SUMMARY
Origin recognition complex (ORC) plays critical roles in the initiation of DNA replication and cell-cycle progression. In metazoans, ORC associates with origin DNA during G1 and with heterochromatin in postreplicated cells. However, what regulates the binding of ORC to chromatin is not understood. We have identified a highly conserved, leucine-rich repeats and WD40 repeat domain-containing protein 1 (LRWD1) or ORC-associated (ORCA) in human cells that interacts with ORC and modulates chromatin association of ORC. ORCA colocalizes with ORC and shows similar cell-cycle dynamics. We demonstrate that ORCA efficiently recruits ORC to chromatin. Depletion of ORCA in human primary cells and embryonic stem cells results in loss of ORC association to chromatin, concomitant reduction of MCM binding, and a subsequent accumulation in G1 phase. Our results suggest ORCA-mediated association of ORC to chromatin is critical to initiate preRC assembly in G1 and chromatin organization in post-G1 cells.
INTRODUCTION
Origin recognition complex (ORC) proteins are critical for the initiation of DNA replication (Bell and Stillman, 1992). Binding of ORC to chromatin serves as a landing pad for the assembly of a multiprotein prereplicative complex at the origins of replication (Bell and Dutta, 2002). Studies from different species have shown that ORC is highly dynamic, with several ORC subunits getting posttranslationally modified by phosphorylation or ubiquitination in a cell cycle-dependent manner (DePamphilis, 2005). ORC binding to origins is ATP dependent, but the sequence requirement for ORC binding shows significant species differences. In S. pombe, Orc4 is known to mediate DNA binding via its multiple AT hooks (Chuang and Kelly, 1999), whereas in Drosophila Orc6 is required for DNA binding by ORC (Balasov et al., 2007). In S. cerevisiae, ORC is known to recognize specific ori elements (Bell and Stillman, 1992). In the early embryonic cell cycles of Xenopus and Drosophila as well as in replication-competent Xenopus egg extracts, the initiation of DNA replication is sequence independent (Blow and Laskey, 1986; Hyrien and Mèchali, 1993; Sasaki et al., 1999; Walter et al., 1998). In human cells, even though certain discrete origins have been identified, no consensus DNA sequence has emerged. Using human ORC purified from insect cells, it has been demonstrated that ORC stimulates replication initiation from any DNA fragment in vitro without any preference for known origin DNA (Vashee et al., 2003). It has been suggested that the binding of ORC to specific origin sequences in vivo is attributed to the chromatin context (Okuno et al., 2001) or to other proteins that provide specificity for chromatin binding, similar to what has been observed for E2F-dependent ORC binding to chorion gene locus or Myb-containing protein complex in site-specific DNA replication in Drosophila (Beall et al., 2002; Royzman et al., 1999) or EBNA-1 dependent ORC loading to EBV origin OriP (Dhar et al., 2001).
In addition to its role in prereplication complex (preRC) assembly, individual subunits of ORC also play vital roles in several non-preRC functions (Chesnokov, 2007; Sasaki and Gilbert, 2007). ORC binds to origins as well as to several other chromosomal regions that represent repressed chromatin in yeast as well as in metazoans. ORC plays key roles in the establishment and spreading of Sir proteins at mating type loci in yeast (Triolo and Sternglanz, 1996). Similarly, ORC plays crucial functions in recruiting HP1 and eventually in its spreading at heterochromatic loci in metazoans (Prasanth et al., 2010; Shareef et al., 2003). Also, in fission yeast S. pombe, ORC interacts with the HP1-related protein Swi6 that is involved in centromere and mating type gene silencing (Matsuda et al., 2007). In all of these events, key chromatin changes accompany ORC binding. These data are indicative of the evolutionarily conserved function of ORC in gene silencing and heterochromatin organization in addition to their conserved function in replication initiation. Even though ORC is highly conserved, how is it recruited to origins as well as to heterochromatic sites in postreplicated cells in metazoans and what additional factor(s) regulate ORC loading remain to be elucidated.
In the present study, we have characterized the function of a WD40 repeat-containing ORC-interacting protein, leucinerich repeats and WD repeat domain-containing protein 1 (LRWD1). We henceforth name it ORC-associated (ORCA) in human cells. ORCA is highly conserved among higher eukaryotes and interacts with ORC, utilizing its WD repeat domain. Further, ORCA associates with chromatin predominantly during G1, with levels diminishing during S phase and reloading during G2. ORCA colocalizes with ORC at specific chromatin structures, including telomeres and centromeres. Tethering of ORCA to an artificially generated chromatin region results in efficient recruitment of ORC to the chromatin site. Finally, depletion of ORCA in human primary diploid cells and embryonic stem cells (hESCs) results in loss of ORC and MCM loading to chromatin, resulting in accumulation in G1. Based on our data, we propose that ORCA is required for the association of ORC on chromatin during G1 to establish preRC and to heterochromatic sites in post-replicated cells.
RESULTS
ORCA Is Conserved among Higher Eukaryotes
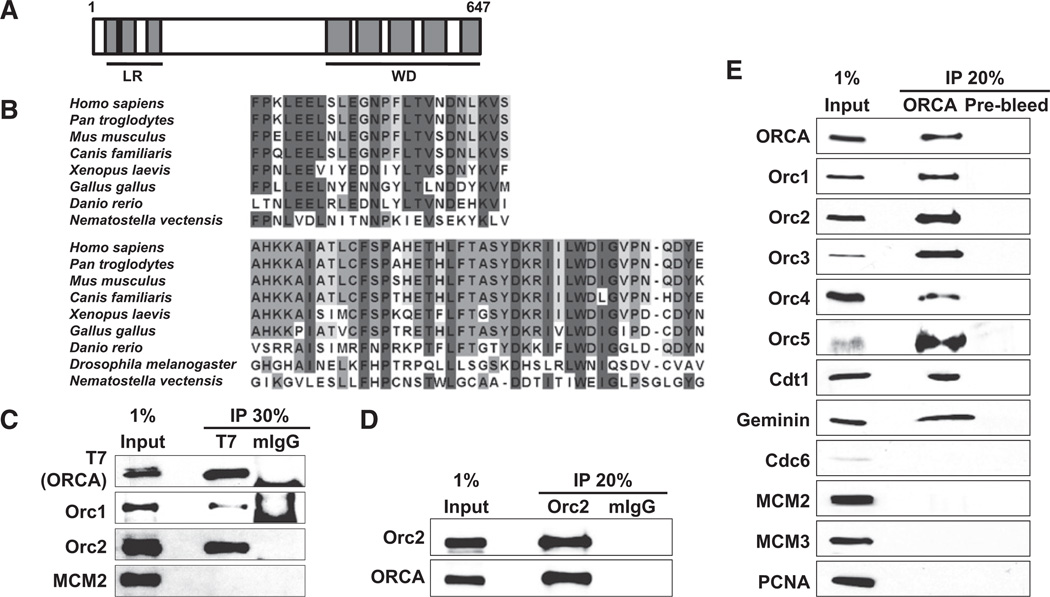
We have performed biochemical analysis in mammalian cells to identify proteins that associate with ORC and play crucial roles in DNA origin specification and/or chromatin organization. Immunoprecipitation (IP) with Orc2 monoclonal antibody (mAb) from nuclear extracts of asynchronously growing human HeLa cells or from nocodazole-arrested mitotic cells was conducted, and the IP material was subjected to mass spectrometric analysis. Similarly, IP with T7 antibody from nuclear extracts of asynchronously growing T7-Orc2-expressing stable cell line followed by mass spectrometry was also conducted. We have identified a protein, ORCA, alias LRWD1 (NP_690852, DKFZp434K1815), as one of the ORC interactors in both IP-mass spectrometric screens. ORCA is composed of 647 aa and motif scan predictions suggest that it contains three leucine rich repeats (LRR) at the N terminus and five WD40 repeats at the C terminus (Figure 1A). Alignment of ORCA from different species, including human, chimpanzee, mouse, dog, zebrafish, chicken, frog, fly, and sea anemone, showed very high levels of conservation among higher eukaryotes, with maximum homology discernible at the LRR- and WD-containing regions (Figure 1B and Figure S1 available online; see the Supplemental Experimental Procedures for accession numbers).
Figure 1. ORCA Is a Highly Conserved ORC-Binding Protein.
(A) Schematic representation of ORCA domain structure. Simple Modular Architecture Research Tool (SMART)-based domain predictions reveal three leucine-rich repeats at the N terminus and five WD repeats at the C terminus.
(B) Sequence alignment shows high conservation of ORCA among higher eukaryotes (see the Supplemental Experimental Procedures for accession numbers). The top block represents alignment of LRR-3 (aa 90–113 human) and the lower block is WD40 repeat-1 (aa 390–430 human). Note that on the basis of bioinformatics prediction, only a conserved WD40-containing protein was found as a possible ortholog in Drosophila.
