Abstract
Titin truncating variants (TTNtv) commonly cause dilated cardiomyopathy (DCM). TTNtv are also encountered in ~1% of the general population where they may be silent, perhaps reflecting allelic factors. To better understand TTNtv we integrated TTN allelic series, cardiac imaging and genomic data in humans and studied rat models with disparate TTNtv. In patients with DCM, TTNtv throughout TTN were significantly associated with DCM. Ribosomal profiling in rat revealed the translational footprint of premature stop codons in Ttn, TTNtv position-independent nonsense-mediated degradation of the mutant allele and a signature of perturbed cardiac metabolism. Heart physiology in rats with TTNtv was unremarkable at baseline but became impaired during cardiac stress. In healthy humans, machine-based analysis of high-resolution cardiac scans showed TTNtv to be associated with eccentric cardiac remodelling. These data show that TTNtv have molecular and physiological effects on the heart across species, with a continuum of expressivity in health and disease.
Dilated cardiomyopathy (DCM) has a prevalence of up to 1:2501, is the commonest indication for heart transplantation and is often associated with titin truncating variants (TTNtv, 15-20% of DCM cases), which are enriched in the titin A-band2,3. A surprising ~1% of the general population has a TTNtv in the absence of DCM, which has stimulated much debate as to the pathogenicity of TTNtv4–7. It has been suggested that TTNtv in the healthy population may be phenotypically silent and that TTNtv that segregate in familial DCM are perhaps modifiers of other DCM-causing variants3. However, it is also known that genetic variation affecting disease genes can be associated with quantitative variation in the physiology of healthy individuals8, which has not been assessed formally for TTNtv.
We showed previously that titin exon characteristics are important determinants of TTNtv pathogenicity. Variants encoded in exons that are not spliced into titin isoforms expressed in the heart (non-cardiac exons with ‘Percent Spliced In’ (PSI) < 15%) are not associated with DCM3, while variants encoded in exons that only incorporate into the N2BA and not into the N2B isoform have weak associations with disease. In studies of human induced pluripotent stem cell (iPSC)-derived cardiomyocytes (iPSC-CMs)9 alternative exon splicing is a major mechanism for reduced penetrance for some I-band TTNtv. When taking only TTNtv that are located in cardiac exons (PSI > 15%) into account, approximately 0.5% of the general population have a TTNtv that might be expected to cause DCM but does not.
In addition to the complexities surrounding TTNtv penetrance, the molecular mechanisms underlying the pathogenesis of TTNtv are uncharacterized. Nonsense-mediated decay (NMD) of the mutated allele by RNA-seq or a reduction in full-length TTN by agarose gel analysis has not been demonstrated, which if documented would support a haploinsufficient disease mechanism3,9,10. Equally, accumulation of a truncated TTN protein molecule is not apparent in human heart samples3 and is rarely present in iPSC-CMs9 suggesting a poison-peptide/dominant negative mechanism to be unlikely.
Here, we undertook studies of DCM patients, rat models of TTNtv and human volunteers from the general population to better understand TTNtv pathogenicity and molecular effect with a specific focus on dissecting a hypothesized position-dependent penetrance effect of TTNtv alleles. To do this, we performed a meta-analysis of TTNtv in DCM patients (n = 2,495) as compared to controls (n = 61,834) and generated two rat models of TTNtv with mutations at opposite ends of the titin molecule. We integrated RNA-sequencing (RNA-seq) and ribosome profiling (Ribo-seq) data across models and species and performed metabolic and signaling studies to outline potential disease mechanisms. To define the effects of TTNtv on the heart in the general population we deeply phenotyped and TTN sequenced healthy volunteers (n = 1,409). We supplemented standard 2D cardiac magnetic resonance imaging (CMR) with high-resolution, high-fidelity 3D CMR, integrating and analysing the imaging and sequencing data using machine learning techniques.
Results
TTNtv in constitutive exons across the TTN molecule from the Z-disc to the M-band are associated with DCM
We and others have shown that A-band TTNtv are associated with DCM2,3,9 but the association of DCM with TTNtv in the I-band, Z-disc or the M-line has not been demonstrated and it is suggested that proximal TTNtv, in particular, may be non-penetrant. To address variant effect across the titin molecule we retrieved TTNtv alleles from DCM patients from available sources, combined these data with novel DCM cases (n = 1,105) and performed a meta-analysis using ExAC11 and other controls (cohort totals: DCM (n = 2,495) controls (n = 61,834); see Methods). This showed that TTNtv in constitutive exons (PSI > 90%) are significantly associated with DCM irrespective of their position in TTN (Table 1).
Table 1. Meta-analysis shows an association of TTNtv in constitutive exons throughout the gene with DCM.
Exon usage are displayed base on the levels of PSI14. Constitutive exons are spliced into titin isoforms with an efficiency of at least 90%. OR, odds ratio. EF, etiologic fraction. P-values indicate significant enrichment for TTNtv in DCM [binomial test].
| DCM Positive | Control Positive | DCM Prevalence (%) | Control Prevalence (%) | OR | OR (hi) | OR (lo) | EF | P-value | |
|---|---|---|---|---|---|---|---|---|---|
| (n=2,495) | (n=61,834) | ||||||||
| Sarcomere domain affected by variant | |||||||||
| A-band (constitutive) | 268 | 149 | 10.74 | 0.24 | 49.8 | 61.1 | 40.6 | 0.98 | 2.4x10-260 |
| I-band (Distal constitutive Post-Cronos) | 9 | 7 | 0.36 | 0.01 | 32.0 | 85.9 | 11.9 | 0.97 | 2.5x10-9 |
| I-band (Distal constitutive Pre-Cronos) | 18 | 23 | 0.72 | 0.04 | 19.5 | 36.2 | 10.5 | 0.95 | 6.6x10-15 |
| I-band (Non-constitutive) | 6 | 102 | 0.24 | 0.17 | 1.5 | 3.3 | 0.6 | 0.31 | 0.46 |
| I-band (Proximal constitutive) | 22 | 29 | 0.88 | 0.05 | 19.0 | 33.0 | 10.9 | 0.95 | 1.1x10-17 |
| M-band (constitutive) | 6 | 40 | 0.24 | 0.07 | 3.7 | 8.8 | 1.6 | 0.73 | 0.01 |
| Z-disc (constitutive) | 7 | 33 | 0.28 | 0.05 | 5.3 | 11.9 | 2.3 | 0.81 | 0.001 |
| Z-disc (Non-constitutive) | 0 | 11 | 0.00 | 0.02 | NA | NA | NA | NA | NA |
While some I-band TTNtv may be rescued by differential splicing9, we observed that I-band TTNtv in constitutive cardiac exons were significantly associated with DCM, which was true for both proximal and distal I-band variants. TTNtv in Z-disc exons were also associated with DCM, although with a much lower odds ratio (OR: 5.3) than A-band TTNtv (OR: 49.8). It has been proposed that a distal internal promoter of the Cronos TTN isoform12, which we confirmed to be present in adult human heart using Cap Analysis of Gene Expression (CAGE)13 data (Fig S1), can rescue proximal TTNtv effect. However, we found that proximal TTNtv are also penetrant and associated with DCM. By splitting regions of titin into protein regions (Table 1), or consecutive uniformly-sized bins (n = 40, (Fig S2)) we were able to show the proportion of truncations that are penetrant (etiological fraction) in regions of TTN upstream of the Cronos promoter is comparable to the proportion in the downstream A-Band. These data show that distal I-band and all A-band TTNtv have larger ORs than very proximal or distal variants, which suggests position-dependent effects for the penetrance of TTNtv in DCM.
Premature stop codons cause nonsense-mediated RNA decay and disrupt translation of full length, sarcomere-spanning Ttn
Our meta-analyses of DCM patients showed that TTNtv throughout the titin molecule are penetrant but have variable, position-related odds ratios. To study putative position-dependent effects in greater detail while controlling for other genetic and non-genetic factors we modelled proximal and distal TTNtv in two independent rat strains on the same genetic background (TTNtvA: with variation in the A-band; TTNtvZ: with variation in the Z-disc; for details see Fig S3). Animals with homozygous mutations were not viable, as previously described for TTNtv in the mouse10 while heterozygous animals were born in normal Mendelian ratios (data not shown).
We bred TTNtv heterozygous rats on a F344 background with Ttn wild type Brown Norway rats to obtain an F1 cross to specifically enable allele-specific analysis of Ttn transcription and translation in the heart of TTNtvA (n=3), TTNtvZ (n=3) and WT (n=4) animals using RNA-seq and Ribo-seq (Fig 1a and Fig S4)15,16. Ribosome protected fragments (RPFs) showed clear trinucleotide periodicity across the translatome, indicating actively translating ribosomes (Fig 1b). Ttn transcripts with a truncation in the A-band were transcribed and translated as far as the premature nonsense codon, which efficiently stopped translation thereafter (Fig 1c,d).
