Abstract
Rice blast is a destructive disease caused by Magnaporthe oryzae, and it has a large impact on rice production worldwide. Compared with leaf blast resistance, our understanding of panicle blast resistance is limited. The japonica landrace Jiangnanwan from Taihu Lake region in China shows highly resistance to panicle and leaf blast. In this study, three generations (F2:5, F2:6, F2:7) consisting of 221 RILs (recombination inbreeding lines), developed from the cross of Jiangnanwan and Suyunuo, a susceptible-blast japonica variety, were evaluated for panicle blast resistance in the fields and leaf blast resistance in greenhouse in Nanjing in 2013, 2014 and 2015. A blast resistance gene Pi-jnw1 referring to panicle blast resistance and leaf blast resistance was identified in the three generations and located in the region of RM27273 and RM27381 in chromosome 11. The RIL18 line harboring Pi-jnw1 was selected to be backcrossed with Suyunuo to develop BC2F2 populations. According to the genotyping of 1,150 BC2F2 individuals and panicle blast and leaf blast resistance evaluation of 47 recombinants between RM27150 and RM27381, Pi-jnw1 was finally mapped to the 282 kb region between markers W28 and BS39. This study revealed that Jiangnanwan harboring a panicle blast and leaf blast resistance gene Pi-jnw1 could be a genetic source for breeding new rice cultivars with panicle blast resistance.
Introduction
Rice blast, caused by the fungus pathogen Magnaporthe oryzae, is one of the most destructive diseases worldwide, and it occurred in all stages of rice growth [1,2]. The disease pathosystem comprises two major interrelated phases: leaf blast and panicle blast [3]. Compared with leaf blast resistance, less is known about the genetic components for panicle blast resistance, which is indispensable for stable rice production. Leaf blast resistant cultivars may be susceptible to panicle blast, and it implies that the genetic mechanisms of blast resistance might vary across the plant growth stages [4–7]. The technical problems as lacking of standard inoculation and evaluation systems, variations in heading date and weather conditions, are obstacles to the exploration of new gene resources of rice panicle blast resistance. Up to date, only Pb1 was cloned from the indica cultivar Modan conferring to the panicle blast resistance [3,8]. It encodes a NBS-LRR protein, and can protect WRKY45 from degradation by ubiquitin proteasome system. The blast resistance of cultivar usually can be lost after few years for the genetic instability and pathogenic variability of M. oryzae [9]. Therefore, to further explore new resistance genes especially panicle blast resistance genes from rice landraces will be the most useful strategy in rice blast resistance breeding.
Up to date, approximately 100 blast resistance loci or genes have been mapped on 12 chromosomes except chromosome 3 [6, 7]. Twenty five blast resistance genes have been cloned [10], and eight of them located in two gene clusters, including three genes Pi2, Pi9 and Piz-t in Pi2 locus and five genes Pik, Pik-m, Pik-p, Pi1 and Pi-ke in Pik locus [11–17]. Among 25 cloned genes, 23 genes encode NBS-LRR (nucleotide-binding site -leucine-rich repeat) proteins, except Pi21 encodes proline-containing protein and Pid2 encodes receptor kinase [18–20]. It has been shown that at least six R genes, Pi1[21], Pi2 [22], Pi9 [23], Pi5 [24], Pi33 [25] and Pigm[26], probably confer broad-spectrum resistance to a number of isolates from different countries respectively. For instance, Pi9 located on the same region with Pi2, showed resistance to 43 isolates from 13 countries [23]. Pi5, a locus associated with resistance to at least 6 blast races from Philippines and 26 isolates from Korea [24], and Pi33 was resistance to more than 2,000 isolates originating from 55 countries [25].
In our previous research, Jiangnanwan, a japonica rice landrace from Taihu Lake region of China, exhibited broad-spectrum resistance to rice blast [27]. Li et al. [28] concluded that two effect genes might be involved in the leaf blast resistance with F2 population deriving from a across between Jiangnanwan and a blast-susceptible variety Suyunuo. In this study, we obtained 221 F2:7 RILs with three generations (F2:5, F2:6 and F2:7), and identified panicle blast and leaf blast resistance genes to the strain Hoku 1 with QTL mapping method. Furthermore, we examine the correlation between the resistance of panicle and leaf blast, and fine mapped the blast resistance gene Pi-jnw1.
