Abstract
Several arenaviruses, chiefly Lassa (LASV) in West Africa, cause hemorrhagic fever (HF) disease in humans and pose important public health problems in their endemic regions. To date, there are no FDA-approved arenavirus vaccines and current anti-arenaviral therapy is limited to the use of ribavirin that has very limited efficacy. In this work we dcument that a recombinant prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) with a codon deoptimized (CD) surface glycoprotein (GP), rLCMV/CD, exhibited wild type (WT)-like growth properties in cultured cells despite barely detectable GP expression levels in rLCMV/CD-infected cells. Importantly, rLCMV/CD was highly attenuated in vivo but able to induce complete protection against a subsequent lethal challenge with rLCMV/WT. Our findings support the feasibility of implementing an arenavirus GP CD-based approach for the development of safe and effective live-attenuated vaccines (LAVs) to combat diseases caused by human pathogenic arenaviruses.
Keywords: Arenavirus, Lymphocytic choriomeningitis virus, Live-attenuated vaccines, Codon deoptimization, Glycoprotein, Codon usage, Reverse genetics
1. Introduction
Several members in the Arenaviridae family cause hemorrhagic fever (HF) disease in humans (Buchmeier et al., 2007; McCormick and Fisher-Hoch, 2002). Thus, Lassa (LASV) in West Africa and Junin (JUNV) in the Argentine Pampas, cause Lassa fever (LF) and Argentine HF, respectively, diseases in humans that are associated with high morbidity and significant mortality, and pose an important public health problem in their endemic areas (Borio et al., 2002; Buchmeier et al., 2007; McCormick and Fisher-Hoch, 2002). Moreover, increased travel has resulted in the importation of LF cases into non-endemic metropolitan regions in the USA, Europe and Japan (Buchmeier et al., 2007; Holmes et al., 1990; Isaacson, 2001). Moreover, novel arenaviruses are being discovered every one to three years (Kunz, 2009), including the recent identification of two novel HF-causing arenaviruses: Chapare in Bolivia in 2003 (Delgado et al., 2008) and Lujo in Southern Africa in 2008 (Briese et al., 2009). It should be also noted that mounting evidence indicates that the worldwide-distributed prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical relevance (Fischer et al., 2006; Palacios et al., 2008; Schafer et al., 2014). In addition, several arenaviruses pose a credible bioterrorism threat and six of them, including LASV and JUNV are classified as Category A agents by the National Institute of Allergy and Infectious Diseases (NIAID) (Borio et al., 2002; Charrel and de Lamballerie, 2003). Despite the significance of arenaviruses in public health and biodefense readiness, to date there are no vaccines approved by the Food and Drug Administration (FDA) and current anti-arenavirus therapy is limited to the off-labeled use of the broad-spectrum nucleoside analog ribavirin that is only partially effective and requires an early and intravenous administration and can also cause significant side effects (Kilgore et al., 1997; McKee et al., 1988; Snell, 1988).
Epidemiological studies indicate that live-attenuated vaccines (LAV) represent the most feasible approach to control HF arenaviruses within their endemic regions, as LAV induce long-term robust cellular and humoral immune responses following a single immunization (Falzarano and Feldmann, 2013; Fisher-Hoch and McCormick, 2004; Lukashevich, 2012; McCormick and Fisher-Hoch, 2002). The JUNV live-attenuated Candid#1 strain developed from a joint effort between the USA Army Medical Research Institute of Infectious Diseases (USAMRIID) and the Argentine Ministry of Health in the early 1990 s, has been shown to be an effective vaccine against Argentine HF (Enria et al., 2008, 2010; Maiztegui et al., 1998; McKee et al., 1992). However, outside Argentina, Candid#1 remains as an investigational new drug (IND). The reassortant ML29 carrying the L segment from the non-pathogenic Mopeia virus (MOPV) and the S segment from LASV has been shown to be safe, immunogenic and capable of provide protection in guinea pig and non-human primate models of LF (Carrion et al., 2007; Lukashevich et al., 2005, 2008). Differences in polymerase activity between LASV and MOPV likely contributed to ML29 attenuation but the mechanisms of attenuation of ML29 remain unknown, which raises concerns about whether the acquisition of additional mutations by the L polymerase of ML29 could result in increased virulence. Likewise, there are concerns that potential reassortants between ML29 and circulating virulent strains of LASV could result in viruses with enhanced virulence (Greenbaum et al., 2012). Therefore, there is an unmet need for novel strategies to develop safe and effective LAV against disease caused by arenavirus infections.
Arenaviruses use an ambisense coding strategy to produce two viral proteins from each of its genome segments (Buchmeier et al., 2007). The large (L) segment encodes for the RNA-dependent-RNA-polymerase (L) (Buchmeier et al., 2007; Lee et al., 2000) and the matrix protein (Z) that mediates viral assembly and budding (Perez et al., 2003; Strecker et al., 2003; Urata and de la Torre, 2011). The small (S) segment encodes for the viral glycoprotein precursor (GPC) and the viral nucleoprotein (NP). GPC is co-translationally cleaved by signal peptidase to produce a stable 58 amino acid Stable Signal Peptide (SSP) and GPC that is post-translationally processed by the cellular Site 1 Protease (S1P) to yield the two mature virion GP1 and GP2 glycoproteins that together with SSP form the GP complex involved in receptor binding and virus cell entry (Beyer et al., 2003; Buchmeier et al., 2007; Pinschewer et al., 2003; Rojek and Kunz, 2008; Rojek et al., 2008), NP, is the most abundant viral protein in infected cells and virions (Buchmeier et al., 2007) and performs multiple roles during infection, including encapsidation of the viral genome RNA species to form, together with L, the viral ribonucleoprotein (vRNPs) complex that directs the biosynthetic processes of RNA replication and gene transcription of the viral genome (Lee et al., 2000; Ortiz-Riano et al., 2012a, 2012b), and inhibition of the cellular type I interferon (IFN-I) (Borrow et al., 2010; Martinez-Sobrido et al., 2006, 2007, 2009, Pythoud et al., 2012, 2015) and inflammatory (Borrow et al., 2010; Lee et al., 2000; Rodrigo et al., 2012) responses.
Codon usage bias refers to the redundancy of the genetic code, where amino acids are determined by synonymous codons at different frequencies (Gustafsson et al., 2004; Ikemura, 1982; Nakamura et al., 1996). The process of codon optimization (CO), where each amino acid is encoded by the most abundant codon, is frequently exploited to improve gene expression in heterologous systems (Kane, 1995; Smith, 1996; Yadava and Ockenhouse, 2003), a strategy that is used to increase immune responses to antigens (Andre et al., 1998; Ramakrishna et al., 2004; Tenbusch et al., 2010). Conversely, codon deoptimization (CD), where each amino acid is encoded by the less abundant codon, is used to decrease gene expression leading to reduced viral protein production (Zhou et al., 1999). Accordingly, several RNA viruses have been effectively attenuated by CD of a single or a limited number of viral gene products (Burns et al., 2006; Cheng et al., 2015b; Le Nouen et al., 2014; Mueller et al., 2006, 2010; Nogales et al., 2014; Yang et al., 2013). We have previously documented the feasibility of generating rLCM viruses with partial, but not fully, CD of the viral NP that were attenuated in vivo but able to induce protective immunity against a lethal challenge with wild type (WT) LCMV (Cheng et al., 2015c). Our inability to rescue an rLCMV with a fully CD NP likely reflected the multiple critical roles played by NP in the arenavirus life cycle. Therefore, the generation of arenavirus LAV based on CD of NP would require extensive testing to identify the degree of NP CD compatible with robust viral growth in cell substrates used for vaccine production, while resulting in complete lack of virulence in vivo while retaining the virus’s ability to induce protective immunity against a virulent strain. In this work we have investigated whether these limitations could be solved by the use of an rLCMV expressing a fully CD GP. We successfully rescued a rLCMV encoding a fully CD GP (rLCMV/CD) that replicated to similar titers as rLCMV/WT in cultured cells despite expression levels of GP CD in infected cell were barely detectable. Notably, rLCMV/CD was highly attenuated in vivo and able to confer full protection, upon a single immunization dose, against a subsequent lethal challenge with rLCMV/WT. These results support the feasibility of using a GP CD-based approach for the development of safe and protective LAV to protect against disease caused by human pathogenic arenaviruses.
2. Results
2.1. Effect of codon deoptimization (CD) on LCMV GP expression levels
We have shown that magnitude of CD of LCMV NP negatively correlates with NP expression levels in transfected or infected cells (Cheng et al., 2015c). To determine whether this was a finding generally applicable to other arenaviral gene products, we synthesized de novo an LCMV GP where each amino acid was substituted by the least frequently used codon in mammalians cells (Gustafsson et al., 2004; Ikemura, 1982; Nakamura et al., 1996) (Supplementary Fig. 1), while preserving the intact LCMV GP WT amino acid sequence (Fig. 1A). The resulting LCMV GP CD contained a total of 431 nucleotide silent mutations out of a total of 1,494 (28.8%) that resulted in the codon-deoptimization of 357 out of 498 amino acids (71.7%) (Fig. 1B). LCMV GP CD was cloned into the pCAGGS plasmid to evaluate GP CD expression levels in transfected human 293 T cells by IFA (Fig. 1C), FACS (Fig. 1D) and WB (Fig. 1E) assays (Cheng et al., 2015c). We observed significantly reduced GP expression levels in cells transfected with LCMV GP CD as compared to cells transfected with the same amount of plasmid encoding LCMV GP WT.
Fig. 1.
CD reduces LCMV GP expression levels. A) Amino acid sequence of LCMV GP CD. CD LCMV GP amino acid residues are indicated in red. Methionine (M) and tryptophan (T) residues in LCMV GP, as well as amino acids already associated with deoptimized codons are indicated in black. Underlined (solid) amino acids represent the stable signal peptide (SSP). No underlined region represents GP1. Underlined (dotted) amino acids represent GP2. Box represents the GP transmembrane domain. Amino acids highlighted in gray represent the GP cytoplasmic tail. B) Mutations in LCMV GP CD. Number of nucleotide mutations and the percentage (%) of CD amino acids are indicated. C-E) LCMV GP CD expression levels in transfected cells. Human 293 T cells were transiently transfected with pCAGGS expression plasmids encoding LCMV GP WT or GP CD and at 48 h p.t. GP expression was assessed by IFA (C), FACS (D) and WB (E) using the LCMV GP mouse monoclonal antibody 83.6 (GP2). Empty (E) plasmid was included as negative control (C–D). IFA, FACS and WB results correspond to representative of three independent transfection experiments. (C) Scale bar=100 μm. (D) Blue line: mock-transfected cells. Black line: cells transfected with LCMV GP WT. Red line: cells transfected with LCMV GP CD. (E) GAPDH expression levels were used as loading controls.
2.2. Generation and characterization of a recombinant LCMV expressing a CD GP (rLCMV/CD)
We have described the generation of attenuated rLCM viruses containing partially CD NP (Cheng et al., 2015c), but we were unable to rescue a rLCMV containing a fully CD NP (Cheng et al., 2015c). In contrast, we were able to rescue rLCMV/CD with high efficiency. The identity of the rLCMV/CD was confirmed by RT-PCR, using RNA extracted from infected BHK-21 cells and primers specific to either LCMV GP WT or LCMV GP CD (Fig. 2), and sequence analysis of the PCR products (data not shown). Next, we compared the growth kinetics of the rLCMV/CD and rLCMV/WT in human A549 (Fig. 3A), rodent BHK-21 (Fig. 3B) and African green monkey kidney Vero cells (Fig. 3C). Complete CD of GP should result in highly reduced expression levels of GP1 and GP2 proteins, which would be expected to affect LCMV multiplication. However, unexpectedly, in BHK-21 and Vero cells rLCMV/CD and rLCMV/WT exhibited similar growth kinetics and peak titers, whereas in A549 cells rLCMV/CD displayed a growth kinetics delay and lower peak titers compared to rLCMV/WT. These differences could be due to species-specific codon usage (CD was based on mammalian codon usage), tRNA availability in these different cell lines, or a decrease in viral fitness in IFN-I competent A549 cells compared to IFN-I deficient BHK-21 and Vero cells (Karki et al., 2012; Martinez-Sobrido et al., 2009).
Fig. 2.
Genetic identity of rLCMV/CD. BHK-21 cells were infected (moi=0.01) with rLCMV/WT or rLCMV/CD. At 72 h p.i., RNA was isolated from LCMV- and mock-infected control cells and subjected to RT-PCR using WT (top) or CD (bottom) LCMV GP specific primers. PCR products were analyzed by agarose (1% wt/vol) gel electrophoresis.
Fig. 3.
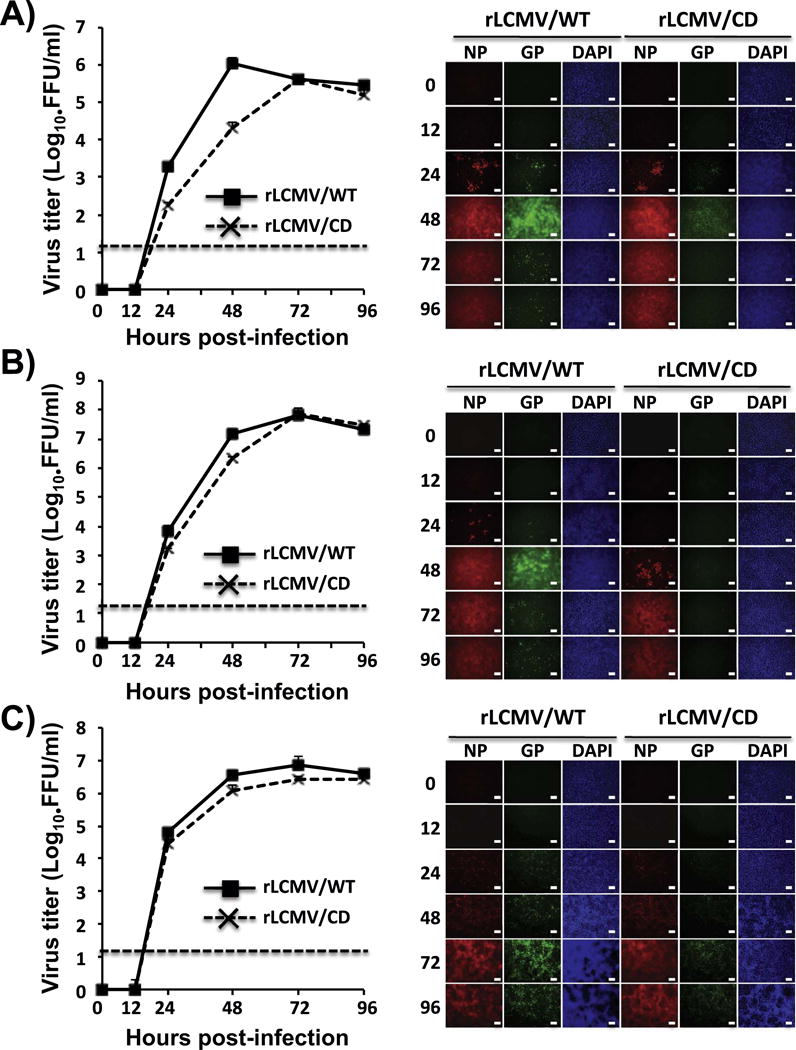
rLCMV/CD growth kinetics in cultured cells. A549 (A), BHK-21 (B), and Vero (C) cells (triplicates) were infected (moi=0.01) with either rLCMV/WT or rLCMV/CD. Viral titers in TCS at the indicated h. p.i were determined by immune-focus assay (FFU/ml) (left). Dotted line indicates the limit of detection (20 FFU/ml). Replicates of cells infected as before were evaluated for NP and GP expression levels by IFA assay (right) using the LCMV GP and NP mouse monoclonal antibodies 83.6 and 1.1.3, respectively. DAPI was used for nuclear staining. Representative images of three independent infection experiments are illustrated. Scale bar=100 μm.
2.3. GP expression levels in rLCMV/CD-infected cells
Our results from cells transfected with WT and CD GPs (Fig. 1) were consistent with the expected negative effect of CD on protein translation efficiency (Zhou et al., 1999). However, CD of GP did not affect significantly growth properties of rLCMV/CD in cultured cells (Fig. 3). This could reflect that translation efficiency of GP CD was not affected in the context of infection, or that reduced expression levels of GP in rLCMV/CD-infected cells were still sufficient to support high levels of LCMV multiplication. To evaluate these possibilities, we infected (moi=0.1) BHK-21 cells with rLMCV/WT and rLCMV/CD and analyzed the kinetics of RNA synthesis, both replication and transcription (Fig. 4A) and GP expression (Fig. 4B–D). Consistent with the results of production of infectious progeny over time (Fig. 3), we did not detect significant differences in the kinetics and levels of viral RNA synthesis, both replication and transcription, between rLCMV/WT- and rLCMV/CD-infected cells (Fig. 4A). In contrast, GP expression levels were significantly lower in rLCMV/CD than in rLCMV/WT-infected BHK-21 cells as determined by WB (Fig. 4B) and FACS (Fig. 4D), even with similar levels of infection, as determined by NP expression (Fig. 4C and D). These results indicated that reduced levels of GP expression caused by CD of GP were still sufficient to promote WT-like growth properties in cultured cells.
Fig. 4.
Viral RNA synthesis (A) and GP expression levels (B–D) in rLCMV/CD-infected BHK-21 cells. A) Cells were infected (moi=0.1) with rLCMV/WT or rLCMV/CD. At the indicated times, RNA was isolated and analyzed by NB hybridization using a double strand DNA NP probe that hybridized to the S genome and NP mRNA species. Signals corresponding to the S and NP mRNA species were quantified and normalized first with respect levels of 18 S rRNA used as loading and transfer control. Corrected S and NP mRNA species signals were then normalized to the signals detected at 24 h p.i. in rLCMV/WT-infected cells. B–D) Cells were mock (M) infected or infected (moi=0.01) with rLCMV/WT or rLCMV/CD. At 48 h p.i., cells lysates were prepared and evaluated for GP expression levels by WB (B) using the LCMV GP mouse monoclonal antibody 83.6. Expression levels of β-actin were used as loading controls. Infection levels at the same times post-infection were evaluated by IFA using the LCMV NP mouse monoclonal antibody 1.1.3 (C). Nuclei were stained with DAPI. Scale bar=100 μm. Cells mock-infected (blue line) or infected with rLCMV/WT (black line) or rLCMV/CD (red line) were subjected to FACS analysis with the LCMV NP monoclonal antibody 1.1.3 or the LCMV GP monoclonal antibody 36.1 (D).
2.4. Morphology of rLCMV/CD virions
We next evaluated if reduced GP expression levels in rLCMV/CD-infected cells (Figs. 3 and 4) resulted in lower incorporation of GP into virions or had an effect on virus morphology. For this, we purified rLCMV/WT and rLCMV/CD virions from the TCS of infected BHK-21 cells. Both rLCMV/WT (Fig. 5A) and rLCMV/CD (Fig. 5B) exhibited similar spherical shape morphology with a heterogeneous (60–100 nm) size in diameter, when observed at low or high magnification, which was consistent with previous findings (Rodrigo et al., 2011). These results indicated that the significantly lower expression levels of GP in rLCMV/CD compared to rLCMV/WT-infected cells (Figs. 3 and 4), did not significantly affect virion morphology. We also evaluated the presence of GP in purified rLCMV/WT and rLCMV/CD by immunostaining with a monoclonal antibody to LCMV GP1 (36.1) (Fig. 5C). We observed that significant higher levels of GP WT than GP CD were incorporated into virion particles as determined by the presence of 12 nm gold particles (Fig. 5C). We quantified the presence of viral gold particles/virion (Fig. 5D), which indicated that purified rLCMV/WT contained about 10-fold higher levels of gold particles than purified rLCMV/CD (Fig. 5D), indicating a lower density of GP in purified rLCMV/CD as compared to rLCMV/WT.
Fig. 5.
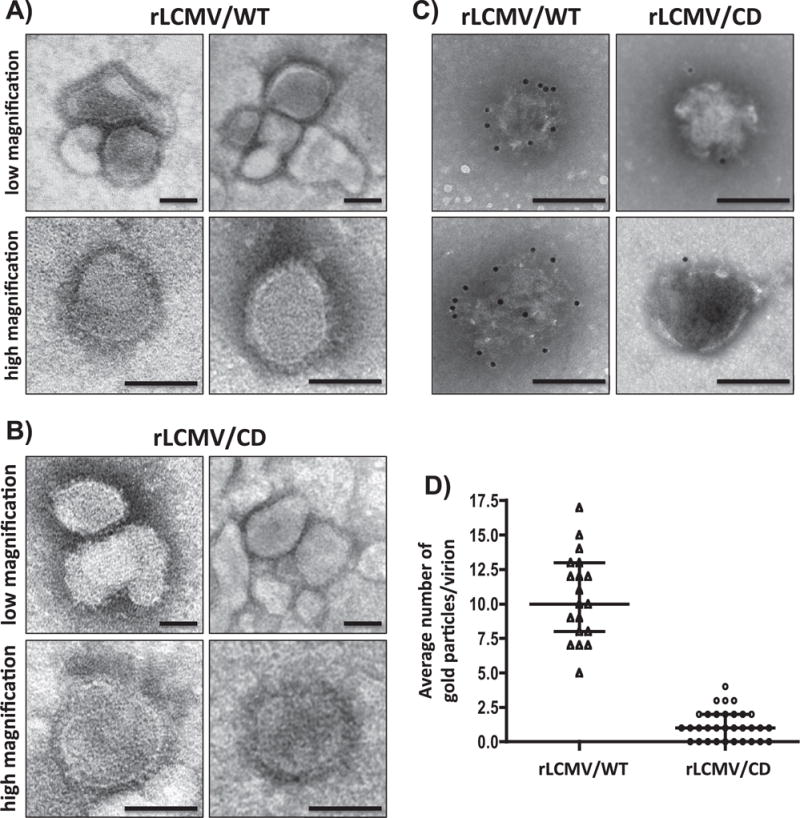
Comparison of rLCMV/WT and rLCMV/CD virion particles. rLCMV/WT and rLCMV/CD virions were purified from infected (moi=0.01) BHK-21 cells at 72 h p.i. by ultracentrifugation through an 20% (wt/vol) sucrose cushion. Morphology of rLCMV/WT (A) and rLCMV/CD (B) was assessed by TEM using negative straining. Same viral preparations were surface stained with the LCMV GP1-specific monoclonal antibody 36.1 and counterstained with 12-nm-gold-labeled goat anti-mouse IgG antibody (C). The number of gold particles/virion was determined for rLCMV/WT (N=19) and rLCMV/CD (N=32) immune stained viruses (D). Representative images are shown. Scale bar=100 μm.
2.5. Virulence and protective efficacy of rLCMV/CD against a lethal challenge with rLCMV/WT in mice
To assess the potential of GP CD-based approach for the development of arenavirus LAV, we examined the safety (Table 1) and protection efficacy (Table 2) of our rLCMV/CD using the well-characterized mouse model of LCMV induced fatal choriomeningitis (Cheng et al., 2015a, 2015c). In this model, six week-old male and female WT B6 mice inoculated i.c. with rLCMV/WT (103 PFU) succumb (100% mortality) between 7 and 8 days p.i. due to a robust immune cellular response to viral antigens present at high levels in the choroid plexus and meninges, which causes a fatal lymphocytic choriomeningitis (LCM). This infection model provides us with a reliable and rapid test to assess the attenuation in vivo of rLCMV/CD. As expected, all mice infected i.c. with rLCMV/WT developed clinical symptoms of LCM disease and died within the first 8 days of infection (Table 1). In contrast, all mice infected with rLCMV/CD survived and remained free of clinical symptoms throughout the duration (12 days) of the experiment. Attenuation of rLCMV/CD correlated with significantly reduced levels of virus multiplication in brains of infected mice, compared to rLCMV/WT-infected mice (Fig. 6). We then assessed whether a single-dose immunization with rLCMV/CD using a standard protocol of immunization (105 FFU, i.p.) was capable of inducing protective immunity against a subsequent lethal challenge with WT LCMV (103 FFU, i.c.) (Table 2). Within this model, mice immunized with rLCMV/CD can be subsequently exposed to a lethal challenge with rLCMV/WT (103 FFU, i.c.) to assess the protective efficacy of rLCMV/CD as candidate LAV. All mice vaccinated with rLCMV/CD were fully protected against the lethal challenge with rLCMV/WT and remained free of clinical symptoms throughout the duration of the experiment (12 days). As expected, all PBS-vaccinated mice developed clinical symptoms and died within the 8 days after the challenge with rLCMV/WT (Table 2) (Cheng et al., 2015a, 2015c). These results demonstrated that rLCMV/CD was highly attenuated in vivo (Table 1) but able to trigger a protective immune response against a subsequent lethal challenge with rLCMV/WT (Table 2).
Table 1.
In vivo attenuation of rLCMV/CD. Six week-old male and female B6 mice (N=8) were infected intracraneallly (i.c) with 103 FFU of rLCMV/WT or rLCMV/CD, or mock-infected (PBS) as control. Mice were monitored daily for morbidity and mortality until the experimental endpoint (12 days p.i.).
| Days p.i. (i.c. 103 PFU) | % Survival (n=8)
|
|||
|---|---|---|---|---|
| 6 | 7 | 8 | 12 | |
| PBS | 100 | 100 | 100 | 100 |
| rLCMV/WT | 100 | 37.5 | 0 | – |
| rLCMV/CD | 100 | 100 | 100 | 100 |
Table 2.
Ability of a single dose of rLCMV/CD to protect against a lethal challenge with rLCMV/WT. Six week-old male and female B6 mice (N=8) were immunized intraperitoneally (i. p.) with 105 FFU of rLCMV/WT, rLCMV/CD, or mock-immunized (PBS). Four weeks after immunization, mice were subjected to a lethal challenge with rLCMV/WT (i.c., 103 FFU) and then monitored daily for morbidity and mortality until the experimental endpoint (12 days p.i.).
| Days post-challenge (i.c. 103 PFU; rLCMV/WT) | % Survival (n=8)
|
|||
|---|---|---|---|---|
| 6 | 7 | 8 | 12 | |
| PBS | 100 | 25 | 0 | – |
| rLCMV/WT | 100 | 100 | 100 | 100 |
| rLCMV/CD | 100 | 100 | 100 | 100 |
Fig. 6.
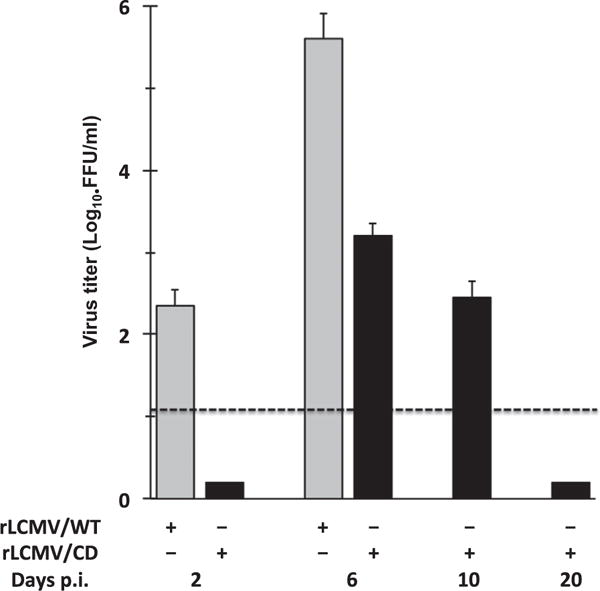
Multiplication of rLCMV/CD in brains of mice infected by the i.c. route. B6 mice (6–8 weeks, females and males) were infected (103 FFU) with either rLCMV/WT (N=6) or rLCMV/CD (N=15). At the indicated days p.i. brain tissue was collected and virus titers determined. Titers correspond to average and SD of three mice. Dotted line indicates the limit of detection (20 FFU/ml).
2.6. Generation of r3LCMV/CD
We have documented the generation of r3 arenaviruses expressing foreign genes of interest (GOI) (Cheng et al., 2013; Emonet et al., 2009; Ortiz-Riano et al., 2013; Popkin et al., 2011). Several r3 arenaviruses tested exhibited attenuation in mice, but still retained a significant degree of pathogenicity upon i.c. inoculation of adult immune mice (Emonet et al., 2009). Therefore, the use of r3 arenaviruses as vaccine vectors would require to increase their degree of attenuation. To evaluate if the GP CD-based approach could be used for the development of safer r3 arenavirus vaccine vectors, we generated a r3LCMV/CD that expressed GFP and Gaussia luciferase (Gluc) from the NP and GPC loci, respectively (Fig. 7). Growth kinetics in A549 (Fig. 7Ai), BHK21 (Fig. 7Bi) and Vero (Fig. 7Ci) cells demonstrated further attenuation of r3LCMV/CD as compared to r3LCMV/WT. Reduced r3LCMV/CD multiplication was further confirmed in each of these cell lines by measuring expression levels of Gluc (Fig. 7A–C, ii) and GFP (Fig. 7A–C, iii).
Fig. 7.
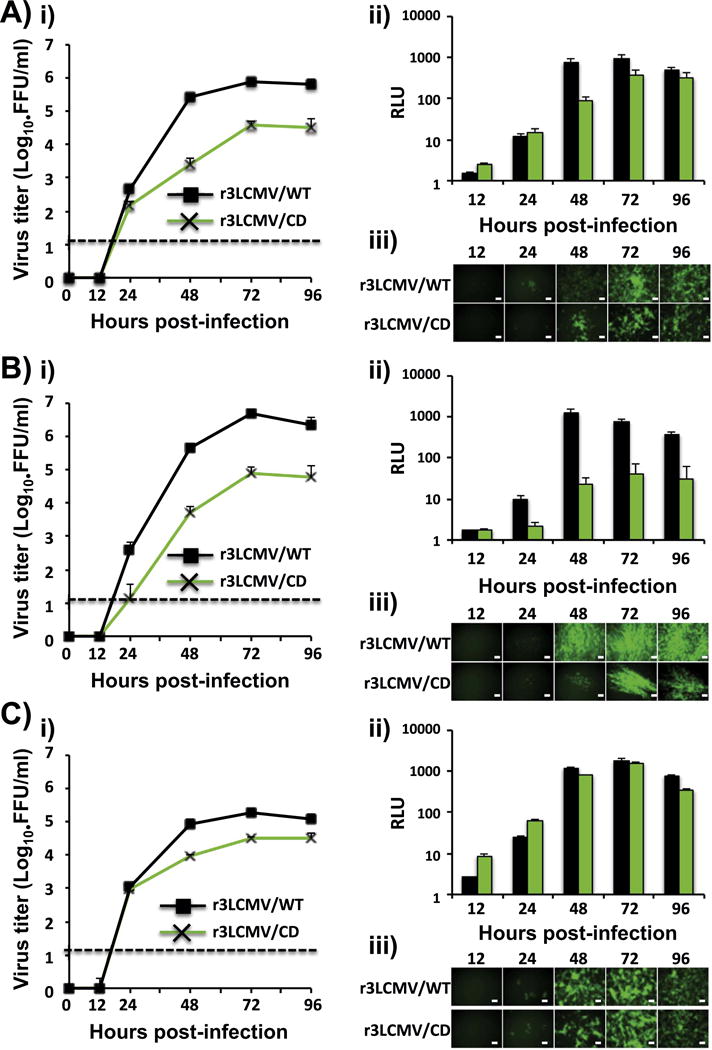
Growth kinetics of r3LCMV/CD in cultured cells. A549 (A), BHK-21 (B), and Vero (C) cells (triplicates) were infected (moi=0.01) with either r3LCMV/WT (black) or r3LCMV/CD (green) and viral titers in TCS at the indicated h. p.i. were determined by fluorescent forming units (FFU/ml) (i). Dotted line indicates the limit of detection (20 FFU/ml). Gluc activity from same TCS was determined by luminescence (ii). Viral replication was also assessed by GFP expression by fluorescence microscopy (iii). Representative images are illustrated. Scale bar=100 μm.
3. Discussion
LF poses a major public health problem in West Africa, and the importation of LF cases, due to traveling, to non-endemic regions across the world illustrates the global public threat posed by LASV, and underscores the need of having in place intervention strategies to control LASV within its endemic regions (Buchmeier et al., 2007; Holmes et al., 1990; Isaacson, 2001). Currently, there are no FDA-approved vaccines to combat human arenavirus infections and current anti-arenaviral therapy is limited to an off-label use of the nucleoside analog ribavirin that is only partially effective (Kilgore et al., 1997; McKee et al., 1988; Snell, 1988). The ML29 reassortant is a promising LASV LAV that has been shown to provide effective protection in different animal models of LASV induced disease (Carrion et al., 2007; Lukashevich et al., 2005, 2008). However, the mechanisms responsible for ML29 attenuation remain unknown (Olschlager and Flatz, 2013), and therefore the incorporation of a limited number of additional mutations into the ML29 genome might result in viruses with enhanced virulence (Greenbaum et al., 2012). Similarly, the phenotypic stability of Candid#1, the JUNV LAV, has not been fully examined and safety concerns have recently arose since a single amino acid substitution (F427I) in the GP of the pathogenic JUNV XJ13 leads to an Candid#1-like attenuated phenotype (Albarino et al., 2011; Droniou-Bonzom et al., 2011), suggesting that a limited number of mutations could result in reversion of Candid#1 into a more virulent strain.
The use of suboptimal codon pair bias has gained attention recently as a novel strategy for the development of viral LAV. This strategy has been used for the generation of LAV candidates for poliovirus (Burns et al., 2009; Mueller et al., 2006), influenza A virus (Mueller et al., 2010; Nogales et al., 2014; Yang et al., 2013), and respiratory syncytial virus (Le Nouen et al., 2014), among others. This approach, however, requires the use of computer algorithms to design viral genomes with appropriate pair-deoptimized codons. The use of different combinations of deoptimized codon pairs results in different degrees of viral attenuation (Burns et al., 2006; Le Nouen et al., 2014; Mueller et al., 2006, 2010; Yang et al., 2013), and therefore this approach needs to be evaluated for each codon pair-deoptimized virus. With the aim of overcoming these difficulties we have explored the alternative approach of using a purely CD-based strategy where each amino acid residue in a given viral protein is encoded by the corresponding least preferred codon in mammalian cells. We provided for the first time evidence supporting the feasibility of this approach for the generation of arenavirus LAV by showing that cells infected with rLCMV containing a CD NP, rLCMV/NPCD, expressed highly reduced levels of NP but was able to grow to high titers (Cheng et al., 2015c). More importantly, rLCMV/NPCD was highly attenuated in mice but able to induce protective immunity, upon a single immunization dose, against a subsequent lethal challenge with rLCMV/WT (Cheng et al., 2015c). These earlier results had the limitation that we were unable to rescue a rLCMV encoding a fully CD NP, and we therefore had to identify the optimal degree of CD for NP to generate a viable recombinant virus that lack virulence in vivo whereas it retained WT-like growth properties in cell substrates used for vaccine production (e.g. Vero cells). This strategy would pose the problem that similar levels of attenuation and protection efficacy might not be achieved by introducing comparable CD regions in other arenavirus NPs. The results provided in this present work have overcome these limitations by showing that a rLCMV with a completely CD GP exhibited WT-like growth properties in cultured cells but was highly attenuated in vivo and able to provide a very effective protective immunity against a lethal challenge with rLCMV/WT. Notably, our results also suggest that rLCMV/WT produces more GP than is required for incorporation into virions and explains the similar replication kinetics of rLCMV/CD infection in vitro. It is worth noting that the lower expression levels of GP CD in both GP CD transfected and rLCMV/CD infected cells may parallel the situation observed during persistent LCMV infection, where GP expression levels have been shown to be significantly lower to GP levels observed during acute LCMV infection (Pasqual et al., 2011). This could explain why drastic lower expression levels of GP in rLCMV/CD infected cells do not have a strong impact in production of infectious LCMV progeny. It should be noted that compared to the acute infection, production of infectious LCMV progeny is significantly reduced. This, however, could reflect additional mechanisms, including production defective interfering genomes, which operate during persistent LCMV infection and down-regulate virus multiplication. Our findings resemble also to those observed with other RNA viruses, including influenza virus for which CD of the viral HA resulted in lower protein expression levels during viral infection without significantly affecting virus multiplication in cultured cells but with significant attenuation in vivo (Mueller et al., 2010; Nogales et al., 2014; Yang et al., 2013). It remains to be determined whether the attenuation observed with rLCMV containing CD genes is entirely due to reduced translation efficiency of the corresponding gene product, or whether altered CpG/UpA frequencies generated as a consequence of CD can play a critical role in attenuation as documented for other CD recombinant viruses (Tulloch et al., 2014).
The generation of LAV candidates using this CD-based approach offers several unique advantages. Current LAVs usually rely on a few amino acid changes to generate the attenuated phenotype, like the current temperature-sensitive live-attenuated influenza vaccines, LAIV (Cox et al., 1988, 2015; Jin et al., 2003; Snyder et al., 1988). In contrast, CD-based LAV candidates would be the result of hundreds of silent nucleotide mutations, thus making viral reversion to a virulent WT phenotype highly unlikely, if possible at all. Accordingly, we have shown that rLCMV/NPCD was very stable during serial passages in Vero cells (Cheng et al., 2015c). This is particularly important for arenaviruses, as their error prone replication machinery confer them with the potential for rapid evolution (Drake and Holland, 1999). Another advantage of a CD-based approach for vaccine development is that the corresponding LAV candidate retains the intact antigenic repertoire of the WT virus and can generate similar B and T cell immune responses against all the viral polypeptides. Moreover, reassortment between a circulating pathogenic arenavirus strain and the corresponding CD-based arenavirus LAV cannot result in a viral reassortant with increased virulence as compared to the already circulating pathogenic strain. It should be also noted that with the implementation of de novo synthesis of genes and already developed robust plasmid-based reverse genetics technologies (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013), generation of CD-based LAV candidates can be rapidly executed. Likewise, as illustrated in the present work, it is feasible to implement the use of a CD-based approach for arenavirus attenuation in combination with our arenavirus tri-segmented platform (Cheng et al., 2013; Emonet et al., 2009; Ortiz-Riano et al., 2013; Popkin et al., 2011) that allows for virus directed expression of additional GOI, including antigens from relevant pathogens, which could facilitate the generation of polyvalent LAV against a given arenavirus and other pathogen of interest.
4. Conclussions
Currently, there are no FDA-licensed vaccines or antiviral drugs to combat arenavirus infections. Here we demonstrate that a recombinant lymphocytic choriomeningitis virus (LCMV) encoding a codon-deoptimized glycoprotein (GP CD) exhibits wild type (WT)-like growth properties in cultured cells, while being highly attenuated in vivo but able to confer protective immunity against a subsequent lethal challenge with WT LCMV. These results provide proof-of-concept support for the feasibility of implementing an arenavirus GP codon deoptimized (CD)-based approach as a strategy to develop safe and effective live-attenuated vaccines (LAVs) to combat diseases caused by human pathogenic arenaviruses.
5. Materials and methods
5.1. Cells and viruses
Human lung adenocarcinoma epithelial A549 (ATCC CCL-185), human embryonic kidney 293T (ATCC CRL-11268), African green monkey kidney epithelial Vero (ATCC CCL-81), and baby hamster kidney BHK-21 (ATCC CCL-10) cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech, Inc.) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, penicillin (100 units/ml) and streptomycin (100 units/ml) and maintained in a 5% CO2 humidified atmosphere at 37 °C (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013). Recombinant wild-type (WT) and CD GP (CD) bi-segmented and tri-segmented (r3) LCM viruses were based on Armstrong (ARM53b) strain of LCMV (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013).
5.2. Plasmids
Fully codon deoptimized (CD) LCMV GP was generated by de novo synthesis (Biomatik). GP WT codons were replaced with the codon least represented in mammalian genomes (Supplementary Fig. 1). LCMV GP CD was amplified by PCR and cloned into pCAGGS using EcoRI and BglII restriction enzymes. The S segment vRNA-expressing plasmid for the rescue of rLCMV/CD was generated by amplifying LCMV GP CD and cloned into the mouse (m)pPol-I BsmBI/NP viral (v) RNA rescue plasmid (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013). Likewise, LCMV GP CD was cloned into a modified mpPol-I S BsmBI/GFP plasmid for the generation of the r3LCMV/CD (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013). Primers used to generate the described plasmid constructs are available upon request. Plasmid were verified by DNA sequencing.
5.3. Immunofluorescence assays (IFA)
Human 293T cells (1.8×105 cells/well; 24-well plate format) were transiently transfected in suspension with 1 μg of the respective pCAGGS GP expression plasmids using lipofectamine 2000 (LPF2000) (Invitrogen). Empty plasmid-transfected cells were included as negative control, whereas LCMV GP WT was included as positive control. At 48 h post-transfection (h p.t.), cells were fixed with 4% (vol/vol) formaldehyde diluted in 1X PBS, permeabilized with 0.1% (vol/vol) Triton X-100, and blocked using 2.5% bovine serum albumin (BSA) diluted in 1X PBS. After blocking, LCMV GP-transfected cells were incubated with the cross-reactive anti-LCMV GP mouse monoclonal antibody clone 83.6 (Cheng et al., 2013, 2015a, 2015c) and probed with a FITC-conjugated rabbit anti-mouse secondary antibody (Dako). Anti-LCMV NP (1.1.3) (Ortiz-Riano et al., 2011, 2012a, 2012b) was used for detection of LCMV in infected cells. Staining with 4′,6-diamindino-2-phenylindole (DAPI) was used to visualize cell nuclei. Protein expression was observed under a Leica fluorescent microscope. Microscope images were colored using Adobe Photoshop CS4 (v11.0) software.
5.4. Fluorescence-activated cell sorting (FACS) analysis
FACS analysis was performed as previously described (jvi2011. w.w. Shanaka et al. use of single-cycle…). Briefly, mock or LCMV infected single-cell preparations were incubated with blocking buffer (2.5% BSA, 1% FBS in 1X PBS) for 30 min on ice and then incubated with the mouse monoclonal antibody clone 36.1 for 40 min on ice. Cells were washed with 1X PBS and incubated with an allophycocyanin (APC)-conjugated goat anti-mouse antibody in blocking buffer for 30 min on ice. Cells were washed with 1X PBS and fixed with 0.5% formaldehyde diluted in 1X PBS for 10 min. For the intracellular staining of NP, after the incubation in blocking solution, cells were fixed and permeabilized with 4% formaldehyde and 0.1% Saponin diluted in 1X PBS. Then cells were stained with the anti-LCMV NP mouse monoclonal antibody clone 1.1.3 diluted in blocking buffer with 0.1% Saponin. Cells were washed with 1X PBS and incubated with APC-conjugated goat anti-mouse antibody in blocking buffer for 30 min on ice and washed twice with 1X PBS. All cells preparations were resuspended in blocking buffer before analysis using an Accuri C6 flow cytometer and the FlowJo software.
5.5. Protein Western blot (WB) analysis
Human 293T cells (3×106 cells/well; 6-well plate format) were transiently transfected in suspension with 1 μg of the LCMV GP CD pCAGGS protein expression plasmid using LPF2000. Empty and LCMV GP WT plasmid-transfected cells were included as negative and positive controls, respectively. At 48 h p.t., cells were collected and lysed with 400 μl of lysis buffer (10 mM Tris-HCl, pH 7.4, 5 mM EDTA, 100 mM NaCl, 1% (vol/vol) NP-40, complete cocktail of protease inhibitors (Roche)). Aliquots (20 μl) of the total cell lysates were separated on a 12% SDS-PAGE and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were incubated with the cross reactive LCMV GP mouse monoclonal antibody 83.6 and probed with a secondary HRP-conjugated rabbit anti-mouse antibody (GE Healthcare). LCMV GP expression was detected using a chemiluminescent kit (Denville Scientific). Cellular GAPDH (Abcam ab9485-100) or β-actin (Sigma clone AC-15, A1978) were used to normalize amount of cell lysates.
5.6. Northern blot (NB) analysis
Total cellular RNA was isolated using Tri-Reagent (Sigma) according to the manufacturer’s instructions. RNA samples were fractionated by 2.2 M formaldehyde-agarose (1.2%) gel electrophoresis followed by transfer in 20×SSC [3 M sodium chloride, 0.3 M sodium citrate] of the RNA to a Magnagraph membrane using the rapid downward transfer system (TurboBlotter). Membrane-bound RNA was cross-linked by exposure to UV. The position and amounts of 28 and 18 S rRNA species transferred and immobilized to the membrane were determined by methylene blue staining. After removal of methylene blue staining by washing with SDS, the membrane was hybridized to a 32P-labeled double strand NP probe that hybridized to both the S genome RNA and NP mRNA.
5.7. Virus rescue and growth kinetics in cultured cells
Rescue of rLCMV/CD (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013) was performed in biosafety level (BSL) 2 conditions, in accordance with the University of Rochester institutional biosafety committee. To rescue rLCMV/CD, BHK-21 cells (1×106 cells/well; 6-well plate format) were co-transfected with 0.8 μg of pCAGGS NP, 1 μg of pCAGGS L, 1.4 μg of the mpPol-I L, and 0.8 μg of the mpPol-I S encoding LCMV GP CD, using LPF2000 (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013). To rescue r3LCMV/CD we followed a similar protocol, but the mpPol-I S plasmid was substituted with the mpPol-I S Gluc/NP and mpPol-I S GP CD/GFP plasmids (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013). At 12 h p.t. media was replaced with 2 ml of post-infection media (1:2 ratio of DMEM 10% FBS and Opti-MEM). At 72 h p.t., cells were trypsinized and scaled up into 10 cm dishes and maintained for 3 days, before tissue culture supernatants (TCS) were collected and virus titers determined using an immunofocus assay (fluorescent forming units [FFU]) (Cheng et al., 2013, 2015a; Ortiz-Riano et al., 2013). Briefly, serial dilutions of TCS were used to infect Vero cells (4×104 cells/well, 96-well plate format, triplicates) and 16–20 h later cells were fixed and NP positive cells identified using an LCMV NP-specific monoclonal antibody (1.1.3). To evaluate viral growth kinetics, BHK-21, A549 and Vero cells (1.25×105 cells/well; 24-well plate format, triplicates) were infected (moi=0.01) and TCS were collected at indicated time points and virus titers (FFU/ml) determined by immunofocus assay. Mean value and standard deviation were calculated using Microsoft Excel software.
5.8. RT-PCR
BHK-21 cells (5×105 cells/well; 6-well plate format) were infected with rLCMV/WT or rLCMV/CD (moi=0.01) and at 72 h post-infection (h p.i.) total cellular RNA was prepared using Trizol (Invitrogen) (Cheng et al., 2015a). Random hexamers were used for cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) (Cheng et al., 2015a). cDNA was amplified by PCR using a combination of LCMV GP WT or GP CD specific 5′ and 3′ primers (Cheng et al., 2015a). Amplified gene products were examined on a 1% (wt/vol) agarose DNA gel and analyzed by sequencing (ACGT) (Cheng et al., 2015a).
5.9. Transmission electron microscopy (TEM) and immunoelectron microscopy (IEM)
WT and CD rLCM viruses were collected from the TCS of BHK-21 cells (10 cm dishes) infected (moi=0.01) with WT and CD rLCM viruses at 72 h p.i. and clarified (SW-32 Ti rotor, 10,000 rpm, 30 min), followed by pelleting the virus at high-speed ultracentrifugation (SW-32 Ti rotor, 25,000 rpm, 2.5 h) through a 20% (wt/vol) sucrose cushion (Rodrigo et al., 2011). Virus pellet was resuspended in PBS and stored at −80 °C. Purified virus was fixed using 2.5% (vol/vol) glutaraldehyde (diluted in filtered water) for 5 min at room temperature, then absorbed onto Formvar-carbon-coated 200-mesh copper grids for 2 min at room temperature (Rodrigo et al., 2011). After 20 s of drying, the grids were negatively stained using 2% (vol/vol) uranyl acetate (pH 4.2–4.5) for 1 min at room temperature before imaging using a Hitachi (Tokyo, Japan) 7650 TEM with an attached Gatan 11-megapixel Erlangshen (Pleasanton, CA) digital camera at 80-kv acceleration voltage, 3.5 s exposures, and magnification ×200k (Rodrigo et al., 2011). For IEM, fixed viruses were absorbed onto Formvar-carbon-coated 200-mesh nickel grids for 30 min and placed on droplets of 2.5% (vol/vol) of BSA for 30 min. Virions were stained with a mouse monoclonal antibody against LCMV GP1 (36.1) diluted 1:1 in 2.5% BSA for 2 h at room temperature. The grids were rinsed with 1X PBS and transferred onto droplets of 12 nm colloidal gold-AffiniPure goat anti-mouse IgG (Jackson ImmunoResearch) diluted 1:40 in 2.5% of BSA for 1 h at room temperature. The grids were rinsed with 1X PBS and post-fixed in 2% phosphate-buffered glutaraldehyde for 15 min, rinsed in distilled water, and negatively stained with an aqueous solution of 2% (vol/vol) uranyl acetate for 1 min at room temperature. The grids were examined and photographed as previously described. Representative micrographs were obtained. The average number of gold particles/virion was calculated by counting the number of gold particles present in 19 (rLCMV/WT) or 32 (rLCMV/CD) immune stained viruses.
5.10. Mouse experiments
LCMV virulence in vivo was assessed using the mouse model of fatal LCM (Cheng et al., 2015a, 2015c). WT B6 mice (six-week old immune competent males and females, N=8) were infected intracranially (i.c.) with 103 FFU of rLCMV/CD or rLMCV/WT and monitored daily for development of clinical symptoms and survival for 12 days (Cheng et al., 2015a, 2015c). To evaluate protection efficacy of rLCMV/CD, mice were immunized with PBS or with either rLCMV/WT or rLCMV/CD intraperitoneally (i.p.) with 105 FFU (N=8). At 4 weeks (h p.i, mice were subjected to a lethal challenge with rLCMV/GPWT (i.c.,103 FFU), and monitored for development of clinical symptoms and survival over 12 days (Cheng et al., 2015a, 2015c). To determine virus titers in brain, mice were perfused i.c. with PBS (30 ml/mouse) and collected brain tissue used to prepare 20% (W/V) homogenates in DMEM. Homogenates were clarified (12,000 g for 10 min at 4 °C) and virus titers determined by immune focus assay in Vero cells. All animal experiments with rLCM viruses were performed under protocol 09–0137 approved by The Scripps Research Institute IACUC.
Supplementary Material
Acknowledgments
Arenavirus research in LM-S laboratory was funded by the NIH/NIAID R21 AI119775-01, R43 AI119775-01, and R21 AI121550. Research in J.C.T. laboratory is supported by grants RO1 AI047140, RO1 AI077719, and RO1 AI079665.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.virol.2016.11.001.
References
- Albarino CG, Bird BH, Chakrabarti AK, Dodd KA, Flint M, Bergeron E, White DM, Nichol ST. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemorrhagic fever is located in the G2 glycoprotein transmembrane domain. J Virol. 2011;85:10404–10408. doi: 10.1128/JVI.00856-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. 2003;77:2866–2872. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borio L, Inglesby T, Peters CJ, Schmaljohn AL, Hughes JM, Jahrling PB, Ksiazek T, Johnson KM, Meyerhoff A, O’Toole T, Ascher MS, Bartlett J, Breman JG, Eitzen EM, Jr, Hamburg M, Hauer J, Henderson DA, Johnson RT, Kwik G, Layton M, Lillibridge S, Nabel GJ, Osterholm MT, Perl TM, Russell P, Tonat K, Working Group on Civilian, B Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA. 2002;287:2391–2405. doi: 10.1001/jama.287.18.2391. [DOI] [PubMed] [Google Scholar]
- Borrow P, Martinez-Sobrido L, de la Torre JC. Inhibition of the type I interferon antiviral response during arenavirus infection. Viruses. 2010;2:2443–2480. doi: 10.3390/v2112443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;(5):e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmeier MJ, Peter CJ, de la Torre JC. Arenaviridae: The viruses and their replication 2007 [Google Scholar]
- Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CC, Campagnoli R, Shaw J, Vincent A, Jorba J, Kew O. Genetic inactivation of poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J Virol. 2009;83:9957–9969. doi: 10.1128/JVI.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrion R, Jr, Patterson JL, Johnson C, Gonzales M, Moreira CR, Ticer A, Brasky K, Hubbard GB, Moshkoff D, Zapata J, Salvato MS, Lukashevich IS. A ML29 reassortant virus protects guinea pigs against a distantly related Nigerian strain of Lassa virus and can provide sterilizing immunity. Vaccine. 2007;25:4093–4102. doi: 10.1016/j.vaccine.2007.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel RN, de Lamballerie X. Arenaviruses other than Lassa virus. Antivir Res. 2003;57:89–100. doi: 10.1016/s0166-3542(02)00202-4. [DOI] [PubMed] [Google Scholar]
- Cheng BY, Ortiz-Riano E, de la Torre JC, Martinez-Sobrido L. Generation of recombinant arenavirus for vaccine development in FDA-approved Vero cells. J Vis Exp: JoVE. 2013 doi: 10.3791/50662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BY, Ortiz-Riano E, de la Torre JC, Martinez-Sobrido L. Arenavirus genome rearrangement for the development of live attenuated vaccines. J Virol. 2015a;89:7373–7384. doi: 10.1128/JVI.00307-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BY, Ortiz-Riano E, Nogales A, de la Torre JC, Martinez-Sobrido L. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J Virol. 2015b;89:3523–3533. doi: 10.1128/JVI.03401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BY, Ortiz-Riano E, Nogales A, de la Torre JC, Martinez-Sobrido L. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J Virol. 2015c doi: 10.1128/JVI.03401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox NJ, Kitame F, Kendal AP, Maassab HF, Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain, A/Ann Arbor/6/60 (H2N2) Virology. 1988;167:554–567. [PubMed] [Google Scholar]
- Cox A, Baker SF, Nogales A, Martinez-Sobrido L, Dewhurst S. Development of a mouse-adapted live attenuated influenza virus that permits in vivo analysis of enhancements to the safety of live attenuated influenza virus vaccine. J Virol. 2015;89:3421–3426. doi: 10.1128/JVI.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, Erickson BR, Agudo R, Blair PJ, Vallejo E, Albarino CG, Vargas J, Comer JA, Rollin PE, Ksiazek TG, Olson JG, Nichol ST. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 2008;(4):e1000047. doi: 10.1371/journal.ppat.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droniou-Bonzom ME, Reignier T, Oldenburg JE, Cox AU, Exline CM, Rathbun JY, Cannon PM. Substitutions in the glycoprotein (GP) of the Candid#1 vaccine strain of Junin virus increase dependence on human transferrin receptor 1 for entry and destabilize the metastable conformation of GP. J Virol. 2011;85:13457–13462. doi: 10.1128/JVI.05616-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emonet SF, Garidou L, McGavern DB, de la Torre JC. Generation of recombinant lymphocytic choriomeningitis viruses with trisegmented genomes stably expressing two additional genes of interest. Proc Natl Acad Sci USA. 2009;106:3473–3478. doi: 10.1073/pnas.0900088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enria DA, Ambrosio AM, Briggiler AM, Feuillade MR, Crivelli E. Candid#1 vaccine against Argentine hemorrhagic fever produced in Argentina. Immunogenicity and safety. Medicina. 2010;70:215–222. [PubMed] [Google Scholar]
- Enria DA, Briggiler AM, Sanchez Z. Treatment of argentine hemorrhagic fever. Antivir Res. 2008;78:132–139. doi: 10.1016/j.antiviral.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D, Feldmann H. Vaccines for viral hemorrhagic fevers–progress and shortcomings. Curr Opin Virol. 2013;3:343–351. doi: 10.1016/j.coviro.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SA, Graham MB, Kuehnert MJ, Kotton CN, Srinivasan A, Marty FM, Comer JA, Guarner J, Paddock CD, DeMeo DL, Shieh WJ, Erickson BR, Bandy U, DeMaria A, Jr, Davis JP, Delmonico FL, Pavlin B, Likos A, Vincent MJ, Sealy TK, Goldsmith CS, Jernigan DB, Rollin PE, Packard MM, Patel M, Rowland C, Helfand RF, Nichol ST, Fishman JA, Ksiazek T, Zaki SR, Team, Li.T.R.I. Transmission of lymphocytic choriomeningitis virus by organ transplantation. N Engl J Med. 2006;354:2235–2249. doi: 10.1056/NEJMoa053240. [DOI] [PubMed] [Google Scholar]
- Fisher-Hoch SP, McCormick JB. Lassa fever vaccine. Expert Rev Vaccines. 2004;3:189–197. doi: 10.1586/14760584.3.2.189. [DOI] [PubMed] [Google Scholar]
- Greenbaum BD, Li OT, Poon LL, Levine AJ, Rabadan R. Viral reassortment as an information exchange between Viral segments. Proc Natl Acad Sci USA. 2012;109:3341–3346. doi: 10.1073/pnas.1113300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Holmes GP, McCormick JB, Trock SC, Chase RA, Lewis SM, Mason CA, Hall PA, Brammer LS, Perez-Oronoz GI, McDonnell MK, et al. Lassa fever in the United States. Investigation of a case and new guidelines for management. N Engl J Med. 1990;323:1120–1123. doi: 10.1056/NEJM199010183231607. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982;158:573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Isaacson M. Viral hemorrhagic fever hazards for travelers in Africa. Clin Infect Dis: Off Publ Infect Dis Soc Am. 2001;33:1707–1712. doi: 10.1086/322620. [DOI] [PubMed] [Google Scholar]
- Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang CF, Kemble G, Greenberg H. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (flumist) derived from cold-adapted A/ann arbor/6/60. Virology. 2003;306:18–24. doi: 10.1016/s0042-6822(02)00035-1. [DOI] [PubMed] [Google Scholar]
- Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- Karki S, Li MM, Schoggins JW, Tian S, Rice CM, MacDonald MR. Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity. PloS One. 2012;7:e37398. doi: 10.1371/journal.pone.0037398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore PE, Ksiazek TG, Rollin PE, Mills JN, Villagra MR, Montenegro MJ, Costales MA, Paredes LC, Peters CJ. Treatment of Bolivian hemorrhagic fever with intravenous ribavirin. Clin Infect Dis: Off Publ Infect Dis Soc Am. 1997;24:718–722. doi: 10.1093/clind/24.4.718. [DOI] [PubMed] [Google Scholar]
- Kunz S. The role of the vascular endothelium in arenavirus haemorrhagic fevers. Thromb Haemost. 2009;102:1024–1029. doi: 10.1160/TH09-06-0357. [DOI] [PubMed] [Google Scholar]
- Le Nouen C, Brock LG, Luongo C, McCarty T, Yang L, Mehedi M, Wimmer E, Mueller S, Collins PL, Buchholz UJ, DiNapoli JM. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc Natl Acad Sci USA. 2014;111:13169–13174. doi: 10.1073/pnas.1411290111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashevich IS. Advanced vaccine candidates for Lassa fever. Viruses. 2012;4:2514–2557. doi: 10.3390/v4112514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashevich IS, Patterson J, Carrion R, Moshkoff D, Ticer A, Zapata J, Brasky K, Geiger R, Hubbard GB, Bryant J, Salvato MS. A live attenuated vaccine for Lassa fever made by reassortment of Lassa and Mopeia viruses. J Virol. 2005;79:13934–13942. doi: 10.1128/JVI.79.22.13934-13942.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashevich IS, Carrion R, Jr, Salvato MS, Mansfield K, Brasky K, Zapata J, Cairo C, Goicochea M, Hoosien GE, Ticer A, Bryant J, Davis H, Hammamieh R, Mayda M, Jett M, Patterson J. Safety, immunogenicity, and efficacy of the ML29 reassortant vaccine for Lassa fever in small non-human primates. Vaccine. 2008;26:5246–5254. doi: 10.1016/j.vaccine.2008.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiztegui JI, McKee KT, Jr, Barrera Oro JG, Harrison LH, Gibbs PH, Feuillade MR, Enria DA, Briggiler AM, Levis SC, Ambrosio AM, Halsey NA, Peters CJ. Protective efficacy of a live attenuated vaccine against Argentine hemorrhagic fever. AHF Study Group. J Infect Dis. 1998;177:277–283. doi: 10.1086/514211. [DOI] [PubMed] [Google Scholar]
- Martinez-Sobrido L, Zuniga EI, Rosario D, Garcia-Sastre A, de la Torre JC. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2006;80:9192–9199. doi: 10.1128/JVI.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sobrido L, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J Virol. 2007;81:12696–12703. doi: 10.1128/JVI.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sobrido L, Emonet S, Giannakas P, Cubitt B, Garcia-Sastre A, de la Torre JC. Identification of amino acid residues critical for the anti-interferon activity of the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2009;83:11330–11340. doi: 10.1128/JVI.00763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JB, Fisher-Hoch SP. Lassa fever. Curr Top Microbiol Immunol. 2002;262:75–109. doi: 10.1007/978-3-642-56029-3_4. [DOI] [PubMed] [Google Scholar]
- McKee KT, Jr, Huggins JW, Trahan CJ, Mahlandt BG. Ribavirin prophylaxis and therapy for experimental argentine hemorrhagic fever. Antimicrob Agents Chemother. 1988;32:1304–1309. doi: 10.1128/aac.32.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee KT, Jr, Oro JG, Kuehne AI, Spisso JA, Mahlandt BG. Candid No. 1 Argentine hemorrhagic fever vaccine protects against lethal Junin virus challenge in rhesus macaques. Intervirology. 1992;34:154–163. doi: 10.1159/000150276. [DOI] [PubMed] [Google Scholar]
- Mueller S, Coleman JR, Papamichail D, Ward CB, Nimnual A, Futcher B, Skiena S, Wimmer E. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28:723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Wada K, Wada Y, Doi H, Kanaya S, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases. Nucleic Acids Res. 1996;24:214–215. doi: 10.1093/nar/24.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales A, Baker SF, Ortiz-Riano E, Dewhurst S, Topham DJ, Martinez-Sobrido L. Influenza A virus attenuation by codon deoptimization of the NS gene for vaccine development. J Virol. 2014;88:10525–10540. doi: 10.1128/JVI.01565-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschlager S, Flatz L. Vaccination strategies against highly pathogenic arenaviruses: the next steps toward clinical trials. PLoS Pathog. 2013;9:e1003212. doi: 10.1371/journal.ppat.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Riano E, Cheng BY, de la Torre JC, Martinez-Sobrido L. The C-terminal region of lymphocytic choriomeningitis virus nucleoprotein contains distinct and segregable functional domains involved in NP-Z interaction and counteraction of the type I interferon response. J Virol. 2011;85:13038–13048. doi: 10.1128/JVI.05834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Riano E, Cheng BY, de la Torre JC, Martinez-Sobrido L. D471G mutation in LCMV-NP affects its ability to self-associate and results in a dominant negative effect in viral RNA. Synth Virus. 2012a;4:2137–2161. doi: 10.3390/v4102137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Riano E, Cheng BY, de la Torre JC, Martinez-Sobrido L. Self-association of lymphocytic choriomeningitis virus nucleoprotein is mediated by its N-terminal region and is not required for its anti-interferon function. J Virol. 2012b;86:3307–3317. doi: 10.1128/JVI.05503-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Riano E, Cheng BY, Carlos de la Torre J, Martinez-Sobrido L. Arenavirus reverse genetics for vaccine development. J Gen Virol. 2013;94:1175–1188. doi: 10.1099/vir.0.051102-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991–998. doi: 10.1056/NEJMoa073785. [DOI] [PubMed] [Google Scholar]
- Pasqual G, Burri DJ, Pasquato A, de la Torre JC, Kunz S. Role of the host cell’s unfolded protein response in arenavirus infection. J Virol. 2011;85:1662–1670. doi: 10.1128/JVI.01782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci USA. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinschewer DD, Perez M, Sanchez AB, de la Torre JC. Recombinant lymphocytic choriomeningitis virus expressing vesicular stomatitis virus glycoprotein. Proc Natl Acad Sci USA. 2003;100:7895–7900. doi: 10.1073/pnas.1332709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popkin DL, Teijaro JR, Lee AM, Lewicki H, Emonet S, de la Torre JC, Oldstone M. Expanded potential for recombinant trisegmented lymphocytic choriomeningitis viruses: protein production, antibody production, and in vivo assessment of biological function of genes of interest. J Virol. 2011;85:7928–7932. doi: 10.1128/JVI.00486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pythoud C, Rodrigo WW, Pasqual G, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKepsilon. J Virol. 2012;86:7728–7738. doi: 10.1128/JVI.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pythoud C, Rothenberger S, Martinez-Sobrido L, de la Torre JC, Kunz S. Lymphocytic Choriomeningitis virus differentially affects the virus-induced type i interferon response and mitochondrial apoptosis mediated by RIG-I/MAVS. J Virol. 2015;89:6240–6250. doi: 10.1128/JVI.00610-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna L, Anand KK, Mohankumar KM, Ranga U. Codon optimization of the tat antigen of human immunodeficiency virus type 1 generates strong immune responses in mice following genetic immunization. J Virol. 2004;78:9174–9189. doi: 10.1128/JVI.78.17.9174-9189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo WW, de la Torre JC, Martinez-Sobrido L. Use of single-cycle infectious lymphocytic choriomeningitis virus to study hemorrhagic fever arenaviruses. J Virol. 2011;85:1684–1695. doi: 10.1128/JVI.02229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo WW, Ortiz-Riano E, Pythoud C, Kunz S, de la Torre JC, Martinez-Sobrido L. Arenavirus nucleoproteins prevent activation of nuclear factor kappa B. J Virol. 2012;86:8185–8197. doi: 10.1128/JVI.07240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek JM, Kunz S. Cell entry by human pathogenic arenaviruses. Cell Microbiol. 2008;10:828–835. doi: 10.1111/j.1462-5822.2007.01113.x. [DOI] [PubMed] [Google Scholar]
- Rojek JM, Perez M, Kunz S. Cellular entry of lymphocytic choriomeningitis virus. J Virol. 2008;82:1505–1517. doi: 10.1128/JVI.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer IJ, Miller R, Stroher U, Knust B, Nichol ST, Rollin PE. Notes from the field: a cluster of lymphocytic choriomeningitis virus infections transmitted through organ transplantation-Iowa, 2013. Am J Transpl. 2014;14:1459. doi: 10.1111/ajt.12802. [DOI] [PubMed] [Google Scholar]
- Smith DW. Problems of translating heterologous genes in expression systems: the role of tRNA. Biotechnol Prog. 1996;12:417–422. doi: 10.1021/bp950056a. [DOI] [PubMed] [Google Scholar]
- Snell N. Ribavirin therapy for lassa fever. Practitioner. 1988;232:432. [PubMed] [Google Scholar]
- Snyder MH, Betts RF, DeBorde D, Tierney EL, Clements ML, Herrington D, Sears SD, Dolin R, Maassab HF, Murphy BR. Four viral genes independently contribute to attenuation of live influenza A/Ann Arbor/6/60 (H2N2) cold-adapted reassortant virus vaccines. J Virol. 1988;62:488–495. doi: 10.1128/jvi.62.2.488-495.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strecker T, Eichler R, Meulen J, Weissenhorn W, Dieter Klenk H, Garten W, Lenz O. Lassa virus Z protein is a matrix protein and sufficient for the release of virus-like particles [corrected] J Virol. 2003;77:10700–10705. doi: 10.1128/JVI.77.19.10700-10705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenbusch M, Grunwald T, Niezold T, Storcksdieck Genannt Bonsmann, M. Hannaman D, Norley S, Uberla K. Codon-optimization of the hemagglutinin gene from the novel swine origin H1N1 influenza virus has differential effects on CD4(+) T-cell responses and immune effector mechanisms following DNA electroporation in mice. Vaccine. 2010;28:3273–3277. doi: 10.1016/j.vaccine.2010.02.090. [DOI] [PubMed] [Google Scholar]
- Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife. 2014;3:e04531. doi: 10.7554/eLife.04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata S, de la Torre JC. Arenavirus budding. Adv Virol. 2011;2011:180326. doi: 10.1155/2011/180326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadava A, Ockenhouse CF. Effect of codon optimization on expression levels of a functionally folded malaria vaccine candidate in prokaryotic and eukaryotic expression systems. Infect Immun. 2003;71:4961–4969. doi: 10.1128/IAI.71.9.4961-4969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Skiena S, Futcher B, Mueller S, Wimmer E. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc Natl Acad Sci USA. 2013;110:9481–9486. doi: 10.1073/pnas.1307473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu WJ, Peng SW, Sun XY, Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J Virol. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