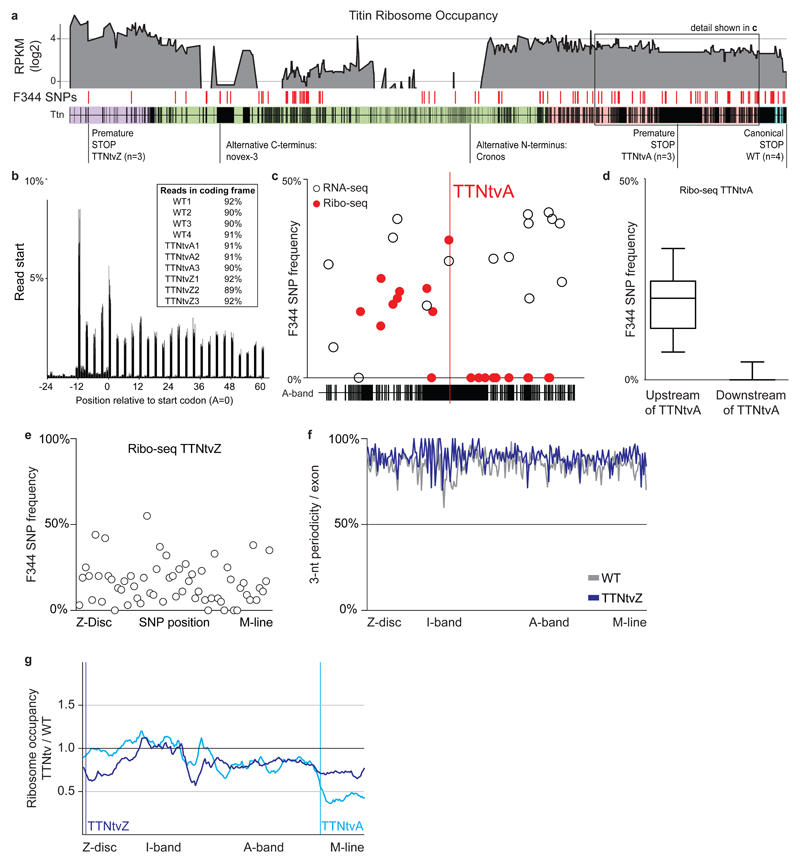
Fig 1. Ribosome profiling reveals the translational footprint of truncating variants in Ttn.
a Ribosome occupancy across Ttn: Average reads per kilobase per million mapped reads (RPKM) per exon for 10 BN/F344 F1 rats. F344 SNPs on the BN background (n=121) allow assessment of allele-specific translation for 2 distinct models of truncations and Ttn wild type animals. Purple indicates Z-disc, green I-band, pink A-band and blue M-line. b Ribo-seq reads (28-mers) show clear 3-nt periodicity across the genome for all replicates indicating that the datasets effectively capture actively translating ribosomes in the heart. c,d Ribosomes occupy the F344 allele in TTNtvA animals upstream of the premature stop codon in the A-band of Ttn exposing the synthesis of truncated Ttn isoforms. The premature stop releases ribosomes from the mutant Ttn message. Titin protein synthesized after TTNtvA is exclusively generated from the healthy BN allele (Median ± 25-75 percentile, Whiskers show 10-90 percentile). Allele frequencies in RNA-seq and Ribo-seq were below 50%, indicative for NMD. e F344 SNPs located after TTNtvZ are occupied by ribosomes, indicating translation after the proximal truncation in Ttn. f Ribo-seq reads located downstream of TTNtvZ do not lower 3-nt periodicity suggesting they actively translate canonical Ttn sequence at similar levels as animals with two wild type Ttn alleles. g TTNtvZ reduces ribosome density initially but translation of Ttn is gradually rescued. Translation of A-band exons, but not of I-band exons, is reduced in mutants compared to WT animals. TTNtvA efficiently reduces translation after the premature stop codon.
The premature stop codon of the TTNtvZ allele did not prevent translation downstream of the non-canonical stop and F344 SNPs encoded on the truncated Ttn allele were detected throughout Ttn (Fig. 1e). Rescue of translation from the truncated F344 allele did not decrease the 3-nt periodicity across Ttn when compared to F1 wild type (WT) animals that synthesize titin from two healthy alleles (Fig. 1f). Exon-level analyses showed that the TTNtvZ variant initially reduces ribosome occupancy at the N-terminus of Ttn (~50% that of WT) but subsequently translation is partially recovered (Fig 1g). This apparent rescue of translation might be explained in parts by internal ribosomal entry sites, transcription start sites that generate Ttn isoforms with alternative N-termini and potentially by factors that maintain ribosomes on the huge Ttn transcript during its translation. In support of this, human CAGE data13 and epigenetic marks17 revealed distal TTN promoters in the human heart (Fig S1), as previously suggested12. In TTNtvA animals, we only detected ribosomes on the healthy BN allele after the premature stop codon (Fig 1c). This led to a decrease of ribosome occupancy by 50% when we compared to WT animals that still translate this section of titin from both alleles (Fig 1g).
In both mutant rats there was a relative increase in exon translation in the I-band (Fig. 1g) that reflects differential Ttn mRNA processing and higher PSI ratios14 of exons in the I-band, as observed in human iPS-derived cardiomyocytes with TTNtv9. This is indicative of an increase in N2BA isoform expression in TTNtv carriers. To translate this finding, we compared cardiac RNA-seq data3 from DCM patients either with (n = 17) or without (n = 91) a TTNtv and observed similar patterns of alternative splicing as seen in the rat models (Fig 2a). Allele-specific RNA-seq data revealed a slight upregulation of the WT allele and profound NMD of the allele carrying the TTNtv in both the TTNtvA and the TTNtvZ models (Fig 2b). These data demonstrate a multi-allelic effect of TTNtv on the overall TTN isoform composition across species (that can cause DCM18,19), and that TTNtv confer position-independent NMD.
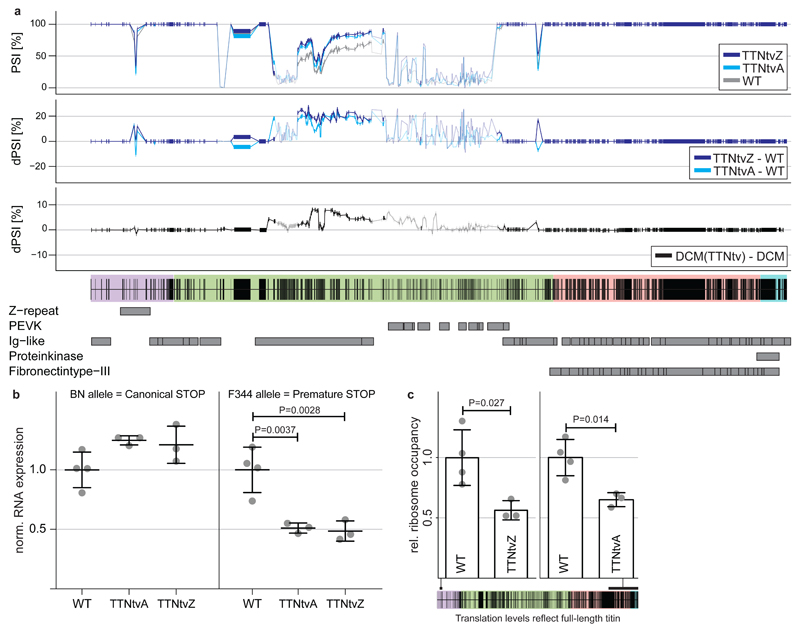
Fig 2. Proximal and distal truncations in titin alter isoform processing and trigger NMD.
a PSI and deltaPSI of titin exons expressed in the heart for TTNtvA, TTNtvZ and WT animals and human DCM. Exons with at least 10 inclusion reads are marked in solid colours. Truncating mutations in Ttn activate splicing in of I-band exons in the TTNtv rat models and also human DCM patients that carry TTN truncations. b RNA-seq reads assigned to either BN or F344 alleles: TTNtv selectively trigger nonsense-mediated decay of truncated Ttn transcripts. [Mean ± SD; Dunnett] c Ribo-seq expression of exons that are exclusively being synthesized in TTNtvA and TTNtvZ animals reveal that both TTNtv models generate 60% of full-length titin compared to WT [Mean ± SD; Student’s t test]. Purple indicates Z-disc, green I-band, pink A-band and blue M-line.
It was noticeable that both the proximal and the distal TTNtv triggered NMD with identical efficiencies irrespective of their location in the Ttn molecule. The fact that the proximal truncation triggers NMD indicates that the premature stop codon close to the N-terminus in the TTNtvZ is functional and that ribosomes cannot clear exon-exon junction complexes20,21. Assessing translation beyond the truncation in the TTNtvA model and within the 4-exon deletion in TTNtvZ revealed that both rat models synthesize lesser amounts (~60% of WT rats) of full-length, sarcomere-spanning Ttn (Fig 2c). In keeping with our published data3, protein gel electrophoresis did not identify a truncated Ttn isoform or a reduction in the Ttn:Mhc ratio in mutant rat heart (Fig S5). This suggests rapid turnover of mutant protein that, for the first time, we document the existence of by showing its synthesis (Fig 1). It also shows that while there are significantly less ribosomes translating full length protein, titin protein changes are not apparent at the resolution of gel analysis. Overall changes in full length titin protein might be compensated not just by an upregulation of the wildtype allele (P = 0.02; Fig 2b), but also via an increase in ribosome translational speed or changes in protein-turnover.