Materials and Methods
Plant materials and growth
Jiangnanwan is a japonica rice landrace from Taihu Lake region in China and has broad spectrum resistance to leaf blast. Suyunuo is a susceptible japonica rice landrace from Taihu Lake region. We developed an F2 population from a cross between Jiangnanwan and Suyunuo, and three generations (F2:5, F2:6 and F2:7) of 221 recombination inbreeding lines (RILs) were generated by a single-seed descent method (Fig 1).
Fig 1. Flowchart showing the development of plant materials used in this study.
The populations of F2:5, F2:6, F2:7 and two parents (Jiangnanwan and Suyunuo) were evaluated for panicle blast resistance in 2013, 2014 and 2015 at Jiangpu Experimental Station of Nanjing Agricultural University (Jiangsu Province, China; 118°50′E, 32°02′N). Twenty plants of each RIL grew in two rows per test plot, and the spaces were 30 cm between rows and 10 cm between plants within rows. Suyunuo and Jiangnanwan were planted adjacent to the test rows as susceptible and resistant controls respectively. Field management was carried out in accordance with the local production process [29].
The populations of F2:5, F2:6, F2:7 and two parents (Jiangnanwan and Suyunuo) were also evaluated for leaf blast resistance in 2013, 2014 and 2015 in the greenhouse. The seeds were sown in plastic trays of 60 × 30 × 5 cm with sieved garden soil as described by Wang et al. [27]. Thirty lines and two parents were sown in each tray, and 6–8 seeds per line were sown for inoculation. Seedlings were grown in greenhouse at 22–30°C with a light and dark cycle of 16 h and 8 h until they were at the four-leaf stage for disease evaluation.
Inoculation and disease evaluation
To evaluate the panicle blast resistance in the field, 221 RILs (F2:5, F2:6, F2:7), forty-seven BC2F2 recombinants and two parents at the mid-booting stage were inoculated with the strain Hoku 1 of M. oryzae by the injecting method as described as Liu et al. [30]. Fifteen booting panicles of each line were injected by 1–2 ml blast isolate Hoku 1 conidial suspension (5×104 conidia/ml). Three weeks after inoculation, phenotypic evaluation was conducted based on visual assessment of diseased grains percentage as described by Koizumi et al.[31] and the scores were ranged from 0 (without diseased grain) to 100% (100% diseased grains).
Four-leaf stage rice seedlings of Jiangnanwan, Suyunuo, 221 RILs (F2:5, F2:6, F2:7), and forty-seven BC2F2 recombinants were inoculated with the strain Hoku1 spore suspension (5×104 conidia/ml) in inoculation chambers as the method described by Wang et al. [27]. After inoculation, the plants were kept in dark at 26°C with relative humidity 95% for 24 h, and then transferred to a greenhouse with 25–28°C and 100% relative humidity by intermittently spraying water for 2 min every three hours. After seven days of inoculation, lesion scores of 0 to 5 were investigated based on lesion type with appropriate reference of the disease area of each plant as described by Shi et al.[32]. Each line was inoculated with three replications in each experiment and three independent experiments were conducted either leaf blast or panicle blast resistance evaluation.
Genetic map construction and identification
221 RILs of F2:6 population were used for genotyping and constructing molecular linkage map 0.2 to 0.5 g of leaves at the four-leaf stage from each line of RILs (F2:6) and parents were collected specifically for DNA extraction by using the CTAB method [33]. 2,300 SSR markers kept in our lab and 108 newly designed InDel markers distributed on 12 chromosomes were screened for polymorphisms. 93 markers with polymorphisms between the two parents were finally used for genetic map construction.
All of the PCR reactions with the markers used a 10 μl reaction mixture containing of 1 μl template DNA, 0.5 μl of each primer, 0.1 μl of Taq (0.01U/μl), 1.6 μl of 10×Buffer, 0.2 μl of dNTP and 6.1 μl of ddH2O. PCR procedures were conducted as follows: Preheating for 5 min at 95°C, 32 cycles (40 sec at 95°C, 40 sec annealing temperature, and 40 sec at 72°C), finally 72°C for 10 min. The PCR products were analyzed on the 8% acrylamide gels.
In order to identify panicle blast and leaf blast resistance genes, QTL mapping method was performed using software IciMapping v4.0 (http://www.isbreeding.net/). The software was set LOD > 2.5 as a threshold which must be operate 1000 times at the p< 0.05 level. In this study, the panicle blast resistance QTLs were named as qPbj-A-B, and the leaf blast resistance QTLs were named as qLbj-A-B, in which A means the chromosome number and B means the sequence of QTL.
