others in their importance in prediction of treatment response; these were OPG, IL-2Rα, urinary protein-to-creatinine ratio, IL-8, and TWEAK. MCP-1, IP-10, age, estimated GFR, C3, IL-1α, C4, cystatin C, and GM-CSF were another distinct set of biomarkers with a mean decrease in Gini Index of ≥2. Of interest, race, anti-dsDNA antibodies, and induction medication were not significant contributors to the model.
Figure 2.
Importance plot for individual biomarkers in the random forest model. A random forest model for prediction of complete response to therapy for lupus nephritis at 1 year was developed using standard clinical markers and clinical markers plus novel urinary biomarkers. The importance of each individual biomarker in the model is plotted as mean decrease in Gini Index (a measure of prediction purity). OPG = osteoprotegerin; IL-2Rα = interleukin-2 receptor α; urine prot/creat = urinary protein-to-creatinine ratio; MCP-1 = monocyte chemotactic protein 1; IP-10 = interferon-inducible protein 10; EGFR = estimated glomerular filtration rate; CysC = cystatin C; GM-CSF = granulocyte–macrophage colony-stimulating factor; NGAL = neutrophil gelatinase–associated lipocalin; MIP-1β = macrophage inflammatory protein 1β; PDGF-BB = platelet-derived growth factor BB; NAG = N-acetyl-β-d-glucosaminidase; IFNγ = interferon-γ; dsDNA = doublestranded DNA; MMF = mycophenolate mofetil.
In a clinical setting, it is ideal to have smaller numbers of biomarkers in a panel. Thus, we ran additional random forest models including 1) only the top 5 most important predictors, 2) the top 13 most important predictors (selected because they had a mean decrease in Gini Index of >2), and 3) the top 4 most important novel biomarkers plus all traditional clinical markers. We then evaluated the predictive performance of these reduced models relative to the full model (including all predictors) (Table 4). These “reduced” models showed equivalent discriminative ability relative to the “full” model and in some cases, had better sensitivity and specificity than the “full” model, suggesting that a more tailored model based on a subset of markers might provide good predictive performance.
Table 4.
Performance of random forest models using different subsets of variables*
| Threshold | |||
|---|---|---|---|
| Model | t = 0.3 | t = 0.4 | t = 0.5 |
| Full model (30 variables) | 0.79 (0.70–0.88) | 0.24 (0.17–0.31) | 0.98 (0.96–1.00) |
| 5 most important predictors | 0.77 (0.70–0.84) | 0.43 (0.35–0.51) | 0.92 (0.87–0.96) |
| 13 most important predictors | 0.80 (0.73–0.87) | 0.35 (0.27–0.43) | 0.95 (0.91–0.99) |
| Clinical variables plus 4 most important biomarkers |
0.79 (0.72–0.86) | 0.51 (0.43–0.59) | 0.91 (0.86–0.96) |
Values are the receiver operating characteristic area under the curve (95% confidence interval). The threshold for prediction when calculating the sensitivities and specificities was set at the default value of ≥50%.
DISCUSSION
In this study, models of complete response at 1 year using panels of biomarkers had superior diagnostic power over single biomarkers or over models developed using traditional clinical biomarkers. The combined model provided improvements in sensitivity compared to traditional models. This improvement provides the rationale for a future randomized implementation study to determine if its use in the clinical setting improves renal response over current standard of care.
Biomarkers in the novel panels represented multiple mechanisms of disease pathogenesis and cellular damage. Thus, the effectiveness of this more inclusive approach to diagnosis (use of biomarker panels rather than individual markers in isolation) likely reflects the multistaged, heterogeneous nature of lupus nephritis. The biomarkers with the greatest importance to the models have biologic plausibility. Both OPG and MCP-1 have been described as markers of renal disease activity (21). OPG is a soluble, decoy receptor to RANKL, thus preventing the RANK–RANKL interaction that can promote inflammation and bone resorption (22). Whether OPG is a marker or mediator of lupus nephritis disease activity is unknown, since OPG levels were higher in nonresponders. Kiani et al have described increased OPG levels in patients with more active lupus nephritis (21). IL-2Rα (CD25) is important in the regulation of immune responses, and mutation of this receptor predisposes to autoimmunity. Serum levels are associated with general lupus disease activity (23). IL-8 is a neutrophil chemoattractant stimulated by CD40 and IL-17 receptor engagement (24). IL-8 polymorphisms were identified in African Americans with lupus nephritis (25), and IL-8 was increased in lupus nephritis (26). TWEAK induced the production of inflammatory chemokines by mesangial and endothelial cells (27) and is a promising therapeutic target for lupus nephritis (28).