Taken together these data show that proximal and distal Ttn truncations disrupt Ttn protein synthesis but have different translational footprints. However, their effect on NMD of the sarcomere-spanning Ttn isoforms is similar and leads to identical reductions in full-length Ttn expression.
Titin truncations cause position-independent perturbation of cardiac metabolism and signaling
To determine whether distinct or similar molecular phenotypes are associated with proximal or distal TTNtv, we performed pathway analysis22 of the genome-wide transcription (RNA-seq) and translation (Ribo-seq) profiles of TTNtvZ and TTNtvA as compared to controls. The gene expression differences observed between TTNtv animals and WT controls were highly correlated for TTNtvA and TTNtvZ both for RNA-seq and also Ribo-seq data, showing both mutant strains to have similarly perturbed transcriptomes (R2 = 0.841; P < 0.0001) and translatomes (R2 = 0.837; P < 0.0001; see also Fig S6 & Table S1). Gene set enrichment analyses showed significantly overlapping KEGG 22,23 terms between TTNtv animals when compared to WT (RNA-seq P value < 10-15; Ribo-seq P value < 10-15; Pearson Chi-square test; Fig 3a), which suggested altered cardiac metabolism that was independent of the position of the TTNtv.
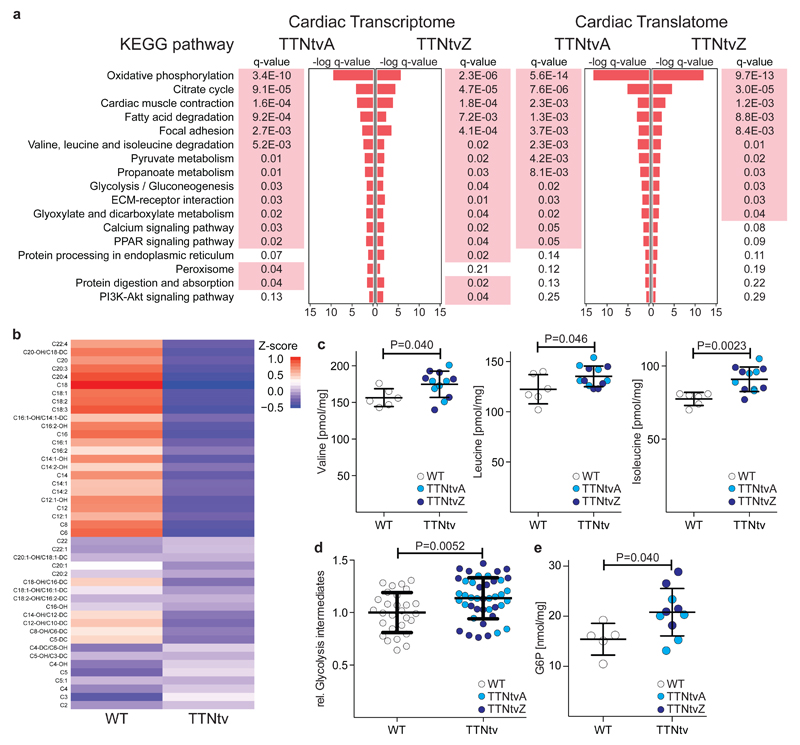
Fig 3. Hearts with proximal and distal truncations in titin undergo metabolic reprogramming.
a Pathway analyses based on RNA-seq and Ribo-seq data suggest perturbed metabolism, structural integrity and mechano-sensation of the TTNtv heart. This molecular signature is strikingly similar in rats with proximal and distal truncations (P value < 10-15; Pearson Chi-square test). Significantly enriched pathways are indicated in red (corrected P<0.05). b Unsupervised clustering by k-means of cardiac acylcarnitine abundance in WT (n=5) and TTNtv (n=10) rats. –OH and –DC designate hydroxylated and dicarboxylic acid acylcarnitine species respectively. Metabolite profiles showing c branched chain amino acids (valine, leucine and isoleucine) [Mean ± SD; Student’s t test], d sum of measured glycolytic intermediates (metabolites are detailed in Table S3) [Mean ± SD; Student’s t test] and e glucose-6-phosphate (G6P) in cardiac tissue of WT (n=6) and TTNtv (n=12) rats [Mean ± SD; Student’s t test]. For individual genotypes (WT vs TTNtvA or TTNtvZ) see Fig S7.
To investigate further the molecular signatures of altered cardiac metabolism in mutant animals, we performed quantitative metabolomic profiling of WT and TTNtv hearts. Liquid chromatography mass spectrometry (LC-MS) showed reduced amounts of medium and long chain fatty acid acyl-carnitines in TTNtv hearts as compared to controls (Fig 3b; Table S2). We also performed capillary electrophoresis mass spectrometry, complementary to LC-MS, and observed accumulation of alternative myocardial substrates (branched-chain amino acid metabolites and glycolytic intermediates; Fig 3c-e; Fig S7; Table S3) in mutants. These changes are similar to those seen in the failing heart and the pressure-loaded non-failing heart24,25 and are associated with a shift in cardiac metabolism away from fatty acids and towards glycolysis, which may be adaptive26–28. There was no change in the major energy substrates (e.g. ATP; Fig S8), which are only diminished in advanced cardiac failure29.
The signalling changes in the heart due to TTNtv are likely many. We found that variation in metabolic proteins interacting with titin, such as FHL2 (Fig S9), are small and their functional role cannot be established based on these data alone. It is known that mTORC1 signalling is activated in familial DCM30 and its activity is detrimental in a mouse model of DCM due to LMNA mutation31. We profiled the mTORC1 pathway in the TTNtv rat hearts, where metabolites that activate mTOR32 are elevated (Fig 3) and observed activation of this pathway (Fig S10). The relative importance of these signalling variations for DCM pathobiology remain to be established.
TTNtv impair cardiac performance during stress in rats and have adverse effects the heart in the general population
In young TTNtv rats (<8 months old) cardiac imaging showed features of concentric remodeling but normal LV mass and systolic function (Fig S11). In older rats (>1 year), TTNtv hearts were similar to controls although with a suggestion of slightly impaired systolic function (Fig 4a, Fig S12). While changes in cardiac morphology and function were mild in young mutant rats it was possible that this represented a compensated state as evidenced by the shift in cardiac metabolism (Fig 3) that is adaptive, at least in the short term26. We examined cardiac function ex vivo and used a volume overload model to test the Frank-Starling response, which may be specifically impaired by genetic variation in TTN33. Under basal conditions (LV end diastolic pressure, 5-10mmHg) TTNtv rat hearts tended to have higher strain rates and LV developed pressures, perhaps reflecting compensatory metabolism (Fig 3) and signalling (Fig S10) but, when subjected to sequential volume overload stress, mutant heart function became increasingly impaired (Fig 4b). As observed previously34–36, cardiac stress ex vivo activated mTORC1 signalling in control animals, which is an adaptive response. However, TTNtv hearts had elevated mTORC1 signalling at baseline and were not able to appropriately increase mTORC1 activity further when stressed (see Fig S10).
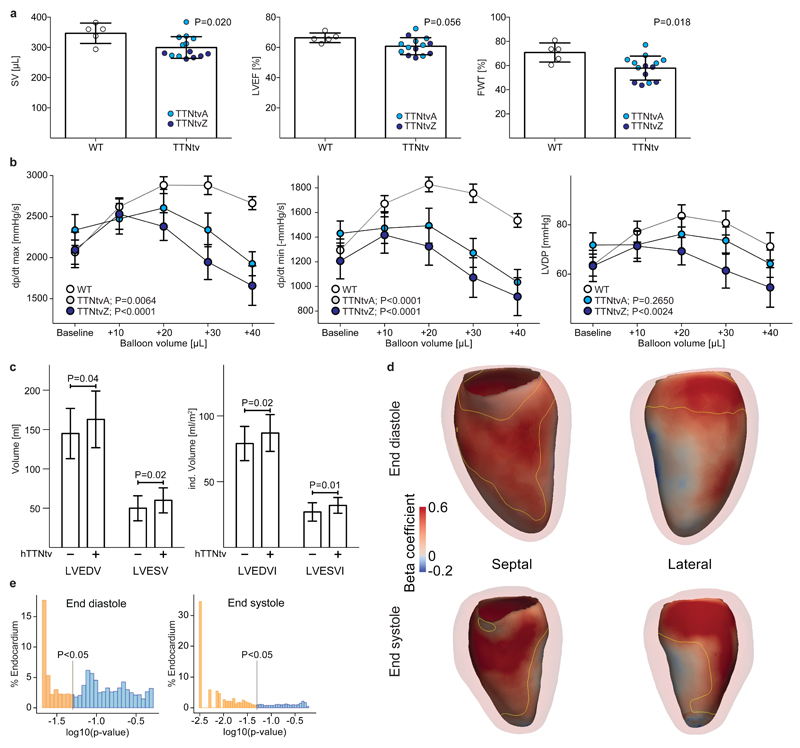
Fig 4. TTNtv in rats and humans adversely affect cardiac geometry and function.
