Abstract
Objective: Phenotyping algorithms applied to electronic health record (EHR) data enable investigators to identify large cohorts for clinical and genomic research. Algorithm development is often iterative, depends on fallible investigator intuition, and is time- and labor-intensive. We developed and evaluated 4 types of phenotyping algorithms and categories of EHR information to identify hypertensive individuals and controls and provide a portable module for implementation at other sites.
Materials and Methods: We reviewed the EHRs of 631 individuals followed at Vanderbilt for hypertension status. We developed features and phenotyping algorithms of increasing complexity. Input categories included International Classification of Diseases, Ninth Revision (ICD9) codes, medications, vital signs, narrative-text search results, and Unified Medical Language System (UMLS) concepts extracted using natural language processing (NLP). We developed a module and tested portability by replicating 10 of the best-performing algorithms at the Marshfield Clinic.
Results: Random forests using billing codes, medications, vitals, and concepts had the best performance with a median area under the receiver operator characteristic curve (AUC) of 0.976. Normalized sums of all 4 categories also performed well (0.959 AUC). The best non-NLP algorithm combined normalized ICD9 codes, medications, and blood pressure readings with a median AUC of 0.948. Blood pressure cutoffs or ICD9 code counts alone had AUCs of 0.854 and 0.908, respectively. Marshfield Clinic results were similar.
Conclusion: This work shows that billing codes or blood pressure readings alone yield good hypertension classification performance. However, even simple combinations of input categories improve performance. The most complex algorithms classified hypertension with excellent recall and precision.
Keywords: phenotyping algorithms, machine learning, random forests, hypertension, natural language processing, electronic health records
Background and Significance
Hypertension is a prototypic intervenable chronic disease with significant longitudinal morbidity when ineffectively treated. Hypertension affects one-third of Americans1,2 and contributes to 1 in 6 adult deaths in the United States.2–5 Thus, identifying hypertension is important as both a primary disease of study and as an important covariate in many clinical and genetic studies, yet a rigorously evaluated algorithm to identify hypertensive individuals from electronic health records (EHRs) is lacking. In this work, we developed and evaluated the performance of different portable algorithms and EHR data types to identify individuals with and without hypertension.
Current clinical guidelines define hypertension as a consistent blood pressure greater than or equal to 140 mmHg systolic and/or 90 mmHg diastolic.6,7 For example, EHR blood pressure measurements do not always correlate well with a diagnosis of hypertension: many conditions can temporarily elevate blood pressure,8 and patients with well-controlled hypertension may display consistently normal values. Prior studies have not rigorously evaluated a general-purpose hypertension algorithm. Studies have leveraged simple thresholds based on a minimum number of hypertension billing code counts9 to classify hypertension for use as covariates in studies of other diseases such as abdominal aortic aneurysm, stroke, chronic kidney disease, heart failure, and atrial fibrillation.2,6 Algorithms have been developed for subtypes of hypertension, such as resistant hypertension.10 Most phenotype algorithm evaluations have typically focused on precision.11 Given that hypertension is both a primary phenotype of interest and an important covariate for other diseases, a phenotyping algorithm that minimizes both false negatives and false positives is desirable.
EHRs contain a diverse set of data types—structured lab values, vital signs, billing codes, narrative clinical documentation, visual data such as X-rays, and semistructured questionnaires, among many others. Using automated phenotyping algorithms, investigators have identified cases and controls for diseases of interest to replicate known phenotype-genotype associations and make novel discoveries,12–17 potentially with decreased cost18 and faster execution than traditional trials.
Phenotyping algorithms can be constructed from sets of nested Boolean logic statements, exclusions, and temporal relationships applied to EHR data elements designed to identify individuals with a given phenotype.19,20 Each data source poses unique challenges, and use of multiple data sources often improves performance.21
Billing code-based phenotyping methods have variable performance with estimates for cardiovascular and stroke risk factors ranging from 0.55 to 0.95 positive predictive value (PPV).22 Similarly, various phenotyping studies have used natural language processing (NLP)-extracted concepts alone, with sensitivities ranging from 72% to 99.6% and PPV between 63% and 100%.23–26 However, due to hypertension’s high prevalence, it is a very common entry within the family history section of clinical notes and may result in many false positives.
Here we show that algorithms that combine multiple EHR data sources achieved the best overall results. We found that machine learning performed the best, but that deterministic algorithms also performed well. Both approaches performed similarly at a replication site.
Methods
Patient selection and review at Vanderbilt University Medical Center
Our starting population consisted of all individuals in the Synthetic Derivative, a de-identified image of the Vanderbilt University Medical Center EHR.27 This study was reviewed by the internal review board and found to be exempt from requiring informed consent. We randomly selected 643 adults with regular outpatient care, defined as at least 2 outpatient visits and 2 blood pressure readings between January 1, 2007, and January 1, 2009. Vitals are routinely recorded at all visits for all patients (including all internal medicine clinics), regardless of hypertension status. Inpatient blood pressures are recorded multiple times a day based on patient acuity, regardless of hypertension status. Authors with a clinical background (R.M.C., W.Q.W., H.M., P.L.T.) manually reviewed an initial cohort (n = 303) with 20% overlap for cases, controls, and unknowns using de-identified notes, billing codes, and vital signs. After determining sufficient interrater agreement (Fleiss’s κ = 0.93), the remaining 340 individuals were reviewed without overlap. A board-certified internist (J.D.) provided guidance and adjudicated any conflicting or undetermined reviews.