Data analysis
Experimental data were analyzed using the IBM SPSS Statistics software 19.0, and bivariate analysis method were used for analyzing the correlation between the panicle blast resistance and leaf blast resistance [33].
Fine mapping of Pi-jnw1
All of the simple sequence repeat (SSR) markers in this study were downloaded from the gramene database (http://www.gramene.org/) [34]. Five InDel markers W26, W28, BS33, BS39 and BS71 were designed on the basis of sequence difference between 93–11 (http://www.genomics.org.cn/) and Nipponbare (http://www.ncbi.nlm.nih.gov/) in the target region by Pairwise BLAST (http://blast.ncbi.nlm.nih.gov/blast.cgi) as described as Wu et al. [35] (S1 table).
The RIL18 line harboring Pi-jnw1 was selected from F2:6 RIL populations and backcrossed with Suyunuo for developing fine mapping populations,.1,150 plants of BC2F2 population generated by 26 resistant BC2F1 individuals were used for constructing fine genetic linkage map and identifying recombinants in the target region of Pi-jnw1. The franking markers RM27150 and RM27381 were used to genotype the 1,150 BC2F2 segregating plants and 47 recombinants were detected. Then the 47 recombinants were inoculated to identify the panicle blast resistance phenotypes and the next generation seeds of 47 recombinants were inoculated to identify the leaf blast resistance phenotypes.
Results
Characterization of resistance to panicle and leaf blast in Jiangnanwan
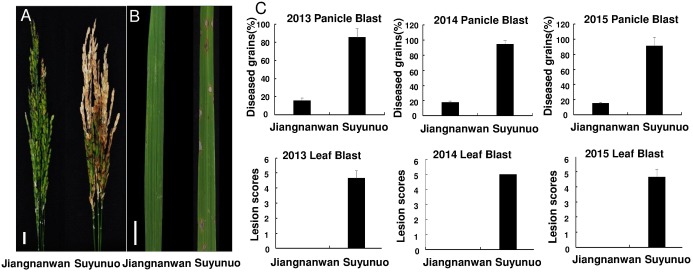
In 2013, 2014 and 2015, Jiangnanwan and Suyunuo were inoculated with the strain Hoku1 in field for panicle blast resistance evaluation and in greenhouse for leaf blast resistance evaluation. The results showed that Jiangnanwan was highly resistance to panicle blast and leaf blast, while Suyunuo was highly susceptible (Fig 2A–2C, Table 1). The frequency distributions of panicle blast and leaf blast resistance in 221 RILs (F2:6, F2:7 and F2:8) were asymmetric and continuous, and they were all predisposed resistance-inclined distribution (Fig 3A–3G). Similar results of frequency distributions of panicle blast and leaf blast resistance were obtained by the IBM SPSS Statistics software 19.0, and the characteristic parameters (Skewness and Kurtosis) showed the frequency distributions of panicle blast and leaf blast resistance in 221 RILs were all predisposed resistance-inclined distribution (Table 1).
Fig 2. The resistant phenotypes of Jiangnanwan and Suyunuo at seedling and heading stage after inoculating by Hoku 1.
A, The phenotypes of panicle blast in Jiangnanwan and Suyunuo. Bar = 1cm. B, The phenotypes of leaf blast in Jiangnanwan and Suyunuo. Bar = 1 cm. C, Characterization of panicle and leaf blast severity distribution of Jiangnanwan and Suyunuo in three years.
Table 1. Phenotypic values of panicle and leaf blast resistance to strain of Hoku 1 in RILs F2:5, F2:6, and F2:7 populations.
| Blast resistance type | Year | Parents | RIL Population c | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Jiangnanwan | Suyunuo | Mean | Max | Min | SD d | Skewness | Kurtosis | ||
| Panicle blast resistance a | 2013 | 15.97±2.64% (R) | 85.67±9.56% (S) | 39.66% | 100% | 0 | 0.32371 | 0.888 | -0.563 |
| 2014 | 18.2±3.45%(R) | 100% (S) | 38.04% | 100% | 0 | 0.31704 | 0.736 | -0.735 | |
| 2015 | 15.46±1.21%(R) | 91.67±11.11% (S) | 33.09% | 100% | 0 | 0.32573 | 1.011 | -0.333 | |
| Leaf blast resistance b | 2013 | 0(R) | 5(S) | 1 | 5 | 0 | 1.29192 | 1.459 | 1.199 |
| 2014 | 0(R) | 5(S) | 1 | 5 | 0 | 1.43337 | 1.486 | 1.27 | |
| 2015 | 0(R) | 5(S) | 1 | 5 | 0 | 0.64204 | 2.329 | 5.321 | |
a means diseased grains (%);
b means lesion score;
c RILs sample size n = 221, replications r = 3;
d standard deviation.