(C) ORCA associates with ORC. Immunoprecipitation with T7 mAb in cells transfected with T7-ORCA followed by immunoblots with antibodies against T7, Orc1, Orc2, and MCM2 reveals specific interaction of T7-ORCA with Orc1 and Orc2. Mouse IgG (mIgG) was used as the control.
(D) Reverse IP with Orc2 mAb shows Orc2 can efficiently pull down endogenous ORCA from human cells.
(E) ORCA interacts with multiple components of preRC. IP with ORCA antibody shows the interaction with ORC subunits, Cdt1, and Geminin. See also Figure S1.
ORCA Is an ORC-Associating Protein
To validate the IP-mass spectrometric data on the interaction of ORCA with ORC, we conducted reverse IP and coIP experiments. An epitope-tagged version of ORCA, T7-ORCA, was transfected into HeLa cells, and IP was carried out with T7 monoclonal antibody (mAb). Both human Orc1 and Orc2 coimmunoprecipitated with T7-ORCA (Figure 1C). Further, T7-ORCA and HA-Orc2 were cotransfected into HeLa cells, and IP with either T7 or HA antibody followed by immunoblot analyses further confirmed their interaction (Figure S1B). IP with Orc2 antibody followed by immunoblot with ORCA polyclonal antibody (pAb) showed the association of Orc2 with endogenous ORCA (molecular weight of ORCA ~70kDa; Figure 1D). Finally, immunoprecipitation with ORCA pAb in human cells clearly demonstrated the association of ORCA with ORC subunits (Figure 1E). Cdt1 and Geminin were also associated with ORCA (Figure 1E). Other preRC components like Cdc6 or MCMs or DNA replication-related proteins PCNA were not part of the ORCA-containing complex (Figure 1E). Thus, immunoprecipitation studies using antibodies against endogenous proteins as well as tagged versions confirmed the interaction between ORCA and ORC.
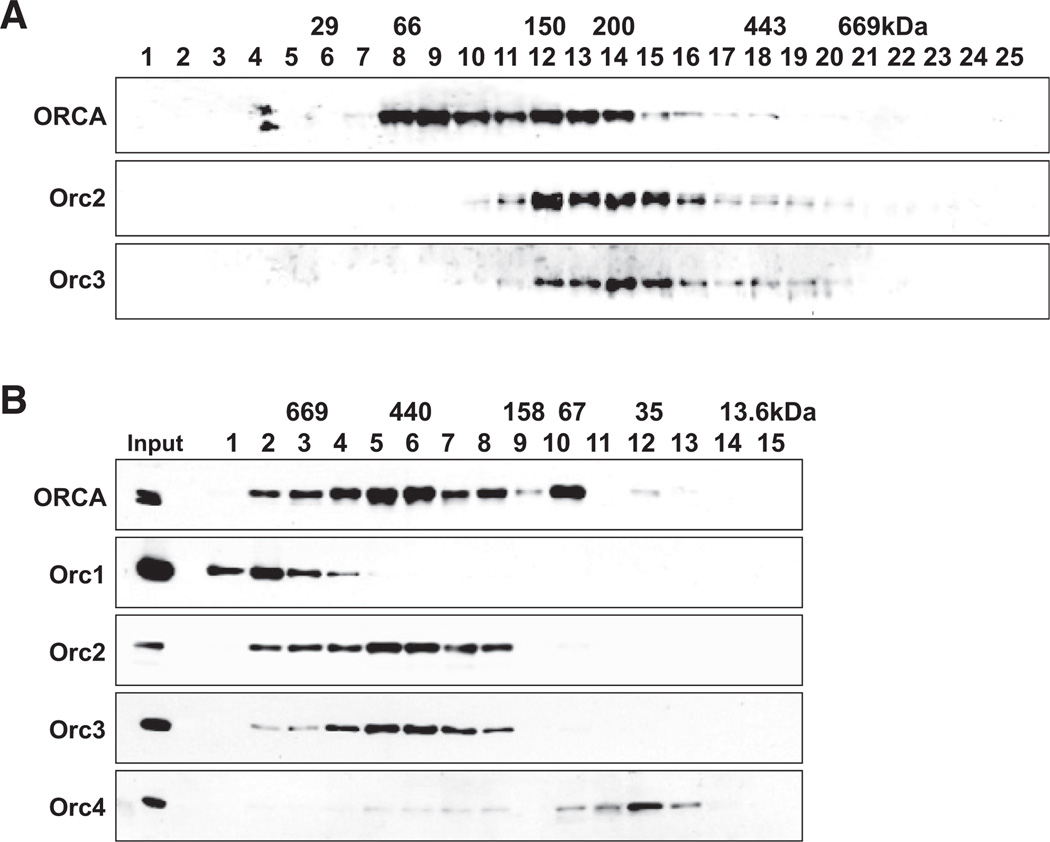
To study the complex assembly of ORCA and ORC, we eluted immunoprecipitates after ORCA IP with specific peptides, subjected the eluate to 10%-50% glycerol gradient sedimentation, and examined the fractions by Orc2 and 3 immunoblots (Figure 2A). Cosedimentation of ORCA and Orc2 and 3 (Figure 2A) to a position >200 kDa suggested that multiple ORC subunits are present in a complex with ORCA. To further investigate the complex assembly of ORCA and multiple ORC subunits, we fractionated HeLa nuclear extract on a Superdex-200 gel filtration column. The fractions were examined by immunoblots with antibodies against ORCA and various ORC subunits (Figure 2B). A significant portion of ORCA was found in fractions 5 and 6 (>440 kDa). As reported previously (Thome et al., 2000), Orc2 (66 kDa) and Orc3 (80 kDa) both coeluted at ~400 kDa (Figure 2B, fractions 5 and 6). Only a small fraction of Orc4 was present in the fraction coeluting with Orc2 and Orc3. The majority of Orc4 was present in apparently monomeric form (Figure 2B, fractions 11 and 12). In addition to fractions 4 and 5, Orc1 and ORCA along with Orc2 and 3 are also present in a higher molecular weight range. The predicted molecular mass of ORCA along with all ORC subunits is ~450 kDa, indicating that ORCA is in complex containing multiple ORC subunits in a high molecular weight (HMW) complex.
Figure 2. ORCA Exists in Multiple Subcomplexes in the Cell.
(A) Glycerol gradient sedimentation analysis of ORCA complex on material immunoprecipitated with ORCA antibodies. The corresponding molecular weight markers are labeled. Note that ORCA, Orc2, and Orc3 cosediment in fractions 12–14.
(B) Association of ORCA with ORC in human cells. HeLa nuclear extract fractionated over a Superdex 200 gel filtration column and fractions analyzed for ORCA and various ORC subunits by immunoblot. Molecular weight markers are labeled on top of the panel. Note the multiple peaks of ORCA corroborating its existence in multiple subcomplexes.
The Binding of ORCA to Chromatin Is Cell-Cycle Dependent
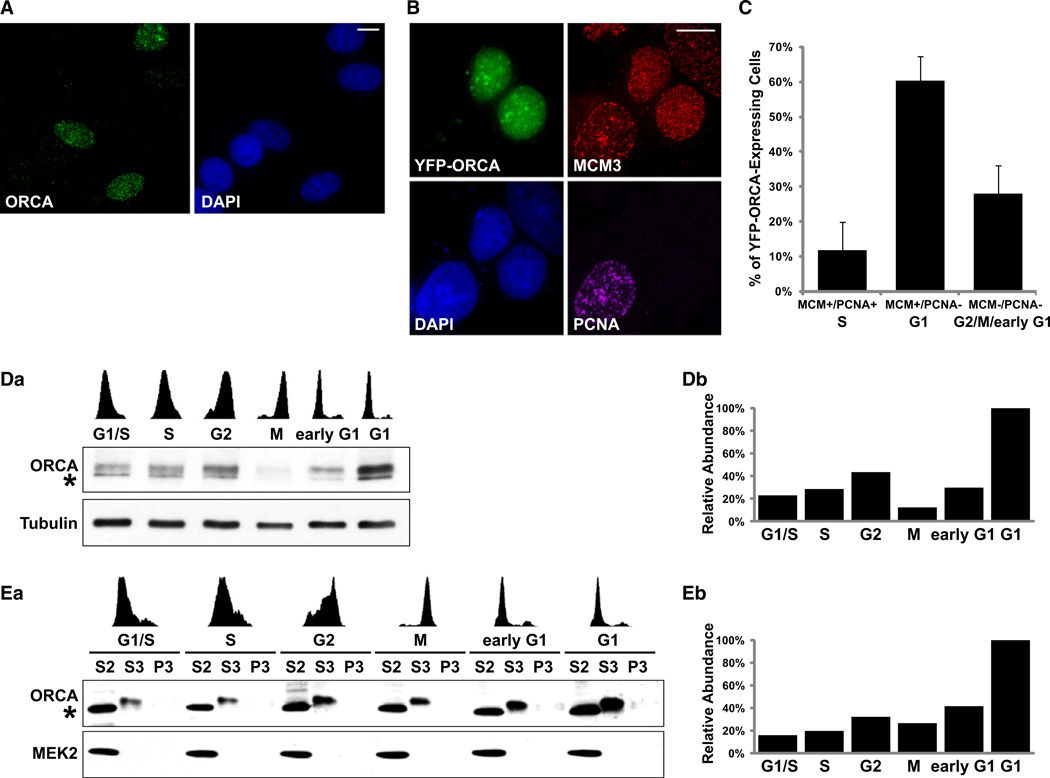
Intracellular localization and cell-cycle distribution of a protein are vital indicators of its cellular function(s). To study the distribution of ORCA in human cells, we performed immunofluorescence (IF) localization analysis using ORCA antibodies in MCF7 (Figure 3A) and U2OS cells (data not shown). ORCA localized predominantly in the nucleus of some cells as punctate foci, and several cells showed differential staining (Figure 3A), suggesting potential cell cycle-regulated expression of ORCA.
Figure 3. ORCA Association with Chromatin Is Cell-Cycle Regulated.
(A) Immunostaining of human cells with ORCA-specific antibody shows differential staining of ORCA, with some cells showing strong staining, while others lacking it.
(B) YFP-ORCA-expressing MCF7 cells were pre-extracted and immunostained with MCM3 and PCNA. Note that the YFP-ORCA-expressing cells show punctate labeling of MCM3 and lack of PCNA, pointing to the presence of ORCA protein in G1 cell population. DNA is counterstained with DAPI (blue). Scale bars in (A) and (B) represent 10 µm.
(C) Statistical analysis of MCM/PCNA profile in YFP-ORCA-expressing MCF7 cells. Note that ORCA is predominantly present in cells that show MCM+/PCNA– (60.3% ± 6.1%), representing G1 cells. ORCA-expressing, MCM+/PCNA+ cells denoting cells in S phase is 11.8% ± 8.2%, and MCM–/PCNA– is 27.9% ± 8.1%. The graph represents mean values plus standard deviation from four independent experiments (n = ~100 cells in each experiment).
(D) Levels of ORCA during the cell cycle. ORCA immunoblot of whole-cell extract from cells synchronized during specific stages of the cell cycle. Note the increase in ORCA during G1 phase.
(E) Biochemical fractionation of ORCA. Chromatin fractionation from cells synchronized during different stages of the cell cycle show accumulation of ORCA in G1 cells (S3 fraction) and decrease during G1/S and S. “*” denotes the cross-reacting band of ORCA pAb. Note that the cross-reacting band appears in the cytosolic fraction and is therefore not found in any of the experiments where nuclear extracts were used (Figures 1 and 2). MEK2 is shown as a control for chromatin fractionation. Flow cytometry profiles are shown on top of the panel. S2 represents cytosolic fraction, S3 nuclear soluble and MNase released nuclear fraction, and P3 the insoluble and MNase resistant nuclear fraction.
Bar diagrams in (Db) and (Eb) represent the relative abundance of ORCA throughout cell cycle (level in G1 is being considered 100%).
To investigate the cell cycle-restricted distribution of ORCA, we immunostained MCM3 and PCNA in YFP-ORCA-expressing MCF7 (Figure 3B) and U2OS (data not shown) cells after pre-extraction with a nonionic detergent, which reflects the fraction of proteins that preferentially associates with chromatin. MCM proteins are associated with chromatin during G1 and S phase, while PCNA is associated with chromatin only during S phase (Prasanth et al., 2004a). Therefore, dual IF of MCM and PCNA (following pre-extraction procedures) serves as a tool to identify the cell-cycle stage of individual cells. Immunolabeling of MCM3 and PCNA in YFP-ORCA-expressing cells demonstrated that ORCA was present predominantly in cells that showed homogenous MCM labeling and were PCNA negative (Figures 3B and 3C), indicating G1 cells.
Biochemical fractionation of cells collected at different stages of the cell cycle further corroborated that ORCA whole-cell levels and chromatin association are cell-cycle regulated (Figures 3D and 3E). ORCA was obtained in the nuclear chromatin fraction (released into S3 after MNase treatment; Figure 3E), with maximum association observed during G1 phase. The levels of ORCA were found to decrease during S phase, subsequently beginning to rise during G2, and peaking at G1 (Figure 3E). Time-lapse live-cell imaging of YFP-ORCA in human cells further confirmed the significant increase of ORCA during G1, after mitotic cell division (Movie S1 and Figure S2). Immunofluorescence and biochemical fractionation experiments have clearly indicated that ORCA distribution is regulated during the cell cycle. The dynamics of ORCA through the cell cycle are reminiscent of ORC dynamics, especially Orc2 (Figure S3B), suggesting that ORCA and ORC might cooperate in chromatin association during specific cell-cycle stages.
ORCA Localizes to Heterochromatic Structures
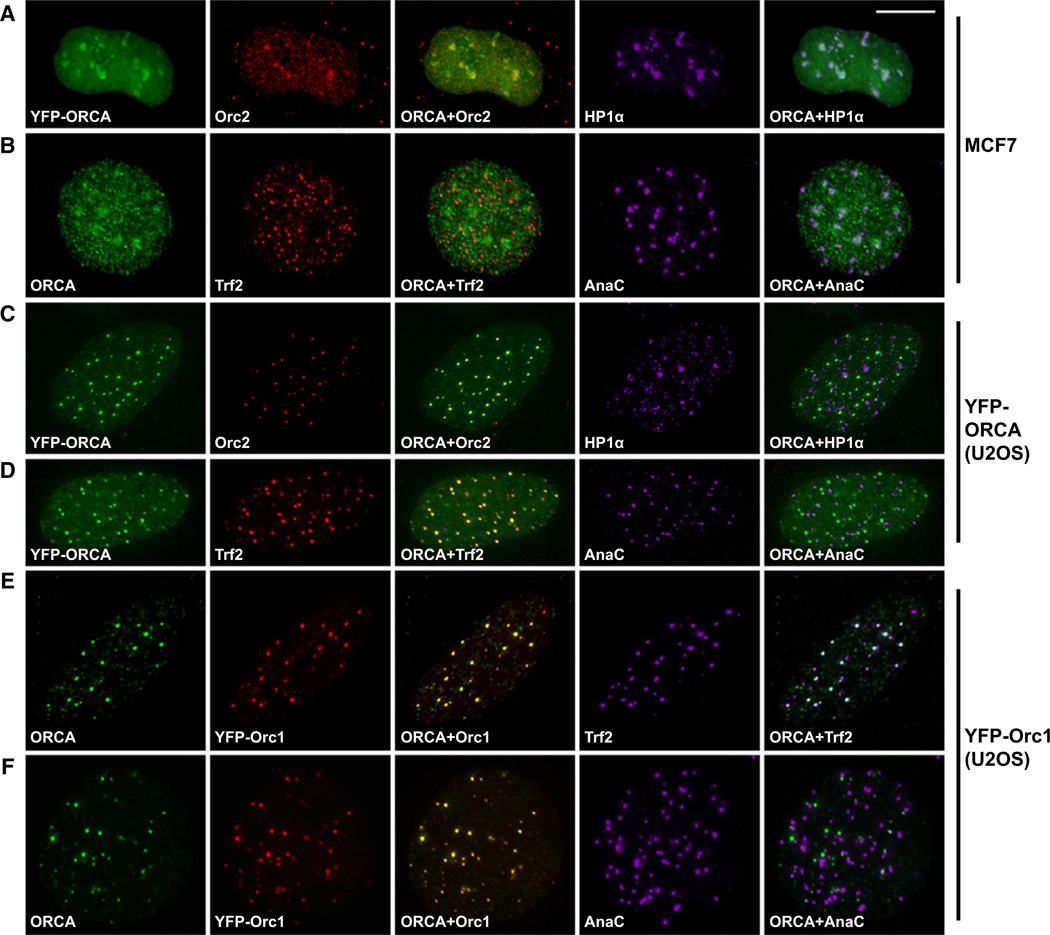
Previous studies have shown that multiple subunits of human ORC associate with heterochromatic structures (including centromeres and telomeres) and interact with the heterochromatin protein, HP1 (Deng et al., 2007; Lidonnici et al., 2004; Prasanth et al., 2004b, 2010). We observed prominent nuclear foci of YFP-ORCA in MCF7 cells (Figure 4A). Pre-extraction with detergent was used to remove soluble fraction of YFP-ORCA in MCF7 and U2OS cells. Immunolabeling of Orc2 and HP1α in YFP-ORCA-expressing MCF7 cells showed colocalization of ORCA and ORC at heterochromatic foci (Figure 4A). Immunofluorescence staining with ORCA antibody or YFP-ORCA localization in MCF7 cells showed distribution of ORCA in heterochromatic foci that overlapped with centromeric heterochromatin (AnaC) but not with telomeric sites (telomere binding protein; Trf2) (Figure 4B and Figure S3A; pearson coefficient of correlation for Figure 4B is provided in Figure S3A). Localization studies of ORCA in U2OS cells showed complete colocalization of YFP-ORCA with Orc2 (Figure 4C) and ORCA pAb with YFP-Orc1 (Figure 4E and 4F) at specific punctate foci. Detailed examination of the punctate foci revealed that in U2OS cells, ORCA overlaps with Orc1 and Orc2 at telomeric heterochromatin specifically in interphase cells (Trf2, Figures 4D and 4E) but not at centromeres as observed in MCF7 cells (AnaC, Figures 4D and 4F). ORCA showed similar cell-cycle dynamics as that of Orc2; it showed punctate homogenous staining during G1 and was restricted to centromeres (MCF7) or telomeres (U2OS, Figure S3B) as cells progressed though S phase (monitored by CAF1, Figure S3B). But as cells entered mitosis, ORCA and Orc2 relocalized to centromeres in all cell lines tested (Figure S3C). The difference in ORCA and ORC distribution between MCF7 and U2OS interphase cells was rather striking. Examining several other cell lines including HeLa and IMR-90 (data not shown) revealed that the association of ORCA to inter-phase cell centromeres was restricted to telomerase-positive cells, whereas ORCA predominantly localized to telomeres in cells utilizing alternative lengthening of telomeres (ALT). Similar observations were made for Orc1 and Orc2 distribution in human cells (Figures 4A, 4C, 4E, and 4F). Our results indicate that ORCA displayed similar localization in cells to that of ORC and also showed complete overlap with ORC at heterochromatic sites, including centromeres and telomeres, implying that the intracellular distribution of ORCA and ORC are coregulated.
Figure 4. ORCA Colocalizes with ORC at Heterochromatic Structures.
(A) IF with Orc2 pAb and HP1α antibodies after pre-extraction procedure in YFP-ORCA-expressing MCF7 cells reveals colocalization of ORCA with Orc2 and HP1α at heterochromatic regions.
(B) Triple immunolabeling with antibodies against ORCA, telomere binding protein Trf2, and centromere protein AnaC shows colocalization of ORCA with centromeric and pericentromeric heterochromatin in MCF7 cells.
(C) Immunostaining of Orc2 and HP1α in YFP-ORCA-expressing stable U2OS cells shows the complete overlap of ORCA and Orc2.
(D) Immunostaining of Trf2 and AnaC in YFP-ORCA-expressing stable U2OS cells shows colocalization of YFP-ORCA at telomeres but not at centromeres.
(E and F) Similar overlap of endogenous ORCA and YFP-Orc1 in U2OS cells at telomeres (E) but not at centromeres (F).
The scale bar represents 10 µm. See also Figure S3.
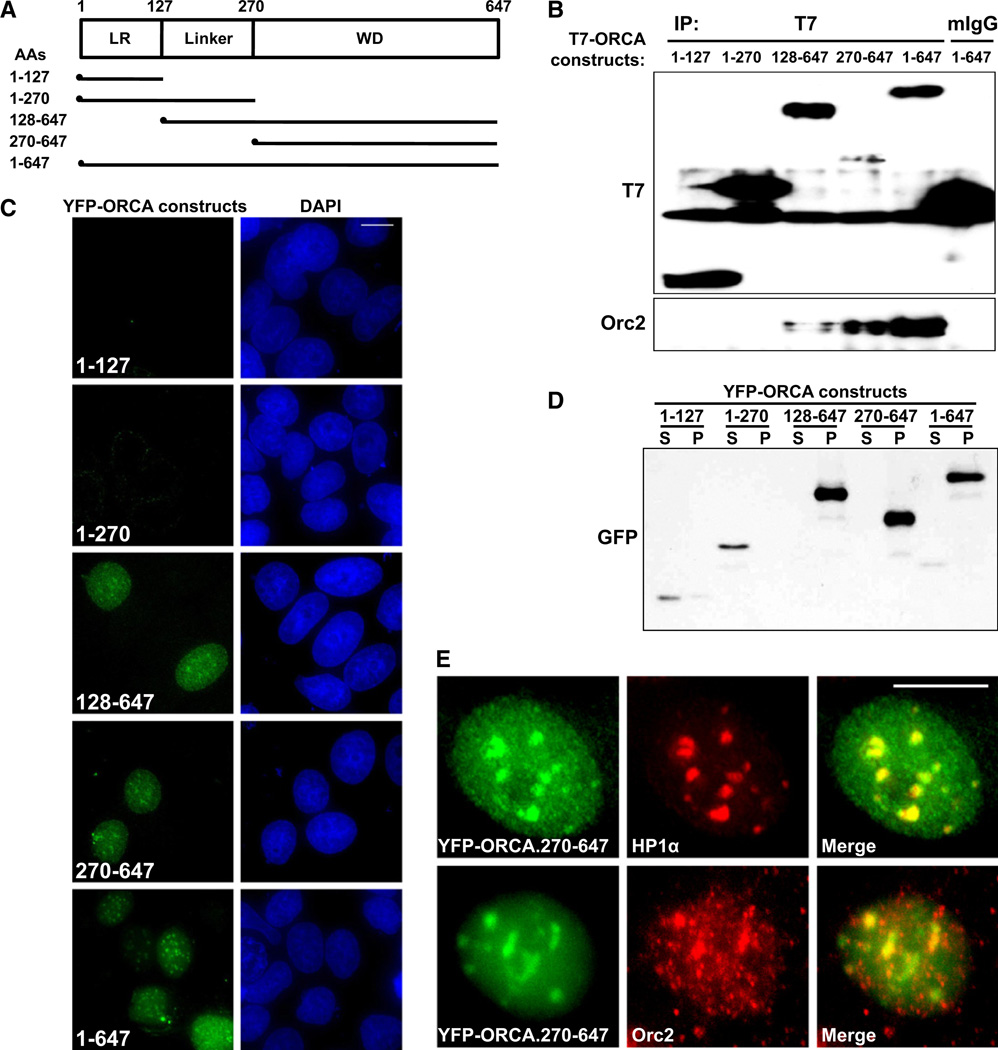
WD Domain of ORCA Is Required for ORC Interaction and Chromatin Association
Leucine-rich repeats and WD repeats are known to mediate protein-protein interactions (Smith, 2008). In order to understand the functional relevance of the structural organization of ORCA and to map the region of ORCA that is responsible for ORC interaction and association with chromatin, we generated T7-and YFP-tagged mutants of ORCA (Figure 5A). T7-tagged full-length and mutant ORCA constructs were transiently transfected into human cells, and IP was conducted from extracts with T7 antibody. Immunoblot analyses with Orc2 antibody revealed that only ORCA constructs that contain the WD repeat domain interacted with Orc2 (Figure 5B, constructs 128–647, 270–647, and 1–647).
Figure 5. WD40 Repeat of ORCA Mediates ORC and Chromatin Association.
(A) Schematic representation of various truncation mutants of ORCA. Each of the mutants contains an epitope tag—T7 or YFP—at their N terminus for IP or IF analyses, respectively.
(B) IP in cells expressing various T7-ORCA mutants with T7 antibody and analysis of Orc2 by immunoblot. Note that only ORCA constructs possessing the WD40 domain efficiently interact with Orc2 (T7-ORCA.128–647; 270–647; 1–647).
(C) YFP-tagged truncated ORCA constructs and their chromatin binding as detected by detergent pre-extraction procedure followed by formaldehyde fixation. Note that only the constructs having WD repeat domain associate with chromatin. DNA was stained with DAPI (blue). The scale bar represents 10 µm.
(D) Immunoblot analysis of YFP-tagged truncated ORCA constructs shows association of FL (1–647) and WD containing fragments (128–647 and 270–647) with chromatin (P). YFP-ORCA mutant constructs lacking WD (1–127 and 1–270) appear in nonchromatin-associated detergent-extracted supernatant (S) fraction.
(E) YFP-ORCA.270–647 localizes to heterochromatin as evident by colocalization with HP1α and Orc2. The scale bar represents 10 µm.
See also Figure S4.
Further, to examine the localization of each of the ORCA mutants, we utilized YFP-tagged ORCA mutants. YFP-ORCA.1–127 (only leucine-rich repeats) showed a homogeneous nuclear and cytoplasmic staining, whereas YFP-ORCA.1–270 showed exclusive nuclear pattern in formaldehyde fixed cells (Figure S4A). However, detergent pre-extraction procedures showed complete loss of YFP-ORCA.1–127 and 1–270 signal, indicating that these ORCA mutants are not associated with chromatin (Figure 5C). This was corroborated by biochemical approach where only ORCA mutants containing WD repeat associated with chromatin (Figure 5D). Similarly, YFP-ORCA.128–647 and YFP-ORCA.270–647 (mutants containing WD domain) showed predominantly nuclear staining (Figure 5C) with prominent heterochromatic foci colocalizing with HP1α and Orc2 upon pre-extraction (Figure 5E and Figure S4B). WD40 repeat domain-containing proteins are known to bind histones and nucleosomes and have been suggested to be important players in chromatin modifying complexes (Suganuma et al., 2008). Our results demonstrate that WD domain of ORCA is critical for ORC interaction and chromatin binding and suggest that ORCA interaction with ORC is critical for their stable association/recruitment onto chromatin. This is corroborated by the fact that ORCA interaction with Orc2 was significantly reduced in DNase I pretreated extracts, demonstrating that the ORC-ORCA interaction is stabilized on the chromatin (Figure S4C).
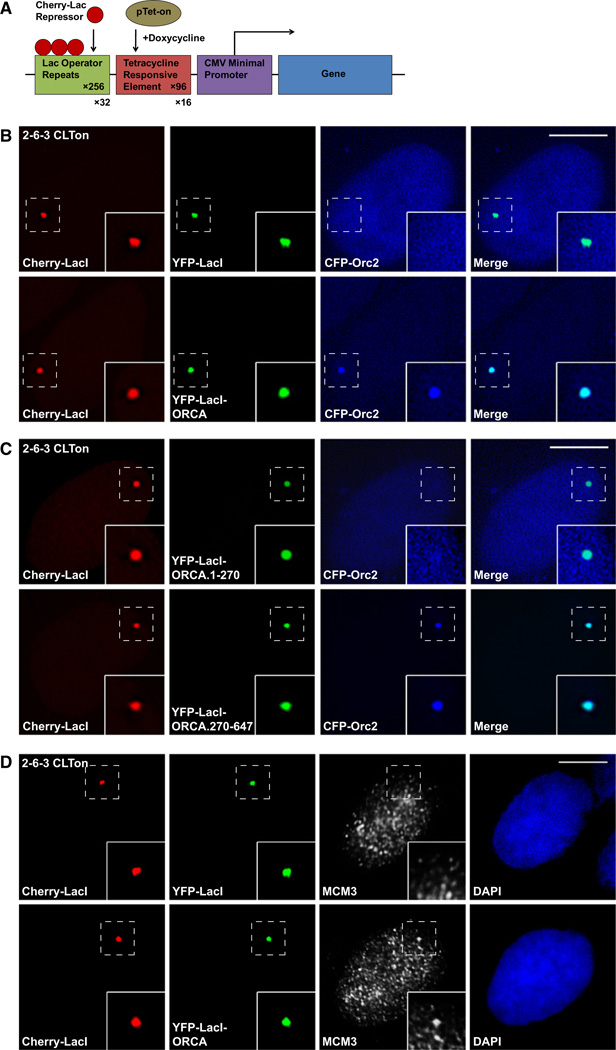
ORCA Tethers ORC to Heterochromatin
Since the ORC-ORCA interaction was stabilized on chromatin, we were interested to address the role of ORCA in ORC recruitment/tethering to chromatin. We have utilized a modified version of the in vivo cell system originally generated by David Spector’s group, where a 200 copy transgene array has been integrated into chromosome 1 in human U2OS cell as a single heterochromatic locus (U2OS-2–6-3) (Janicki et al., 2004). Each transgene copy has 256 copies of Lac operator. We have stably integrated Cherry-LacI (Lac operator repressor) and rtTa (tet-activator) in 2–6-3 cells, so that the locus containing the transgene array can be readily visualized in living cells by the presence of Cherry-LacI (U2OS-2–6-3 CLTon (Figure 6A, Figure S5A). A triple-fusion protein expressing YFP-LacI-ORCA was generated so that ORCA could be directly targeted to the stably integrated chromatin locus in 2–6-3 CLTon via a Lac operator-repressor interaction. By targeting ORCA to the locus, we examined the recruitment of various ORC subunits to the chromatin locus using CFP-tagged versions of ORC. Targeting of YFP-LacI-ORCA to the locus resulted in the specific recruitment of all the ORC subunits to the chromatin locus, with Orc2 (Figure 6B) and Orc3 (Figure S5C) being recruited most efficiently in majority of the cells. Similar results were obtained for Orc5 (Figure S5D) as well as Orc4 and Orc6 (data not shown). Interestingly, CFP-Orc1 is recruited in only ~63% of YFP-LacI-ORCA-positive cells (n = 650, three independent experiments; Figure S5B). Immunolabeling of these cells with either MCM or PCNA suggested that ORCA brings Orc1 to the locus specifically during G1 phase or at the G1/S boundary of the cell cycle (YFP-LacI-ORCA + CFP-Orc1; whenever CFP-Orc1 is at the locus, 100% cells show MCM-positive staining; n = 50; data not shown).
Figure 6. ORCA Tethers ORC to Chromatin.
(A) Schematic representation of the 2-6-3 CLTon locus in human U2OS cells (adapted and modified from Janicki et al. [2004]). The chromatin locus is visualized by Cherry-LacI staining.
(B) Cells were cotransfected with YFP-LacI and CFP-Orc2 or with YFP-LacI-ORCA and CFP-Orc2. YFP-LacI does not recruit CFP-Orc2 to the locus, whereas YFP-LacI-ORCA can efficiently recruit CFP-Orc2 to the chromatin locus.
(C) Cells cotransfected with YFP-LacI-ORCA.1-270 or YFP-LacI-ORCA.270-647 and CFP-Orc2 show that only YFP-LacI-ORCA.270-647 recruits Orc2 to the locus.
(D) Cells transfected with YFP-LacI-ORCA and not YFP-LacI alone show accumulation of MCM3 to the locus, demonstrating that ORC binding to this locus is functional.
Scale bars represent 10 µm. See also Figure S5.
Similar recruitment assays with YFP-LacI-ORCA mutants, including YFP-LacI-ORCA.1–127, YFP-LacI-ORCA.1–270, YF-P-LacI-ORCA.128–647, and YFP-LacI-ORCA.270–647, demonstrated that only the mutants that possess the WD domain recruit ORC to chromatin (Figure 6C and Figure S5E). YFP-LacI alone does not recruit any of the ORC subunits to the chromatin locus (Figure 6B and Figures S5B–S5D). These data clearly demonstrate that ORCA can recruit ORC or stabilize the association of ORC to a chromatin region and WD repeat is essential for the ORC recruitment to chromatin.
Further, when YFP-LacI-ORCA was tethered to the locus, robust accumulation of MCM was observed at the locus (44.4% ± 0.8% cells showed MCM3 at the locus), demonstrating that the ORC bound to the locus is functional (Figure 6D and Figure S5F). These results are similar to what we see for Orc1 recruitment, suggesting that Orc1 loading may be critical for MCM loading and that only in the cells where Orc1 is recruited, MCM could load.
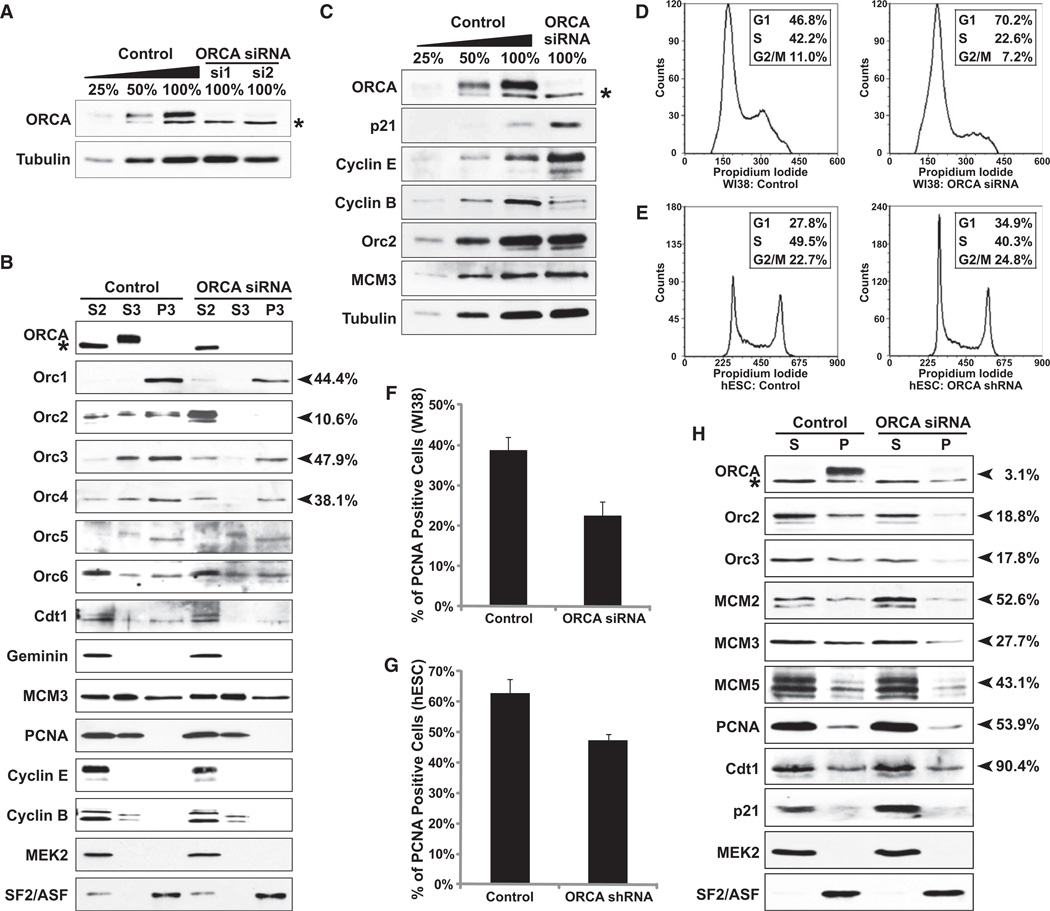
Depletion of ORCA Results in Loss of ORC Binding to Chromatin
To gain an insight into the functional significance of the ORCA-ORC interaction and to unravel the cellular function of ORCA, we used RNA interference (RNAi)-mediated knockdown of ORCA with two small interfering RNA (siRNA) oligonucleotides and two short hairpin RNAs (shRNAs; Figure S6Aa) in human Osteosarcoma U2OS cells and primary diploid fibroblast WI38 cells (Elbashir et al., 2001). Transfection of ORCA siRNA (two independent siRNA oligonucleotides, ORCA-si1 and ORCA-si2) twice with a gap of 24 hr resulted in significant loss of ORCA in U2OS cells, YFP-ORCA stable cell line, and primary fibroblasts (Figures 7A and 7C and Figures S6Ab, S6B, S6C, and S6D). Immunoblots on whole-cell extracts demonstrated that total levels of ORC proteins were unaltered in cells lacking ORCA (Figure 7C and Figure S6Ab). Immunoprecipitation with Orc2 antibodies in ORCA-depleted cells showed that even though negligible ORCA is left behind, it is in complex with ORC, suggesting that ORCA-ORC complex is highly stable (Figure S6E). Interestingly, chromatin fractionation after ORCA depletion in U2OS cells clearly demonstrated that ORC proteins, including Orc1, Orc2, Orc3, and Orc4 (see arrowheads), showed significant reduction in chromatin fraction (P3) and redistribution to the cytosolic fraction (S2), with most pronounced effects on Orc2 (Figure 7B). Similar results were obtained from chromatin fractionation of U2OS cells that stably express shRNAs against ORCA (Figure S6F). However, no significant changes in the chromatin association of other preRC proteins, including MCMs, were discernible in ORCA depleted human cancer cell lines. Similarly, previous results have demonstrated that depletion of human Orc2 and Orc3 in human cancerous cells results in increase in mitotic population without any obvious defects in loading of MCM proteins onto the chromatin (Prasanth et al., 2004b). We believe that after ORCA depletion, even the low levels of ORCA or ORC associated with chromatin in human cancerous cells are sufficient to load MCMs.
Figure 7. ORCA Is Required for Loading ORC to Chromatin.
(A) Immunoblot analysis after siRNA depletion of ORCA with two independent siRNA oligonucleotides shows >90% depletion in U2OS cells. “*” denotes the cross-reacting band. Tubulin was used as the loading control.
(B) Chromatin fractionation in ORCA depleted U2OS cells and immunoblot analysis. Note that in the ORCA-siRNA treated cells, there is significant reduction of Orc2 from P3 fraction and a concomitant accumulation in S2. Similar reduction is apparent for Orc1, Orc3, and Orc4. MEK2 was used as a control for chromatin fractionation. SF2/ASF, a splicing factor, is used as loading control.
(C) Immunoblot analysis after depletion of ORCA in human primary diploid fibroblasts WI38. Note the increase in levels of p21 and Cyclin E and decrease in Cyclin B upon ORCA depletion.
(D and E) Cell-cycle profile by flow cytometry of WI38 cells and hESCs depleted of ORCA shows increased accumulation in G1 phase.
(F and G) Percentage of PCNA positive cells (by immunofluorescence) in control and ORCA-depleted WI38 and hESCs. The graphs represent mean values plus standard deviation from three independent experiments (n = ~400).
(H) Chromatin fractionation in ORCA depleted WI38 cells and immunoblot analysis. Note the significant reduction of Orc2 and 3 and MCM2,3, and 5 on chromatin (P) and the increase in p21 in ORCA-depleted cells.
“%” on the right side of (B) and (H) denotes the amount of protein left on chromatin after ORCA depletion. See also Figure S6.
To address this, we performed siRNA depletion of ORCA in primary diploid fibroblasts, WI38 (46XX normal diploid female). Greater than 90% knockdown of ORCA was achieved via the siRNA approach (Figure 7C). In ORCA-depleted primary cells, significant accumulation of p21 and Cyclin E and reduction of Cyclin B was observed, all indicative of G1 arrest (Figure 7C). Proliferation assay in ORCA-depleted primary cells showed significant proliferation defect (Figure S6G). Further, FACS analysis showed prominent G1 accumulation upon ORCA depletion (Figure 7D; 70.2% in G1 compared to 46.8% in control cells). Immunofluorescence staining with PCNA antibodies showed significant decrease in PCNA labeling (Figure 7F and Figure S6H; 38.8% ± 3.1% in control versus 22.5% ± 3.5% in ORCA-depleted cells), corroborating the FACS data. Chromatin fractionation in ORCA-depleted primary cells showed reduction in ORC subunits (Figure 7H and Figure S6H; seen both with cell biological and biochemical approaches) and also reduced MCM on chromatin (Figure 7H). In summary, depletion of ORCA in human primary cells results in reduced ORC loading to chromatin with a concomitant defect in chromatin loading of MCM, resulting in cells accumulating in G1 phase.
We have also conducted stable shRNA-mediated depletion of ORCA in hESCs (Figures 7E and 7G and Figures S6I–S6L). The key difference between somatic and hESCs is the short G1 phase in hESCs, with most of the cells being in active proliferation phase (S phase) (Becker et al., 2006). hESCs are phenotypically and karyotypically normal cells that can proliferate in culture almost indefinitely. They are similar to pluripotent cell lines derived from the postimplantation epiblast, making studies on them physiologically relevant (Brons et al., 2007; Tesar et al., 2007). Most strikingly, the hESCs like primary cells lacking ORCA proliferated very slowly compared to the control (Figure S6I), and FACS analyses clearly showed increase in G1 population as also evident by an increase in total Cyclin E levels (Figure 7E and Figure S6K). ORCA-depleted hESCs also showed fewer PCNA-positive cells, indicating reduced number of S phase cells (Figure 7G). Oct4, a well-established marker for pluripotency of hESCs, remained unaltered in ORCA-depleted hESCs, suggesting that ORCA depletion reduces proliferation without impairing pluripotency (Figure S6K). Further, immunofluorescence studies in detergent pre-extracted cells revealed loss of ORC association to chromatin in ORCA-depleted hESCs (Figure S6J). Similar observations on the loss of Orc1 and Orc2 binding to chromatin in ORCA-depleted hESCs (P3) were seen by chromatin fractionation procedures (Figure S6L). This observation was similar to what we found in human cancerous cell lines as well as in primary cells.
Taken together, our data demonstrate that ORCA is an essential protein that is required for loading or stabilizing ORC to chromatin. Future studies will focus on the mechanistic insights on how ORCA recruits ORC specifically to origins and to hetero-chromatic regions.
DISCUSSION
ORC is critical for the establishment of preRCs during G1 phase of the cell cycle (Bell and Dutta, 2002). Apart from preRC function, recent evidence in different model organisms has pointed to the role of ORC in transcriptional silencing, heterochromatin formation, and position effect variegation (Sasaki and Gilbert, 2007; Shimada and Gasser, 2007). In yeast, the differential binding affinity of ORC to HMRa locus determines their function either for DNA replication or for heterochromatin establishment (Palacios DeBeer et al., 2003). In yeast, ORC binds adjacent to replication start point, suggesting that ORC directs the preRC to a distinct chromatin site (Das-Bradoo and Bielinsky, 2009). While ORC binds specific DNA sequences in yeast, metazoans do not show sequence-specific association of ORC at origins (Cvetic and Walter, 2005). Though the replication initiation events are nonrandom in metazoans, the mechanism or factors that recruits ORC or specifies ORC binding to specific chromatin sites remains elusive. In the present study, we have identified an ORC-interacting protein, ORCA, that associates and colocalizes with ORC. While this manuscript was in press, another study also reported that ORCA (LRWD1) is an ORC interactor (Vermeulen et al., 2010). We demonstrate that ORCA is critical for loading ORC to the chromatin and/or stabilizing the complex on chromatin. Furthermore, by using an artificial gene locus system, we have shown that ORCA regulates the association of ORC to chromatin sites. Recent work has demonstrated that recruitment of ORC or Cdc6 to DNA is sufficient to create an artificial origin of replication in mammalian cells using preRC components fused to the GAL4 DNA-binding sites (Takeda et al., 2005). Similar to their observations, we now show that tethering ORCA to a chromatin locus results in efficient recruitment of ORC to chromatin and robust accumulation of MCM, suggesting that this system like the artificial origin developed by Dutta and coworkers is functional and provides a useful tool to study preRC function.