Input feature development
We developed 67 different features for consideration alone and as part of different algorithms. We hypothesized that billing codes, medications, vital readings, and clinic note content provide broad coverage, thus enabling accurate identification of hypertension cases and controls despite problems within each data source. We aggregated general information (document counts, maximum age, total International Classification of Diseases, Ninth Revision [ICD9] code counts, etc.) and hypertension-specific elements (hypertensive ICD9 code count, hypertensive medication count, hypertensive blood pressure reading count, hypertensive note-item count, etc.). Supplemental Table 1 includes the full feature list and descriptions. We curated a set of hypertension-related billing codes (Supplemental Table 2). Medications were available from structured electronic prescribing records and also extracted from narrative documents using MedEx.28,29 Hypertension medications were determined using medication strings with indications determined as part of Medication-Indication resource-High Performance Subset (MEDI-HPS), which lists on- and off-label indications of medications (Supplemental Table 3).30–33 We used the hypertensive blood pressure guideline thresholds of 140 mmHg systolic and 90 mmHg diastolic. We separated vital readings into outpatient and inpatient only and collapsed multiple daily readings to their median values.
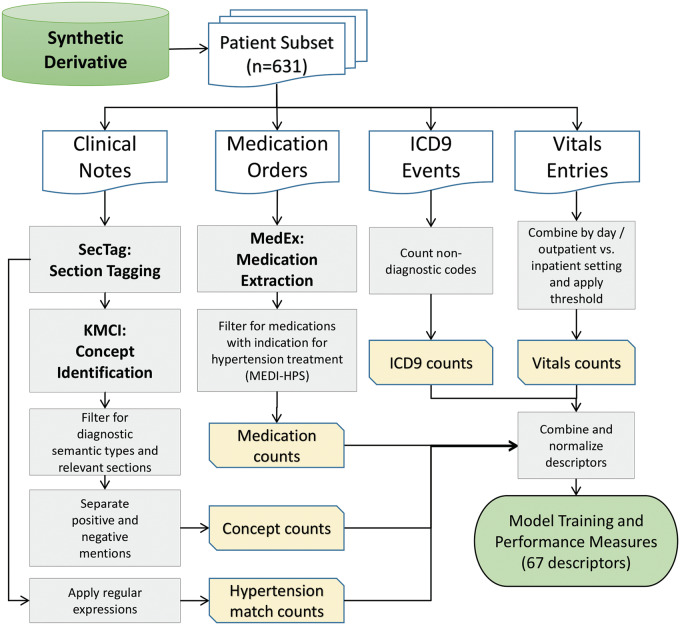
We restricted narrative documents to problem lists, clinic notes, discharge summaries, and admission history and physical notes. We identified sections using SecTag34 and used only high-yield sections to reduce NLP false positives, including but not limited to the “history of present illness,” “past medical history,” and “assessment and plan.” We extracted concepts from these sections using the KnowledgeMap Concept Identifier with a SNOMED-CT focused subset as the vocabulary (Supplemental Table 4).26,35 We used non-negated patient-related concepts. From the set of KnowledgeMap Concept Identifier-extracted UMLS concepts, we identified 12 hypertension concepts (Supplemental Table 5). We also calculated hypertension-related counts from full notes using regular expressions. Regular expression matches targeted “hypertension” not preceded by “pulmonary” or “HTN” with word boundaries (to avoid matching strings such as “tightness”; regular expression included in Supplemental Information). Figure 1 depicts the processing pipeline. The full protocol is available on PheKB (http://phekb.org/phenotypes/hypertension).
Figure 1.
Algorithm dataset generation flowchart. We randomly sampled 631 adults for the initial population. We limited sampling to concepts that were in high-yield sections, which included “history of present illness,” “past medical history,” and “assessment and plan.” Billing codes were available as structured data, and hypertension-related codes were physician-curated. We also separated inpatient and outpatient vitals using Current Procedural Terminology (CPT) codes.
We assembled the final features for each category (billing codes, medications, vital signs, hypertension regular expression matches, and hypertension concepts) by taking the following individually and in combination: total counts of each item, all hypertensive elements (blood pressures above the threshold, medications with hypertension as an indication), counts of unique items, and normalized versions of each. We normalized by dividing hypertension-related counts by total category counts or total unique item counts. We normalized inputs to account for the variable number of observations per individual. In addition, we added unique elements for ICD9 and medication data to compensate for high-frequency concepts found in clinical notes due to copy and paste.36 Several different medications or billing codes seemed more likely to identify a case correctly.
Algorithm development
We developed several simple algorithms as easier-to-implement alternatives to those that include NLP. There were 2 types of simple algorithms. The first summed features, 1 per category. The second type summed the number of categories with a nonzero feature, where each category contained a single representative feature. The sum of category counts included an integer threshold (n = 1–4) to predict case vs control. We also used permutations that included normalization—normalizing by total occurrences, unique items, documents, or total concepts as appropriate. In addition to simple expert-defined algorithms, we also used random forests, which used the 67 input features developed above. We chose random forests because they are robust, computationally cheap to train, and able to handle large numbers of descriptors well.
Bootstrap analysis
To compare random forest models, individual features, and several simple algorithms, we used a version of the 0.632+ bootstrap37,38 method and then applied each model, feature, or simple algorithm to the same test set (200 individuals). Briefly, this method samples N elements with replacement from a population of size N, which results in mean coverage of 0.632N of the population. Sampling with replacement exposes the model to more varied and potentially representative weightings of the different possible populations that could have been sampled. We repeated this sampling 1000 times and used the 2.5th percentile and the 97.5th percentile based on the sorted results from the entire bootstrap to empirically establish the 95% confidence interval (CI). Bootstraps were run for random forest models trained across each category of features individually (e.g., ICD9 codes, medications, vitals) as well as with increasingly complex combinations (e.g., Boolean or count combinations of different features). We ran bootstraps for each set across training set sizes of 25, 50, 100, 150, 200, 250, 300, 350, 400, 450, 500, 550, and 600 to test the effect of training set size on algorithm performance. For each iteration, the validation set was a random subset of 200 individuals not sampled for the training set. We calculated the area under the receiver operating characteristic curve (AUC), sensitivity, and PPV for each test set. We used the randomForest package in R to train models and the ROCR package to calculate performance metrics.39,40
Best random forest model performance evaluation
To evaluate the random forest predictions per individual, we used the 1000 models generated in the bootstrap run along with the 1000 accompanying validation sets. For the best-performing random forest model by AUC—using ICD9, medications, all vitals, and NLP-derived concepts—we aggregated independent test set predictions across all 1000 runs and calculated the mean prediction for each individual. We applied the same approach for calculating a log-based score,41 although in the case of the normalized sum model we used the training set to calculate a logistic regression that mapped the sums to the 0–1 range necessary. We then plotted a histogram of the mean predictions to determine the counts of individuals with different prediction ranges and identified misclassified individuals using a threshold of 0.5. We then reviewed a subset of these sets of false positives and false negatives as part of an error analysis.