MCP-1 (CXCL2) acted as a chemoattractant for monocytes, memory T cells, and dendritic cells (29,30). MCP-1 levels were increased in the urine of patients with lupus nephritis (31), preceded flares, and were associated with nephritis activity (21,32). More recently, MCP-1 within a biomarker panel has been associated with interstitial inflammation and chronicity in lupus nephritis (33). This latter approach is advocated by Rovin et al (34) and is similar to the use of a composite biomarker panel rather than a single biomarker in this study. The novel contribution of the present study is the use of a broader biomarker panel and machine learning techniques to optimize models of disease outcome. The importance of urinary protein-to-creatinine ratio in the model is not surprising given that the outcome is dependent on this measure.
This is also the first study to use artificial intelligence or machine learning modeling to develop models of outcome using multiple biomarkers that can be measured using commercial kits. Others have used machine learning modeling to improve the diagnostic accuracy of traditional clinical markers over that of expert diagnosis (35). Our group used artificial neural network modeling of proteomic data to identify glycosylation variants of common proteins that were predictive of class, a finding later confirmed by others (36). However, a commercial assay for these glycosylation variants is not available.
This study has several potential limitations. The input variable of biopsy class was determined by multiple pathologists. However, the International Society of Nephrology/Renal Pathology Society classification system has proven to have reduced interobserver variability (37,38) when compared to the World Health Organization classification. The models were created using only findings in urine samples obtained at the start of induction therapy. As we have previously determined with measures of systemic nitric oxide production (39), markers evaluated after the initiation of induction therapy were more indicative of outcome than baseline measures. However, models created in that study using change in biomarkers from baseline to 3 months did not yield superior predictive power over simple baseline measures. Markers in the present study were not normalized to creatinine level. While normalizing to creatinine level may reduce variability and improve the predictive power of the model, data on creatinine level were not available for all patient samples from the clinical trials. While the models have not been validated in an external cohort of patients, a separate study of the same data set, using separate training and validation sets, demonstrated that the random forest modeling did not over fit. Similar model performance to that seen in this study was observed in the validation set in the separate study (17).
The variation in choice of induction therapy at each site is accounted for in the models (induction therapy was included as a variable). However, differences in induction therapy could affect the outcome after baseline urine is collected for modeling and introduce bias in the model. Contrary to this notion, Figure 2 illustrates that biomarkers are more important in the model than choice of induction therapy. This study did not test why induction therapy was not important in the models. However, biomarkers may have contributed more to models because they were measured in every patient, while therapies other than mycophenolate mofetil monotherapy did not occur in sufficient frequency to contribute to the model (Table 1 and Figure 2). The same is true for the abatacept and rituximab add-on therapies. While there were numerical differences in the percentage of responders and nonresponders who were receiving each biologic agent, the study was not powered to study differences between therapies. Specifically, 52-week complete response outcome used in this study is predicted to be sufficient to detect differences in treatments with 50 or more subjects in each group (15).
The study cohort could similarly affect outcomes through different practice patterns or protocols. However, the prediction performance of the model that included study cohort was similar to that of the random forest model presented above. Additionally, the cohort variable was found to be of low importance (ranked 24th of 30 predictor variables). Cumulative prednisone dose could have affected outcomes but was not measured in this study. Finally, the lack of full representation of patients at each of the contributing sites could introduce a bias. Specifically, only patients who had complete follow-up and baseline data were selected. It is possible that those patients who did not have complete assessments represented a population more at risk for treatment failure.
This study provides support for the use of commercially available assay biomarker panels to develop models of lupus nephritis outcome. The heterogeneity of lupus nephritis is reflected in the finding that panels of multiple biomarkers provided superior diagnostic power compared to single biomarkers. This raises the possibility of using panels such as this with interpretive modeling as a clinical decision tool in future studies. Prospective trials would need to be performed to determine if their use in aiding the choice of induction therapy affects outcomes.