Fig 3. Characterization of panicle and leaf blast severity distribution in RILs F2:5, F2:6, and F2:7.
The frequency distributions of panicle blast resistance in three generations of 221 RILs (F2:5, F2:6 and F2:7) were asymmetric and continuous, and they were all predisposed resistance-inclined distribution (Fig 2A–2C). The frequency distributions in the three tested generations under the experimental paddy field and greenhouse were not bimodal, suggesting that multiple loci are involved in the panicle blast and leaf blast resistance of Jiangnanwan (Fig 2D–2F).
Identification of Pi-jnw1
With a linkage map which covering 1,690.76 cM on the 12 chromosomes and an average distance 18.18 cM between two connected markers, a blast resistance gene Pi-jnw1 referring to panicle blast and leaf blast resistance was detected in the same region of RM27273 and RM27381 on chromosome 11 by QTL IciMapping 4.0 in three generations (F2:5, F2:6 and F2:7) (Table 2, Fig 4).
Table 2. Identification of panicle and leaf blast resistance genes by QTL mapping method in RILs F2:5, F2:6, and F2:7 populations.
| Traits | Generation | QTL Names | Chr. | Left Marker | Right Marker | LOD | PVE (%) | Add |
|---|---|---|---|---|---|---|---|---|
| Panicle blast | F2:5 | qPbj-7-1 | 7 | RM3186 | RM346 | 4.8629 | 6.9138 | 0.0849 |
| qPbj-11-1 (Pi-jnw1) | 11 | RM27273 | RM27381 | 23.2412 | 39.9217 | -0.2045 | ||
| F2:6 | qPbj-7-2 | 7 | RM3186 | RM346 | 2.8482 | 2.8882 | 0.0536 | |
| qPbj-11-2 (Pi-jnw1) | 11 | RM27273 | RM27381 | 34.7518 | 53.6856 | -0.2321 | ||
| F2:7 | qPbj-6-1 | 6 | RM276 | AP5659-5 | 3.2884 | 5.2431 | 0.0766 | |
| qPbj-9-1 | 9 | RM3164 | RM2144 | 2.5526 | 3.9206 | -0.0644 | ||
| qPbj-11-3 (Pi-jnw1) | 11 | RM27273 | RM27381 | 24.7161 | 42.4911 | -0.2130 | ||
| Leaf blast | F2:5 | qLbj-11-1 (Pi-jnw1) | 11 | RM27273 | RM27381 | 9.7449 | 18.8430 | -0.5623 |
| F2:6 | qLbj-11-2 (Pi-jnw1) | 11 | RM27273 | RM27381 | 13.1284 | 23.6070 | -0.6635 | |
| F2:7 | qLbj-11-3 (Pi-jnw1) | 11 | RM27273 | RM27381 | 5.3520 | 10.9116 | -0.3286 |
Fig 4. Identification of panicle and leaf blast resistance genes in Jiangnanwan by QTL mapping method.
Marker names and their positions were showed on the left linkage group. The color lines indicated LOD scores.
For the panicle blast resistance, Pi-jnw1 was detected in the same region of RM27273 and RM27381 on chromosome 11 (qPbj-11-1, qPbj-11-2 and qPbj-11-3) in 2013, 2014 and 2015, respectively (Fig 4). They could explain 39.92%, 53.68% and 42.49% of phenotypic variation for panicle blast resistance in three generations (Table 2). Other four minor resistant loci, including qPbj-7-1, qPbj-7-2, qPbj-6-1 and qPbj-9-1 detected in 2013, 2014, and 2015, respectively could explain only 2.89%-6.91% of phenotypic variation (Table 2). Among these four minor resistant loci, qPbj-7-1 and qPbj-7-2 were located in the same region of RM3186 and RM346 on chromosome 7, and qPbj-6-1 and qPbj-9-1 were located in the region of RM276 and AP5659.5 on chromosome 6 and in the region of RM3164 and RM2144 on chromosome 9, respectively (Fig 4).