KNIME module development
We developed a Konstanz Information Miner (KNIME)42 module to provide ease implementation of the algorithm by other users. KNIME provides a graphical user interface to simplify interpretability and editing. The package takes raw inputs with dates and encapsulates data processing, normalization, and analysis, outputting case/control determinations for individuals. The module can also take subsets of available inputs such as coded data only. Given labeled cases and controls, the module outputs aggregate performance statistics (counts, prevalence, sensitivity, specificity, and PPV). The module includes some of the best-performing simple algorithms and random forest models trained with our entire reviewed dataset using the following category combinations: (1) ICD9s, medications, and all vitals; (2) ICD9s, medications, and all vitals including separate outpatient and inpatient vitals; (3) all elements from the second set plus regular expression matches; (4) all elements from the second set plus NLP-derived concepts; and (5) all data including regular expression matches and concepts.
Replication at Marshfield Clinic
The Marshfield Personalized Medicine Research Project is a population-based study in which participants consented and provided DNA, plasma, and serum samples and access to their medical records for genetic research. The cohort consists of approximately 20 000 participants from central Wisconsin with primarily northern European ancestry. Marshfield Clinic provides most of the primary, secondary, and tertiary care for this cohort and the data is stored electronically in an internally-developed EHR with medical information dating back to the early 1960s.43
Participants (n = 15 183) with 2 or more blood pressure measurements between January 1, 2007, and December 31, 2008, were selected from the Personalized Medicine Research Project for this study. One hundred patients were randomly selected from this sample and manually classified by 1 of 2 authors (R.A.D., A.M.N.) as cases (having hypertension) or controls (absence of hypertension), then used to test the KNIME workflow hypertension prediction module. ICD9 codes, medications, pulse, outpatient CPTs, blood pressure measurements, and hypertension concepts indexed using MetaMap44 with negation were provided as input to the module; however, concepts were not restricted to specific note types or note sections as they were at Vanderbilt. Regular expression matches for hypertension mentions within clinical notes were not tested at Marshfield, as they were unable to extract raw text and run the regular expressions (UMLS concepts had been previously extracted).
Results
Table 1 includes the summary information for the populations studied at Vanderbilt and Marshfield Clinic. Both sites had a prevalence of hypertension of almost 60%. Reviewers demonstrated high inter-rater agreement for the subset classified by multiple reviewers at Vanderbilt (Fleiss’s κ = 0.93). Median age was lower for controls (47, IQR = 37–59.75) compared to hypertensive individuals (65, IQR = 56–75) (P < .00001). Median age across the entire population was 59 with an interquartile range of 46–70. Both sites have 1.5-fold more total females. The majority of individuals were white—84% at Vanderbilt and 95% at Marshfield Clinic. There were nonzero counts of hypertension-related ICD9 codes and medications for controls at both sites. We found 101 hypertension ICD9 codes (401.*) for 19 controls; thus, 7.3% of Vanderbilt controls had hypertension ICD9 codes but were judged to be controls. Similarly, 104 (39.7%) of Vanderbilt controls had at least 1 medication with hypertension as a potential indication. Median follow-up was similar between both cases and controls at Vanderbilt—6.6 years or 30 visits for hypertensive individuals and 5.7 years or 17 visits for controls. Median follow-up at Marshfield Clinic was longer at 18.6 years as compared to 6.1 years at Vanderbilt. Supplemental Table 6 lists median and IQR between cases and controls.
Table 1.
Study population demographics and clinical information.
|
Vanderbilt (n = 631) |
Replication (Marshfield, n = 100) |
|||
|---|---|---|---|---|
| Description | Hypertensive | Control | Hypertensive | Control |
| Unique individuals | 369 | 262 | 59 | 41 |
| Median age (IQR) | 65 (56–75) | 47 (37–59.75) | 70.9 (56.9–80.2) | 53.8 (44.4–58.9) |
| Female (%) | 198 (54) | 177 (68) | 32 (54) | 28 (68) |
| Race | ||||
| White | 306 | 227 | 55 | 40 |
| Black | 49 | 16 | 0 | 0 |
| Asian | 2 | 3 | 0 | 0 |
| Indian American | 0 | 2 | 0 | 0 |
| Unknown | 12 | 14 | 4 | 1 |
| Medication (counts per individual and IQR) | ||||
| Median | 462 (207–1015) | 135 (61.25–346) | 1161 (867–2291) | 408 (214–829) |
| Median hypertension-related | 61 (22–173) | 0 (0–5) | 221 (72–538) | 2 (1–7) |
| Median unique | 84 (44–147) | 41.5 (20–76) | 135 (110–204) | 86 (58–121) |
| Median unique hypertension-related | 7 (3–13) | 0 (0–2) | 16 (9–32) | 1 (1–3) |
| Billing codes | ||||
| Any hypertension ICD9 Code | 4951 | 101 | 1650 | 19 |
| Essential hypertension 401.a | 4936 | 101 | 1579 | 19 |
| Secondary hypertension 405.a | 15 | 0 | 71 | 0 |
| EHR Follow-upaand IQR | ||||
| Median follow-up | 6.6 (5.0–8.8) | 5.7 (3.3–7.7) | 19.1 (15.5–19.8) | 18.2 (17.0–19.6) |
| Number of visits with vitals | 30 (16–52) | 17 (9–30) | 86 (66–120) | 52 (35–61) |
aMedian with (IQR = interquartile range) in years, calculated as first vitals reading to last.