Acknowledgments
The authors thank Sally Self, MD, Serena Bagnasco, MD, Lorraine Racusen, MD, and the Genentech and BMS study pathologists for their work in classifying biopsies using the International Society of Nephrology/Renal Pathology Society criteria. The authors also thank Jay Garg, MD, Romeo Maciuca, and Paul Brunetta, MD at Genentech, the LUNAR investigators, Drs. Ralph Small and Sean Connolly at Bristol-Myers Squibb, and the participants who generously contributed to all of the studies from which data and samples came.
Supported in part by a VA Merit Award (1I01CX000218-01), NIH/National Institute of Arthritis and Musculoskeletal and Skin Diseases grant R21-AR-051719, the Medical University of South Carolina General and Clinical Research Center (through NIH grant M01-RR-001070), the South Carolina Clinical and Translational Research Institute, with an academic home at the Medical University of South Carolina (through NIH/National Center for Research Resources grant number UL1-RR-029882), and the Multidisciplinary Clinical Research Center at the Medical University of South Carolina (through NIH grant P60-A-R062755). Support also came from a VA Research Enhancement Award Program award. Subject material (urine samples for biomarker analysis and clinical data) were provided by Genentech (from the Lupus Nephritis Assessment of Rituximab [LUNAR] study; ClinicalTrials.gov identifier: NCT00282347) and Bristol-Myers Squibb (from the Abatacept in Lupus Nephritis study; ClinicalTrials.gov identifier: NCT00430677) and their investigators. Dr. Janech’s work was supported by a NephCure Kidney International Young Investigator Career Development Award. Dr. Petri’s work was supported by the Hopkins Lupus Cohort (through NIH grant AR-43727) and NIH/National Center for Research Resources grant UL1-RR-025005.
Footnotes
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or the US Department of Veterans Affairs.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Oates had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design. Wolf, Arthur, Janech, Oates.
Acquisition of data. Arthur, Petri, Oates.
Analysis and interpretation of data. Wolf, Spainhour, Oates.
REFERENCES
- 1.Wallace DJ, Hahn BH, Klippel JH. Clinical and laboratory features of lupus nephritis. In: Wallace DJ, Hahn B, Dubois EL, editors. Dubois’ lupus erythematosus. 6th. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 1077–1078. [Google Scholar]
- 2.Dooley MA, Hogan S, Jennette C, Falk R for the Glomerular Disease Collaborative Network. Cyclophosphamide therapy for lupus nephritis: poor renal survival in black Americans. Kidney Int. 1997;51:1188–1195. doi: 10.1038/ki.1997.162. [DOI] [PubMed] [Google Scholar]
- 3.Hahn BH, McMahon MA, Wilkinson A, Wallace WD, Daikh DI, Fitzgerald JD, et al. American College of Rheumatology guidelines for screening, treatment, and management of lupus nephritis. Arthritis Care Res (Hoboken) 2012;64:797–808. doi: 10.1002/acr.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajimehr R, Farsiu S, Kouhsari LM, Bidari A, Lucas C, Yousefian S, et al. Prediction of lupus nephritis in patients with systemic lupus erythematosus using artificial neural networks. Lupus. 2002;11:485–492. doi: 10.1191/0961203302lu226oa. [DOI] [PubMed] [Google Scholar]
- 5.Kelley VR, Wuthrich RP. Cytokines in the pathogenesis of systemic lupus erythematosus. Semin Nephrol. 1999;19:57–66. [PubMed] [Google Scholar]
- 6.Petri M, Kasitanon N, Lee SS, Link K, Magder L, Bae SC, et al. for the Systemic Lupus International Collaborating Clinics. Systemic Lupus International Collaborating Clinics renal activity/response exercise: development of a renal activity score and renal response index. Arthritis Rheum. 2008;58:1784–1788. doi: 10.1002/art.23456. [published erratum appears in Arthritis Rheum 2008;58:2823] [DOI] [PubMed] [Google Scholar]
