Abstract
Astrocytes are the most abundant cell type in the mammalian brain. Interest in astrocyte function has increased dramatically in recent years because of their newly discovered roles in synapse formation, maturation, efficacy, and plasticity. However, our understanding of astrocyte development has lagged behind that of other brain cell types. We do not know the molecular mechanism by which astrocytes are specified, how they grow to assume their complex morphologies, and how they interact with and sculpt developing neuronal circuits. Recent work has provided a basic understanding of how intrinsic and extrinsic mechanisms govern the production of astrocytes from precursor cells and the generation of astrocyte diversity. Moreover, new studies of astrocyte morphology have revealed that mature astrocytes are extraordinarily complex, interact with many thousands of synapses, and tile with other astrocytes to occupy unique spatial domains in the brain. A major challenge for the field is to understand how astrocytes talk to each other, and to neurons, during development to establish appropriate astrocytic and neuronal network architectures.
Neurons are not alone in the nervous system; the vast majority of all cells in the adult human brain are glia. Glial cell types in the CNS include astrocytes, oligodendrocytes, microglia, and chondroitin sulfate proteoglycan NG2–positive cells. Together these cells perform a dynamic range of functions essential for nervous system development and physiology, from simple trophic support of neurons to wrapping axons to allow for rapid nerve impulse conduction to modulating synaptic connectivity and efficacy. Astrocytes are the most abundant cell type in the brain; they are intimately associated with synapses and govern key steps in synapse formation and plasticity. However, we understand surprisingly little about the molecular underpinnings of astrocyte development. How are astrocytes specified at the appropriate developmental time from neural precursor cells (NPCs)? What gene expression profile distinguishes astrocytes from other glia or from neurons? How do newly born astrocytes interact with each other or with neurons to promote astrocyte maturation? This review will focus on recent advances in our understanding of how astrocytes are generated by NPCs and of astrocyte morphogenesis during nervous system assembly. Understanding these developmental events in molecular terms will provide insights into the mechanisms by which glia regulate brain development and function, as well as how changes in astrocyte function affect neurological disease.
Making Astrocytes—Nature and Nurture
During mammalian nervous system development, NPCs generate neurons first, followed by glia. Proceeding through the neuron-to-glia switch at the proper time is critical for determining how many neurons or glia are ultimately made in each brain region. The mechanisms regulating this transition are complex and not well understood (1). However, a number of recent studies indicate that this fate switch is governed by both extrinsic environmental cues that promote astrogenesis in NPCs and NPC-intrinsic mechanisms that decrease neurogenic and increase astrogenic competence over developmental time.
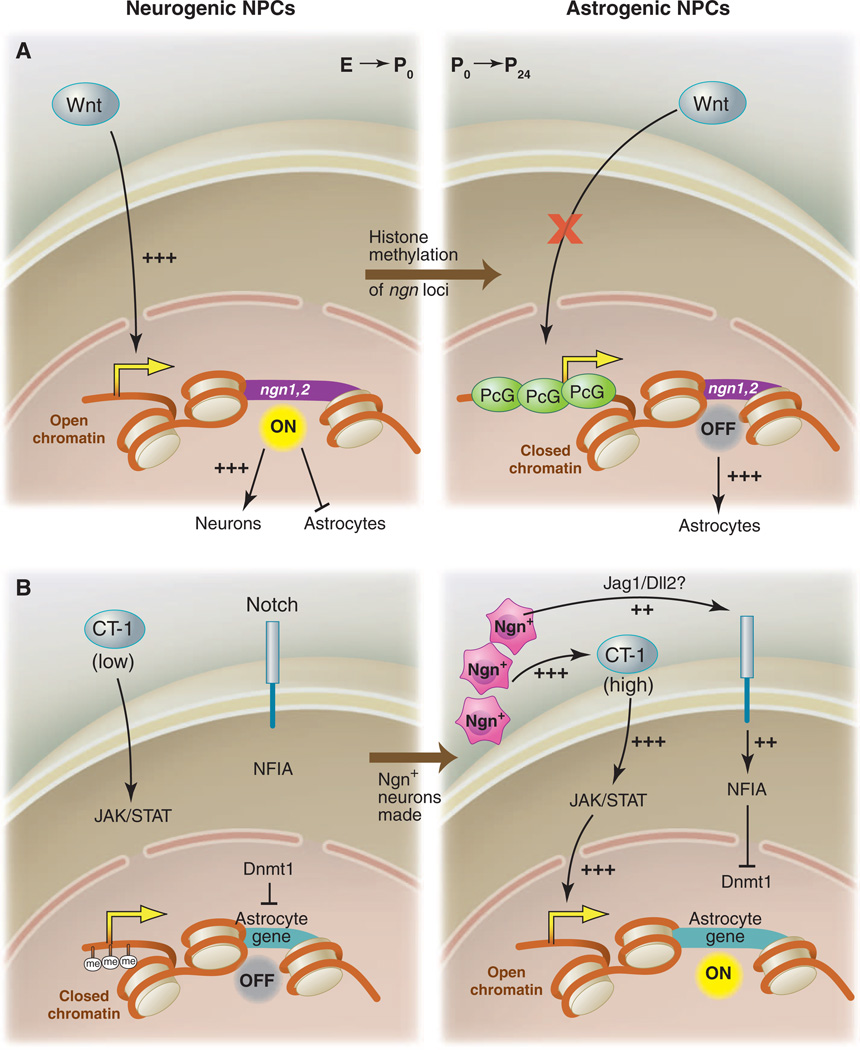
Wnt signaling is required for the activation of the proneural genes neurogenin1 (ngn1) and neurogenin2 (ngn2) in NPCs, where, at an early stage, they act to promote neuronal differentiation (2–4). However, Wnt ligands continue to be expressed in the nervous system even after the neuron-to-glia transition, during which time they fail to induce ngn expression in NPCs. How are glia made when Wnts are still expressed? Hirabayashi et al. (5) found key intrinsic changes in histone H3 acetylation and trimethylation at the ngn1 and ngn2 promoters in NPCs, such that, at early stages, they observed high acetylation and low methylation, but at later stages, low acetylation and high methylation. These would correspond to the open or closed chromatin conformations, respectively. Treatment with histone deacetylase (HDAC) inhibitors resulted in an increase in ngn expression in older, but not younger, NPCs, which indicates that HDAC activity negatively regulates ngn expression in late NPCs. Consistent with a closed chromatin conformation at ngn loci, it was also observed that RNA polymerase II associated at relatively low rates with ngn promoters in late-stage NPCs. This epigenetic change in NPC competence is somehow mediated by the Polycomb group complex (PcG). Knockout of key components of the complex (e.g., Ring1B or Ezh2) prolongs the NPC neurogenic phase and delays the production of astrocytes (5). Thus, despite the continued presence of Wnts, NPCs are able to make the neuron-to-glia switch in part because of intrinsic epigenetic changes that lead to PcG-dependent suppression of expression of the ngns (Fig. 1A).
Fig. 1.
Intrinsic epigenetic mechanisms converge with extrinsic signals to promote astrocyte fate from NPCs. (A) In early NPCs, chromatin is open at the neurogenin1 and neurogenin2 (ngn1 and 2) loci, Wnt signals can activate their expression, and neurogenins promote neuronal fate and inhibit astrocyte fate. In postnatal NPCs, the ngn1 and 2 loci have been methylated and bound by PcG, chromatin is closed, Wnt-dependent ngn1 and 2 expression is suppressed, and astrocyte fate is in turn derepressed. (B) Astrocyte genes are methylated during embryonic stages, which closes local chromatin; this is maintained by DNMT1, and astrocyte fate is suppressed, despite the presence of low levels of the proastrocyte factor CT-1. At the switch from NPC production of neurons to astrocytes, Ngn+ neurons release additional CT-1, which activates JAK-STAT signaling, and also activates Notch signaling (likely through JAG1 or DLL2), which activates NFIA, an inhibitor of Dnmt1 activity. Positive JAK-STAT signaling, coupled with opening of chromatin at astrocyte gene loci, promotes astrocyte fate.
Extrinsic signals also potently influence the fate of NPC progeny. This was demonstrated by culturing embryonic NPCs on either embryonic or postnatal cortical slices: NPCs made neurons when grown on embryonic slices, but glia when grown on postnatal slices (6). Thus, NPCs are competent to make astrocytes during embryonic stages, but normally do not, potentially because of the lack of some extracellular astrogenic cue. The Janus kinase–signal transducers and activators of transcription (JAK-STAT) pathway is a well-known activator of astrocyte cell fate and can be stimulated in NPCs by cytokines including ciliary neurotrophic factor (CNTF), leukemia inhibitory factor (LIF), and cardiotrophin-1 (CT-1) (1). Namihira et al. used CT-1 to stimulate embryonic NPCs at either early or mid-gestational stages and found that CT-1 could promote astrocyte production only in mid-gestational or older NPCs. However, if Notch signaling was also activated in early NPCs, then treatment with CT-1 was sufficient to drive astrocyte production. How does Notch change NPC competence to respond to CT-1? Previous work had shown that methylation of the promoters of key astrocyte genes including glial fibrillary acidic protein (gfap) and S100β suppresses STAT binding and activation of these genes in NPCs. Moreover, blocking DNA methyltransferase activity in NPCs results in early induction of astrocyte genes (7). Thus, changes in DNA methylation status of astrogenic loci was a candidate mechanism. Expressing activated Notch (NICD) resulted in demethylation of STAT binding sites in the promoters and activation of expression of gfap and S100β. The mechanism by which Notch drives this event appears twofold: First, it promotes the dissociation of the DNMT1 maintenance methyltransferase [DNA (cytosine-5-)-methyltransferase 1] from the gfap promoter; second, it leads to increased nuclear factor IA (NFIA) expression (8). NFIA is a key regulator of the neuron-to-glia switch (9). NFIA is necessary and sufficient for induction of glial markers in NPCs like GLAST, is required for Notch to promote the transition to gliogenesis, and actively inhibits neurogenesis by binding to the gfap promoter and protecting it from DNMT1-dependent methylation (8, 9).
Positive Notch signaling is therefore sufficient to make NPCs prematurely responsive to CT-1 for activation of JAK-STAT signaling. But what drives normal activation of Notch in NPCs and, in turn, astrogenesis at the appropriate developmental time? The answer seems to be early-born neurons. Newly born Ngn1+ neurons, which are adjacent to NPCs, express the Notch ligands JAG1 and DLL1 and are thus a likely source for signals activating Notch signaling in NPCs (8). At the same time, CT-1 is secreted by embryonic neurons beginning at mid-gestation, which is thought to boost overall levels of CT-1 (10). It therefore appears that early-born neurons costimulate Notch and, through CT-1 release, JAK-STAT signaling pathways. Notch signaling shifts the chromatin state of NPCs to an astrogenesis-competent configuration, whereby CT-1 can activate astrocytic genes (Fig. 1B). These studies have revealed much about how NPCs shift from neurogenesis to gliogenesis and have led to many exciting new questions including: How are the initial epigenetic marks laid down so that NPCs make neurons, then glia, and not vice versa? What promotes PcG-dependent repression of ngn loci over developmental time? Why doesn't early Notch signaling in NPCs (where it promotes maintenance of NPC fate) promote astrocyte production? Once Notch allows for STAT-dependent activation of astrocyte fate, what genes are turned on to promote astrocyte fate?
Astrocyte Diversity—Location Matters
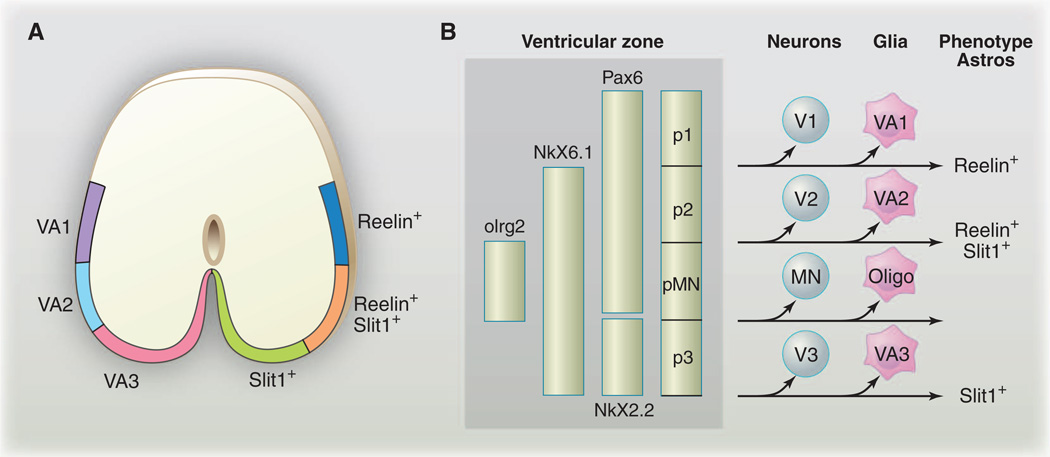
Whether there is molecular and functional diversity among newly born astrocytes remains poorly defined. In one (extreme) scenario, all newly born astrocytes could be essentially identical, with their mature phenotypes being shaped by interactions with their environment. Alternatively, astrocyte fates could be hard-wired, with astrocyte gene expression patterns and functional specializations that are largely predetermined at birth. Some intermediate scenario remains the most likely, as astrocytes are highly responsive to neuron-derived cues (below), and there is now good evidence that at least some aspects of vertebrate astrocyte fates are prepatterned. Positional identity along the dorsoventral (DV) and anteroposterior (AP) axis is a critical determinant of neuronal fates in the mammalian CNS. The ventricular zone is subdivided along the DV and AP axes by a combinatorial code of homeodomain transcription factors and precursors that generate unique pools of neuronal progeny that arise from distinct spatial expression domains (11). Recent work has identified three subpopulations of spinal cord white-matter astrocytes in the chick, which are organized along the dorsoventral axis (12). Each of these astrocyte populations is derived from a unique progenitor domain in the spinal cord (either VA1, VA2, or VA3 from dorsal to ventral) and has a unique pattern of expression of the markers Reelin and Slit1, and their fates appear to be regulated by homeodomain transcription factors which also govern diversification of neuronal fates (Fig. 2). For example, Pax6 is expressed in Reelin+ astrocytes, is required for Reelin expression, and, in the absence of Pax6, the border of Slit1 expression expands dorsally. Reciprocally, overexpression of Pax6 (even after the neurogenic phase is complete) is sufficient to expand the domain of Reelin expression ventrally and to suppress Slit1 expression (12). These types of cell fate switches are reminiscent of those seen in neuronal lineages when the expression domain of Pax6 is manipulated (13). However, the functional significance of VA1, VA2, and VA3 white matter astrocyte diversity remains unclear. Moreover, these represent only a subpopulation of white matter astrocytes (which are themselves a subpopulation of all astrocytes), and it remains to be determined whether the homeodomain code governs the specification of additional populations of astrocytes in a similar way.
Fig. 2.
Astrocyte diversity is generated by production from unique spatial domains. (A) A schematic illustrating VA1, VA2, and VA3 domains of the spinal cord white matter astrocytes and expression domains of Reelin and Slit1. (B) A homeodomain code that controls the generation of neuronal diversity is reused during astrocyte specification to promote white matter astrocyte diversity. [(A) and (B) are adapted from (12)]
Beyond global DV-AP patterning mechanisms there is also evidence for more local regulation of astrocyte production from NPCs. The basic helix-loop-helix transcription factor stem cell leukemia (Scl) is required for distinguishing between astrocyte and oligodendrocyte fates, but only at the border of the pMN and p2 domains of the spinal cord and not in other regions of the spinal cord. This likely represents a patterning step (i.e., binary choice mechanism), rather than a mechanism for astrocyte specification, because Scl also modulates v2a versus v2b interneuron fates in the same region of the spinal cord (14). It is plausible that several such astrocyte subpopulations exist whose production from NPCs is modulated in a regionally restricted way, but such mechanisms await description. Positional identity is thus an important organizing feature of astrocyte populations, with mechanisms like homeodomain codes patterning globally and Scl ensuring production of subpopulations in binary choice scenarios. Because location matters, a model whereby astrocyte phenotype is primarily determined by astrocyte interactions with the environment seems increasingly unlikely.
The Black Box of Astrocyte Specification
Although the above-described pathways potently modulate the neuron-to-glia switch in NPCs (i.e., when and where NPCs will make astrocytes), they do not seem to specify astrocyte fate. PcG genes act in the termination of neurogenesis (rather than induction of gliogenesis), JAK-STAT and Notch signaling makes NPCs competent to make astrocytes, and DV-AP patterning mechanisms determine where NPCs will make astrocytes. COUP-TFI and II (chicken ovalbumin upstream promoter-transcription factors I and II) are also critical regulators of the induction of gliogenesis, but like the above-described pathways, they appear to regulate the timing of NPC competence to make glia (15), rather than specifying glial fates per se. Moreover, all of these pathways affect not only astrocyte development but also that of other glia subtypes (e.g., oligodendrocytes) and/or neurons. It should also be noted that these studies relied primarily on the activation of GFAP or S100β as markers for astrocyte fates—these are known to be imperfect markers—and none traced these cells to the point where they had acquired their mature morphology in the brain. As such, it remains unclear how astrocyte-like the cells of interest did, or did not, become. To date, no mutants exist where astrocytes are completely lacking, and no astrocyte-specific transcription factors have been identified. Thus, genes that direct an immature cell in the nervous system to acquire the characteristics of a mature astrocyte remain to be discovered.
How will we move forward? The above study of DV-AP patterning of astrocytes was possible because Reelin and Slit1 allowed for the molecular discrimination of unique subsets of cells. New markers are needed to understand the development and diversity of these astrocyte subtypes. A recent large-scale screen of the spatial and temporal expression of more than 1400 mammalian transcription factors led to the identification of 12 with expression in spinal cord white matter in either oligodendrocyte or astrocyte lineages. Two of these genes, Scl and Klf15, were strongly enriched in expression in astrocytes, and their misexpression promoted precocious production of GFAP+ cells (16). A number of genomic approaches have been used to define the expression profiles of astrocytes and other major cell populations in the nervous system including GeneChip arrays (17, 18) and translating ribosome affinity purification (TRAP) (19). Comprehensive GeneChip arrays on highly purified astrocytes, oligodendrocytes, and neurons from multiple developmental stages provide a resource to explore how astrocytes change expression patterns over developmental time and have led to the identification of improved astrocyte markers like Aldh1L1 (17). Interestingly, microarray and TRAP studies have both called into question the classification of “glia” as a single brain cell class; expression analysis indicates that different glial subpopulations, such as astrocytes and oligodendrocytes, are as different from one another with respect to gene expression profiles as they are from neurons (17, 19). Both approaches have yielded treasure troves of interesting genes for future functional studies, but the next important steps include generating new markers to define the diversity and development of astrocyte populations, producing a new battery of astrocyte gene promoter-driven Cre lines to manipulate all or selected subsets of astrocytes, and ultimately defining which of these genes are key regulators of astrocyte specification, growth, and function.
The Mystery of Astrocyte Morphogenesis—Blossoms in the Brain
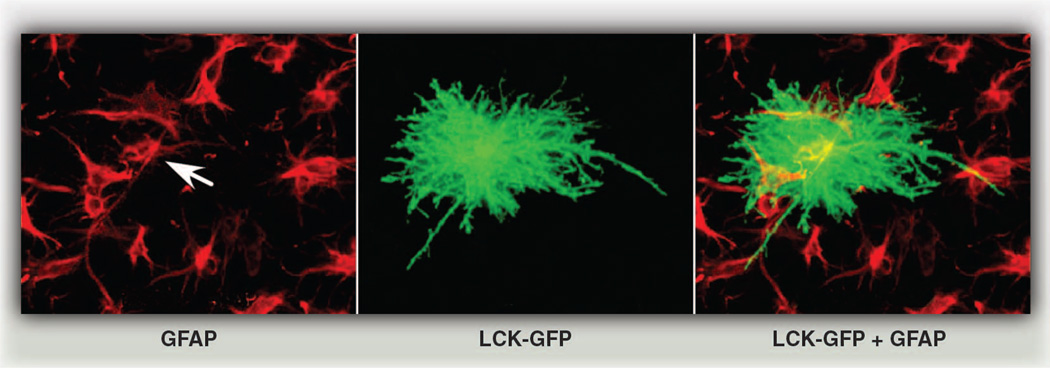
The morphology of a mature mammalian astrocyte is spectacular. From the cell soma radiate primary branches that gradually divide into finer and finer processes to generate a dense network of delicate terminal processes, which associate very closely with synapses. Until recently, most studies used GFAP immunostains to characterize astrocyte morphology, but this marker only reveals the structure of primary branches, which represent a meager ~15% of the total volume of the astrocyte (20) (Fig. 3). Astrocytes are more morphologically complex than was initially appreciated. Impressively, depending upon the particular brain region, a single mature rodent astrocyte can cover a spatial domain in the brain that ranges between 20,000 and 80,000 µm3 (20–22), wrap multiple neuronal somata (21), associate with 300 to 600 neuronal dendrites (21), and contact ~100,000 individual synapses (20, 23). In humans, these numbers increase dramatically, with a single astrocyte occupying a volume in the brain that is almost 30 times the volume in rodents and associating with ~2,000,000 synapses (23).
Fig. 3.
Astrocyte morphology is far more complex than initially appreciated. Astrocytes in an organotypic slice preparation labeled with the common marker GFAP (red) overlaid with a single astrocyte transfected with the membrane marker Lck-GFP (green fluorescent protein). Arrow indicates GFAP label of Lck-GFP–marked astrocyte. [From (53) with permission from Elsevier]
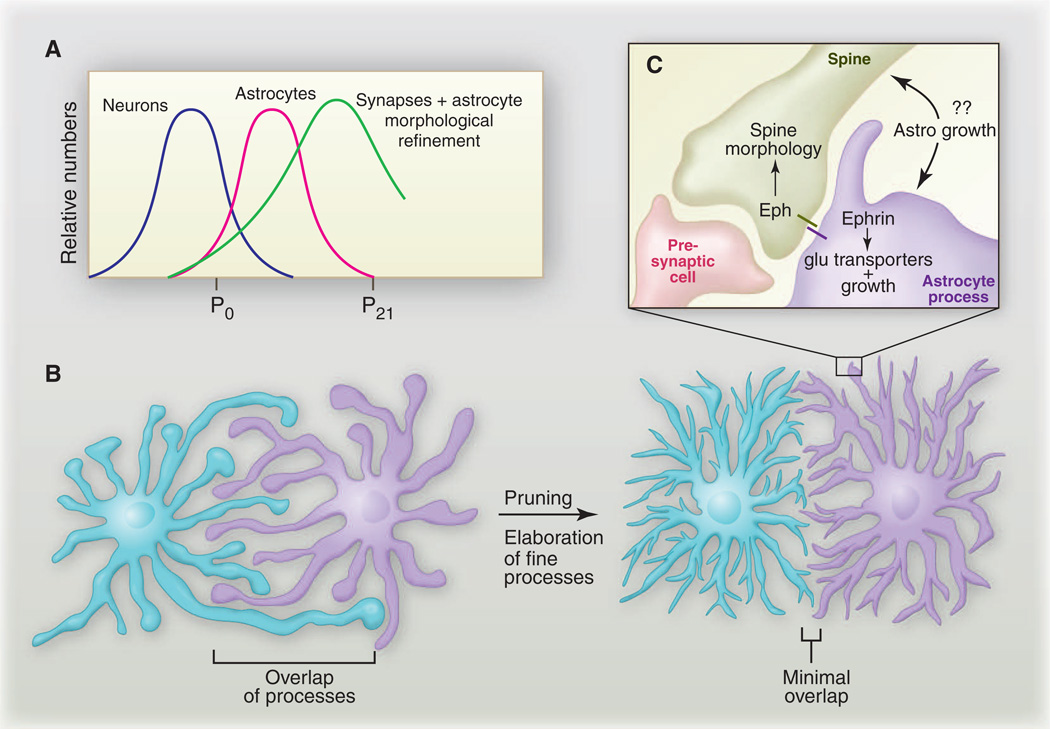
We know remarkably little about signaling pathways that direct immature astrocytes generated by NPCs to transform ultimately into mature astrocytes in vivo. Astrocytes sprout cellular processes as early as the first week of postnatal development, and most processes appear filopodial (i.e., actively growing) in nature. At this developmental time point, the borders of astrocytes are quite ragged, and long processes that extend well beyond the astrocyte’s border are commonly observed. However, during weeks 3 and 4 of postnatal development, astrocyte processes ramify to an increasing degree, distal processes become much thinner and spongiform in morphology, and fine astrocytic processes densely infiltrate the brain tissue (24). Thus, astrocyte morphology appears to be “mature” by week 3 to 4. In two landmark studies using dye injections to differentially label neighboring astrocytes, it was shown that each astrocyte, in fact, occupies a unique spatial domain and forms discrete borders with neighboring astrocytes (20, 22). How these borders form remains an intriguing and unexplored question. Early in development in the hippocampus (at postnatal day 7), which corresponds to the active growth stage of astrocyte development, neighboring astrocytes exhibit significant overlap of processes. However, overlapping processes are “pruned” back, and discrete borders develop by postnatal day 14, and this resolution of spatial domains becomes even more evident by postnatal day 21 (24). It is now believed that astrocytes “tile” with one another, through a mechanism akin to dendritic tiling, to accomplish complete coverage of the brain space (Fig. 4, A and B). How these domains are established and whether they are predefined or generated stochastically remain to be determined. For example, do initially overlapping astrocytic processes simply retract, or are they, in fact, pruned by larger-scale degradative mechanisms (e.g., overlapping processes are severed, degrade, and are cleared from the CNS) as has been observed with pruned dendrites and axons (25)? Is the extreme gap junction coupling observed in mature astrocytes a mechanism for communication among astrocytes during the establishment or maintenance of these domains? Astrocytic domains have been proposed to function as “synaptic islands,” whereby a single astrocyte could coordinate the activity of all synapses within its unique spatial domain, but their functional relevance remains to be determined (21). The integrity of astrocytic domains is compromised in mouse models of epilepsy within a week after injury, with astrocytes exhibiting a dramatic decrease in fine processes and an increase in the overlap of thicker processes, and these changes persist for 6 months after injury (26). The breakdown of unique astrocytic domains may occur in an injury-specific manner in the CNS because reactive astrocyte overlap was negligible when multiple brain regions were subjected to denervation (27).
Fig. 4.
Coordination of astrocyte morphological growth and refinement with synapse formation. (A) Although most neurons are made during embryonic stages, the major waves of synaptogenesis follow and depend on astrocyte production. The timing of astrocyte growth and morphological refinement overlaps significantly with this window of synaptogenesis. (B) Astrocytes initially extend large, filopodial processes that overlap significantly with neighboring astrocytes; however, by postnatal day 21, astrocytes refine their morphology to occupy unique spatial domains and elaborate fine processes that closely associate with synapses. (C) A complex interrelation exists between synapses and astrocyte processes. Eph or ephrin signaling can bidirectionally control astrocyte features, as well as spine morphology (see text).
What governs the branching and elaboration of astrocyte processes? Because astrocytes are sensitive to neuronal activity, it would be surprising if their morphology was not shaped during development in some way by local neural activity. Astrocyte membranes are extremely dynamic in live preparations of the mature brain and apparently more so near synapses than neuronal cell bodies (28), consistent with a correlation of motility with synaptic activity. In a number of physiological settings, astrocytes appear to respond morphologically to changes in neural activity (29, 30). For example, a wide range of studies have reported dynamic and activity-dependent changes in Ca2+ signaling in astrocytes (29); these include events that depend upon the major mammalian neurotransmitter glutamate (31, 32), and glial ensheathment of synapses appears to be increased after neuronal stimulation (33–35). Specific behaviors can also elicit coordinated Ca2+ signaling in glial networks in vivo (36). Astrocytes express an array of neurotransmitter receptors and transporters, as well as ion channels through which they likely sense neuronal activity and direct morphological changes (37, 38). However, the extent to which astrocyte growth during development depends on neuronal activity, or neuron-astrocyte signaling via other mechanisms, remains poorly defined.
Synaptogenesis and Astrocyte Maturation—Reciprocal Signaling During Brain Assembly?
How astrocyte growth is regulated such that initially simple cells can transform into a field of mature astrocytes with tiled and morphologically complex architecture remains a major question for the field. The close association of astrocytes with synapses, which is observed from even the earliest stages of development, is probably providing us with important clues. Astrocytes are now appreciated as important regulators of synapse formation, maintenance, efficacy, and elimination, but whereas increasing attention has been focused on how the synaptogenic activity of astrocytes sculpts neural connectivity, an equally interesting question is, do these newly formed synapses reciprocally shape astrocyte morphology? Not only is there is an intriguing coordinate timing between synapse formation and astrocyte birth (i.e., synapses form in earnest only after astrocytes appear), but the maturation of fine astrocytic processes and the establishment of astrocyte spatial domains seems to immediately follow the formation of new synapses (Fig. 4A). The major waves of rodent CNS synaptogenesis occur during the first 2 to 3 weeks of postnatal life (39–41). This is the time when astrocytes are dynamically growing. However, by weeks 3 to 4 (i.e., after the major wave of synapse formation), astrocytes have taken on their mature morphology, with fine processes closely associating with synapses and astrocytic spatial domains having been tiled out in the brain space. The expression of thrombospondins (TSP1 and TSP2), which are key astrocyte-secreted molecules that induce synapse formation, is high in the developing brain during week 1, but ceases by postnatal week 3 (42). This correlates with reduced synaptogenic potential of neurons and suggests that astrocytes down-regulate pathways that strongly promote synapse formation (41). Because astrocyte processes are broadly responsive to neural activity (30), it is easy to image how the initiation of synaptic activity could immediately begin to draw fine glial processes to newly formed synaptic contacts. For example, glutamate receptors (mGluRs) and transporters (EAAT1 and GLT-1) appear to be expressed in astrocytes during the first week of postnatal life (43, 44), which suggests that astrocytes are primed even at early stages to be responsive to neurotransmission. Accumulating evidence argues for an exciting and central role for Eph receptor–ephrin signaling in sculpting astrocyte-dendritic spine interactions. Astrocytic ephrin-A3 modulates dendritic spine morphology through neuronal EphA4, with activation of EphA4 negatively regulating spine growth (45), and there is an intriguing correlation between astrocyte contact and the maturation and lifetime of dendritic spines (46). Eph or ephrin signaling activates bidirectional signaling between the engaged cells, which would offer an elegant mechanism for simultaneously coordinating astrocyte process growth and spine morphology through a single pathway. Indeed, EphA4 and/or ephrin-A3 can modulate expression of glutamate transporters in glia (47), which appears critical for synaptic plasticity (48) (Fig. 4C). However, the role of Eph or ephrin signaling in astrocyte growth control has only been explored in the mature brain in vivo, despite the fact that ephrins can stimulate filopodial outgrowth from astrocytes within minutes (49).
In Closing—It’s Wide Open
The future looks very bright for the field of astrocyte biology. We have a new appreciation for the fact that astrocytes are far more than simple support cells. They are active participants in brain assembly and signaling, and it is abundantly clear that we have only scratched the surface of the depth of their biology. Interest in glial cells has been increasing dramatically over the past two decades, and this has led to the inevitable development of new sophisticated genetic tools with which to label and manipulate astrocytes in vivo. New tools that allow for temporally controlled deletion of genes, specifically in mouse astrocytes, along with improved high-resolution imaging techniques, are enabling researchers to address fundamental questions in astrocyte biology for the first time. These tools need to be expanded and exploited to pry deeply into astrocyte biology in vivo. There is also growing interest in the use of genetic model organisms like Drosophila, Caenorhabditis elegans, and zebrafish in the study of glial biology. Drosophila have glial subtypes that are morphologically and molecularly similar to mammalian astrocytes (50, 51), C. elegans glia share a range of developmental and functional properties with mammalian glia (52), and these organisms are amenable to rapid forward genetic screens. Now is the time to harness the powerful forward genetic approaches in these organisms to explore astrocyte development and function. From a developmental perspective, central questions for the field include the following: What gene expression profile and key genes make an astrocyte? How do astrocytes grow into intricate morphologies and associate with synapses? How dynamic is this association? Precisely how do astrocytes govern synaptogenesis or other aspects of neural circuit formation? How do synapses reciprocally govern astrocyte maturation? Finally, how are developmental mechanisms reused in the adult during plasticity or changed after injury or in disease?
Acknowledgments
M.R.F. is supported by NIH grant NS053538 and is a Howard Hughes Medical Institute Early Career Scientist.
References and Notes
- 1.Miller FD, Gauthier AS. Neuron. 2007;54:357. doi: 10.1016/j.neuron.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Hirabayashi Y, et al. Development. 2004;131:2791. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 3.Israsena N, Hu M, Fu W, Kan L, Kessler JA. Dev. Biol. 2004;268:220. doi: 10.1016/j.ydbio.2003.12.024. [DOI] [PubMed] [Google Scholar]
- 4.Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuroscience. 2006;142:1119. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Hirabayashi Y, et al. Neuron. 2009;63:600. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 6.Morrow T, Song MR, Ghosh A. Development. 2001;128:3585. doi: 10.1242/dev.128.18.3585. [DOI] [PubMed] [Google Scholar]
- 7.Takizawa T, et al. Dev. Cell. 2001;1:749. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- 8.Namihira M, et al. Dev. Cell. 2009;16:245. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Deneen B, et al. Neuron. 2006;52:953. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Barnabé-Heider F, et al. Neuron. 2005;48:253. doi: 10.1016/j.neuron.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 11.Shirasaki R, Pfaff SL. Annu. Rev. Neurosci. 2002;25:251. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 12.Hochstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson DJ. Cell. 2008;133:510. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi M, Osumi N. Development. 2002;129:1327. doi: 10.1242/dev.129.6.1327. [DOI] [PubMed] [Google Scholar]
- 14.Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Nature. 2005;438:360. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- 15.Naka H, Nakamura S, Shimazaki T, Okano H. Nat. Neurosci. 2008;11:1014. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- 16.Fu H, et al. J. Neurosci. 2009;29:11399. doi: 10.1523/JNEUROSCI.0160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahoy JD, et al. J. Neurosci. 2008;28:264. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovatt D, et al. J. Neurosci. 2007;27:12255. doi: 10.1523/JNEUROSCI.3404-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle JP, et al. Cell. 2008;135:749. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bushong EA, Martone ME, Jones YZ, Ellisman MH. J. Neurosci. 2002;22:183. doi: 10.1523/JNEUROSCI.22-01-00183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halassa MM, Fellin T, Takano H, Dong JH, Haydon PG. J. Neurosci. 2007;27:6473. doi: 10.1523/JNEUROSCI.1419-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata K, Kosaka T. Neuroscience. 2002;113:221. doi: 10.1016/s0306-4522(02)00041-6. [DOI] [PubMed] [Google Scholar]
- 23.Oberheim NA, Wang X, Goldman S, Nedergaard M. Trends Neurosci. 2006;29:547. doi: 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Bushong EA, Martone ME, Ellisman MH. Int. J. Dev. Neurosci. 2004;22:73. doi: 10.1016/j.ijdevneu.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Luo L, O’Leary DD. Annu. Rev. Neurosci. 2005;28:127. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- 26.Oberheim NA, et al. J. Neurosci. 2008;28:3264. doi: 10.1523/JNEUROSCI.4980-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilhelmsson U, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17513. doi: 10.1073/pnas.0602841103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirrlinger J, Hülsmann S, Kirchhoff F. Eur. J. Neurosci. 2004;20:2235. doi: 10.1111/j.1460-9568.2004.03689.x. [DOI] [PubMed] [Google Scholar]
- 29.Nimmerjahn A. J. Physiol. 2009;587:1639. doi: 10.1113/jphysiol.2008.167171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theodosis DT, Poulain DA, Oliet SH. Physiol. Rev. 2008;88:983. doi: 10.1152/physrev.00036.2007. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, et al. Nat. Neurosci. 2006;9:816. doi: 10.1038/nn1703. [DOI] [PubMed] [Google Scholar]
- 32.Petzold GC, Albeanu DF, Sato TF, Murthy VN. Neuron. 2008;58:897. doi: 10.1016/j.neuron.2008.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lushnikova I, Skibo G, Muller D, Nikonenko I. Hippocampus. 2009;19:753. doi: 10.1002/hipo.20551. [DOI] [PubMed] [Google Scholar]
- 34.Wenzel J, Lammert G, Meyer U, Krug M. Brain Res. 1991;560:122. doi: 10.1016/0006-8993(91)91222-m. [DOI] [PubMed] [Google Scholar]
- 35.Genoud C, et al. PLoS Biol. 2006;4:e343. doi: 10.1371/journal.pbio.0040343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nimmerjahn A, Mukamel EA, Schnitzer MJ. Neuron. 2009;62:400. doi: 10.1016/j.neuron.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verkhratsky A, Steinhäuser C. Brain Res. Brain Res. Rev. 2000;32:380. doi: 10.1016/s0165-0173(99)00093-4. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Rothstein JD. In: Astroctyes in (Patho)physiology of the Nervous System. Parpura V, Haydon PG, editors. Boston: Springer; 2009. pp. 69–105. [Google Scholar]
- 39.Lund RD, Lund JS. Brain Res. 1972;42:1. doi: 10.1016/0006-8993(72)90038-8. [DOI] [PubMed] [Google Scholar]
- 40.Warton SS, McCart R. Synapse. 1989;3:136. doi: 10.1002/syn.890030205. [DOI] [PubMed] [Google Scholar]
- 41.Ullian EM, Christopherson KS, Barres BA. Glia. 2004;47:209. doi: 10.1002/glia.20082. [DOI] [PubMed] [Google Scholar]
- 42.Christopherson KS, et al. Cell. 2005;120:421. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 43.Regan MR, et al. J. Neurosci. 2007;27:6607. doi: 10.1523/JNEUROSCI.0790-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schools GP, Kimelberg HK. J. Neurosci. Res. 1999;58:533. doi: 10.1002/(sici)1097-4547(19991115)58:4<533::aid-jnr6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 45.Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Nat. Neurosci. 2003;6:153. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- 46.Nishida H, Okabe S. J. Neurosci. 2007;27:331. doi: 10.1523/JNEUROSCI.4466-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carmona MA, Murai KK, Wang L, Roberts AJ, Pasquale EB. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12524. doi: 10.1073/pnas.0903328106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filosa A, et al. Nat. Neurosci. 2009;12:1285. doi: 10.1038/nn.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nestor MW, Mok LP, Tulapurkar ME, Thompson SM. J. Neurosci. 2007;27:12817. doi: 10.1523/JNEUROSCI.2442-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doherty J, Logan MA, Taşdemir OE, Freeman MR. J. Neurosci. 2009;29:4768. doi: 10.1523/JNEUROSCI.5951-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawai H, Arata N, Nakayasu H. Glia. 2001;36:406. doi: 10.1002/glia.1126. [DOI] [PubMed] [Google Scholar]
- 52.Shaham S. Curr. Opin. Neurobiol. 2006;16:522. doi: 10.1016/j.conb.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 53.Benediktsson AM, Schachtele SJ, Green SH, Dailey ME. J. Neurosci. Methods. 2005;141:41. doi: 10.1016/j.jneumeth.2004.05.013. [DOI] [PubMed] [Google Scholar]