a SV (Stroke Volume), LVEF (Left Ventricular Ejection Fraction) and FWT (Fractional Wall Thickening) measured with CMR in 13-16 month old male WT (n=5) and TTNtv (TTNtvA, n=8; TTNtvZ, n=6) rats [Mean ± SD; Student’s t test]. For individual genotypes see Fig S12. b Measurements of ex vivo myocardial function during volume overload stress in 4 month old WT (n=9) and TTNtv (TTNtvA, n=8; TTNtvZ, n=8) rats. TTNtv hearts have mildly increased dp/dt max/min and LVDevP at baseline. Myocardial contraction rate, dp/dt max (mmHg/s); myocardial relaxation rate, dp/dt min (-mmHg/s); Left ventricle developed pressure, LVDevP (mmHg). [Mean ± SEM; Two-way ANOVA]. Human data: c Univariate analyses of left ventricular end diastolic volume (LVEDV), LEVEDV indexed to body surface area (LVEDVI), left ventricular end systolic volume (LVESV) and LVESV indexed to body surface area (LVESVI). Healthy human individuals without a TTNtv (TTNtv-) are compared to healthy humans with a TTNtv (TTNtv+) [Mean ± SD; Mann-Whitney]. d Computational modeling of cardiac geometry in healthy humans using 3D CMR. Positive standardised beta coefficients indicate where TTNtv genotype status is associated with enlargement of the LV cavity at end-diastole and end-systole. Septal and lateral en face projections are shown with an outline of the LV myocardium. The area enclosed by the yellow contour has a corrected P <0.05 [mass univariate linear regression]. e The distribution of corrected P values as a proportion of the endocardial surface [mass univariate linear regression].
In the rat, TTNtv had mild effects on heart function irrespective of their position in the Ttn molecule but did not cause DCM. To explore the possibility that TTNtv in cardiac exons similarly affect the heart in human subjects, irrespective of disease status, we recruited 1,409 healthy individuals for detailed CMR studies of the heart in combination with TTN sequencing. We specifically focused our studies on the cardiac parameters of LV end diastolic volume (LVEDV) and ejection fraction (EF) that are used to define DCM37 and also LV end systolic volume (LVESV) that is elevated in pre-DCM and predicts heart failure onset38. In this cohort we identified 15 TTNtv (see Table S4; prevalence = 1.0%) in TTN exons (PSI > 15%) in keeping with our previous findings3 and the data from the ExAC Consortium11. The individuals carrying these truncations in cardiac exons are labelled as TTNtv+ in Fig 4. Truncations in TTN in our cohort and in the ExAC dataset were equally distributed across the TTN molecule (Fig S13).
After genotype-blinded analysis of CMR data, we found that no individual from the general population with a TTNtv had imaging criteria for DCM, similar to our observations in the rat models and previous studies3. However, in univariate analyses TTNtv conferred a significant increase in absolute LV volumes and had more pronounced effects on volumes indexed to body surface area (Fig 4c). There was a non-significant trend for a lower LVEF in TTNtv+ individuals (LVEF (%): TTNtv+, 66 ± 5; TTNtv-, 63 ± 5; P = 0.06, Mann-Whitney). Given the multiple clinical and anthropometric variables that predict cardiac morphology and function, we built regression models for LVEDV, LVESV and LVEF (see Methods and Table S5) and tested whether addition of TTNtv status improved model performance, which turned out to be the case (absolute beta values: LVEDV, +11.8mls (8.1%); LVESV +7.7 mls (15%); EF = -2.8% (P<0.03 for all)). Of note, the effect size of TTNtv on cardiac parameters was much greater than the effect sizes of cardiovascular GWAS loci (e.g. systolic blood pressure; combined effect of all GWAS loci ~3%)39,40.
To complement our 2D studies we collected an independent dataset of high-resolution, single-breath hold 3D CMR images and, blinded to genotype, performed atlas- and machine-based analyses of LV geometry with respect to TTNtv status41,42. The 3D data show that TTNtv were associated with eccentric cardiac remodeling in healthy individuals. This was defined by outwards displacement of the endocardial border of the heart (Fig 4d,e and Supplementary video 1) in both systole (79% of total surface, P < 0.05) and diastole (47% of total surface, P < 0.05), in consensus with the 2D data that showed larger LV volumes at these phases of the cardiac cycle.
Discussion
Here we studied the effects of TTNtv in patients with DCM, in the rat and in healthy humans to better understand these variants that represent the commonest genetic cause of DCM, yet are prevalent in the general population. At the molecular level, we found that TTNtv cause altered I-band splicing and position-independent NMD, which attenuates the synthesis of sarcomere-spanning Ttn isoforms. The NMD of the TTNtv allele that we observed in the F1 rat cross is the first demonstration of this effect and something we were not able to show previously using unphased human RNA-seq3. Distal truncations are associated with synthesis of carboxy-terminus truncated Ttn isoforms, whereas proximal TTNtv lead to translation of additional Ttn isoforms with alternative amino-termini. None of these additional isoforms were detected on protein gels, which suggests rapid degradation of these species (Fig S5). It was notable that, irrespective of their position in the Ttn molecule, both proximal and distal premature stop codons in the rat caused highly similar gene expression and translational signatures. While these molecular phenotypes are sufficient to impact heart function, it does not exclude additional, position-dependent effects that modify TTNtv penetrance in DCM where distal I-band and all A-band TTNtvs have the highest ORs.
The molecular phenotype due to TTNtv, which is reminiscent of cardiac adaptation to heart failure stimuli with a shift away from fatty acid metabolism26–28, was associated with activation of the mTORC1 pathway that is also activated in familial DCM30. We suggest that the metabolic (Fig 3) and signalling (Fig S10) changes in the TTNtv heart represent adaptive mechanisms26–28 and that the heart is maintained in a compensated state and therefore inflexible to further stress. This may explain why hemodynamic stress associated with pregnancy or possibly a second genetic factor may reveal TTNtv effects43. It will be interesting to see what other environmental triggers for DCM (e.g. alcohol, viral infection or chemotherapy) combine with TTNtv to cause disease and equally to ascertain why some individuals with TTNtv and an interacting stimulus such as pregnancy, do not develop disease.
We found that TTNtv in constitutive exons throughout the TTN molecule, from the Z-disc to the M-line, are significantly associated with DCM. This has implications for interpretation of TTNtv in patients with DCM although A-band and distal I-band TTNtv have higher ORs than variants in other TTN domains, for reasons that remain unclear. In the general population, we show that TTNtv that were previously thought to be of limited consequence44 are associated with higher left ventricular volumes in 2D CMR analyses, which reflects underlying eccentric remodelling that was revealed using advanced 3D CMR techniques. These data demonstrate the benefits of combining high-resolution phenotyping and machine-based data analysis for imaging genetic studies of the heart, which we suggest may be applied at scale in large cohorts with cardiac imaging and genetics, such as the UK Biobank. It is apparent that the magnitude of effect of TTNtv on cardiac geometry in the general population may be large enough to adversely influence future cardiac events in some individuals, which requires further study.
We note that TTNtv in exons that are expressed in the heart (PSI > 15%) are present in about 0.5% of individuals across all ethnicities3,11 and it may be that this variant class is of clinical relevance for up to 35 million people, particularly if they are exposed to additional genetic or environmental cardiac stresses. Future studies that pinpoint those at greatest risk from the interaction between a TTNtv and a secondary trigger, genetic or environmental, are needed to move the field forward.
Online Methods
Animal studies
Rat TTNtv models were generated by SAGE Laboratories using zinc-finger nuclease–mediated gene targeting. Animals were maintained on a F344 background. For A-band (C-terminus) truncating variant (TTNtvA), 12bp were deleted and 2bp inserted (TA) at 228608-228619 to introduce a stop codon in exon 303 (Genomic NCBI Ref Seq: NC_005102.3) corresponding to exon 327 in the human sequence. For Z-disc (N-terminus) truncating mutation (TTNtvZ) was generated by deletion of exons 2 to 6 (5286bp deletion, coordinates 2323-7608) to introduce a frame-shift. For details see supplementary figure 1. Genotypes were detected by polymerase chain reaction (PCR) and products confirmed by Sanger sequencing. Animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee (IACUC, 2013/SHS/844) at the Duke-National University of Singapore Medical School.
To enable allele-specific expression analyses with RNA-seq and Ribo-seq, we crossed WT, TTNtvA and TTNtvZ F344 animals with healthy BN animals. These F1 animals were only used for sequencing experiments. The WT strain carries a non-truncated BN Ttn allele and one non-truncated F344 Ttn allele. The TTNtvA and TTNtvZ strains carry a truncated F344 Ttn allele and one non-truncated BN Ttn allele.