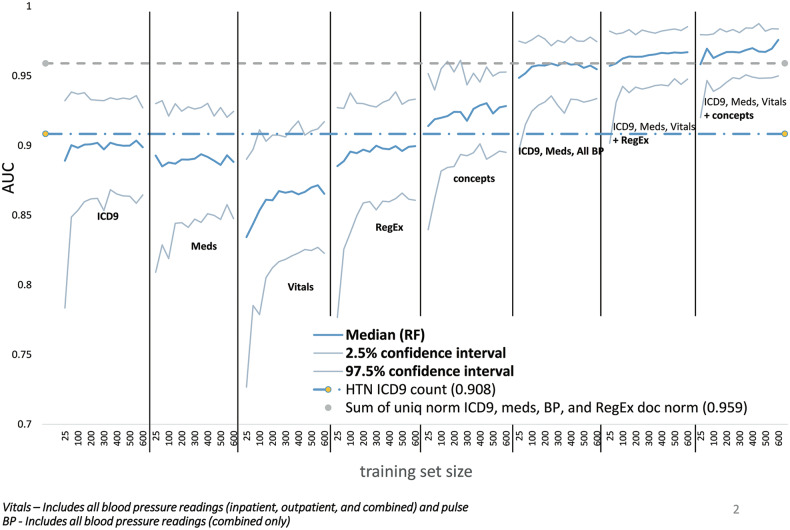
Bootstrap performance for random forest models trended upwards as training set size increased and CI narrowed (Figure 2). The best-performing model was the random forest trained on all features for ICD9 codes, medications, vitals, and UMLS concepts (AUC 0.976). Of the individual category random forests, vitals performed the most poorly (0.865) and models trained on the UMLS concept features performed best (0.928). However, the difference between them was comparable to their CIs.
Figure 2.
Random forests trained on combinations of categories perform best. We did 1000-iteration bootstrap runs for each category of features as well as increasingly comprehensive combinations of categories for successively larger training set sizes from 25 to 600. Labels indicate the set of categories used for each learning curve. Other combinations were tested but were similar to the included examples. The graph below includes the median AUC for each learning curve in addition to the upper and lower bounds of the 95% confidence interval. For reference, lines representing the median AUC for 2 simple methods are included—hypertension (HTN) ICD9 counts and the sum of unique normalized ICD9 codes, medications, blood pressure (BP) readings, and regular expression (RegEx) matches normalized by document counts.
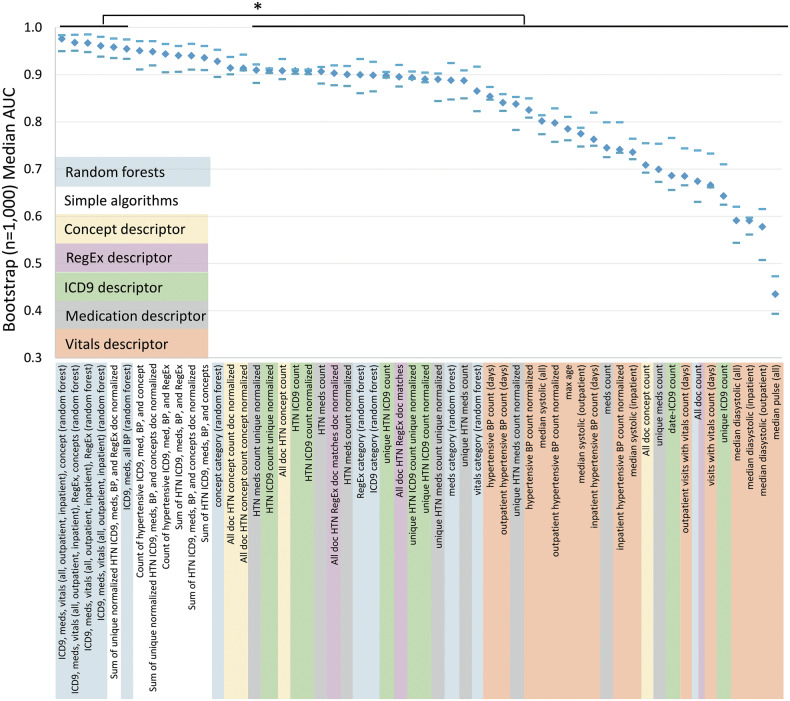
Random forests using combinations of feature categories generally performed better than simple algorithms (Figure 3). The simple algorithms performed well both with and without normalization although there was a trend toward better performance for simple algorithms that sum the individual normalized counts of each category. After the top 3 random forests, the fourth highest median AUC (0.959) was achieved by summing the unique normalized values of hypertension-related ICD9s, medications, blood pressure readings, and regular expression matches normalized by the number of documents. The top 6 algorithms, which were all random forest–based except 1, were statistically better than all individual features except the hypertension concept counts across all notes that were normalized by either the total number of concepts or documents. Only NLP-derived hypertension concepts approached the combined methods’ performance (AUC = 0.914). The worst-performing algorithms used pulse or diastolic blood pressure alone (AUCs of 0.435–0.591). Systolic blood pressure algorithms were better but still underperformed other categories of data (AUCs of 0.775–0.854). Full results, including sensitivities and PPVs at various thresholds, are included in Supplemental Table 7. The normalized sum and random forest using all categories outperformed all other approaches with AUCs of 0.959 and 0.976. Hypertension ICD9 code, concept, and medication counts performed similarly at AUCs of 0.908, 0.908, and 0.907 (Figure 4).
Figure 3.
Algorithm performance. Median AUC and 95% confidence intervals (CI) for the 1000-iteration bootstrap are depicted across all random forests, representative simple algorithms, and representative individual features. Diamonds indicate the AUC and dashes indicate the upper and lower bounds of the 95% CI, respectively. The top 6 by median AUC are statistically significantly better than the lower 41 of the 56 total included—comparing 95% CI.
Figure 4.
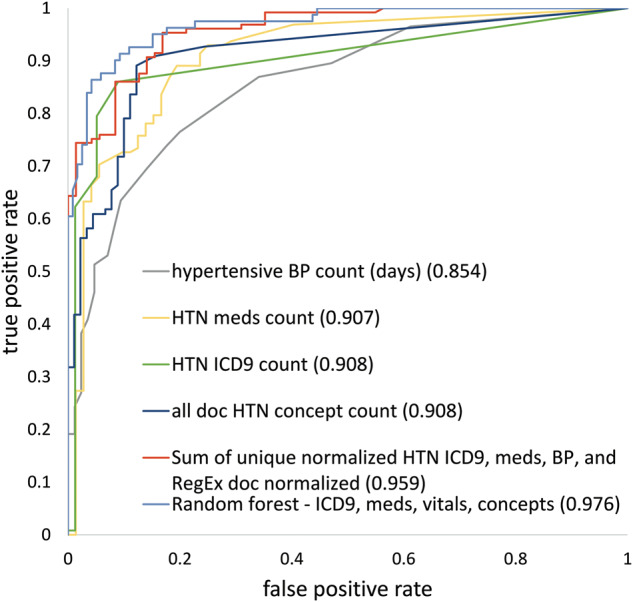
Combination methods achieve the highest AUC. We include the ROC representative of the 50th percentile 1000 iteration bootstrap run below. Numbers in parentheses represent the median AUCs from the bootstrap model. The random forest model represented here is the best-performing RF model from Figure 2. The best simple algorithm is the sum of unique normalized hypertension ICD9, medications, blood pressures, and regular expression matches normalized by the number of documents.
The best random forest model’s per-individual predictions effectively separated cases from controls (Figure 5). For scores > 0.9 (264 total), the random forests correctly classified 97.7% of cases. Similarly, the random forests correctly classified 96.8% of the controls with median predictions < 0.1 (156 total). Performance degraded as predictions approached 0.5 from either extreme. Assuming a threshold of 0.5, the random forests only correctly classified 33.3% of the 0.5–0.6 bin as cases and 52% of the controls for the 0.4–0.5 bin. Overall, the random forests correctly classified 88.9% of the individuals with 36 false negatives and 34 false positives by median bootstrap prediction. Using a log-based scoring metric,41 we also calculated the performance for the best random forest compared to a non–machine learning approach using only structured data—the normalized sum of hypertension-related ICD9 codes, medications, and blood pressure readings. The scores were −0.109 and −0.164 for the random forest and normalized sum model respectively (closer to zero is better).
Figure 5.
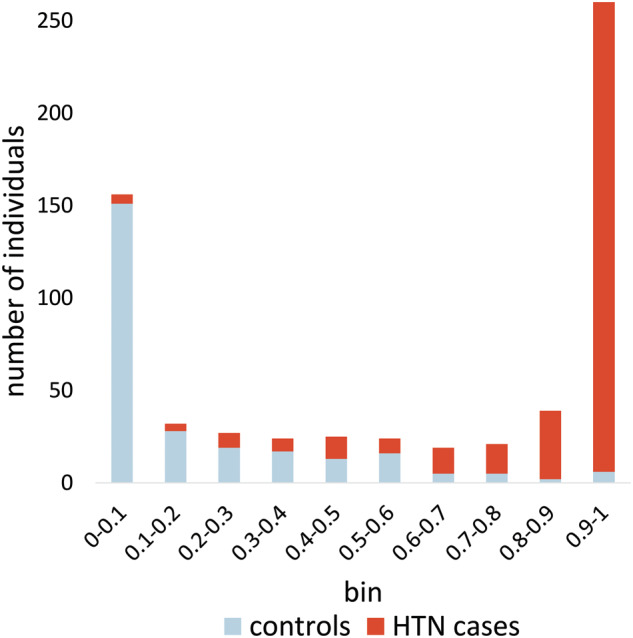
Histogram showing prediction separation between cases and controls. The top column segments, biased toward the right (1.0) are the counts of hypertensive individuals with a mean random forest prediction (each taken from a test set not used for training) within the bin range listed along the x-axis. The bottom column segments represent the counts of controls in each bin range. Individuals with an unexpected score (< 0.5 for cases, > 0.5 for controls) were reviewed.
Comparing the true positives and negatives with false positives and negatives across all features revealed many that were systematically different (Supplemental Table 8). For example, the number of hypertension concepts across all notes was 840 times higher for true positives than for false negatives. When one normalizes for the document count, this increased to a 2335-fold difference.
Finally, we examined the portability of the best random forest models trained on Vanderbilt data as well as the simple algorithms at the Marshfield Clinic. Table 2 includes the AUCs, sensitivities, and PPVs for the 5 random forests trained, 3 simple category count algorithms with integer thresholds, and 2 summing algorithms. Of note, the AUCs at Marshfield for methods that included concepts or required 2 or fewer categories were lower than their counterparts at Vanderbilt. Summing algorithms achieved slightly higher performance than category count algorithms at both sites. All other AUCs were comparable between sites.
Table 2.
Portability evaluation across various algorithms at Vanderbilt and Marshfield Clinic.
|
Vanderbilt (n = 631) |
Replication Marshfield (n = 100) |
|||||
|---|---|---|---|---|---|---|
| Model with expected features | AUC (CI) | Sens. | PPV | AUC | Sens. | PPV |
| ICD9, meds, all BP (random forest) | 0.955 (0.934–0.975) | 0.844 | 0.954 | 0.922 | 0.966 | 0.919 |
| ICD9, meds, all vitals (random forest) | 0.961 (0.938–0.980) | 0.858 | 0.954 | 0.910 | 0.966 | 0.905 |
| ICD9, meds, all vitals, RegEx (random forest)a | 0.967 (0.948–0.985) | 0.866 | 0.954 | 0.934 | 0.966 | 0.934 |
| ICD9, meds, all vitals, concept (random forest) | 0.976 (0.95–0.984) | 0.902 | 0.952 | 0.873 | 0.966 | 0.864 |
| ICD9, meds, all vitals, RegEx, concepts (random forest)a | 0.968 (0.951–0.985) | 0.877 | 0.954 | 0.898 | 0.966 | 0.891 |
| Positive category count ICD9, med, and BP 2 of 3 | 0.833 (0.788–0.868) | 0.952 | 0.822 | 0.646 | 1.000 | 0.670 |
| Positive category count ICD9, med, and BP 3 of 3 | 0.877 (0.849–0.914) | 0.798 | 0.967 | 0.914 | 0.949 | 0.918 |
| Positive category count ICD9, med, BP, and concept 3 of 4 | 0.910 (0.868–0.936) | 0.925 | 0.924 | 0.711 | 0.983 | 0.716 |
| Sum of normalized hypertension ICD9, meds, and BP | 0.915 (0.888–0.942) | 1.000 | 0.673 | 0.949 | 1.000 | 0.702 |
| Sum of normalized hypertension ICD9, meds, BP, and concept | 0.929 (0.897–0.955) | 1.000 | 0.663 | 0.949 | 1.000 | 0.702 |
aMarshfield Clinic inputs to random forest models did not include regular expression (RegEx) information.
The best AUC and model for each site-category combination are bolded.
Discussion
In this work, we evaluated ICD9 codes, medications, vitals, and narrative documents as data sources for hypertension phenotyping algorithms. We also showed that combinations of multiple categories of information result in the best performance, with AUC rising in tandem with the number of categories used. Blood pressure measurements, despite being the basis for determining hypertension clinically, performed worst of all categories for the identification of hypertensive individuals from EHR data, even when restricted to outpatient measurements. This is likely due to issues such as treatment reducing blood pressure to within the normal range, treatment often starting outside of our EHR dataset, and the many nonhypertension causes of high blood pressure readings within the EHR. Medications and ICD9 codes alone achieved reasonable performance. Individually, concepts perform best of all 4 categories. The best-performing algorithm used random forest-based models and identified hypertensive individuals with a median AUC of 0.976. Multicategory random forest models also performed well at Marshfield, with AUCs 0.873–0.934. Thus, using more than just vitals and ICD9 codes individually improved EHR-based hypertension phenotyping.
Combining multiple information sources yielded an increase in performance regardless of method. Confidence intervals overlapped substantially between “count” and “sum” simple algorithm types. Normalized sum algorithms, which include more granularity and information reflecting the total occurrence of categories, performed better at Marshfield than the count algorithm types. This may have been due to different lengths of observation between the 2 sites. Random forests trended higher than these simple aggregations, but implementation may be more difficult than a simple algorithm that combines hypertension ICD9 codes, medications, blood pressures, and regular expression matches, whose performance does not differ greatly (AUC of 0.976 vs 0.959). Easing implementation issues, random forest models required relatively few training cases. As few as 25–50 cases resulted in near peak performance for most random forest models.
Random forests are not necessarily the best possible method, but we have used them because they are an easy way to include nonlinear interactions. If one desires an algorithm that does not require regular expression or UMLS concepts, then the sum of unique normalized hypertension ICD9, meds, and blood pressures is the best algorithm that does not leverage narrative text (AUC 0.948). Interestingly, using readings from both inpatient and outpatient readings consistently outperformed approaches limited to readings taken in the outpatient setting. This may be due to improved coverage, as outpatient-only counts provide far less data, accounting for only 56% of the 21 537 total per-day median vitals readings. Thus, including inpatient data and leveraging the median to reduce the influence of outliers and multiple daily blood pressure readings favors the more inclusive approach.
Individual features are limited in their ability to distinguish cases from controls. Many conditions and circumstances result in abnormal blood pressure readings in nonhypertensive individuals, and many medical encounters tend toward such stressful conditions. In addition, individuals with successfully managed hypertension have normal blood pressure readings. For controls, 55.7% had at least 1 blood pressure reading above the hypertension threshold, and 3.8% had a median systolic or diastolic above threshold. For cases, 4.3% had no blood pressures above the threshold and 1.6% had a median diastolic and systolic below the threshold. Our set was initially selected to have dense records; thus, a population with sparser EHR data is likely to have worse vitals-only performance.
Manual review of random forest misclassifications noted several trends. Random forest models were more likely to miss recently diagnosed hypertensive patients, patients without a hypertension ICD9 code, or individuals with very few notes and only a few hypertension concepts. Controls predicted to be hypertensive by the random forest models were most likely to have been missed during review. These individuals often had well-controlled blood pressures, few if any ICD9 codes, and relatively few notes with complex or severe diagnoses (e.g., cancer and severe Crohn’s disease).
Most algorithms trained on Vanderbilt data successfully replicated on data from the Marshfield Clinic. All random forest–based models achieved AUCs in the range of 0.873–0.934. Algorithms that included NLP-derived concepts did not perform as well at Marshfield. Marshfield data included concepts extracted by MetaMap and a different pipeline that did not limit input to high-yield note subsections, which may have had worse performance. Finally, regular expression matches were not included at Marshfield. Although performance may have been improved by its inclusion, the performance achieved without such data on models trained with regular expression information highlighted the robustness of the random forest models.
For sites that wish to optimize their hypertension phenotyping performance, we provide a KNIME module that will automate many of the normalization and feature creation steps. The module includes a number of the better-performing deterministic algorithms and random forest models trained on the full Vanderbilt dataset. We generated the inputs in our database with 9 relatively simple queries after concept indexing was complete. We have included algorithms that do not require narrative information. We have provided a complete description and protocol as well as example data files on PheKB. The training pipeline for the KNIME module can use occurrence counts of ICD-10-CM codes if provided. ICD-10-CM codes for hypertension are relatively simpler than ICD9 codes (I10 maps all codes in 401.*). However, it is not clear that one could aggregate ICD-10-CM with ICD9 codes without further study. Future work will need to investigate the impact of inclusion of ICD-10-CM codes into ICD9-based algorithms.
Several limitations caution the interpretation of these results. We evaluated the portability at only a single additional site. Other institutions may differ from both Vanderbilt and Marshfield Clinic. While we attempted to standardize the record review between Marshfield and Vanderbilt, there may be systematic differences between the hypertensive and normotensive populations at each site. We limited to ICD9 codes, medications, vitals, and narrative text to achieve broad coverage with simple but readily available information. We focused on the total counts of elements in each category and hypertension-specific counts of each. However, other concepts or lab values for comorbid conditions may prove useful for hypertension classification. More complex NLP—perhaps taking into account temporal patterns—would likely be valuable but would also increase implementation difficulty. While there are significant differences in hypertension prevalence between different demographic groups, we have not included features for sex or ethnicity. Many of our features and relevant codes were expert-curated, thus development of similar phenotyping algorithms is not easily scalable. Medications with an indication of hypertension were determined using MEDI-HPS and as such are imperfect and include some medications that are for pulmonary hypertension. Our algorithm also did not detect the date of onset of hypertension, which could be clinically interesting in a number of circumstances. Anecdotally, we found this challenging to accurately determine for many of the records. Pediatric populations and other subspecialty clinics with higher secondary causes may see different performance.
Conclusion
Our results demonstrated that we can identify hypertensive individuals with high recall and precision by combining EHR data sources. Even simple combinations of elements from different categories are statistically significantly better than current simple ICD9 code count thresholds and within the confidence intervals of the best—random forest—methods. Random forests required relatively few training cases to achieve near peak performance. Models and features based on structured EHR fields are more portable than text-based features, especially regular expression–based features. The best phenotyping algorithms have broad potential applicability.
Funding
This work was supported by Public Health Service award T32 GM07347 from the National Institute of General Medical Studies (NIGMS) for the Vanderbilt Medical-Scientist Training Program and other grants from the National Library of Medicine (R01-LM010685, R21LM011664) and NIGMS (R01 GM105688). The Synthetic Derivative is supported in part by Vanderbilt CTSA grant 1 UL1 RR024975 from the National Center for Advancing Translational Sciences. Replication at the Marshfield Clinic was supported by U01HG006389 from the National Human Genome Research Institute given to Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University.
Competing Interests
The authors have no competing interests to declare.
Contributors
P.L.T., T.A.L., J.P.V., and J.C.D. designed the study. Analysis was performed by P.L.T., E.L., P.L.P., and J.C.D. The literature search was performed by P.L.T. and J.C.D. P.L.T., W.Q.W., R.M.C., H.M., J.C.D., A.M.N., and R.A.D. reviewed cases. Data retrieval was performed by P.L.T., E.L., and J.C.D. The data were interpreted by all authors. The initial document was drafted by P.L.T. and J.C.D. The figures were designed and created by P.L.T. and J.C.D. The tables were created by P.L.T. and J.C.D. Supplementary materials were provided by P.L.T. and J.C.D. All authors revised the document and gave final approval for publication.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
Supplementary Material
REFERENCES
- 1. Yoon SS, Gu Q, Nwankwo T, Wright JD, Hong Y, Burt V. Trends in blood pressure among adults with hypertension: United States, 2003 to 2012. Hypertension. 2015;65(1):54–61. http://hyper.ahajournals.org.proxy.library.vanderbilt.edu/content/65/1/54. Accessed July 8, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mozaffarian D, Benjamin EJ, Go AS, et al. Heart disease and stroke statistics-2015 update: a report from the American Heart Association. Circulation. 2014;131(4):e29–e322. http://www.ncbi.nlm.nih.gov/pubmed/25520374. Accessed December 19, 2014. [DOI] [PubMed] [Google Scholar]
- 3. Cutler JA, Sorlie PD, Wolz M, Thom T, Fields LE, Roccella EJ. Trends in hypertension prevalence, awareness, treatment, and control rates in United States adults between 1988-1994 and 1999-2004. Hypertension. 2008;52(5):818–827. http://www.ncbi.nlm.nih.gov/pubmed/18852389. Accessed October 1, 2013. [DOI] [PubMed] [Google Scholar]
- 4. WHO ISH Writing Group. 2003. World Health Organization (WHO) and Internal Society of Hypertension (ISH) statemnt on management of hypertension - WHO, ISH Writing Group 2003.pdf. 2003. [Google Scholar]
- 5. Myers MG. A proposed algorithm for diagnosing hypertension using automated office blood pressure measurement. J Hypertens. 2010;28(4): 703–708. [DOI] [PubMed] [Google Scholar]
- 6. James PA, Oparil S, Carter BL, et al. 2014. Evidence-based guideline for the management of high blood pressure in adults: report from the panel members appointed to the Eighth Joint National Committee (JNC 8). JAMA. 2014;311(5):507–520. http://jama.jamanetwork.com/article.aspx?article id= 1791497. Accessed July 9, 2014. [DOI] [PubMed] [Google Scholar]
- 7. Wei W-Q, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41 http://genomemedicine.com/content/7/1/41. Accessed May 10, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klabunde RE. Cardiovascular physiology concepts. Heart Failure. 2005:235. [Google Scholar]
- 9. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31(12): 1102–1110. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid= 39 69265&tool=pmcentrez&rendertype=abstract. Accessed October 6, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20(e1):e147–e154. http://www.ncbi.nlm.nih.gov/pubmed/23531748. Accessed October 1, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20(e1):e147–e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hebbring SJ, Rastegar-Mojarad M, Ye Z, Mayer J, Jacobson C, Lin S. Application of clinical text data for phenome-wide association studies (PheWASs). Bioinformatics. 2015;31(12):1981–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics (Oxford, England). 2010;26(9):1205–1210. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2859132&tool=pmcen trez& re n d ertype =abstract. Accessed October 29, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denny JC, Bastarache L, Ritchie MD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013; 31(12): 1102–1110. http://www.pubmedcentral.nih.gov/articlerender.fcgi?ar t id = 3969265&tool=pmcentrez&rendertype=abstract. Accessed May 28, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crosslin DR, Carrell DS, Burt A, et al. Genetic variation in the HLA region is associated with susceptibility to herpes zoster. Genes Immunity. 2015;16(1):1–7. 10.1038/gene.2014.51. Accessed July 10, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crawford DC, Crosslin DR, Tromp G, et al. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5:184 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4060012&tool= pmcentrez & rendertype=abstract. Accessed May 31, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mo H, Pacheco JA, Rasmussen LV, et al. A Prototype for executable and portable electronic clinical quality measures using the KNIME analytics platform. AMIA Jt Summits Transl Sci Proc AMIA Summit on Transl Sci. 2015;2015:127–131. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4525225&tool=pmcentrez&rendertype=abstract. Accessed December 23, 2015. [PMC free article] [PubMed] [Google Scholar]
- 18. Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6(234):234cm3 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=422641 4&tool =pmcentrez&rendertype=abstract. Accessed April 23, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Conway M, Berg RL, Carrell D, et al. Analyzing the heterogeneity and complexity of Electronic Health Record oriented phenotyping algorithms. AMIA Annual Symp Proc/AMIA Symp. AMIA Symposium. 2011;2011:274–283. [PMC free article] [PubMed] [Google Scholar]
- 20. Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc. 2015;22(6):1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wei W-Q, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J Am Med Inform Assoc. 2015:ocv130. http://jamia.oxfordjournals.org/content/early/2015/09/02/jamia.ocv130.abstract. Accessed September 5 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005;43(5):480–485. [DOI] [PubMed] [Google Scholar]
- 23. Savova GK, Fan J, Ye Z, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annual Symp Proc/AMIA Symp. 2010;2010:722–726. [PMC free article] [PubMed] [Google Scholar]
- 24. Penz JFE, Wilcox AB, Hurdle JF. Automated identification of adverse events related to central venous catheters. J Biomed Inform. 2007;40(2):174–182. [DOI] [PubMed] [Google Scholar]
- 25. Friedlin J, Overhage M, Al-Haddad MA, et al. Comparing methods for identifying pancreatic cancer patients using electronic data sources. AMIA Annual Symp Proc/AMIA Symp. 2010;2010:237–241. [PMC free article] [PubMed] [Google Scholar]
- 26. Denny JC, Miller RA, Waitman LR, Arrieta MA, Peterson JF. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inform. 2009;78 (Suppl 1):S34–S42. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2728459 &tool=p m c entrez&rendertype=abstract. Accessed August 22, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Therapeutics. 2008;84(3):362–369. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3763939&tool=pmcentrez&render type=abstract. Accessed February 20, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17(1):19–24. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2995636&tool=pmcentrez&rendertype=abstract. Accessed January 26, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc. 2011;18(4):387–391. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3128409&tool=pmcentrez&rendertype =abstract. Accessed August 23, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei W-Q, Cronin RM, Xu H, Lasko TA, Bastarache L, Denny JC. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc. 2013;20(5):954–961. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3756263&tool=pmcen trez&renderty pe =abstract. Accessed March 12, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bejan CA, Wei W-Q, Denny JC. Assessing the role of a medication-indication resource in the treatment relation extraction from clinical text. J Am Med Inform Assoc. 2015;22(e1):e162–e176. http://jamia.oxfordjournals.org/content/early/2014/11/07/amiajnl-2014-002954.abstract. Accessed July 7, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shang N, Xu H, Rindflesch TC, Cohen T. Identifying plausible adverse drug reactions using knowledge extracted from the literature. J Biomed Inform. 2014;52:293–310. http://www.sciencedirect.com/science/article/pii/S1532046414001580. Accessed July 30, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Khare R, Li J, Lu Z. LabeledIn: cataloging labeled indications for human drugs. J Biomed Inform. 2014;52:448–456. http://www.sciencedirect.com/science/article/pii/S1532046414001853. Accessed July 30, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Denny JC, Spickard A, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. 2014;16(6):806–815. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3002123&tool=pmcentrez&rendertype= abstract. Accessed September 8, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Denny J, Smithers J. “Understanding” medical school curriculum content using KnowledgeMap. Am Med. 2003;10(4):351–363. http://jamia.bmjjournals.com/content/10/4/351.short. Accessed August 6, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wrenn JO, Stein DM, Bakken S, Stetson PD. Quantifying clinical narrative redundancy in an electronic health record. J Am Med Inform Assoc. 2010;17(1):49–53. http://jamia.oxfordjournals.org/content/17/1/49.abstract. Accessed June 4, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Efron B, Tibshirani R. Improvements on cross-validation: The 632+ bootstrap method. J Am Statis Assoc. 1997;92(438):548–560. http://www.tandfonline.com/doi/abs/10.1080/01621459.1997.10474007. Accessed October 31, 2014. [Google Scholar]
- 38. Ohno-machado L. Cross-validation and bootstrap ensembles, bagging, boosting. Harvard-MIT Division of Health Sciences and Technology HST.951J: Medical Decision Support, Fall 2005 Instructors: Professor Lucila Ohno-Machado and Professor Staal Vinterbo, 6.873/HST.951 Medical Decision Support (Spring 2005). http://ocw.mit.edu/courses/health-sciences-and-technology/hst-951j-medical-decision-support-fall-2005/lecture-notes/hst951_6.pdf. [Google Scholar]
- 39. Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–3941. [DOI] [PubMed] [Google Scholar]
- 40. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 41. Bickel JE. Some comparisons among quadratic, spherical, and logarithmic scoring rules. Decision Analysis. 2007;4(2):49–65. [Google Scholar]
- 42. Berthold MR, Cebron N, Dill F, et al. KNIME: The Konstanz information miner (pp. 319–326). Berlin, Heidelberg: Springer, 2008. [Google Scholar]
- 43. McCarty CA, Peissig P, Caldwell MD, Wilke RA. The Marshfield Clinic Personalized Medicine Research Project: 2008 scientific update and lessons learned in the first 6 years. Personalized Med. 2008; 5(5):529–542. [DOI] [PubMed] [Google Scholar]
- 44. Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings of the AMIA Symposium. 2001;17. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.