Abstract
Many enveloped RNA viruses utilize lipid rafts for the assembly of progeny virions, but the role of cholesterol, a major component of rafts, on paramyxovirus budding and virion formation is controversial. In this study, we analyzed the effects of FDA-approved cholesterol-reducing agents, gemfibrozil and lovastatin, on raft formation and assembly of human parainfluenza virus type 1 (hPIV1) and Sendai virus (SeV). Treatment of the human airway epithelial A549 cells with the agents, especially when combined, significantly decreased production of infectious hPIV1 and SeV. Mechanistic analysis indicated that depletion of cellular cholesterol reduced cell surface accumulation of envelope glycoproteins and association of viral matrix and nucleocapsids with raft membrane, which resulted in impaired virus budding and release from the cells. These results indicate that cellular cholesterol is required for assembly and formation of type 1 parainfluenza viruses and suggest that cholesterol could be an attractive target for antiviral agents against hPIV1.
Keywords: Type I parainfluenza virus, assembly, cholesterol, lipid rafts, antiviral therapy
1. Introduction
Acute respiratory infection (ARI) is one of the leading causes of mortality among infants and young children (Liu et al., 2015; Mao et al., 2012). A recent study reports that among 6.3 million children who died before the age of five in 2013, 51.8% died of infectious diseases, and pneumonia was the leading cause of these deaths (Liu et al., 2015). Viruses are the major cause of ARI, but vaccines or effective antivirals are available only for a limited number of viral infections. There are no vaccines or drugs to combat infections caused by major respiratory viruses, such as parainfluenza viruses (PIV) and respiratory syncytial virus (RSV). The available therapeutics against respiratory viruses, such as influenza neuraminidase inhibitors are effective, but the drugs target viral proteins and quick emergence of resistant viruses restrict their usage (Govorkova, 2013). Therefore, antivirals that target host factors that are critical for virus spread are desirable for the control of respiratory RNA viruses.
Human parainfluenza virus type 1 (hPIV1) is one of the major causes of croup in the younger population, and posses the threat of occasional outbreak in immunocompromised patients (Harvala et al., 2012; Sydnor et al., 2012). As a prototype virus, Sendai virus (SeV), a murine counterpart of hPIV1, has been studied extensively to determine the significant molecular and biological properties of PIVs (Henrickson, 2003). Parainfluenza viruses are enveloped and contain non-segmented negative sense RNAs as viral genomes, which are encapsidated with nucleoprotein (NP) and associated with the polymerase complex composed of phosphoprotein (P) and large polymerase (L) proteins to form viral nucleocapsid (vRNP). Viral membrane is derived from the host plasma membrane and is associated with two surface glycoproteins hemagglutinin-neuraminidase (HN) and fusion (F) proteins at the surface of the virion, and matrix (M) protein inside the membrane (Henrickson, 2003). Viral genome replication and transcription takes place entirely in the cytoplasm, and newly synthesized viral components are transported and assembled at the budding sites in the plasma membrane, where progeny virions are produced (Takimoto and Portner, 2004). Specific interactions among viral glycoproteins, M and vRNP are required for efficient infectious virus production (Ali and Nayak, 2000; Harrison et al., 2010). Particularly, the cytoplasmic tail of F protein plays a major role in virus assembly at the plasma membrane (El Najjar et al., 2014; Stone and Takimoto, 2013; Takimoto and Portner, 2004).
Virus assembly is considered to take place at lipid raft microdomains on the plasma membrane. Lipid rafts are small microdomains ranging from 10–200 nm in size, enriched in sphingolipids and cholesterol (Brown and London, 2000). Lipid rafts are resistant to detergent solubilization at 4°C, and viral protein association with lipid rafts has been shown by utilizing this unique property (Brown and London, 2000). Previous studies on various paramyxoviruses suggested that viral proteins preferentially localize at lipid rafts, and this association was required for efficient infectious virus production (Brown et al., 2002a; Brown et al., 2004; Brown et al., 2002b; Gosselin-Grenet et al., 2006; Imhoff et al., 2007; Laliberte et al., 2006; Manie et al., 2000; Ravid et al., 2010; Vincent et al., 2000). It has been shown that many enveloped RNA viruses commonly utilize lipid rafts for virus assembly and budding (Chang et al., 2012; El Najjar et al., 2014; Harrison et al., 2010; Lyles, 2013). Cellular cholesterol is an essential component of lipid rafts, but the requirement of cholesterol for virus assembly has not been fully elucidated. For example, extraction of cholesterol from Newcastle Disease Virus (NDV)-infected cells by methyl-β-cyclodextrin (MBCD) did not inhibit the release of virus particles from cells, although progeny virions were abnormal in protein composition and particle density (Laliberte et al., 2006). Similarly, cholesterol extraction from RSV-infected cells did not inhibit assembly or release of virion particles, but the progeny virions with low cholesterol levels in the viral membranes were poorly infectious (Chang et al., 2012). These studies suggest that, in the case of NDV and RSV, cholesterol is not required for virus budding and release, but affected infectivity of the released virions.
In this study, we have analyzed the role of cholesterol in infection, replication and assembly of type I parainfluenza viruses. We utilized the FDA approved cholesterol reducing agents, gemfibrozil (Gem) and lovastatin (Lov) for our experiments. Gem is a lipid lowering fibrate which causes intracellular cholesterol efflux by activating the nuclear hormone receptor peroxisome proliferators-activated receptor-α (PPAR-α) decreasing intracellular storage of cholesterol (Roy and Pahan, 2009). Lov lowers cholesterol by inhibiting the 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGCR) enzyme, which is essential for the cholesterol biosynthesis process (Maron et al., 2000). We determined the effect of the cholesterol reducing agents on infectious virus production from human alveolar epithelial (A549) cells, and analyzed the mechanism of action. Our data indicate that these drugs, when combined, are effective in reducing both cellular cholesterol and release of parainfluenza virions. Interestingly, unlike RSV or NDV, membrane cholesterol is required for viral budding and release from infected cells, indicating that PIV1 relies on cholesterol and raft membranes for virus assembly, budding and release.