- 7.Han WK, Wagener G, Zhu Y, Wang S, Lee HT. Urinary biomarkers in the early detection of acute kidney injury after cardiac surgery. Clin J Am Soc Nephrol. 2009;4:873–882. doi: 10.2215/CJN.04810908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petri M. Musculoskeletal complications of systemic lupus erythematosus in the Hopkins Lupus Cohort: an update. Arthritis Care Res. 1995;8:137–145. doi: 10.1002/art.1790080305. [DOI] [PubMed] [Google Scholar]
- 9.Weening JJ, D’Agati VD, Schwartz MM, Seshan SV, Alpers CE, Appel GB, et al. The classification of glomerulonephritis in systemic lupus erythematosus revisited. J Am Soc Nephrol. 2004;15:241–250. doi: 10.1097/01.asn.0000108969.21691.5d. [J Am Soc Nephrol 2004;15:835–6] [DOI] [PubMed] [Google Scholar]
- 10.Rovin BH, Furie R, Latinis K, Looney RJ, Fervenza FC, Sanchez-Guerrero J, et al. for the LUNAR Investigator Group. Efficacy and safety of rituximab in patients with active proliferative lupus nephritis: the Lupus Nephritis Assessment with Rituximab study. Arthritis Rheum. 2012;64:1215–1226. doi: 10.1002/art.34359. [DOI] [PubMed] [Google Scholar]
- 11.Furie R, Nicholls K, Cheng TT, Houssiau F, Burgos-Vargas R, Chen SL, et al. Efficacy and safety of abatacept in lupus nephritis: a twelve-month, randomized, double-blind study. Arthritis Rheumatol. 2014;66:379–389. doi: 10.1002/art.38260. [DOI] [PubMed] [Google Scholar]
- 12.Hochberg MC for the Diagnostic and Therapeutic Criteria Committee of the American College of Rheumatology. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus [letter] Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 13.Chaturvedi S, Farmer T, Kapke GF. Assay validation for KIM-1: human urinary renal dysfunction biomarker. Int J Biol Sci. 2009;5:128–134. doi: 10.7150/ijbs.5.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor TP, Janech MG, Slate EH, Lewis EC, Arthur JM, Oates JC. Overcoming the effects of matrix interference in the measurement of urine protein analytes. Biomark Insights. 2012;7:1–8. doi: 10.4137/BMI.S8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wofsy D, Hillson JL, Diamond B. Comparison of alternative primary outcome measures for use in lupus nephritis clinical trials. Arthritis Rheum. 2013;65:1586–1591. doi: 10.1002/art.37940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vervoort G, Willems HL, Wetzels JF. Assessment of glomerular filtration rate in healthy subjects and normoalbuminuric diabetic patients: validity of a new (MDRD) prediction equation. Nephrol Dial Transplant. 2002;17:1909–1913. doi: 10.1093/ndt/17.11.1909. [DOI] [PubMed] [Google Scholar]
- 17.Wolfe BJ, Spainhour JCG, Fleury TJ, Oates JC. Technical report: qualification of a set of novel biomarkers when limited by small sample size for prediction of response to treatment in lupus nephritis. Medical University of South Carolina. 2015 URL: people.musc.edu/~wolfb/TechnicalReport_Oates2.docx. [Google Scholar]
- 18.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 20.Tape TG. The area under an ROC curve. Interpreting diagnostic tests. 2009 URL: http://gim.unmc.edu/dxtests/roc3.htm. [Google Scholar]
- 21.Kiani AN, Johnson K, Chen C, Diehl E, Hu H, Vasudevan G, et al. Urine osteoprotegerin and monocyte chemoattractant protein-1 in lupus nephritis. J Rheumatol. 2009;36:2224–2230. doi: 10.3899/jrheum.081112. [DOI] [PubMed] [Google Scholar]
- 22.Saidenberg Kermanac’h N, Bessis N, Cohen-Solal M, De Vernejoul MC, Boissier MC. Osteoprotegerin and inflammation. Eur Cytokine Netw. 2002;13:144–153. [PubMed] [Google Scholar]
- 23.Swaak AJ, Hintzen RQ, Huysen V, van den Brink HG, Smeenk JT. Serum levels of soluble forms of T cell activation antigens CD27 and CD25 in systemic lupus erythematosus in relation with lymphocytes count and disease course. Clin Rheumatol. 1995;14:293–300. doi: 10.1007/BF02208342. [DOI] [PubMed] [Google Scholar]