Patient and control cohorts and rare TTNtv burden testing
Data were collated from previously published case-series2,3,45,46 alongside a novel cohort of 1,105 DCM patients and 571 healthy controls. Overall, TTN sequencing was performed in a total of 2,495 cases and 61,834 reference samples. Case cohorts included 689 probands previously published3, 241 cases previously published2 (a UK cohort from this study, originally denoted DCM-B, was excluded due to overlap with the Roberts et al series), 156 probands referred to the Partners Healthcare Laboratory of Molecular Medicine for molecular diagnostics45,46 and 304 probands similarly referred to the Oxford Medical Genetics Laboratory (OMGL), UK46. Further prospective unselected patients with a diagnosis of DCM, confirmed using cardiac imaging with reference to established cardiac MRI or echocardiographic diagnostic criteria as previously described3, were recruited via the NIHR Royal Brompton Cardiovascular Biomedical Research Unit (n=542), the German Center for Cardiovascular Research, Berlin (n=386) and the National Heart Centre, Singapore (n=177) and are reported here for the first time. These latter cohorts were sequenced on the Illumina MiSeq, Illumina NextSeq, or Life Technologies SOLiD 5500xl platform after target enrichment using in solution hybridisation (Illumina Nextera47 or Agilent SureSelect3), with analysis using standard pipelines as previously described3. The research studies were approved by a research ethics comittee and all participants gave written informed consent. Informatics pipelines were used to define TTNtvs as previously described3. TTNtvs included frameshifts, non-canonical stops and essential splice site variation.
Reference samples comprised the Exome Aggregation Consortium (ExAC, n=60,706)11 [version 0.3, Jan 2015], alongside 557 controls from our previous studies2,3 after exclusion of control cohorts that overlap with ExAC and 571 unpublished controls recruited at the National Heart Centre Singpore.
Rare variants that were predicted to truncate full-length TTN were included in analyses (rare = ExAC MAF < 1x10-4; predicted truncating = nonsense, frameshift insertion or deletion, or disrupting canonical splice donor/acceptor sequence, TTN reference = inferred complete meta-transcript LRG391_t1).
Variants were first grouped according to their location within the Titin structure and the expression of the exon in which they were found (PSI). We considered exons to be constitutive, if they were spliced into at least 90% of the TTN transcripts present in the heart (PSI >= 90%). For each region the prevalence of TTNtv in cases & controls were compared using the binomial test in R.
Constitutive TTN exons (plus 2bp canonical splice sequences) were divided into 40 uniform bins. For each bin, the total number of TTNtv alleles was compared in cases and controls, an odds ratio was computed, and the etiological fraction (EF: the probability that an individual rare variant, found in a proband, was responsible for the disease) was calculated using as previously described46.
RNA- and Ribo-sequencing library preparation
To assess the translational status of wild type and Ttn mutant rats, ribosome profiling libraries were generated as previously described 16. Briefly, ~70mg of cardiac tissue were used for each animal and strain (WT: n=4, TTNtvA: n=3, TTNtvZ: n=3) and subjected to cryolysis in 1 ml lysis buffer (1× TruSeq mammalian polysome buffer (Illumina), 1% Triton X100, 0.1% NP-40, 1 mM DTT, 10 U ml−1 DNase I and nuclease-free H2O) supplemented with 0.1mg/ml CHX (Sigma). The tissue was further homogenized using a 21-gauge syringe needle, incubated on ice for 10 min and centrifuged at 20,000g. 400μl lysate per sample were used to obtain footprints (or Ribosome protected fragments, RPFs) upon incubation with TruSeq Ribo Profile Nuclease (3 U per OD260 / ml of lysate). RPF purification was performed using MicroSpin S-400 columns (GE Healthcare) followed by phenol-chloroform RNA extraction. Mammalian rRNA was subsequently removed following procedures described for the RiboZero Magnetic Gold kit (Epicentre). Ribosome footprints are expected to be 28-30 nt long. To recover this exact size window, purified fragments were resolved under denaturing PAGE. Following adapter ligation, reverse transcription and a second denaturing PAGE purification, samples were circularized. The amplification of Ribo-seq libraries was performed by PCR (12 cycles) using the circularized products as templates. PCR amplified libraries were purified on native 8% polyacrylamide gels and quantified using the Qubit fluorometer. The quality and average fragment size of the libraries was instead assessed with the Bioanalyzer High Sensitivity Assay (Agilent). In order to reduce technical biases, samples were barcoded and pooled to perform multiplex sequencing on the Hiseq2000 platform (single end, 50bp sequencing chemistry).
To assess transcript abundance, stranded polyA+ RNA-sequencing was performed following standard manufacturer’s instructions (TruSeq stranded mRNA-seq library prep kit), using 1μg total RNA as input. Libraries were pooled and sequenced on a HiSeq2000 instrument (paired end, 100bp sequencing chemistry).
Read mapping
We used the Illumina CASAVA 1.8 software to demultiplex all reads and convert bcl to fastq files. We then clipped adapters from the Ribo-seq cDNA inserts using the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html). Adapters were clipped using the command:
fastx_clipper -a AGATCGGAAGAGCACACGTCT -l 20 -n -v -Q33 | fastx_trimmer -Q33 -f 1
We then also processed the RNA-seq reads in a similar manner and trimmed them to a length of 29 bp, the most abundant read length found in Ribo-seq:
fastx_trimmer -l 29 -Q33
Trimming the RNA-seq data prior to mapping ensures that we can compare both methodologies and draw conclusions regarding translational regulation. This step avoids differences in expression estimates that arise for technical differences of both methods. We then removed mtRNA and rRNA sequences from all libraries using bowtie48,49:
bowtie -l 20 --un clean.fastq /abundant_sequences input.fastq -S abundant.sam
Abundant sequences were stored in a bowtie index derived from fastq files of the rat mitochondrial genome and rRNA sequences annotated in the Ensembl database50. Sequences that did not align (clean.fastq) were then further mapped to the genome using Tophat 2.0.1351 against the rn5 genome:
tophat2 --read-realign-edit-dist 0 -p 6 -z0 -M -j HiQual.juncs -G Rattus_norvegicus.Rnor_5.0.79_TTN.gtf rn5 R1.fastq
To improve the mapping of these short reads, we introduced splicing junctions as described previously16. We also supplied the Ensembl50 rat genome annotation v79 to the mapping pipeline. Titin is not well annotated in the rat, so we decided to lift over the human titin to the rat genome. We used the Lift-Over tool of the Galaxy52,53 platform with standard parameters to transfer the titin transcript model3 to rn5. Split reads in RNA-seq data were used to confirm exon boundaries in the rat. Titin regions and domains are shown as reported previously in the human3. The Ensembl gene IDs representing parts of titin were removed from the annotation and replaced with our custom titin annotation prior the mapping step and then used for all analyses further downstream.
To analyze alternative splicing, we did not trim the RNA-seq reads but utilized the full-length 2x100bp paired-end data to improve the estimation of splice site usage in the rat:
tophat2 --read-realign-edit-dist 0 -p 6 --no-coverage-search -M -j HiQual.juncs -G Rattus_norvegicus.Rnor_5.0.79_TTN.gtf rn5 R1.fastq R2.fastq
Read periodicity
To determine the quality of the Ribo-seq data, we calculated the percentage of reads starting at a position that is indicative for ribosomes that are located on a codon of the open reading frame. We first selected 29-mers and the extracted the position of the A of the start codon ATG. This position is indicated by the location of 5’ UTRs annotated by Ensembl50 Biomart. Plotting the start position of 29-mers in a window around the ATG of known genes reveals a peak of read starts with an offset of 12 bp upstream of the start codon. This is indicative for ribosomes located at the translation start site. Subsequently, read starts are preferentially located periodically every 3 base pairs, representing actively translating ribosomes reading the genetic message while synthesizing new proteins. All libraries generated showed very high periodicity with more than 90% of reads located in frame.
To determine whether Ribosomes localized on titin after the truncation in TTNtvZ rats are translating canonical titin sequence, we first determined the coding frame for each exon in the WT animals. We then compared the percentage of reads located in this frame between WT and TTNtvZ animals and did not detect a difference across the locus. This indicates that ribosomes associated to the truncated titin isoforms are located in frame. We only determined the periodicity in cardiac exons that were covered by at least 10 read starts to avoid noise due to low coverage.
Gene expression analyses
To assess gene transcription and translation levels, we counted uniquely mapping reads that were unambiguously assigned to one gene with htseq-count:
htseq-count --stranded=no --type=CDS --quiet --idattr=gene_id - Rattus_norvegicus.Rnor_5.0.79_TTN.gtf
We considered only reads mapping to the coding sequence of genes annotated in the Ensembl database v7950 for both Ribo-seq and RNA-seq datasets. To detect differentially expressed genes between genotypes, we used the DESeq2 package54 with standard parameters (FDR <= 0.05) for comparisons between both TTNtv models to WT animals. All genes that were detected in any of the comparisons are reported in the supplementary data.
Gene and exon expression levels are given as “reads per kilobase per million mapped reads” (RPKM). Reads overlapping the feature were normalised to the length of the feature and the number of uniquely mapping reads (i.e. sequencing depth). Unlike read counts, these values can be used to compare RNA expression levels between different features within the same sample.
To assess allele-specific transcription and translation, we first determined all F344 SNPs on the BN background annotated by the rat genome database55 overlapping with the titin locus at http://rgd.mcw.edu/. We then counted reads overlapping these positions and assigned them to either the BN or the F344 depending on their sequence. The allele frequency across these positions indicates the ratio of titin transcripts originating from either non-truncated titin (BN) or truncated titin (F344) in the TTNtv models.