For the leaf blast resistance, Pi-jnw1 was also detected in the same region of RM27273 and RM27381 on chromosome 11 (qLbj-11-1, qLbj-11-2 and qLbj-11-3) in 2013, 2014 and 2015, respectively (Fig 4). They could explain 18.84%, 23.60%, and 10.91% phenotypic variation for leaf blast resistance in three generations.
To determine whether leaf blast resistance was related with panicle blast resistance in Jiangnanwan, the correlation of panicle blast resistance and leaf blast resistance of 221 F2:5, F2:6, F2:7 RILs was examined. The results showed that the correlation coefficients between panicle blast resistance and leaf blast resistance were 0.49 in the F2:5 RILs, 0.371 in the F2:6 RILs, and 0.551 in the F2:7 RILs respectively, all with a significantly positive relationship (Table 3).
Table 3. The correlations between panicle and leaf blast resistance reaction.
“**” P<0.01,
“*” P<0.05.
Fine mapping of Pi-jnw1
Five InDel markers W26, W28, BS33, BS39 and BS71 between RM27273 and RM27381 with polymorphisms between two parents Jiangnanwan and Suyunuo were used to fine map the Pi-jnw1. 1,150 BC2F2 plants were genotyped by those markers, and 28, 10, 1, 0, 2, 2, 6 and 19 recombinants were identified by RM27150, RM27273, W28, W26, BS39, BS71, BS33 and RM27381, respectively. Through panicle blast and leaf blast resistance phenotype assays of the forty seven recombinants between RM27150 and RM27381, Pi-jnw1 was mapped in the region of W28 and BS39 with the physical distance of 282 Kb (Fig 5).
Fig 5. Fine mapping of Pi-jnw1.
A. A total of 47 recombinant plants were screened from 1150 BC2F2 segregating plants. B. High resolution analysis of phenotypes and genotypes. Serial numbers represent partial key recombinant plants. The black regions indicated Jiangnanwan genotypes, the white regions indicated Suyunuo genotypes and the blue regions indicated hybrid subtype of Jiangnanwan/Suyunuo.
Discussion
Panicle blast usually caused more loss of yielding than leaf blast in rice production. However, fewer genetic analyses of rice panicle blast resistance have been reported compared with leaf blast resistance. More field works, complex phenotype evaluation system and the influence of environmental conditions are obstacles to study rice panicle blast resistance. There are various ways for evaluating the panicle blast resistance: (1) Injecting inoculation, injecting 2–3 ml of spore suspension into one booting panicle [30]; (2) Inducing inoculation, controlling the field conditions to be suitable for development and epidemic of blast disease [3]; (3) In vitro inoculation, 6 cm rice panicle necks containing 1–3 rachis nodes were put on the filter paper in petri dishes, then the nodes were inoculated with 2 ml spore suspension by a micropipette [5]. The resistance genes Pi-64 and Pi-25 were identified by vitro inoculation [35,36], and Pb1 was identified under suitable field conditions for blast disease development [3]. We use the modified injecting method to inoculate the 221 RILs and forty-seven BC2F2 recombinants in the field. In our study, the frequency distributions of diseased grains percentages in the three tested generations in different fields were relatively stable and a blast resistance gene Pi-jnw1 referring to panicle blast resistance in chromosome 11 were detected in three years and two minor resistant loci (qPbj-7-1 and qPbj-7-2) in chromosome 7 were both detected in two years. It indicated that the injecting inoculation method might be more appropriate for identifying the panicle blast resistance.
Jiangnanwan, one japonica rice landrace from Taihu Lake region in China, exhibited broad-spectrum resistance to leaf blast and highly resistance to panicle blast [37]. Li et al. [28] studied the genetic patterns of leaf blast resistance to Hoku1 in Jiangnanwan with P1, P2, F1 and F2 population deriving from a across between Jiangnanwan and a blast-susceptible variety Suyunuo and concluded that two genes might be involved. In our results, only one blast resistance gene Pi-jnw1 could be detected in 221 F2:5, F2:6, F2:7 RILs, respectively. The possible reason could be due to the different populations and different analysis methods.