2. Results
2.1. Cholesterol reducing agents inhibit infectious virus production
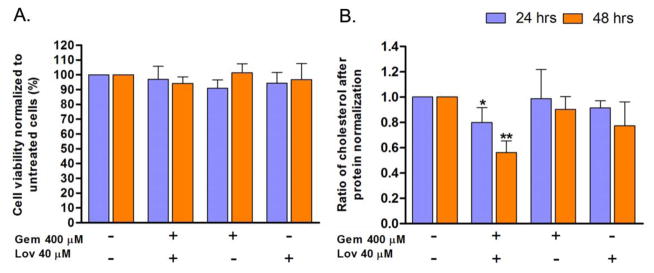
To determine the role of cholesterol on parainfluenza virus growth, we used FDA-approved cholesterol reducing agents Gem and Lov to deplete cellular cholesterol in human airway epithelial A549 cells. First, we evaluated the toxicity of these drugs by treating the cells with various doses of the drugs. The 25% lethal concentration (LC25) for Gem and Lov were 1,392 μM and 146 μM, respectively. Treatment of cells with Gem 400 μM and Lov 40 μM, individually or in combination did not affect viability of the cells by more than 10% in 48 h as determined by trypan blue exclusion assay (Fig. 1A). We also determined the cellular cholesterol level after drug-treatment. Individual treatment of the cells with Gem at 400 μM or Lov at 40 μM did not cause significant reduction in the total cellular cholesterol level (Fig. 1B). However, treatment of the cells with both drugs significantly reduced cellular cholesterol levels by 20% and 44% at 24 and 48 h, respectively (Fig. 1B). These results indicate that the drugs are capable of reducing total cellular cholesterol levels in cultured human airway A549 cells, especially when the drugs are combined. It also suggests that treatment of cells with two drugs exhibiting different mechanisms of action is more effective for efficient reduction of total cellular cholesterol.
Figure 1.
Effect of cholesterol reducing agents on cell viability and cholesterol level. A549 cells were treated with Gem 400 μM and/or Lov 40 μM for 24 or 48 h. (A) Cell viability was compared to untreated cells as measured by trypan blue cell viability assay (n=3). (B) Cellular cholesterol levels were determined by Amplex Red Cholesterol assay. Data were normalized to protein levels as determined by bicinchoninic acid assay (n=3). Gem: gemfibrozil, Lov: lovastatin. *; P<0.05, **; P<0.01 (Student t-test).
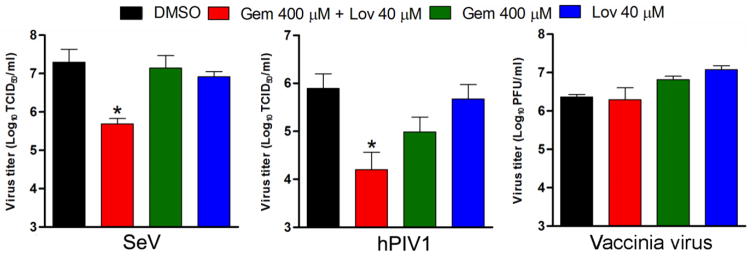
Next, we analyzed the production of infectious SeV and hPIV1 virus from cells treated with the drugs (Gem 400 μM and Lov 40 μM) that effectively reduced cellular cholesterol without affecting cell viability (Fig. 1). The cells were pretreated with the drugs individually or in combination for 24 h and infected with SeV or hPIV1. The infected cells were cultured in the presence of the drugs for an additional 24 h, and the released infectious virions were quantitated. We found that treatment of cells with both Gem and Lov significantly reduced infectious SeV production (97.5% reduction), while treatment with individual drugs showed only a subtle reduction in infectious virus production (Fig. 2). Similarly, treatment of cells with both drugs resulted in a 98% reduction in hPIV1 virus production. While treatment with Gem alone resulted in 87.5% reduction, Lov alone did not significantly reduce infectious virus production (Fig. 2). The inhibitory effect of the drugs on SeV and hPIV1 production correlated with the reduced cellular cholesterol levels (Fig. 1B). As a control, we tested the effect of the drugs on production of infectious intracellular or extracellular vaccinia virus. In contrast to the results of PIV1s, cholesterol reducing agents did not affect production of infectious vaccinia virus, suggesting a specific effect on the release of infectious PIV1s (Fig. 2). Taken together, these results strongly suggest the critical role of cellular cholesterol in the production of infectious SeV and hPIV1.
Figure 2.
Cholesterol is required for infectious virus production of SeV and hPIV1. Cells untreated or treated with the drugs for 24 h were infected with either SeV, hPIV1 or vaccinia virus and cultured in the presence of drugs for an additional 24 h. Titers of released viruses are shown (n=3). *; P<0.05 (Student t-test).
2.2. Reduction of cholesterol levels does not affect cellular susceptibility to PIV1 infection
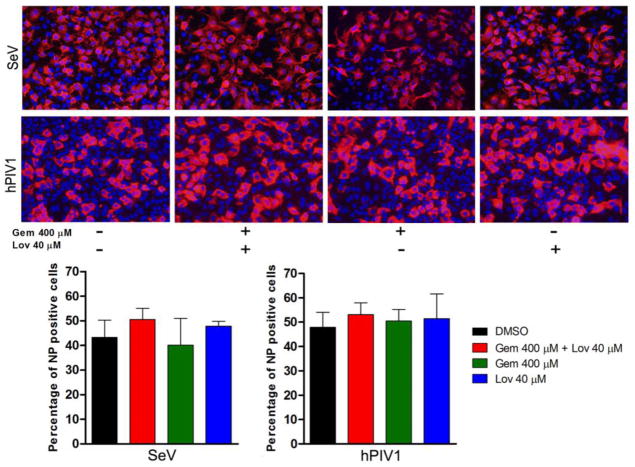
Next, we systematically analyzed the role of cholesterol in the life cycle of SeV and hPIV1. For some enveloped viruses such as HIV, association of cellular receptors to membrane rafts has been shown to be required for viral entry (Chazal and Gerlier, 2003). We first determined if cellular cholesterol is required for the viral entry process. A549 cells were treated with Gem 400 μM and Lov 40 μM for 24 h and infected with the viruses at a multiplicity of infection (MOI) of 0.5 for 24 h. Cells were fixed and the infectivity of the viruses was determined by immunostaining using specific anti-NP monoclonal antibodies (mAb). We detected no difference in virus infectivity between cells untreated or treated with Gem and Lov (Fig. 3). These results indicate that the 20 % depletion of cholesterol achieved at 24 h post treatment (Fig. 1B) has no effect on the entry process of hPIV1 and SeV.
Figure 3.
Cholesterol depletion does not prevent SeV or hPIV1 entry. A549 Cells untreated or treated with the drugs were infected with the viruses at a MOI of 0.5 and cultured for 24 h in medium without trypsin. Cells were fixed and viral NP was detected by IF assay using specific antibodies. The infected NP positive cells were counted and presented as bar graphs at the bottom (n=3).
2.3. Effect of cholesterol reducing agents on virus protein synthesis
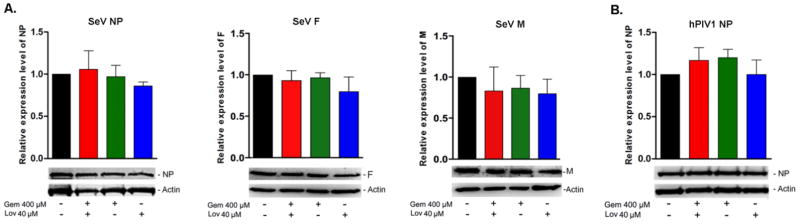
Since cholesterol reducing agents did not inhibit virus entry in A549 cells, we determined whether reduced infectious virus production was due to decreased viral protein synthesis. Cells treated with the drugs were infected with the viruses at a MOI of 1, and the quantities of viral NP, F and M in cell lysates were determined by Western blotting analysis. Treatment of cells with either Gem 400 μM or Lov 40 μM or in combination did not reduce NP, F and M synthesis during SeV infection (Fig. 4A). Similarly, reduction of cellular cholesterol did not affect hPIV1 NP synthesis (Fig. 4B). We were unable to analyze hPIV1 M and F due to the lack of antibodies suitable for Western blot analysis. These results indicate that reduction of infectious virus production from treated cells was not due to reduced virus entry or protein synthesis.
Figure 4.
Cholesterol reducing agents do not affect protein synthesis of SeV or hPIV1. A549 cells untreated or treated with the drugs were infected with SeV (A) or hPIV1 (B) at a MOI of 3, and cultured in the presence of drugs for 24 h. SeV NP, F and M and hPIV1 NP in the cell lysates were detected by Western blot analysis using specific antibodies. Relative expression levels of the proteins are presented in bar graphs (n=3).
2.4. Cholesterol is required for virion production from infected cells
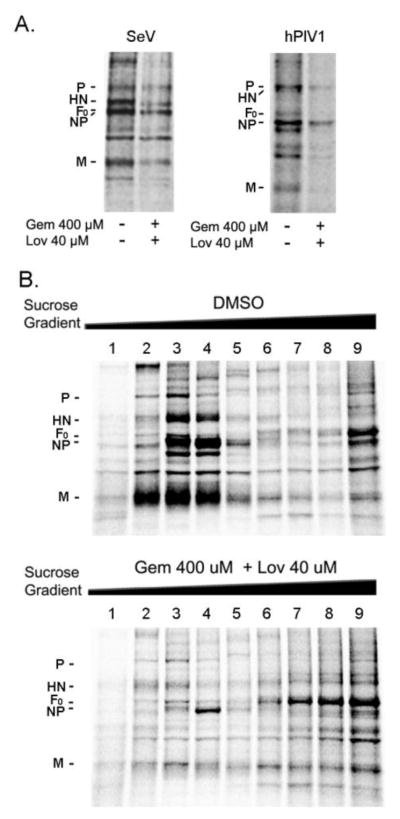
Next, we examined the formation and release of progeny virions from drug-treated cells. A549 cells treated with both Gem and Lov were infected with SeV or hPIV1, and labeled with 35S-Met/Cys for 16 h. Labeled virions released into culture medium were purified and analyzed by SDS-PAGE. The release of virions from drug-treated cells was significantly reduced in both SeV-and hPIV1-infected cells (Fig. 5A). The quantification of NP revealed a 66.5% and 88.5% decrease in total SeV and hPIV1 production from the treated cells, respectively. To further characterize the virions released from drug-treated cells, the labeled culture supernatants were ultracentrifuged over a continuous 5–40% sucrose gradient, and viral proteins in each fraction were analyzed by SDS-PAGE. From untreated cells, SeV particles containing all the major structural proteins were recovered from fractions 3 and 4 of low sucrose density (1.02 g/cm3 – 1.05 g/cm3) (Fig. 5B upper panel). However, few virus specific proteins were detected in the same fractions released from the cells treated with both drugs (Fig. 5B lower panel). These results indicate that, unlike those reported for NDV and RSV, depletion of cholesterol significantly impairs assembly and release of SeV and hPIV1 from infected cells.
Figure 5.
Cholesterol is required for assembly and release of progeny virions. (A) Cells either untreated or treated with the drugs were infected with SeV or hPIV1 and labeled with 35S-Met/Cys for 16 h. Released SeV and hPIV1 virions were purified and analyzed by SDS-PAGE. (B) Labeled virions were ultracentrifuged through a continuous (5–50%) sucrose gradient. Viral proteins in each fraction were analyzed by SDS-PAGE and autoradiography. Fractions 2–4 include progeny virions.
2.5. Depletion of cellular cholesterol reduces cell surface accumulation of viral glycoproteins
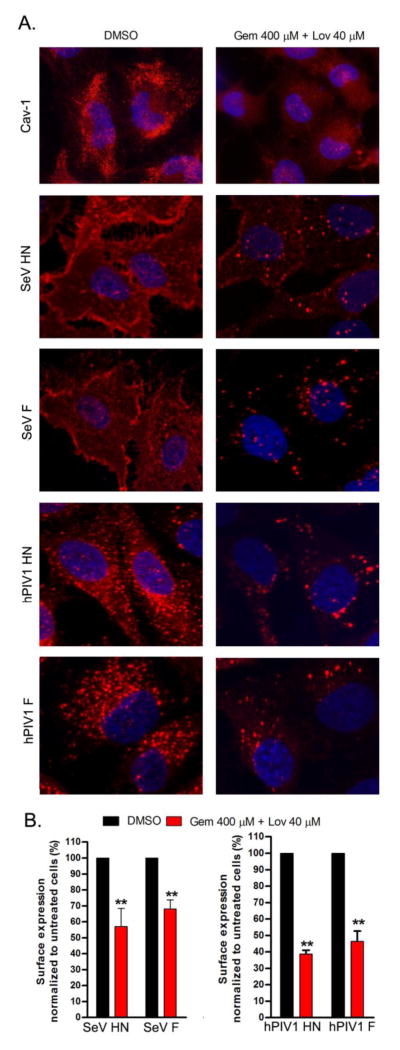
Our previous studies of SeV and hPIV1 showed that viral glycoproteins play a crucial role in parainfluenza virus assembly (Takimoto and Portner, 2004). Notably, the cytoplasmic tail of the F protein plays a major role for accumulation of viral structural components at plasma membrane assembly sites (Stone and Takimoto, 2013). Since paramyxoviruses are assembled at the plasma membrane raft structures, we next analyzed the effect of cholesterol depletion on cell surface expression of viral HN and F proteins by immunofluorescence (IF) assay. First, we determined the effect of cholesterol depletion on cell surface distribution of Cav-1, a lipid raft marker protein. We detected punctate localization of Caveolin-1 (Cav-1) on the surface of untreated cells, which was less evident in cells treated with the drugs (Fig. 6A). These results suggest that drug treatment prevents the proper localization of Cav-1 by disrupting the integrity of lipid rafts. Similarly, the surface expression of SeV and hPIV1 glycoproteins was significantly decreased in the presence of cholesterol reducing agents (Fig. 6). Viral HN and F proteins were also quantified by ELISA. Consistent with the results of the IF analysis, drug treatment significantly reduced cell surface expression of SeV HN and F by 42% and 32%, respectively (Fig. 6B). Similarly, surface expression of hPIV1 HN and F was reduced by 61% and 54% in the treated cells, respectively (Fig. 6B). These results suggest that cholesterol is a critical component for accumulation of viral glycoproteins at the surface of infected cells.
Figure 6.
Reduced expression of viral glycoproteins at the surface of drug-treated cells. (A) Cells untreated or treated with drugs were infected or mock infected (for the detection of cav-1), and analyzed for surface distribution of cav-1 and viral HN and F expression. Fixed and unpermeabilized cells were utilized for IF analysis to detect indicated proteins. (B) The amounts of viral glycoproteins at the surface of infected cells were quantitated by ELISA. (n=3). **; P<0.01 (student t test).
2.6. Depletion of cellular cholesterol reduces viral M proteins and nucleocapsid association with the lipid rafts
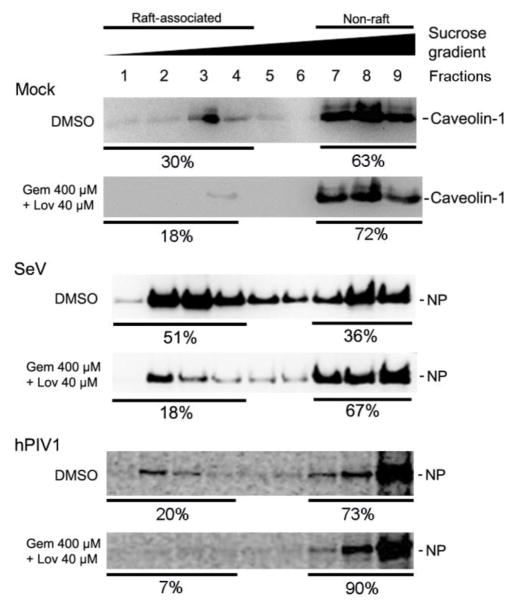
Next, we determined the effect of cholesterol drugs on lipid raft association of vRNPs by raft flotation assay. Lipid rafts were isolated based on their unique property of resistance to detergent solubilization at 4°C and buoyancy at low-sucrose densities (Chazal and Gerlier, 2003; Manie et al., 2000). In untreated cells, 30% of Cav-1 was associated with lipid rafts. Drug treatment decreased raft associated Cav-1 to 18% (Fig. 7), indicating that depletion of cholesterol by drug treatment resulted in disruption of lipid raft integrity. In untreated cells, 51% of SeV NP was detected in raft-associated fractions (Fig 7). In contrast, only 18% of NP was associated with lipid raft fractions in drug treated cells, showing reduced association of vRNP with raft membranes. Similarly, drug treatment reduced raft association of hPIV1 NP from 20% to 7% (Fig. 7).
Figure 7.
Reduced viral nucleocapsid association with lipid rafts after cholesterol depletion. Cells untreated or treated with drugs were infected with mock, SeV or hPIV1. Membrane floatation assay was performed to quantitate the raft-associated NP in each fraction. Caveolin-1 and SeV NP were detected by immunoblotting. Cells infected with hPIV1 were radiolabeled with 35S-Met/Cys and labeled NPs in each fraction were immunoprecipitated and resolved by SDS-PAGE. The numbers show percentage of the proteins recovered from raft (1–4) and non-raft (7–9) fractions.
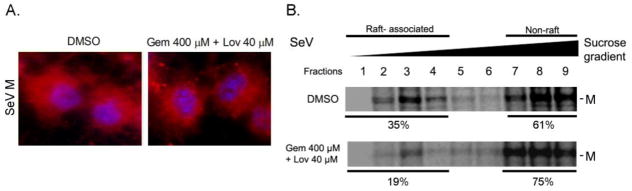
Previous studies have suggested that M plays a major role in the assembly of paramyxoviruses (El Najjar et al., 2014). M interacts with glycoproteins and is required for concentration of NP in the plasma membrane at specific assembly sites. To analyze whether cholesterol depletion affects M mediated virus assembly process, we next determined M distribution in infected cells by IF assay. The diffused M distribution detected in untreated cells was also observed in drug-treated cells (Fig. 8A). However, similar to NP, M association with the raft membrane was decreased from 35% to 19% in cholesterol-depleted cells (Fig. 8B). These results indicate that cholesterol is required for the formation of virus assembly site to accumulate structural components for ordered assembly and production of type I parainfluenza viruses.
Figure 8.
Reduced M association with lipid rafts in cholesterol depleted cells. (A) Cells untreated or treated with drugs were infected with SeV and intracellular M distribution (red) was determined by IF. Cells were counterstained with DAPI. (B) Cells untreated or treated with the drugs were infected with SeV and radiolabeled with 35S-Met/Cys. Membrane floatation assay was performed and M in each fraction was immunoprecipitated using specific mAb. The numbers show percentage of the proteins recovered from raft (1–4) and non-raft (7–9) fractions.
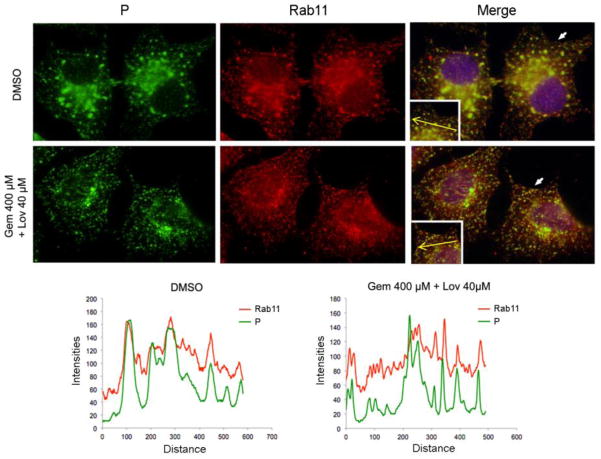
2.7. Reduced cellular cholesterol does not affect vRNP interaction with Rab11
Our lab has previously shown that Rab11-mediated recycling endosomes play a critical role in trafficking and transport of SeV and hPIV1 vRNPs to the plasma membrane assembly sites (Chambers and Takimoto, 2010; Stone et al., 2016). Reduced accumulation of vRNP at the plasma membrane could be explained by impaired interaction with Rab11, since depletion of cholesterol has been reported to result in redistribution of Rab11 (Holtta-Vuori et al., 2002; Takahashi et al., 2007). Therefore, we next determined whether cholesterol depletion affects vRNP association with Rab11 by IF assay (Fig. 9). Punctate staining of SeV vRNP and Rab11 were detected throughout the cytoplasm of infected cells, with a strong perinuclear staining pattern in both untreated and drug treated cells (Fig. 9). Consistent with our previous published data, vRNP and Rab11 co-localized in the untreated cells. Similarly, co-localization of Rab11 and P was detected in drug-treated cells. These results suggest that depletion in cellular cholesterol did not have a strong effect on vRNP association with Rab11 and their distribution in infected cells. The overall data described above suggests that lipid raft integrity at the plasma membrane is required for accumulation and interaction between viral glycoproteins, M and vRNPs, and is essential for efficient assembly and release of progeny PIV1 virions.
Figure 9.
Depletion of cholesterol does not affect vRNP interaction with Rab11. (A) Cells untreated or treated with the drugs were infected with SeV. Viral P proteins and cellular Rab11 were detected by IF assay using specific antibodies. Histograms indicate the fluorescence intensities of Rab11 and P in the area represented by the yellow arrow in the images.
3. Discussion
In this study, we evaluated the role of cholesterol in infection, replication and assembly of type I parainfluenza viruses using FDA-approved cholesterol reducing agents. Cholesterol is a major component of lipid raft structures, which are known to be involved in various stages of enveloped viral infections, including entry, replication, assembly and budding (Chazal and Gerlier, 2003; Takahashi and Suzuki, 2011). Our data indicate that cholesterol depletion significantly inhibits the release of infectious SeV and hPIV1 from infected A549 cells. Further mechanistic analysis revealed that these drugs did not affect viral entry (Fig. 3) or viral protein synthesis (Fig. 4), but prevented virion formation and release from infected cells (Fig. 5). Because depletion of cholesterol disrupts the integrity of lipid rafts, which are the assembly sites of paramyxovirus and other enveloped RNA viruses (Ono and Freed, 2005), it is likely that accumulation of viral structural components at raft structures is essential for budding and formation of progeny PIV1 virions. Previous studies with paramyxoviruses, including NDV, RSV, MeV, and SeV showed that major viral structural proteins associate with lipid rafts in infected cells (Ali and Nayak, 2000; Brown et al., 2004; Chang et al., 2012; Gosselin-Grenet et al., 2006; Laliberte et al., 2006; Manie et al., 2000; Pohl et al., 2007; Sanderson et al., 1995; Vincent et al., 2000). Interestingly, unlike SeV or hPIV1, budding and release of NDV and RSV from cholesterol-depleted cells were not inhibited, although they had irregular protein composition, abnormal particle density, and reduced infectivity (Chang et al., 2012; Laliberte et al., 2006). These results reflect the differences in raft dependency for virus budding/release between parainfluenza viruses and RSV or NDV. It is likely that SeV and hPIV1 strictly depend on lipid raft integrity for viral protein interactions to bud and form progeny virions, which may not be the case for NDV or RSV.
It is unclear what triggers paramyxovirus budding and formation of progeny virions. Using expressed proteins, we previously showed that SeV M and F, but not HN are capable of producing virus-like particles when expressed alone (Takimoto et al., 2001). SeV HN and F can be detected in detergent resistant membrane (DRM) fractions of flotation gradients even when expressed alone (Sanderson et al., 1995). However, SeV M protein, which is the major driving force for virus budding is associated with DRM only when it was co-expressed with either HN or F (Ali and Nayak, 2000). It was also shown that specific interaction with F is required for recruitment of M to DRM (Ali and Nayak, 2000). Consistent with these findings, our lab has shown that the cytoplasmic tail sequence of F protein is critical for accumulation of HN and F at the plasma membrane, and in turn essential for M and NP interaction at the plasma membrane (Stone and Takimoto, 2013). Our results indicate that depletion of cholesterol significantly reduces surface expression of HN and F proteins (Fig. 6) and association of vRNP and M with raft membrane (Figs. 7 and 8). These data stress the role of cholesterol and raft integrity for accumulation of structural components to a confined area of the plasma membrane to form progeny virions. In the case of SeV and hPIV1, these interactions between structural components likely trigger the final steps in virus assembly and release, including budding and scission of virus particles.
Along with previous reports, this study highlights the importance of lipid rafts and the major raft component, cholesterol, on paramyxovirus infection and spread. Interestingly, some reports suggest that viruses modulate the biosynthesis of cholesterol during infection. A global transcriptional analysis of measles virus infected cells showed that most genes associated with the cholesterol biosynthesis pathway along with lipid rafts in the plasma membrane were down-regulated in persistently infected cells as compared to acutely infected cells (Robinzon et al., 2009). Similarly, it was reported that HMGCR and the low-density lipoprotein receptor that is involved in cholesterol homeostasis, are up-regulated during RSV infection, suggesting RSV infection may modulate cholesterol levels during virus infection for optimal growth (Yeo et al., 2009). In addition, up-regulation of the genes encoding enzymes in the cholesterol biosynthesis pathway was reported in cells infected with HIV and influenza viruses (van’t Wout et al., 2003), suggesting a possibility that viral regulation of sterol biosynthesis may affect the life cycle of many viruses.
In addition to its role in the cellular membrane, cholesterol is also known to be a major component of viral membranes. Quantitative analysis of the lipidomes of influenza virus envelope indicate that over 50% of influenza viral membrane lipids are cholesterol (Gerl et al., 2012). Cholesterol depletion from influenza viral membranes markedly reduces virus fusion activity and infection, showing that cholesterol in the viral membrane is crucial for virus stability and infectivity (Sun and Whittaker, 2003). Similarly, NDV or RSV released from cells depleted with cholesterol were reported to be unstable and irregular in structural components (Chang et al., 2012; Laliberte et al., 2006), stressing the essential role of cholesterol in virus assembly and stability.
Accumulating evidence supports the efficacy of statins and fibrates against different enveloped viral infections, such as hepatitis C virus (Ikeda et al., 2006), Andes virus (Petersen et al., 2014), RSV (Gower and Graham, 2001), influenza virus (Budd et al., 2007), and HIV (del Real et al., 2004), thereby exhibiting potential as broadly-reactive antivirals. Correspondingly, our PIV1 data support the idea that FDA-approved cholesterol reducing agents could be re-evaluated for drug repurposing as broadly-reactive antivirals. Our functional analysis described here unveiled the mechanism of how cholesterol drugs inhibit PIV1 growth. We showed that cholesterol reducing agents, gemfibrozil and lovastatin, disrupt lipid raft integrity and prevent specific viral protein interactions required for efficient virus assembly. Together with previous findings of other enveloped viruses, our data strongly suggest that cholesterol could be an attractive target for antiviral therapeutics against various clinically important respiratory RNA viruses.
4. Materials and Methods
4.1. Cells, viruses, and reagents
LLC-MK2 (ATCC, CCL-7) and A549 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Corning) supplemented with 8% fetal calf serum (FCS; Life Technologies), 1% GlutaMAX (Life Technologies) and 0.1% Gentamicin (Life Technologies). SeV (strain Enders) and hPIV1 (strain C-35) were grown in LLC-MK2 cells in DMEM supplemented with 0.15% bovine serum albumin and acetylated trypsin at 2 μg/ml for SeV or 1.5 μg/ml for hPIV1. Vaccinia virus (vA4-YFP) was grown in BSC-40 cells in DMEM containing 2.5% FCS. Gemfibrozil (Sigma) and lovastatin (Sigma) were dissolved in dimethyl sulfoxide (DMSO, Sigma) to make 200 mM and 20 mM stock solutions.
4.2. Assays to measure cytotoxicity, cholesterol level and protein concentrations
Cytotoxicity of cholesterol reducing agents was measured by trypan blue exclusion assay (Strober, 2001). Total cellular cholesterol and protein were measured by Amplex Red Cholesterol assay (Molecular Probes) and bicinchoninic acid (BCA) protein assay (PIERCE), respectively, according to the manufactures’ protocols.
4.3. Titration of released viruses
A549 cells were untreated or pretreated with Gem at 400 μM and/or Lov at 40 μM for 24 h, and infected with SeV and hPIV1 at a MOI of 3 or vaccinia virus at a MOI of 5. Cells were then cultured for 24 h in the presence of drugs at the same concentration. Supernatant containing released SeV and hPIV1 virions were treated with TPCK-treated trypsin and titrated by measuring TCID50 in MK2 cells. Titer of vaccinia virus was measured after lysing cells by freezing and thawing. Virus titer was measured by plaque assay using BSC-40 cells (Ausubel, 1995).
4.4. Immunofluorescence (IF) assay
A549 cells were untreated or pretreated with the drugs for 24 h and infected with SeV or hPIV1 at a MOI of 0.5 for the analysis of virus infectivity, and at a MOI of 3 for the analysis of surface expression of glycoproteins. Similarly, cells were infected with SeV at a MOI of 3 to determine the intracellular distribution of M and co-localization between Rab11 and P. After 24 h infection, cells were fixed with 4% paraformaldehyde (PFA) for 15 min. For the analysis of cell surface expression of viral glycoproteins and caveolin-1, fixed cells were incubated with anti-SeV HN (a cocktail of S16, M27 and M41), anti-SeV F (M49 and M38), anti-hPIV1 HN (P18, P24 and P37), anti-hPIV1 F (P12 and P38) mAbs, or anti-caveolin-1 rabbit antibody (abcam), followed by anti-mouse or anti-rabbit IgG conjugated with Texas Red (Life Technologies). For the analysis of virus infectivity and M distribution, fixed cells were first permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature (RT), and then reacted with anti-SeV NP (M52, WS16) or anti-hPIV1 NP (P19, P27, P35) mAbs, or anti-SeV M (M1) mAb, followed by anti-mouse IgG conjugated with Texas Red. To detect Rab11 and vRNP in infected cells, permeabilized cells were reacted with anti-Rab11 rabbit antibody (Cell Signaling) and anti-SeV P (M56) mAb, followed by anti-rabbit IgG conjugated with Texas Red and anti-mouse IgG-FITC. Fluorescent images were taken using an Olympus FV1000 confocal microscope or Olympus IX50 inverted fluorescence microscope.
4.5. Western blot analysis
To determine the effect of the drugs on viral protein synthesis, A549 cells were untreated or pretreated with Gem 400 μM and/or Lov 40 μM for 24 h, and infected with SeV and hPIV1 at a MOI of 1 for 24 h. Cells were lysed with Triton-X lysis buffer (20 mM HEPES [pH7.5], 1.5 mM MgCl2, 500 mM NaCl, 0.2 mM EDTA, 1% Triton-X 100 and 20% glycerol), and viral and cellular proteins were analyzed by Western blot analysis using PVDF membranes. Anti-SeV NP (M52, WS16), anti-SeV M (M1), anti-F (M16), anti-hPIV1 NP (P19, P27, P35) mAbs or anti-β-actin mAb (Cell Signaling) were used to detect the proteins (Chambers and Takimoto, 2010; Stone and Takimoto, 2013). Band intensities were quantitated using BioRad Quantity One software.
4.6. Virion production from cells
A549 cells were untreated or pretreated with the drugs for 12 h, and infected with SeV and hPIV1 at a MOI 3 for 1 h. At 12 h post infection, cells were cultured in labeling medium containing 50 μCi of 35S-Met/Cys (PerkinElmer) and the drugs at the same concentrations as previously used for an additional 16 h. Labeled progeny virions were purified by ultracentrifuge over 20% sucrose for SeV and 30% glycerol in PBS for hPIV1 and analyzed by SDS-PAGE. Fixed and dried gels were exposed onto a phosphor screen and visualized using Personal Molecular Image (Bio-Rad).
4.7. Sucrose gradient analysis of released virions
One ml of radiolabeled virions was applied over 10 ml of 5 to 40% sucrose gradient in PBS and ultracentrifuged at 27,600 rpm for 18 h at 4°C in a SW44Ti rotor (Beckman). Nine equal fractions were collected from the top and an aliquot of each fraction was removed for density measurement by hand held refractometer (R-5000, Atago, U.S.A). The remainder of each fraction was diluted with 3 volumes of PBS and centrifuged at 39,500 rpm for 16 h at 4°C in a SW55 rotor. The pellet was resuspended in NuPAGE sample buffer and analyzed by SDS-PAGE.
4.8. Cell surface expression of viral glycoproteins
Mock or drug-treated A549 cells in 24-well plates were infected with SeV or hPIV1 at a MOI of 3 for 24 h. Cells were fixed with 4% PFA in PBS for 10 min at RT, and blocked with PBS containing 0.1% BSA for 30 min. Cells were incubated with anti-SeV or hPIV1 mAbs against HN and F for 1 h at RT. Plates were washed and incubated with goat anti-mouse antibody conjugated with horseradish peroxidase (1:1000, Abcam) for 1 h at RT. After washing, cells were incubated with 250 μl of ABTS (2,2′-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt) (ThermoFisher) substrate for 10 min followed by 250 μl of 1% SDS solution in water to terminate the reaction. The absorbance of the solution at 405 nm was measured using a microplate spectrophotometer (Molecular Devices).
4.9. Raft flotation assay
Raft flotation assay was performed as previously described (Bialas et al., 2012) with several modifications. Briefly, cells pretreated with the drugs for 12 h were infected with SeV or mock for 1 h and cultured for an additional 28 h in the presence of drugs. Cells were harvested by scraping and pelleted by low speed centrifugation in an Eppendorf minispin centrifuge (3,000 rpm for 3 min). Following re-suspension in 400 μl of cold hypotonic TE buffer (10 mM Tris HCl [pH 7.5], 4 mM EDTA), cells were passaged 30 times through a 27 ½ gauge hypodermic needle and centrifuged at 2,000 rpm for 3 min. Then, 100 μl of cold TNE buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 4 mM EDTA) containing 1% Triton-X 100 was added to 300 μl of lysate and incubated on ice for 40 min. Each lysate (300 μl) was mixed with 700 μl of 70% sucrose in TNE with 1% Triton-X 100 and overlaid with 2 ml 30% sucrose and 1 ml 2.5% sucrose in TNE containing 1% Triton-X 100. After centrifugation at 28,000 rpm for 18 h at 4°C in an SW55 rotor, 400 μl fractions were collected from the top. Proteins in the samples were precipitated with Trichloroacetic acid (TCA), re-suspended with NuPAGE sample buffer (life technologies) and resolved by SDS-PAGE, followed by immunoblot analysis using anti-SeV NP (M52, WS16) or anti-caveolin 1 mAbs. To detect raft association of hPIV1 NP and SeV M, cells pretreated with the drugs for 12 h were infected with hPIV1 or SeV at a MOI of 3 for 12 h, and then labeled with 35S-Met/Cys for 16 h in the presence of drugs as previously described. Cell lysates were subjected to membrane floatation assay, and NP or M in each fraction was immunoprecipitated by anti hPIV1-NP (P19, P27, P35) or anti-SeV M (M1) mAbs and resolved by SDS-PAGE. The fixed and dried gels were exposed on a phosphor screen and visualized using Personal Molecular Image (Bio-Rad). Band intensities were quantitated using BioRad Quantity One software.
4.10. Statistical analysis
All statistical analysis were performed using a two-tailed t-test assuming equal variance. All quantitative data is represented as the mean±standard deviation (SD) of the indicated number of experiments. Significant differences between groups of data are represented as *(P<0.05) or **(P<0.01).
Highlights.
Cholesterol-reducing agents inhibit production of infectious hPIV1 and SeV.
The drugs block virus assembly and release from infected cells.
Cholesterol is required for assembly and formation of parainfluenza viruses.
Acknowledgments
This work was supported by National Institutes of Health Grant R01AI081779. For the microscopy data, we would like to thank Linda Callahan for the use of the URMC Confocal and Conventional Microscopy Core. We also thank Raychel Stone for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali A, Nayak DP. Assembly of Sendai virus: M protein interacts with F and HN proteins and with the cytoplasmic tail and transmembrane domain of F protein. Virology. 2000;276:289–303. doi: 10.1006/viro.2000.0556. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. Short protocols in molecular biology: a compendium of methods from Current protocols in molecular biology. 3. Wiley; New York; Chichester: 1995. [Google Scholar]
- Bialas KM, Desmet EA, Takimoto T. Specific Residues in the 2009 H1N1 Swine-Origin Influenza Matrix Protein Influence Virion Morphology and Efficiency of Viral Spread In Vitro. PLoS One. 2012;7:e50595. doi: 10.1371/journal.pone.0050595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid-and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- Brown G, Aitken J, Rixon HW, Sugrue RJ. Caveolin-1 is incorporated into mature respiratory syncytial virus particles during virus assembly on the surface of virus-infected cells. J Gen Virol. 2002a;83:611–621. doi: 10.1099/0022-1317-83-3-611. [DOI] [PubMed] [Google Scholar]
- Brown G, Jeffree CE, McDonald T, Rixon HW, Aitken JD, Sugrue RJ. Analysis of the interaction between respiratory syncytial virus and lipid-rafts in Hep2 cells during infection. Virology. 2004;327:175–185. doi: 10.1016/j.virol.2004.06.038. [DOI] [PubMed] [Google Scholar]
- Brown G, Rixon HW, Sugrue RJ. Respiratory syncytial virus assembly occurs in GM1-rich regions of the host-cell membrane and alters the cellular distribution of tyrosine phosphorylated caveolin-1. J Gen Virol. 2002b;83:1841–1850. doi: 10.1099/0022-1317-83-8-1841. [DOI] [PubMed] [Google Scholar]
- Budd A, Alleva L, Alsharifi M, Koskinen A, Smythe V, Mullbacher A, Wood J, Clark I. Increased survival after gemfibrozil treatment of severe mouse influenza. Antimicrob Agents Chemother. 2007;51:2965–2968. doi: 10.1128/AAC.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. PLoS One. 2010;5:e10994. doi: 10.1371/journal.pone.0010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Segovia J, Sabbah A, Mgbemena V, Bose S. Cholesterol-rich lipid rafts are required for release of infectious human respiratory syncytial virus particles. Virology. 2012;422:205–213. doi: 10.1016/j.virol.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazal N, Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol Mol Biol Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Real G, Jimenez-Baranda S, Mira E, Lacalle RA, Lucas P, Gomez-Mouton C, Alegret M, Pena JM, Rodriguez-Zapata M, Alvarez-Mon M, Martinez AC, Manes S. Statins inhibit HIV-1 infection by down-regulating Rho activity. J Exp Med. 2004;200:541–547. doi: 10.1084/jem.20040061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Najjar F, Schmitt AP, Dutch RE. Paramyxovirus glycoprotein incorporation, assembly and budding: a three way dance for infectious particle production. Viruses. 2014;6:3019–3054. doi: 10.3390/v6083019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerl MJ, Sampaio JL, Urban S, Kalvodova L, Verbavatz JM, Binnington B, Lindemann D, Lingwood CA, Shevchenko A, Schroeder C, Simons K. Quantitative analysis of the lipidomes of the influenza virus envelope and MDCK cell apical membrane. J Cell Biol. 2012;196:213–221. doi: 10.1083/jcb.201108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin-Grenet AS, Mottet-Osman G, Roux L. From assembly to virus particle budding: pertinence of the detergent resistant membranes. Virology. 2006;344:296–303. doi: 10.1016/j.virol.2005.09.035. [DOI] [PubMed] [Google Scholar]
- Govorkova EA. Consequences of resistance: in vitro fitness, in vivo infectivity, and transmissibility of oseltamivir-resistant influenza A viruses. Influenza and other respiratory viruses. 2013;7(Suppl 1):50–57. doi: 10.1111/irv.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower TL, Graham BS. Antiviral activity of lovastatin against respiratory syncytial virus in vivo and in vitro. Antimicrob Agents Chemother. 2001;45:1231–1237. doi: 10.1128/AAC.45.4.1231-1237.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MS, Sakaguchi T, Schmitt AP. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol. 2010;42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvala H, Gaunt E, McIntyre C, Roddie H, Labonte S, Curran E, Othieno R, Simmonds P, Bremner J. Epidemiology and clinical characteristics of parainfluenza virus 3 outbreak in a Haemato-oncology unit. J Infect. 2012;65:246–254. doi: 10.1016/j.jinf.2012.04.011. [DOI] [PubMed] [Google Scholar]
- Henrickson KJ. Parainfluenza viruses. Clin Microbiol Rev. 2003;16:242–264. doi: 10.1128/CMR.16.2.242-264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtta-Vuori M, Tanhuanpaa K, Mobius W, Somerharju P, Ikonen E. Modulation of cellular cholesterol transport and homeostasis by Rab11. Mol Biol Cell. 2002;13:3107–3122. doi: 10.1091/mbc.E02-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Abe K, Yamada M, Dansako H, Naka K, Kato N. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology. 2006;44:117–125. doi: 10.1002/hep.21232. [DOI] [PubMed] [Google Scholar]
- Imhoff H, von Messling V, Herrler G, Haas L. Canine distemper virus infection requires cholesterol in the viral envelope. J Virol. 2007;81:4158–4165. doi: 10.1128/JVI.02647-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberte JP, McGinnes LW, Peeples ME, Morrison TG. Integrity of membrane lipid rafts is necessary for the ordered assembly and release of infectious Newcastle disease virus particles. J Virol. 2006;80:10652–10662. doi: 10.1128/JVI.01183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE. Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–440. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- Lyles DS. Assembly and budding of negative-strand RNA viruses. Adv Virus Res. 2013;85:57–90. doi: 10.1016/B978-0-12-408116-1.00003-3. [DOI] [PubMed] [Google Scholar]
- Manie SN, de Breyne S, Vincent S, Gerlier D. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J Virol. 2000;74:305–311. doi: 10.1128/jvi.74.1.305-311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao N, Ji Y, Xie Z, Wang H, Wang H, An J, Zhang X, Zhang Y, Zhu Z, Cui A, Xu S, Shen K, Liu C, Yang W, Xu W. Human parainfluenza virus-associated respiratory tract infection among children and genetic analysis of HPIV-3 strains in Beijing, China. PLoS One. 2012;7:e43893. doi: 10.1371/journal.pone.0043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron DJ, Fazio S, Linton MF. Current perspectives on statins. Circulation. 2000;101:207–213. doi: 10.1161/01.cir.101.2.207. [DOI] [PubMed] [Google Scholar]
- Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. doi: 10.1016/S0065-3527(05)64010-9. [DOI] [PubMed] [Google Scholar]
- Petersen J, Drake MJ, Bruce EA, Riblett AM, Didigu CA, Wilen CB, Malani N, Male F, Lee FH, Bushman FD, Cherry S, Doms RW, Bates P, Briley K., Jr The major cellular sterol regulatory pathway is required for Andes virus infection. PLoS Pathog. 2014;10:e1003911. doi: 10.1371/journal.ppat.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Duprex WP, Krohne G, Rima BK, Schneider-Schaulies S. Measles virus M and F proteins associate with detergent-resistant membrane fractions and promote formation of virus-like particles. J Gen Virol. 2007;88:1243–1250. doi: 10.1099/vir.0.82578-0. [DOI] [PubMed] [Google Scholar]
- Ravid D, Leser GP, Lamb RA. A role for caveolin 1 in assembly and budding of the paramyxovirus parainfluenza virus 5. J Virol. 2010;84:9749–9759. doi: 10.1128/JVI.01079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinzon S, Dafa-Berger A, Dyer MD, Paeper B, Proll SC, Teal TH, Rom S, Fishman D, Rager-Zisman B, Katze MG. Impaired cholesterol biosynthesis in a neuronal cell line persistently infected with measles virus. J Virol. 2009;83:5495–5504. doi: 10.1128/JVI.01880-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Pahan K. Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacol Immunotoxicol. 2009;31:339–351. doi: 10.1080/08923970902785253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson CM, Avalos R, Kundu A, Nayak DP. Interaction of Sendai viral F, HN, and M proteins with host cytoskeletal and lipid components in Sendai virus-infected BHK cells. Virology. 1995;209:701–707. doi: 10.1006/viro.1995.1308. [DOI] [PubMed] [Google Scholar]
- Stone R, Hayashi T, Bajimaya S, Hodges E, Takimoto T. Critical role of Rab11a-mediated recycling endosomes in the assembly of type I parainfluenza viruses. Virology. 2016;487:11–18. doi: 10.1016/j.virol.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone R, Takimoto T. Critical role of the fusion protein cytoplasmic tail sequence in parainfluenza virus assembly. PLoS One. 2013;8:e61281. doi: 10.1371/journal.pone.0061281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W. Trypan blue exclusion test of cell viability. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.ima03bs21. Appendix 3, Appendix 3B. [DOI] [PubMed] [Google Scholar]
- Sun X, Whittaker GR. Role for influenza virus envelope cholesterol in virus entry and infection. J Virol. 2003;77:12543–12551. doi: 10.1128/JVI.77.23.12543-12551.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydnor ER, Greer A, Budd AP, Pehar M, Munshaw S, Neofytos D, Perl TM, Valsamakis A. An outbreak of human parainfluenza virus 3 infection in an outpatient hematopoietic stem cell transplantation clinic. Am J Infect Control. 2012;40:601–605. doi: 10.1016/j.ajic.2011.11.011. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Murate M, Fukuda M, Sato SB, Ohta A, Kobayashi T. Cholesterol controls lipid endocytosis through Rab11. Mol Biol Cell. 2007;18:2667–2677. doi: 10.1091/mbc.E06-10-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Suzuki T. Function of membrane rafts in viral lifecycles and host cellular response. Biochem Res Int. 2011;2011:245090. doi: 10.1155/2011/245090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. Role of matrix and fusion proteins in budding of Sendai virus. J Virol. 2001;75:11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106:133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- van’t Wout AB, Lehrman GK, Mikheeva SA, O’Keeffe GC, Katze MG, Bumgarner RE, Geiss GK, Mullins JI. Cellular gene expression upon human immunodeficiency virus type 1 infection of CD4(+)-T-cell lines. J Virol. 2003;77:1392–1402. doi: 10.1128/JVI.77.2.1392-1402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S, Gerlier D, Manie SN. Measles virus assembly within membrane rafts. J Virol. 2000;74:9911–9915. doi: 10.1128/jvi.74.21.9911-9915.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo DS, Chan R, Brown G, Ying L, Sutejo R, Aitken J, Tan BH, Wenk MR, Sugrue RJ. Evidence that selective changes in the lipid composition of raft-membranes occur during respiratory syncytial virus infection. Virology. 2009;386:168–182. doi: 10.1016/j.virol.2008.12.017. [DOI] [PubMed] [Google Scholar]