The sum of reads assigned to the BN or F344 allele was calculated for each animal , normalized by library size and then compared between WT and TTNtvA or TTNtvZ animals. To avoid BN or F344 specific differences in Ttn expression to disturb the signal, we normalised the expression levels of both alleles to the levels in WT animals.
To assess the protein synthesis levels of full-length titin, we considered all Ribo-seq reads that we could assign to regions that were exclusive to the non-truncated alleles in TTNtv animals and compared these counts to WT animals. For the TTNtvA rats, all Ribo-seq reads located after the premature stop codon are derived from the healthy BN allele. This is indicated by the complete absence of F344 SNPs in the data due to the efficient stop of translation after the premature stop codon. The proximal truncation in the TTNtvZ animal does not completely stop translation and Ribo-seq reads downstream are not necessarily representative of full-length titin expression. These reads partly derive from isoforms with alternative N-termini that are not truncated by the TTNtvZ mutation. Anticipating this, we chose to generate a stop by introducing a large deletion that encompasses several exons. These exons (3-5) are only present in the healthy BN allele and thus reads mapping to this location are indicative for full-length titin expression in the absence of a truncation in the TTNtvZ animals.
We also assessed the translation of each exonic part of titin across all animal models. We only considered exons with an expression level of RPKM >1 in the rat heart (n = 237). FPKM values of all 3 replicates for each TTNtv model where compared to WT animals and outliers were removed (ROUT Q=1%). We then calculated the average ratio for each exon across all biological replicates and performed polynomial smoothing of the 2nd order using 10 neighbors to average. Smoothing was required to visualize a trend of translation rates compared to healthy animals across the entire titin locus.
To assess allele frequency in the RNA-seq and Ribo-seq data, we required the location of the F344 SNP (n=121) to be covered by at least 2 reads on average across all animals.
Detection of alternative transcription start site expression in human heart
In order to identify transcription start sites in Titin two sources of data were used; Firstly capped analysis of gene expression data (CAGE) from heart tissue in the FANTOM5 consortium dataset13 to identify the location of active transcription start sites (TSSs). Secondly ChipSeq data from heart tissue of two histone marks that are associated with the presence of an active promoter, namely H3K4me3 and H3K9ac, in the epigenome roadmap dataset17. Specifically, the FANTOM dataset heart samples were CNhs11758, CNhs11757, CNhs10621, CNhs10653, CNhs12855, CNhs12856 and CNhs12857 and were downloaded using CAGEr56. These were then processed using CAGEr (standard parameters) in order to identify TSSs by first normalising the tag counts using a power law and then clustering the reads in order to find CAGE peaks that correspond to active TSSs. This identified three TSSs within the Titin region. In order to further validate these TSSs H3K4me3 narrow peaks from sample E095 (adult heart left ventricle) and H3K9ac narrow peaks and H3K4me3 narrow peaks from sample E083 (fetal heart) were downloaded from the epigenome roadmap data portal and overlaid onto Titin. This showed corresponding histone modifications for two out of the three TSSs identified using CAGEr. All of these datasets were then plotted using circlize57.
PSI
To assess splicing of titin in TTNtv hearts, we calculated the ‘Percent Spliced In’ (PSI) ratio for all exons in titin across species. The PSI is a measure of how efficiently an exon is spliced into the final isoform population of a gene. First we generated an exonic matrix based on our custom titin annotation using the prepare_annotation tool58. Then we executed the PSI.sh14 script to calculate PSI ratios with the following parameters:
PSI.sh StartPSIStrictFilter TTN_exonic.gtf 100 accepted_hits.bam junctions.bed PSI_result
We slightly altered the script and included a more stringent filter for exclusion reads to avoid mapping artifacts of split alignments. Every exclusion read was required to start and end at a known exon boundary in order to be considered in the final PSI calculation (StrictFilter). We also calculated PSI values for human, cardiac DCM samples with and without TTNtv for patients reported previously3. We then compared patterns of TTN isoform regulation between DCM patients carrying truncating variants in TTN versus those who do not, by means of the deltaPSI (dPSI): mean(PSI(DCMTTNtv)) - mean(PSI(DCMcontrols)).
Exons with a PSI in that heart of at least 15% are considered cardiac and at least 90% are considered constitutive exons throughout the manuscript.
Pathway analysis
To better understand the effects of TTNtvs on the genome-wide molecular landscape of the heart, we identified overrepresented pathways in differentially transcribed and translated genes. We used counts normalized by DESeq254 and performed a KEGG23 pathway enrichment analysis with GAGE22:
res <- gage(counts,gsets = kegg.gs,ref = 4:7,samp = 1:3,compare = “unpaired”)
where columns 4-7 represent normalized counts of WT animals and 1-3 counts from TTNtv rats. Ensembl IDs were converted to Entrez IDs prior to this analysis using BiomaRt50. GAGR does not rely on a list of significant differential genes, but rather performs a statistical test whether fold changes in a certain pathway are more different between groups than expected. This enabled use to detect affected pathways in between groups with great sensitivity.
Rat MRI imaging
Rats (13 to 16 month old) were anaesthetized using 1 to 3% isoflurane and maintained at 0.5 to 1.5% isoflurane during the imaging session. Heart rate, blood oxygen saturation, respiratory rate, temperature and ECG were monitored (SA Instruments, NY, USA). Imaging was performed on a 7T ClinScan small animal MRI (Bruker Ettlingen, Germany) equipped with a rat cardiac array surface coil. 2D cine gradient echo sequences were acquired in long-axis, four–chamber and short axis (SAX) views of the left ventricle59. Left ventricle chamber volumes were quantified using Segment v2.0 R4377 software60 (Medviso, Sweden), and left ventricle wall parameters were measured as previously described61.
Rat echocardiography
4 to 8 month old rats were anesthetized initially with 2% to 2.5% isoflurane and maintained at 1.6% to 2.0% isoflurane during image acquisition. Transthoracic echocardiographic measurements were performed on a Vevo 2100 system with MS250linear array transducer, 13–24 MHz (VisualSonics, Canada). Standard two dimensional (2D) and M-mode short axis at mid papillary muscle level were acquired, and an average of 10 cardiac cycles were stored in cine loops for subsequent offline analysis using the same system62,63.
Ex vivo Langendorff studies
Rats (13-16 weeks old) were anesthetized with Ketamine (80mg/kg) and Xylazine (10mg/kg) via intraperitoneal injection. Heparin (1000U) was administered subcutaneously and the heart excised and perfused with modified Krebs-Henseleit buffer solution in a retro-grade fashion on a Langendorff apparatus64 (ADInstruments). An incision was made on the left atrium and a fluid-filled latex balloon placed in the left ventricular cavity. End-diastolic pressure was set at between 5 to 10mmHg for all hearts for baseline measurements by adjustment of the volume of buffer in the latex balloon. Hearts were paced at 300 beats per minute. To mimic volume overload, the volume of buffer in the balloon in the left ventricular cavity was sequentially increased by 10uL every 5 minutes up to a final 40uL incremental volume. Myocardial contractility (dP/dtmax) and relaxation rate (dP/dtmin) were derived from left ventricular developed pressure (LVDevP) using the MLT844 physiological pressure transducer. Data was acquired and analysed using LabchartPro software (ADInstruments).
Metabolomic analysis
Rat hearts were snap frozen in liquid nitrogen and stored at -80°C prior to analysis. Acylcarnitine profiling was performed as described65. Briefly, 50 to 100 mg of tissue was homogenised in 50 % Acetonitrile, 0.3 % formic acid. Tissue homogenates were extracted using methanol and derivatised using 3M Hydrochloric acid in methanol (Sigma Aldrich, USA), dried and reconstituted in methanol for analysis by Liquid chromatography-mass spectrometry (LC-MS) using the Agilent 6430 Triple Quadrupole LC/MS system (Agilent Technologies, CA, USA). 2µL sample was injected at 0.4ml/min with 80/20 methanol/water as mobile phase. Raw data analysis were performed on Agilent MassHunter Workstation B.06.00 Software.
CE-TOFMS (capillary electrophoresis-time of flight mass spectrometry) was performed to quantify glycolytic intermediates, energy substrates and amino acids. Heart tissue were mixed in 50% acetonitrile in water (v/v) containing internal standards (40 μM for cation and 10 μM for anion measurement) and homogenized (1,500rpm, 120 sec x 5 times) before addition of 50% acetonitrile in water (v/v). The supernatant was then filtered through 5-kDa cut-off filter (Human Metabolome Technologies, Yamagata, Japan) to remove macromolecules and the filtrate was centrifugally concentrated and resuspended in ultrapure water before metabolite measurement. The compounds were analysed using Agilent capillary electrophoresis system equipped with an Agilent 6210 TOFMS, a 1100 isocratic HPLC pump, a G1603A CE-MS adapter kit and a G1607A CE-electrospray ionization-mass spectrometry (ESI-MS) sprayer kit (Agilent Technologies, Germany). The system was controlled using G2201AA ChemStation software (Agilent).
Cationic metabolites were analysed using a fused silica capillary (i.d. 50 μm x 80 cm length) with Cation buffer solution (H3301-1001; Human Metabolome Technologies). The samples were injected at a pressure of 50 mbar for 10 sec and the positive applied voltage was 27 kV. ESI –MS was conducted in a positive ion mode with the capillary voltage set at 4,000V. Sample were scanned from 50 to 1,000 m/z66. Anionic metabolites were analysed using a fused silica capillary (i.d. 50 μm x 80 cm length) with Anion buffer solution (H3302-1021; Human Metabolome Technologies). The samples were injected at a pressure of 50 mbar for 25 sec and the positive applied voltage was 30 kV. ESI –MS was conducted in a negative ion mode with the capillary voltage set at 3,500V. Sample were scanned from 50 to 1,000 m/z67.
Raw data obtained by CE-TOFMS were processed using automatic integration software (MasterHands ver. 2.17.1.11, Keio University) to obtain peak information68. Peak area was converted to relative peak area and peak detection limit was determined based on signal-noise ratio; S/N = 3. Putative metabolites were then assigned from HMT’s standard library and known-unknown peak library on the basis of m/z and migration time. Absolute quantification was obtained by single-point (100 μM) calibrations and further normalized by sample weight.
Glucose-6-phosphate (G6P) assay
Rat hearts were snap frozen in liquid nitrogen and stored at -80°C prior to analysis. Heart tissue were homogenized in ice-cold PBS and samples were deproteinized following manufacturer’s instructions (ab204708, Abcam). Deproteinized samples were added in duplicate wells of a 96-well plate and G6P content determined following manufacturer’s instructions (ab83426, Abcam).
Titin protein quantification
SDS-PAGE (polyacrylamide gel electrophoresis) was performed on 2.5 % polyacrylamide/1% agarose gels and total protein visualised by Coomassie blue staining. Ttn protein was visualised at approximately 3 MDa and normalised to MyHC (205 KDa) as a loading control. For immunoblotting quantification, total protein were separated on 1.8 % polyacrylamide/1% agarose gels, transferred to PVDF membrane and probed with T12 antibody against the N-terminus of Ttn and Novex3 as previously described9.
Western blotting
Rat left ventricle tissues were homogenised in primary lysis buffer [50 mM Sodium Phosphate pH 7.4, 150 mM NaCl, 1 % Triton X-100 with protease and phosphatase inhibitor cocktail (Sigma)] and then centrifuged. The supernatant was collected and a secondary extraction was performed on the pellet containing nuclear, structural and membrane proteins using secondary lysis buffer [1 % SDS, 5 mM EDTA, 10 mM DTT, 15 U/ml DnaseI and protease and phosphatase inhibitors]. The primary and secondary extracts were combined and protein concentration was determined by BCA protein assay (Pierce, Thermo Scientific). Equal amounts of protein were separated by SDS-PAGE, transferred to PVDF membranes and probed with antibodies to mTOR pSer2448 (#2971), 4EBP1 pThr37/46 (#2855), S6K pThr389 (#9205), mTOR (#2972), S6K (#2708), 4EBP1 (#9644), GAPDH (#2118) or β-tubulin (#2146) from Cell Signalling Technology.
Healthy Volunteer MRI imaging
Healthy volunteers without self-reported cardiovascular disease or a family history of disease were recruited prospectively via advertisement to the UK Digital Heart Project (https://digital-heart.org/) at the MRC-CSC, Imperial College, London69. The study was approved by a research ethics comittee and all participants gave written informed consent.
Conventional two dimensional and high-resolution three dimensional cine balanced steady-state free precession (b-SSFP) CMR imaging was performed on a 1.5T Philips Achieva system (Best, Netherlands)41. Analysis of the CMR scans was carried out by 2 experienced cardiologists, blinded to genotyping data, using commercially available semi-automated software (CMRtools, Cardiovascular Imaging Solutions, London, UK). Measurements followed standard methodology70,71 with papillary muscles included in the LVM and valve position tracked in the long axis images. Three dimensional CMR data were segmented and co-registered to provide a model of phenotypic variation in the population42.
Statistical analysis
GraphPad Prism6 software was used for statistical analysis. For two factors, trend testing was performed with two-way ANOVA. Comparison of two means was tested by 2-tailed t-test. When several conditions were each compared to one reference condition, we performed the Dunnett’s test to correct for multiple testing. Data is reported as the mean ± standard error / deviation as described42, with a significance level of * = P < 0.05, ** = P < 0.01, *** = P < 0.001. Using RStudio Server version 0.98 (Boston, Massachusetts) standard linear regressions were performed to evaluate the relationship between TTNtv genotype and cardiovascular phenotypes, as previously described3. Multivariate models were generated using known clinical covariates and optimized to minimize Bayesian Information Criterion. The relationships between morphologic parameters and TTN genotype were assessed by ANOVA between nested linear models. To test for position-independent association of TTNtv across titin, we performed a binomial tests. Three dimensional phenotypic regression modelling applied threshold-free cluster enhancement and permutation testing to derive the P values associated with each regression coefficient following adjustment to control the false discovery rate72,73.
Data availability
The RNA-seq and Ribo-seq data used in the manuscript can be obtained from http://www.ebi.ac.uk/ena/data/view/ERP015402.
Supplementary Material
Acknowledgements
We thank all the patients and healthy volunteers for taking part in this research, and our team of research nurses across the hospital sites. We also thank M. von Frieling-Salewsky for technical support. The research was supported by the Medical Research Council Clinical Sciences Centre UK to J.S.W. & S.A.C. & D.P.O., NIHR Biomedical Research Unit in Cardiovascular Disease at Royal Brompton & Harefield NHS Foundation Trust and Imperial College London to J.S.W. & S.A.C., NIHR Imperial Biomedical Research Centre, British Heart Foundation UK (SP/10/10/28431, PG/12/27/29489 to S.A.C. & D.P.O. & C.B.), Wellcome Trust UK (107469/Z/15/Z to J.S.W., 087183/Z/08/Z, 092854/Z/10/Z, WT095908), Wellcome Trust Fellowship (100211/Z/12/Z & P43579_WMET to T.J.W.D.), Fondation Leducq to J.S.W., Tanoto Foundation to S.A.C., CORDA, National Institutes of Health (NHLBI: 2R01HL080494 to J.G.S. & C.E.S.), National Medical Research Council (NMRC) Singapore (CIRG13nov024 & STaR13nov002 to D.P.V.d.K.), SingHealth Duke-NUS Institute of Precision Medicine, Rosetrees Trust, Health Innovation Challenge Fund (HICF-R6-373 to J.S.W.) funding from the Wellcome Trust and Department of Health, UK, Howard Hughes Medical Institute, the European Union EURATRANS award (HEALTH-F4-2010-241504 to N.H.), the Helmholtz Alliance ICEMED to N.H., European Union FP7 (CardioNeT-ITN-289600 to F.M.), Deutsche Forschungsgemeinschaft (SFB1002, TPA08 to W.A.L., Forschergruppe 1054 HU 1522/1-1 to N.H. & TP1 to V.R.Z.) and an EMBO Long-Term Fellowship (ALTF 186-2015 to S.v.H.) and Marie Curie Actions (LTFCOFUND2013, GA-2013-609409 to S.v.H.). This publication includes independent research commissioned by the Health Innovation Challenge Fund (HICF), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this work are those of the authors and not necessarily those of the Department of Health or Wellcome Trust. The RNA-seq and Ribo-seq data used in the manuscript can be obtained from http://www.ebi.ac.uk/ena/data/view/ERP015402.
Footnotes
Author Contributions
S.A.C. conceived, managed and arranged funding for the project. A.d.M., E.A., L.R.F., B.N., E.K., S.v.H., C.J.P., U.T., S.K.P., T.J.W.D., N.S.J.K., D.S., L.L.H.C., C.W.L.C., P.J.B., D.P.V.d.K., T.T., C.B., N.T., V.R.Z., J.G.S., C.E.S. and W.A.L. performed experiments and contributed clinical data. S.S., A.d.M., O.J.L.R., M.K. R.W., F.M., F.K., D.R., V.S., A.F., J.P.K., D.P.O., J.S.W., N.H. and S.A.C. performed data analysis and interpretation. S.S., B.N. and S.A.C. prepared the manuscript with input from co-authors.
Competing Financial Interests
S.A.C. consults for Illumina.