So far, more than 20 blast resistance genes were reported on rice chromosome 11, four of them locate near Pi-jnw1 region. The Pb1 locus was mapped in the Modan-derived chromosomal region in the middle part of the long arm of chromosome 11, located closet with the RFLP marker of S723[3]. The rice blast resistance gene Pik, which confers high and stable resistance to many Chinese rice blast isolates, encoded two coiled-coil nucleotide binding site leucine-rich repeat (NBS-LRR) proteins[17]. The Pi34 locus was located in the 54.1 kb region on the genomic sequence of Nipponbare and acted partial resistance to blast in Chubu 32[38]. Pi-hk1 was identified on chromosome 11 of Heikezijing, located between the SSR markers of RM7654 and RM27381[20]. According to the fine mapping results, Pi-jnw1 was not in the same region of Pb1. In our further study, we will confirm the fine mapping results and use more markers to detect whether Jiangnanwan harbors Pik, Pi34 and Pi-hk1 genes in the Pi-jnw1 region.
In our study, the blast resistance gene Pi-jnw1 was identified both in panicle blast resistance and leaf blast resistance of the three generations (F2:5, F2:6 and F2:7), suggesting that there was a positive relationship between panicle blast and leaf blast resistance detected in Jiangnanwan. It is consistent with the common viewpoint that panicle blast resistance is correlated with leaf blast resistance in many rice cultivars[39]. However, there were also four minor panicle blast resistance specific loci qPbj-6-1, qPbj-7-1, qPbj-7-2 and qPbj-9-1, and it indicates that some loci might be only influence the panicle blast resistance. Interestingly, qPbj-7-1, qPbj-7-2 and qPbj-6-1 were contributed by Suyunuo indicated that there were some genes in Suyunuo against panicle blast which could be detected under specific conditions. In this study, 93 genetic markers with polymorphisms between two parents were used for genetic map construction, and the frequency of polymorphisms between Jiangnanwan and Suyunuo was not as high as the indica/japonica crosses. In further research, more genetic markers between W28 and BS39 will be designed and larger segregation populations will be constructed for fine mapping the Pi-jnw1. The recombinants harboring Pi-jnw1 will be further used for breeding new cultivars with back crossing with the elite cultivars and marker associated selection method (MAS).
Breeding new blast resistant cultivars is considered as an effective and economical way to control this disease. However, among the cloned 25 resistance genes, 24 of them were referring to the leaf blast resistance and few of them have been widely applied in rice breeding. Many cultivars show different levels of partial resistance to leaf and panicle blast. This implies that the genetic mechanisms of host resistance might vary across growth stages. In this study, Jiangnanwan showed broad spectrum resistance to leaf blast and highly resistance to panicle blast, and the panicle blast resistance showed a positive correlation with leaf blast resistance. The mapping results also showed that Pi-jnw1 could be detected with panicle blast resistance phenotypic data and leaf blast resistance phenotypic data in three years and located in the same region of RM27273 and RM27381 on chromosome 11 in the three generations (F2:5, F2:6 and F2:7). It indicated that Jiangnanwan might be a good resource for application in rice breeding programs, and further cloning and functional analysis of Pi-jnw1 could be necessary for clarifying the molecular basis of panicle blast resistance and leaf blast.
Supporting Information
(DOCX)
Acknowledgments
This research has been supported by grants from the National Key Project for Transgenic Crops (2016ZX08009-003-001, 2014ZX08009-001B), the Natural Science Foundation of China (30900888, 31171516) and the Fundamental Research Funds for the Central Universities (KYZ201302), Jiangsu Agriculture science and technology innovation fund (CX(12)1003-3, CX(15)1054). We would like to thank Prof. Chen Zhi-Yi and Dr. Liu Yong-Feng for their kindly providing all the blast isolates and Mr. Chen Hao for his help in field panicle inoculation.
Data Availability
All relevant data are within the paper.