- 24.Woltman AM, de Haij S, Boonstra JG, Gobin SJ, Daha MR, van Kooten C. Interleukin-17 and CD40-ligand synergistically enhance cytokine and chemokine production by renal epithelial cells. J Am Soc Nephrol. 2000;11:2044–2055. doi: 10.1681/ASN.V11112044. [DOI] [PubMed] [Google Scholar]
- 25.Rovin BH, Lu L, Zhang X. A novel interleukin-8 polymorphism is associated with severe systemic lupus erythematosus nephritis. Kidney Int. 2002;62:261–265. doi: 10.1046/j.1523-1755.2002.00438.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsai CY, Wu TH, Yu CL, Lu JY, Tsai YY. Increased excretions of beta2-microglobulin, IL-6, and IL-8 and decreased excretion of Tamm-Horsfall glycoprotein in urine of patients with active lupus nephritis. Nephron. 2000;85:207–214. doi: 10.1159/000045663. [DOI] [PubMed] [Google Scholar]
- 27.Campbell S, Burkly LC, Gao HX, Berman JW, Su L, Browning B, et al. Proinflammatory effects of TWEAK/Fn14 interactions in glomerular mesangial cells. J Immunol. 2006;176:1889–1898. doi: 10.4049/jimmunol.176.3.1889. [DOI] [PubMed] [Google Scholar]
- 28.Michaelson JS, Wisniacki N, Burkly LC, Putterman C. Role of TWEAK in lupus nephritis: a bench-to-bedside review. J Autoimmun. 2012;39:130–142. doi: 10.1016/j.jaut.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr MW, Roth SJ, Luther E, Rose SS, Springer TA. Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proc Natl Acad Sci U S A. 1994;91:3652–3656. doi: 10.1073/pnas.91.9.3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu LL, Warren MK, Rose WL, Gong W, Wang JM. Human recombinant monocyte chemotactic protein and other C-C chemokines bind and induce directional migration of dendritic cells in vitro. J Leukoc Biol. 1996;60:365–371. doi: 10.1002/jlb.60.3.365. [DOI] [PubMed] [Google Scholar]
- 31.Noris M, Bernasconi S, Casiraghi F, Sozzani S, Gotti E, Remuzzi G, et al. Monocyte chemoattractant protein-1 is excreted in excessive amounts in the urine of patients with lupus nephritis. Lab Invest. 1995;73:804–809. [PubMed] [Google Scholar]
- 32.Rovin BH, Song H, Birmingham DJ, Hebert LA, Yu CY, Nagaraja HN. Urine chemokines as biomarkers of human systemic lupus erythematosus activity. J Am Soc Nephrol. 2005;16:467–473. doi: 10.1681/ASN.2004080658. [DOI] [PubMed] [Google Scholar]
- 33.Zhang X, Nagaraja HN, Nadasdy T, Song H, McKinley A, Prosek J, et al. A composite urine biomarker reflects interstitial inflammation in lupus nephritis kidney biopsies. Kidney Int. 2012;81:401–406. doi: 10.1038/ki.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rovin BH, Zhang X. Biomarkers for lupus nephritis: the quest continues. Clin J Am Soc Nephrol. 2009;4:1858–1865. doi: 10.2215/CJN.03530509. [DOI] [PubMed] [Google Scholar]
- 35.Rajimehr R, Farsiu S, Kouhsari LM, Bidari A, Lucas C, Yousefian S, et al. Prediction of lupus nephritis in patients with systemic lupus erythematosus using artificial neural networks. Lupus. 2002;11:485–492. doi: 10.1191/0961203302lu226oa. [DOI] [PubMed] [Google Scholar]
- 36.Oates JC, Varghese S, Bland AM, Taylor TP, Self SE, Stanislaus R, et al. Prediction of urinary protein markers in lupus nephritis. Kidney Int. 2005;68:2588–2592. doi: 10.1111/j.1523-1755.2005.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furness PN, Taub N. Interobserver reproducibility and application of the ISN/RPS classification of lupus nephritis: a UK-wide study. Am J Surg Pathol. 2006;30:1030–1035. doi: 10.1097/00000478-200608000-00015. [DOI] [PubMed] [Google Scholar]
- 38.Markowitz GS, D’Agati VD. The ISN/RPS 2003 classification of lupus nephritis: an assessment at 3 years. Kidney Int. 2007;71:491–495. doi: 10.1038/sj.ki.5002118. [DOI] [PubMed] [Google Scholar]
- 39.Oates JC, Shaftman SR, Self SE, Gilkeson GS. Association of serum nitrate and nitrite levels with longitudinal assessments of disease activity and damage in systemic lupus erythematosus and lupus nephritis. Arthritis Rheum. 2008;58:263–272. doi: 10.1002/art.23153. [DOI] [PMC free article] [PubMed] [Google Scholar]