References
- 1.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–47. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 2.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–28. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts AM, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci Transl Med. 2015;7:270ra6. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Norton N, et al. Exome sequencing and genome-wide linkage analysis in 17 families illustrate the complex contribution of TTN truncating variants to dilated cardiomyopathy. Circ Cardiovasc Genet. 2013;6:144–53. doi: 10.1161/CIRCGENETICS.111.000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chauveau C, Rowell J, Ferreiro A. A Rising Titan: TTN Review and Mutation Update. Human Mutation. 2014;35:1046–1059. doi: 10.1002/humu.22611. [DOI] [PubMed] [Google Scholar]
- 6.Akinrinade O, Koskenvuo J, Alastalo T-P. Prevalence of Titin Truncating Variants in General Population. PLOS ONE. 2015;10:e0145284. doi: 10.1371/journal.pone.0145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinrinade, Alastalo T-P, Koskenvuo JW. Relevance of truncating titin mutations in dilated cardiomyopathy. Clinical Genetics. 2016 doi: 10.1111/cge.12741. n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 8.Robinson EB, et al. Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet. 2016 doi: 10.1038/ng.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hinson JT, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–6. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gramlich M, et al. Stress-induced dilated cardiomyopathy in a knock-in mouse model mimicking human titin-based disease. J Mol Cell Cardiol. 2009;47:352–358. doi: 10.1016/j.yjmcc.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2015 doi: 10.1038/nature19057. 030338. http://biorxiv.org/content/early/2015/10/30/030338.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou J, et al. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin. eLife. 2015;4 doi: 10.7554/eLife.09406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest A, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer S, et al. Alternative Splicing Signatures in RNA-seq Data: Percent Spliced in (PSI) Curr Protoc Hum Genet. 2015;87:11.16.1–11.16.14. doi: 10.1002/0471142905.hg1116s87. [DOI] [PubMed] [Google Scholar]
- 15.Ingolia N, Ghaemmaghami S, Newman J, Weissman J. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schafer S, et al. Translational regulation shapes the molecular landscape of complex disease phenotypes. Nature communications. 2015;6:7200. doi: 10.1038/ncomms8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo W, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–73. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maatz H, et al. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–30. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hir L, Izaurralde, Maquat, Moore The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. Embo J. 2000;19:6860–9. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hir L, Gatfield, Izaurralde, Moore The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. Embo J. 2001;20:4987–97. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai L, et al. Energy metabolic reprogramming in the hypertrophied and early stage failing heart: a multisystems approach. Circ Heart Fail. 2014;7:1022–31. doi: 10.1161/CIRCHEARTFAILURE.114.001469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibayama J, et al. Metabolic remodeling in moderate synchronous versus dyssynchronous pacing-induced heart failure: integrated metabolomics and proteomics study. PLoS ONE. 2015;10:e0118974. doi: 10.1371/journal.pone.0118974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doenst T, Nguyen TD, Abel ED. Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res. 2013;113:709–24. doi: 10.1161/CIRCRESAHA.113.300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stanley WC, Recchia FA, Lopaschuk GD. Myocardial substrate metabolism in the normal and failing heart. Physiol Rev. 2005;85:1093–129. doi: 10.1152/physrev.00006.2004. [DOI] [PubMed] [Google Scholar]
- 28.Schisler JC, et al. Cardiac energy dependence on glucose increases metabolites related to glutathione and activates metabolic genes controlled by mechanistic target of rapamycin. J Am Heart Assoc. 2015;4 doi: 10.1161/JAHA.114.001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neubauer S. The failing heart--an engine out of fuel. N Engl J Med. 2007;356:1140–51. doi: 10.1056/NEJMra063052. [DOI] [PubMed] [Google Scholar]
- 30.Yano T, et al. Clinical impact of myocardial mTORC1 activation in nonischemic dilated cardiomyopathy. Journal of molecular and cellular cardiology. 2016;91:6–9. doi: 10.1016/j.yjmcc.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 31.Ramos F, et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Science translational medicine. 2012;4:144ra103. doi: 10.1126/scitranslmed.3003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neishabouri, Hutson, Davoodi Chronic activation of mTOR complex 1 by branched chain amino acids and organ hypertrophy. Amino acids. 2015;47:1167–82. doi: 10.1007/s00726-015-1944-y. [DOI] [PubMed] [Google Scholar]
- 33.Ait-Mou Y, et al. Titin strain contributes to the Frank-Starling law of the heart by structural rearrangements of both thin- and thick-filament proteins. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2306–11. doi: 10.1073/pnas.1516732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen S, et al. Glucose regulation of load-induced mTOR signaling and ER stress in mammalian heart. J Am Heart Assoc. 2013;2:e004796. doi: 10.1161/JAHA.113.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shende P, et al. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123:1073–82. doi: 10.1161/CIRCULATIONAHA.110.977066. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D, et al. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120:2805–16. doi: 10.1172/JCI43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mestroni, et al. Guidelines for the study of familial dilated cardiomyopathies. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. Eur Heart J. 1999;20:93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 38.Vasan RS, Larson MG, Benjamin EJ, Evans JC, Levy D. Left ventricular dilatation and the risk of congestive heart failure in people without myocardial infarction. N Engl J Med. 1997;336:1350–5. doi: 10.1056/NEJM199705083361903. [DOI] [PubMed] [Google Scholar]
- 39.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Marvao A, et al. Population-based studies of myocardial hypertrophy: high resolution cardiovascular magnetic resonance atlases improve statistical power. J Cardiovasc Magn Reson. 2014;16:16. doi: 10.1186/1532-429X-16-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai W, et al. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Med Image Anal. 2015;26:133–45. doi: 10.1016/j.media.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 43.Ware J, et al. Shared Genetic Predisposition in Peripartum and Dilated Cardiomyopathies. New Engl J Medicine. 2016;374:233–241. doi: 10.1056/NEJMoa1505517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watkins H. Tackling the achilles’ heel of genetic testing. Sci Transl Med. 2015;7:270fs1. doi: 10.1126/scitranslmed.aaa4276. [DOI] [PubMed] [Google Scholar]
- 45.Pugh TJ, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014;16:601–8. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 46.Walsh, Thomson, Ware, Funke, Woodley Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. 2016 doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pua CJ, et al. Development of a Comprehensive Sequencing Assay for Inherited Cardiac Condition Genes. J Cardiovasc Transl Res. 2016;9:3–11. doi: 10.1007/s12265-016-9673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nature Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunningham F, et al. Ensembl 2015. Nucleic Acids Res. 2015;43:D662–9. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goecks J, Nekrutenko A, Taylor J, Team T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giardine B, et al. Galaxy: A platform for interactive large-scale genome analysis. Genome Research. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimoyama M, et al. The Rat Genome Database 2015: genomic, phenotypic and environmental variations and disease. Nucleic Acids Res. 2015;43:D743–50. doi: 10.1093/nar/gku1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haberle V, Forrest A, Hayashizaki Y, Carninci P, Lenhard B. CAGEr: precise TSS data retrieval and high-resolution promoterome mining for integrative analyses. Nucleic Acids Research. 2015;43:e51–e51. doi: 10.1093/nar/gkv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu, Gu, Eils, Schlesner, Brors circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 58.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Research. 2012;22:2008–2017. doi: 10.1101/gr.133744.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price AN, et al. Cardiovascular magnetic resonance imaging in experimental models. Open Cardiovasc Med J. 2010;4:278–92. doi: 10.2174/1874192401004010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heiberg E, et al. Design and validation of Segment--freely available software for cardiovascular image analysis. BMC Med Imaging. 2010;10:1. doi: 10.1186/1471-2342-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ross AJ, et al. Serial MRI evaluation of cardiac structure and function in mice after reperfused myocardial infarction. Magn Reson Med. 2002;47:1158–68. doi: 10.1002/mrm.10166. [DOI] [PubMed] [Google Scholar]
- 62.Tortoledo FA, Quinones MA, Fernandez GC, Waggoner AD, Winters WL. Quantification of left ventricular volumes by two-dimensional echocardiography: a simplified and accurate approach. Circulation. 1983;67:579–84. doi: 10.1161/01.cir.67.3.579. [DOI] [PubMed] [Google Scholar]
- 63.Sahn DJ, DeMaria A, Kisslo J, Weyman A. Recommendations regarding quantitation in M-mode echocardiography: results of a survey of echocardiographic measurements. Circulation. 1978;58:1072–83. doi: 10.1161/01.cir.58.6.1072. [DOI] [PubMed] [Google Scholar]
- 64.Sutherland FJ, Hearse DJ. The isolated blood and perfusion fluid perfused heart. Pharmacol Res. 2000;41:613–27. doi: 10.1006/phrs.1999.0653. [DOI] [PubMed] [Google Scholar]
- 65.Muoio DM, et al. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012;15:764–77. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soga T, Heiger DN. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2000;72:1236–41. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 67.Soga T, et al. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2002;74:2233–9. doi: 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- 68.Sugimoto M, et al. Differential metabolomics software for capillary electrophoresis-mass spectrometry data analysis. Metabolomics. 2010;6:27–41. [Google Scholar]
- 69.de Marvao A, et al. Precursors of Hypertensive Heart Phenotype Develop in Healthy Adults: A High-Resolution 3D MRI Study. JACC Cardiovasc Imaging. 2015;8:1260–9. doi: 10.1016/j.jcmg.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bellenger NG, Grothues F, Smith GC, Pennell DJ. Quantification of right and left ventricular function by cardiovascular magnetic resonance. Herz. 2000;25:392–9. doi: 10.1007/s000590050031. [DOI] [PubMed] [Google Scholar]
- 71.Grothues F, et al. Comparison of interstudy reproducibility of cardiovascular magnetic resonance with two-dimensional echocardiography in normal subjects and in patients with heart failure or left ventricular hypertrophy. Am J Cardiol. 2002;90:29–34. doi: 10.1016/s0002-9149(02)02381-0. [DOI] [PubMed] [Google Scholar]
- 72.Smith SM, Nichols TE. Threshold-free cluster enhancement: addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 73.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq and Ribo-seq data used in the manuscript can be obtained from http://www.ebi.ac.uk/ena/data/view/ERP015402.