Funding Statement
This research has been supported by grants from the National Key Project for Transgenic Crops (2016ZX08009-003-001, 2014ZX08009-001B), the Natural Science Foundation of China (30900888, 31171516) and the Fundamental Research Funds for the Central Universities (KYZ201302), Jiangsu Agriculture science and technology innovation fund (CX(12)1003-3, CX(15)1054). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sesma A, Osbourn AE (2004) The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431: 582–586. 10.1038/nature02880 [DOI] [PubMed] [Google Scholar]
- 2.Valent B, Chumley FG (1991) Molecular genetic analysis of the rice blast fungus, magnaporthe grisea. Annu Rev Phytopathol 29: 443–467. 10.1146/annurev.py.29.090191.002303 [DOI] [PubMed] [Google Scholar]
- 3.Fujii K, Hayano-Saito Y, Saito K, Sugiura N, Hayashi N, Tsuji T, et al. (2000) Identification of a RFLP marker tightly linked to the panicle blast resistance gene, Pb1, in rice. Breeding Sci 50: 183–188. [Google Scholar]
- 4.Sirithunya P, Tragoonrung S, Vanavichit A, Pa-In N, Vongsaprom C, Toojinda T (2002) Quantitative trait loci associated with leaf and neck blast resistance in recombinant inbred line population of rice (Oryza sativa). DNA Res 9: 79–88. [DOI] [PubMed] [Google Scholar]
- 5.Zhuang JY, Ma WB, Wu JL, Chai RY, Lu J, Fan YY, et al. (2002) Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica 128: 363–370. [Google Scholar]
- 6.Puri KD, Shrestha SM, Chhetri GB, Joshi KD (2009) Leaf and neck blast resistance reaction in tropical rice lines under green house condition. Euphytica 165: 523–532. [Google Scholar]
- 7.Ishihara T, Hayano-Saito Y, Oide S, Ebana K, La NT, Hayashi K, et al. (2014) Quantitative trait locus analysis of resistance to panicle blast in the rice cultivar Miyazakimochi. Rice (N Y) 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi K, Yasuda N, Fujita Y, Koizumi S, Yoshida H (2010) Identification of the blast resistance gene Pit in rice cultivars using functional markers. Theor Appl Genet 121: 1357–1367. 10.1007/s00122-010-1393-7 [DOI] [PubMed] [Google Scholar]
- 9.Manandhar HK, Lyngs Jorgensen HJ, Mathur SB, Smedegaard-Petersen V (1998) Suppression of rice blast by preinoculation with avirulent Pyricularia oryzae and the nonrice pathogen Bipolaris sorokiniana. Phytopathology 88: 735–739. 10.1094/PHYTO.1998.88.7.735 [DOI] [PubMed] [Google Scholar]
- 10.Zhu D, Kang H, Li Z, Liu M, Zhu X, Wang Y, et al. (2016) A genome-wide association study of field resistance to Magnaporthe Oryzae in rice. Rice (N Y) 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu S, Liu G, Zhou B, Bellizzi M, Zeng L, Wang Y, et al. (2006) The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics 172: 1901–1914. 10.1534/genetics.105.044891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou B, Qu SH, Liu GF, Dolan M, Sakai H, Lu G, et al. (2006) The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol Plant Microbe In 19: 1216–1228. [DOI] [PubMed] [Google Scholar]
- 13.Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu JZ, Matsumoto T, et al. (2008) Two adjacent nucleotide-binding Site-Leucine-Rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics 180: 2267–2276. 10.1534/genetics.108.095034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhai C, Lin F, Dong Z, He X, Yuan B, Zeng X, et al. (2011) The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication. New Phytol 189: 321–334. 10.1111/j.1469-8137.2010.03462.x [DOI] [PubMed] [Google Scholar]
- 15.Yuan B, Zhai C, Wang WJ, Zeng XS, Xu XK, Hu H, et al. (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet 122: 1017–1028. 10.1007/s00122-010-1506-3 [DOI] [PubMed] [Google Scholar]
- 16.Hua L, Wu J, Chen C, Wu W, He X, Lin F, et al. (2012) The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor Appl Genet 125: 1047–1055. 10.1007/s00122-012-1894-7 [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Peng P, Tian JS, He YG, Zhang LP, Liu Z, et al. (2015) Pike, a rice blast resistance allele consisting of two adjacent NBS-LRR genes, was identified as a novel allele at the Pik locus. Mol Breeding 35. [Google Scholar]