DNA replication must occur once and only once in each cell cycle, and preRC components are key players in this regulation. In S. cerevisiae, ORC remains associated with the chromatin throughout the cell cycle, although phosphorylation plays a key role in regulating ORC activity (Diffley et al., 1994; Liang and Stillman, 1997; Nguyen et al., 2001). Recent work in yeast has demonstrated that ORC, Cdc6, and Cdt1 are required for loading but not for maintaining MCM at the origins of replication (Tsakraklides and Bell, 2010). In S. pombe, ORC association to chromatin increases during mitosis and peaks at M/G1 (Wu and Nurse, 2009). In Xenopus, ORC is not associated with chromatin during S phase and is absent from the mitotic chromatin (Hua and Newport, 1998; Romanowski et al., 1996; Rowles et al., 1999; Sun et al., 2002). In Drosophila, Orc1 is degraded at the end of M phase by the anaphase-promoting complex activated by Fzr/Cdh1 (Araki et al., 2005). In contrast, in human cells, Orc1 dissociates from chromatin, is ubiquitinated, and then is degraded during the G1 to S phase transition and reloaded at the M-G1 transition when new preRCs are formed (Kreitz et al., 2001; Mèndez et al., 2002; Siddiqui and Stillman, 2007; Tatsumi et al., 2003). Human Orc2 and Orc3 associate predominantly with centric and pericentromeric heterochromatin during the end of S phase, G2, and mitosis (Prasanth et al., 2004b). A key molecule regulating replication licensing is Cdt1, which accumulates during M and G1 and is degraded during S phase by SCFSkp2-mediated proteolysis through Cdk phosphorylation and replication-coupled proteolysis by Cullin4-Ddb1Cdt2 ubiquitin ligase and PCNA (Fujita, 2006; Tada, 2007). In addition, Gem-inin, a protein expressed in cells from S phase through early mitosis, controls the levels of Cdt1 (McGarry and Kirschner, 1998; Wohlschlegel et al., 2000). We demonstrate that ORCA modulates ORC loading and shows cell-cycle dynamics similar to that of human ORC with maximum expression during G1 and then sequential release from most sites, except at heterochromatic sites during the post-G1 part of the cell cycle. Our data suggest that ORCA gets modified (presumably ubiquitinated), since immunoblots with ORCA antibody that were exposed for longer duration show a smear in the high molecular weight region (molecular weight of ORCA ~70 kDa, Figures S6B and S6C). It is a possibility that once ORCA accomplishes the loading of ORC, it is released from the chromatin, gets ubiquitinated, and finally is degraded during the G1/S boundary. Interestingly, epitope-tagged ORCA (YFP as well as T7) also shows this modification, suggesting that this is a highly coordinated posttranslational event. The association of ORCA with ORC, Cdt1, and Geminin suggests that this might be a key component that regulates timely association of each of these components to chromatin to ensure “once per cell cycle” replication.
The association of ORCA and ORC to centromeres and telomeres in postreplicated cells suggests that this complex plays a key role in chromatin organization at heterochromatic structures. Recent work has established that ORC interacts with the Shelterin complex, as well as the RNA component, TERRA, and plays important roles in structural integrity of telomeres and chromatin structure therein (Deng et al., 2009; Tatsumi et al., 2008). Proper organization of ORC and its associated proteins at heterochromatic sites are critical for cell-cycle progression (Pflumm and Botchan, 2001; Prasanth et al., 2004b). ORCA and ORC bind to centromeres throughout the cell cycle in human MCF7, HeLa, and IMR-90 cells but associate predominantly with telomeres during interphase and to centromeres in mitotic cells in the Osteosarcoma U2OS cells. These data imply that ORCA and ORC colocalize to heterochromatic structures in all human cell lines examined. In support of this observation, a recent proteomic screen has also identified ORCA and ORC as components of human telomeres (Déjardin and Kingston, 2009). MCF7 breast epithelial cells and HeLa cells use telomerase to perform RNA-dependent DNA replication at telomeres to maintain telomere length (Counter et al., 1992), whereas U2OS cells use the mechanism called ALT, which is mediated by DNA recombination (Reddel, 2003). The specific association of ORCA and ORC at telomeres only in ALT+ cells implies that these proteins might also be involved in DNA repair/recombination. Interestingly, ORCA has recently been identified as a target of ATM and ATR in a screen to identify substrates modified by these kinases (Matsuoka et al., 2007). Further, transcription microarray data as well as other recent data have provided evidence for significant enrichment of ORCA in adult testis, a tissue that is highly recombinogenic (Teng et al., 2010). The binding of ORCA to telomeres in cells that use recombination-mediated ALT supports this hypothesis. Recent evidence has also pointed out that centromeres are highly recombinogenic, and recombination occurs at higher frequency at centromeres than at telomeres, implicating the multiple roles of ORCA at heterochromatic sites (Jaco et al., 2008).
Work in yeast has demonstrated that ORC is critical to maintain the nucleosomal configuration adjacent to origins (Lipford and Bell, 2001). Interestingly, disruption of nucleosomal arrangement at an origin has been shown to affect replication initiation, suggesting that ORC is critical not only for replication initiation, but also for altering the local chromatin environment to facilitate origin function (Lipford and Bell, 2001). In yeast, origin sequences have been shown to be sufficient for maintenance of nucleosome-free origin, whereas ORC is needed for accurate nucleosome position flanking origin (Eaton et al., 2010). In Drosophila, open chromatin seems to be a critical feature that governs ORC binding (Macalpine et al., 2010). Recent work has indicated that chromatin proteins like HMGa can specifically target ORC to specific sites on DNA, which in turn is crucial for specification of origins (Thomae et al., 2008). Our data provide evidence that the binding of ORCA stabilizes ORC binding to chromatin, presumably to origins in G1 and to heterochromatin in postreplicated human cells. Based on our data, we propose that the WD40-repeat containing ORCA is a critical factor that is required for loading/stabilizing ORC to chromatin.
EXPERIMENTAL PROCEDURES
Cell Culture
WI38, HeLa, MCF7, and U2OS cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing high glucose, supplemented with penicillin-streptomycin and 10% fetal bovine serum (FBS; Hyclone). For transient transfection, FuGENE 6 (Roche) and Lipofectamine 2000 (Invitrogen) were used according to the manufacturers’ protocols. For generation of stable cell lines, Lipofectamine 2000-mediated transfection was carried out in U2OS, followed by G418 selection (600 µg/ml). The U2OS-2–6-3 cells were transfected with a plasmid containing pVITRO2.Cherry.Lac Repressor (LacI) and Tetracyline activator (Tet-On) (kind gift from Tetsuya Nakamura) and selected with Hygromycin and G418 to generate 2–6-3 CLTon cells.
Transfection
U2OS-2–6-3 CLTon cells were grown in DMEM with 10% Tet system approved FBS (Clontech). Cells grown on coverslips to 50%–60% confluency were cotransfected with 100 ng pEYFP-LacI vector or pEYFP-LacI-ORCA construct along with 500 ng various CFP-tagged ORC subunits with Lipofectamine 2000 reagent. Sixteen to twenty-four hours after transfection, cells were fixed in 2% (w/v) formaldehyde.
Raising ORCA Antibodies
Antibodies against ORCA (S2853, S2854) were raised in rabbits by immunization with peptide LSARLLMQRGRPKSDRLGKIR (aa 4–24) and peptide LAALKRPDDVPLSLSPSKRA (aa 228–247), respectively (Proteintech Group). S2854-1 and S2854-2 were used for immunoprecipitation, S2854-1 affinity purified (AP; 1:1000) for immunofluorescence, and S2853-2 AP (1:1500) for immunoblots.
siRNA and shRNA Knockdown
ThesiRNAstargetingORCA(ORCA-si1CCAACCAGGACUACGAAUU,ORCA-si2 GUGAAGUGGAAUUCGUCUU) and control luciferase (Elbashir et al., 2001) were synthesized by Dharmacon. siRNAs were delivered into cells at a final concentration of 100 nM, mediated by Oligofectamine or Lipofectamine RNAimax (Invitrogen), and were delivered twice at a gap of 24 hr. shRNAs for ORCA (Sigma-Aldrich; Clone ID, shRNA1 NM_152892.1–465s1c1 and shRNA2 NM_152892.1–510s1c1) and scramble (plasmid 1864, Addgene) were in pLKO.1. For viral packaging, pLKO-shRNA, pCMV-dR8.91, and pCMV-VSV-G were cotransfected into 293T cells with FuGENE 6. Viruses were obtained by collecting media at 24 and 48 hr after transfection. For knockdown of ORCA, cells were infected with the viruses in the presence of 8 µg/ml polybrene (Sigma-Aldrich) for 24 hr and then subjected to puromycin selection.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Prasanth laboratory for discussions and suggestions. We thank X. Cao, J. Chen, J. Cook, M. Dundr, A. Dutta, Y. Ge, P. Leiber-man, M. Li, C. Mizzen, T. Nakamura, B. Pilas, D. Spector, B. Stillman, and P. Yao for providing reagents and suggestions. We would like to thank P. Newmark and D. Rivier for critical reading of the paper. This work was supported by a special fellowship from Leukemia and Lymphoma Society, startup funds, University of Illinois at Urbana-Champaign (UIUC), and a National Science Foundation award (0843604) to S.G.P. and by start-up funds, UIUC to K.V.P.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one movie and can be found with this article online at doi: 10.1016/j.molcel.2010.09.021.
REFERENCES
- Araki M, Yu H, Asano M. A novel motif governs APC-dependent degradation of Drosophila ORC1 in vivo. Genes Dev. 2005;19:2458–2465. doi: 10.1101/gad.1361905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M, Huijbregts RP, Chesnokov I. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol. Cell. Biol. 2007;27:3143–3153. doi: 10.1128/MCB.02382-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 2006;209:883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chesnokov IN. Multiple functions of the origin recognition complex. Int. Rev. Cytol. 2007;256:69–109. doi: 10.1016/S0074-7696(07)56003-1. [DOI] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc. Natl. Acad. Sci. USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetic C, Walter JC. Eukaryotic origins of DNA replication: could you please be more specific? Semin. Cell Dev. Biol. 2005;16:343–353. doi: 10.1016/j.semcdb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Das-Bradoo S, Bielinsky AK. Replication initiation point mapping: approach and implications. Methods Mol. Biol. 2009;521:105–120. doi: 10.1007/978-1-60327-815-7_6. [DOI] [PubMed] [Google Scholar]
- Déjardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr. Biol. 2007;17:1989–1995. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML. Cell cycle dependent regulation of the origin recognition complex. Cell Cycle. 2005;4:70–79. doi: 10.4161/cc.4.1.1333. [DOI] [PubMed] [Google Scholar]
- Dhar SK, Yoshida K, Machida Y, Khaira P, Chaudhuri B, Wohlschlegel JA, Leffak M, Yates J, Dutta A. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106:287–296. doi: 10.1016/s0092-8674(01)00458-5. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fujita M. Cdt1 revisited: complex and tight regulation during the cell cycle and consequences of deregulation in mammalian cells. Cell Div. 2006;1:22. doi: 10.1186/1747-1028-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J. Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Mèchali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaco I, Canela A, Vera E, Blasco MA. Centromere mitotic recombination in mammalian cells. J. Cell Biol. 2008;181:885–892. doi: 10.1083/jcb.200803042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitz S, Ritzi M, Baack M, Knippers R. The human origin recognition complex protein 1 dissociates from chromatin during S phase in HeLa cells. J. Biol. Chem. 2001;276:6337–6342. doi: 10.1074/jbc.M009473200. [DOI] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidonnici MR, Rossi R, Paixão S, Mendoza-Maldonado R, Paolinelli R, Arcangeli C, Giacca M, Biamonti G, Montecucco A. Subnuclear distribution of the largest subunit of the human origin recognition complex during the cell cycle. J. Cell Sci. 2004;117:5221–5231. doi: 10.1242/jcs.01405. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Macalpine HK, Gordan R, Powell SK, Hartemink AJ, Macalpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;24:748–753. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Makise M, Sueyasu Y, Takehara M, Asano T, Mizushima T. Yeast two-hybrid analysis of the origin recognition complex of Saccharomyces cerevisiae: interaction between subunits and identification of binding proteins. FEMS Yeast Res. 2007;7:1263–1269. doi: 10.1111/j.1567-1364.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mèndez J, Zou-Yang XH, Kim SY, Hidaka M, Tansey WP, Stillman B. Human origin recognition complex large subunit is degraded by ubiquitin-mediated proteolysis after initiation of DNA replication. Mol. Cell. 2002;9:481–491. doi: 10.1016/s1097-2765(02)00467-7. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Okuno Y, McNairn AJ, den Elzen N, Pines J, Gilbert DM. Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. EMBO J. 2001;20:4263–4277. doi: 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios DeBeer MA, Muller U, Fox CA. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 2003;17:1817–1822. doi: 10.1101/gad.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflumm MF, Botchan MR. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development. 2001;128:1697–1707. doi: 10.1242/dev.128.9.1697. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Mèndez J, Prasanth KV, Stillman B. Dynamics of pre-replication complex proteins during the cell division cycle. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004a;359:7–16. doi: 10.1098/rstb.2003.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004b;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc. Natl. Acad. Sci. USA. 2010;107:15093–15098. doi: 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddel RR. Alternative lengthening of telomeres, telomerase, and cancer. Cancer Lett. 2003;194:155–162. doi: 10.1016/s0304-3835(02)00702-4. [DOI] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr. Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J. Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Gilbert DM. The many faces of the origin recognition complex. Curr. Opin. Cell Biol. 2007;19:337–343. doi: 10.1016/j.ceb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shareef MM, Badugu R, Kellum R. HP1/ORC complex and heterochromatin assembly. Genetica. 2003;117:127–134. doi: 10.1023/a:1022963223220. [DOI] [PubMed] [Google Scholar]
- Shimada K, Gasser SM. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell. 2007;128:85–99. doi: 10.1016/j.cell.2006.11.045. [DOI] [PubMed] [Google Scholar]
- Siddiqui K, Stillman B. ATP-dependent assembly of the human origin recognition complex. J. Biol. Chem. 2007;282:32370–32383. doi: 10.1074/jbc.M705905200. [DOI] [PubMed] [Google Scholar]
- Smith TF. Diversity of WD-repeat proteins. Subcell. Biochem. 2008;48:20–30. doi: 10.1007/978-0-387-09595-0_3. [DOI] [PubMed] [Google Scholar]
- Suganuma T, Pattenden SG, Workman JL. Diverse functions of WD40 repeat proteins in histone recognition. Genes Dev. 2008;22:1265–1268. doi: 10.1101/gad.1676208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WH, Coleman TR, DePamphilis ML. Cell cycle-dependent regulation of the association between origin recognition proteins and somatic cell chromatin. EMBO J. 2002;21:1437–1446. doi: 10.1093/emboj/21.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S. Cdt1 and geminin: role during cell cycle progression and DNA damage in higher eukaryotes. Front. Biosci. 2007;12:1629–1641. doi: 10.2741/2175. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J. Biol. Chem. 2003;278:41528–41534. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- Tatsumi Y, Ezura K, Yoshida K, Yugawa T, Narisawa-Saito M, Kiyono T, Ohta S, Obuse C, Fujita M. Involvement of human ORC and TRF2 in pre-replication complex assembly at telomeres. Genes Cells. 2008;13:1045–1059. doi: 10.1111/j.1365-2443.2008.01224.x. [DOI] [PubMed] [Google Scholar]
- Teng YN, Liao MH, Lin YB, Kuo PL, Kuo TY. Expression of lrwd1 in mouse testis and its centrosomal localization. Int. J. Androl.. in press. Published online February. 2010;17:2010. doi: 10.1111/j.1365-2605.2009.01038.x. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomae AW, Pich D, Brocher J, Spindler MP, Berens C, Hock R, Hammerschmidt W, Schepers A. Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc. Natl. Acad. Sci. USA. 2008;105:1692–1697. doi: 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome KC, Dhar SK, Quintana DG, Delmolino L, Shahsafaei A, Dutta A. Subsets of human origin recognition complex (ORC) subunits are expressed in non-proliferating cells and associate with non-ORC proteins. J. Biol. Chem. 2000;275:35233–35241. doi: 10.1074/jbc.M005765200. [DOI] [PubMed] [Google Scholar]
- Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- Tsakraklides V, Bell SP. Dynamics of pre-replicative complex assembly. J. Biol. Chem. 2010;285:9437–9443. doi: 10.1074/jbc.M109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 2003;17:1894–1908. doi: 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wu PY, Nurse P. Establishing the program of origin firing during S phase in fission Yeast. Cell. 2009;136:852–864. doi: 10.1016/j.cell.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.