- 18.Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, et al. (2009) Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325: 998–1001. 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- 19.Kouzai Y, Kaku H, Shibuya N, Minami E, Nishizawa Y (2013) Expression of the chimeric receptor between the chitin elicitor receptor CEBiP and the receptor-like protein kinase Pi-d2 leads to enhanced responses to the chitin elicitor and disease resistance against Magnaporthe oryzae in rice. Plant Mol Biol 81: 287–295. 10.1007/s11103-012-9998-7 [DOI] [PubMed] [Google Scholar]
- 20.Wu Y, Bao Y, Xie L, Su Y, Chu R, He W, et al. (2013) Fine mapping and identification of blast resistance gene Pi-hk1 in a broad-spectrum resistant japonica rice landrace. Phytopathology 103: 1162–1168. 10.1094/PHYTO-02-13-0044-R [DOI] [PubMed] [Google Scholar]
- 21.Mackill D, Bonman J (1992) Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82: 746–749. [Google Scholar]
- 22.Chen D, Zeigler R, Ahn S, Nelson R (1996) Phenotypic characterization of the rice blast resistance gene Pi-2 (t). Plant Dis 80: 52–56. [Google Scholar]
- 23.Liu G, Lu G, Zeng L, Wang GL (2002) Two broad-spectrum blast resistance genes, Pi9 (t) and Pi2 (t), are physically linked on rice chromosome 6. Mol Genet Genomics 267: 472–480. 10.1007/s00438-002-0677-2 [DOI] [PubMed] [Google Scholar]
- 24.Jeon JS, Chen D, Yi GH, Wang GL, Ronald PC (2003) Genetic and physical mapping of Pi5(t), a locus associated with broad-spectrum resistance to rice blast. Mol Genet Genomics 269: 280–289. 10.1007/s00438-003-0834-2 [DOI] [PubMed] [Google Scholar]
- 25.Berruyer R, Adreit H, Milazzo J, Gaillard S, Berger A, Dioh W, et al. (2003) Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor Appl Genet 107: 1139–1147. 10.1007/s00122-003-1349-2 [DOI] [PubMed] [Google Scholar]
- 26.Deng Y, Zhu X, Shen Y, He Z (2006) Genetic characterization and fine mapping of the blast resistance locus Pigm (t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor Appl Genet 113: 705–713. 10.1007/s00122-006-0338-7 [DOI] [PubMed] [Google Scholar]
- 27.Wang JF, He XJ, Zhang HS, Chen ZY (2002) [Genetic analysis of blast resistance in japonica rice landrace heikezijing from Taihu region]. Yi Chuan Xue Bao 29: 803–807. [PubMed] [Google Scholar]
- 28.Li PF, Shi XL, Wang JF, Zhang HS (2007) [Genetic analysis of resistance to rice blast in four Japonica landraces from Taihu Lake region]. Yi Chuan 29: 1249–1255. [DOI] [PubMed] [Google Scholar]
- 29.Cheng JP, Wang L, Du WL, Lai YY, Huang X, Wang Z, et al. (2014) Dynamic quantitative trait locus analysis of seed dormancy at three development stages in rice. Mol Breed 34: 501–510. [Google Scholar]
- 30.Liu SF, Yang XR, Sun SQ, Liu CY, Wang Y, Zhang CX, et al. (2007) Identification technique of rice resistance to Magnaporthe Grisea. Tianjin Agricultural Sciences 13(4): 55–58. [Google Scholar]
- 31.Koizumi S, Tani T (1998) A method for evaluating field resistance to panicle blast in rice cultivars using cut panicles. Annual Report of the Society of Agricultural Chemicals of North Japan 1998: 27–32. [Google Scholar]
- 32.Shi X, Wang J, Bao Y, Li P, Xie L, Huang J, et al. (2010) Identification of the quantitative trait loci in japonica rice landrace Heikezijing responsible for broad-spectrum resistance to rice blast. Phytopathology 100: 822–829. 10.1094/PHYTO-100-8-0822 [DOI] [PubMed] [Google Scholar]
- 33.Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, et al. (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9: 199–207. [DOI] [PubMed] [Google Scholar]
- 35.Ma J, Lei C, Xu X, Hao K, Wang J, Cheng Z, et al. (2015) Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol Plant Microbe In 28: 558–568. [DOI] [PubMed] [Google Scholar]
- 36.Wu JL, Fan YY, Li DB, Zheng KL, Leung H, Zhuang JY, et al. (2005) Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor Appl Genet 111: 50–56. 10.1007/s00122-005-1971-2 [DOI] [PubMed] [Google Scholar]
- 37.Li P, Shi X, Wang J, Liu C, Zhang H (2007) Molecular mapping of rice blast resistance gene in a japonica landrace Heikezijing from the Taihu lake area, China. Chinese Journal of Rice Science 21(6): 579–584. [Google Scholar]
- 38.Zenbayashi-Sawata K, Fukuoka S, Katagiri S, Fujisawa M, Matsumoto T, Ashizawa T, et al. (2007) Genetic and physical mapping of the partial resistance gene, pi34, to blast in rice. Phytopathology 97: 598–602. 10.1094/PHYTO-97-5-0598 [DOI] [PubMed] [Google Scholar]
- 39.Bonman JM (1991) Durable resistance to rice blast disease-environmental influences. Euphytica 63: 115–123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper.