Abstract
Gold nanoparticles (AuNPs) have been increasingly integrated in biological systems, making it imperative to understand their interactions with cell membranes, the first barriers to be crossed to enter cells. Herein, liposomes composed of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) as a model membrane system were treated with citrate stabilized AuNPs from 5 to 30 nm at various concentrations. The fluorescence shifts of Laurdan probes reveal that AuNPs in general made liposomes more fluidic. The increased fluidity is expected to result in an increased surface area, and thus liposome shape changes from circular to less circular, which was further confirmed with fluorescence microscopy. The localized stress in lipids induced by electrostatically adsorbed AuNPs was hypothesized to cause the dominant long-range effect of fluidization of unbound lipid membranes. A secondary effect of the AuNP-induced lateral pressure is the membrane rupture or formation of pores, which was probed by AFM under fluid. We found in this study a nanoparticle-mediated approach of modulating the stiffness of lipid membranes: by adsorption of AuNPs, lipids at the binding sites are stiffened whereas lipids afar are fluidized. Understanding the factors that modulate lipid packing is important for the discovery of alternative therapeutic methods for diseases linked to membrane integrity such as high blood pressure and cancer metastasis.
The size resemblance of nanoparticles (NPs) to biological macromolecules coupled with the NPs' high surface energy leads to intricate interactions between the two entities. Myriads of study have been conducted to gain insights into the bio-nano interface for biomedical applications.1,2 Gold nanoparticles (AuNPs) are the most extensively studied NPs owing to their tunable sizes, well-defined surface chemistry, and the quantum size effects.3,4 Being biocompatible and inert,4,5 AuNPs are very attractive for biomedical and pharmaceutical applications,6,7 such as drug and gene delivery,8–10 medical diagnostics,11,12 and therapeutics.13,14 The increasing integration of AuNPs in the biological systems makes it imperative to understand how they interact with cell membranes, the first barriers to be crossed to enter cells. We show in this paper how unmodified AuNPs of various sizes affect the lipid packing in a simplified model membrane system. The extent of lipid packing is one of the key physicochemical features of biological membranes and plays an important role in providing anchorage platform for the orchestrated function of receptors and enzymes in cell signaling.15 Some drugs, e.g., Losartan, are known to take effect by altering membrane viscosity.16 The goal of this study is to identify key factors on how AuNPs affect the lipid packing.
Lipid vesicles, or liposomes, are spherical constructs of lipid bilayers and provide a simplified, repeatable, and stable model for cell membranes. Devoid of the complexity of embedded proteins and the large variability in different cellular lines and culture media,17 liposomes allow for the physicochemical characterization of the AuNP-lipid interaction.7,18–21 In this study, the sizes of AuNPs are chosen from 5 to 30 nm to be comparable to those of bio-macromolecules and other components of cell membranes.
We prepared liposomes by thin lipid film extraction (supplementary material). A chloroform solution of DMPC (1,2-dimyristoyl-sn-glycero-3-phosphocholine, Sigma-Aldrich, Figure 1(b)) and Laurdan (6-dodecanoyl-2-dimethylamino-naphthalene, Invitrogen) was first dried and then hydrated in phosphate buffered saline (1× PBS). Repeated incubation at 37 °C and vortexing were applied followed by sonication. A room temperature (∼22 °C) incubation for 48 h led to a suspension of large unilamellar vesicles (LUVs) and giant unilamellar vesicles (GUVs). Then, 25 μl of DMPC liposomes (2 mg/ml) was mixed with AuNPs (citrate stabilized in 0.1 mM PBS, Sigma-Aldrich) of various sizes (5, 10, 20, and 30 nm) and different amounts (25, 50, 75, and 100 μl) into 1 ml of 1× PBS. Spectrofluorometry (K2 Spectrofluorometer, ISS, Inc.) was later performed at 22 (±1) °C with excitation at 340 nm and emission recorded from 360 to 500 nm.
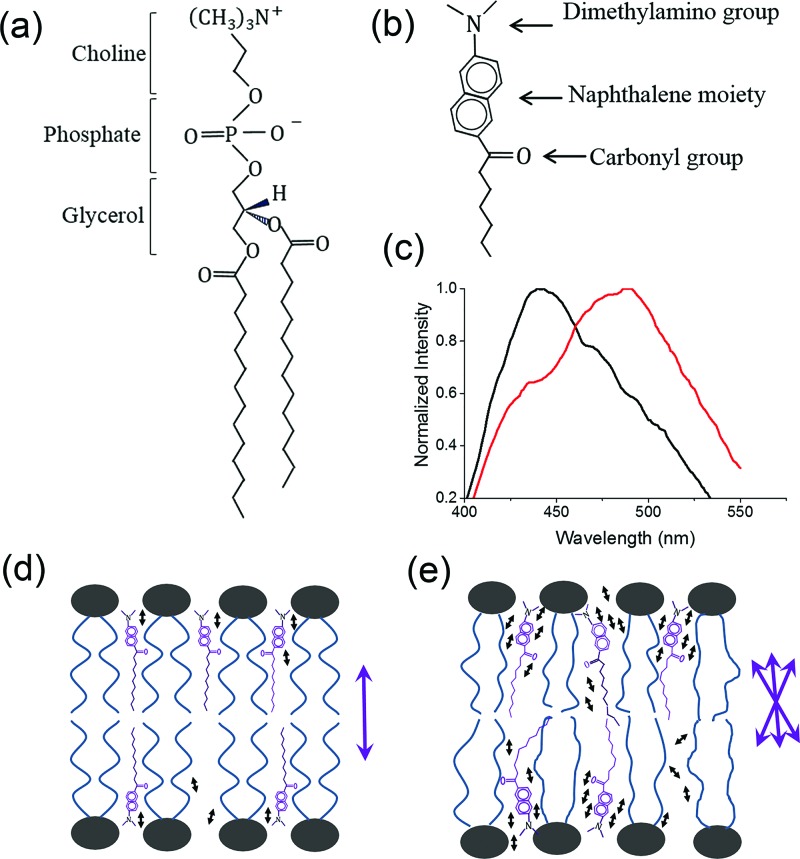
FIG. 1.
The structural formula of DMPC (a) and Laurdan (b). Fluorescence red shift (c) of Laurdan when partitioned between ordered (d) and disordered (e) DMPC bilayers. In (d) and (e), the Laurdan naphthalene moieties anchor at the glycerol backbone of DMPC but alignments differ. The purple double-headed arrows next to bilayer diagrams represent the possible alignments of Laurdan dipole moments. Water molecules are represented as tiny black double-headed arrows.
Laurdan, a hydrophobic fluorescent probe for evaluation of lipid packing, is made up of a dimethylamino group, a carbonyl group, and a naphthalene moiety (Figure 1(b)).22 When lodged between the acyl chains of a lipid bilayer, Laurdan probes the polarity changes associated with the concentration and molecular dynamics of surrounding water molecules, a direct measure of lipid packing or membrane phases.23 The emission maximum of Laurdan is 440 nm for lipids in the ordered gel phase and shifts to 490 nm in the disordered fluid phase (Figure 1(c)), a red shift caused by the increased dipolar relaxation of the fluorescent naphthalene moiety of Laurdan. In the gel phase (Figure 1(d)), the lipids are tightly packed so the reorientation of water molecules around the excited dipole of Laurdan can hardly occur. Whereas in the liquid disordered phase (Figure 1(e)), the increased concentration and mobility of water molecules around increase Laurdan's dipolar relaxation and therefore a red shift in emission. The dual emission of Laurdan is quantified by the generalized polarization (GP) function as , where I440 and I490 are the emission intensities at 440 and 490 nm, respectively.24,25 An increase in GP function suggests a transition toward the gel phase whereas a decrease represents fluidization.26
Multiple fluorescence images (Olympus IX-71) of Laurdan-labeled liposomes were recorded for size and circularity analysis with ImageJ software package (National Institutes of Health). Circularity, calculated as , is a measure of how circular each particle is. Particles in ImageJ are modelled as ellipses. An ellipse with a circularity of 0 is a straight line, while a circularity of 1 represents a perfect circle.
A multimode AFM (Bruker, CA) study was carried out in a fluid cell filled with 1× PBS. Supported DMPC bilayers deposited on freshly cleaved mica surface incubated with AuNPs (5 to 50 nm) were imaged (supplementary material). The peak force quantitative nanomechanical (PF-QNM) characterization, including thickness, deformation, and Young's modulus, was performed using soft calibrated MSCT cantilever on bilayers spanning their break-through strength.27 Multiple force curves were recorded to determine the elastic limit, and the Young's modulus was derived from the Hertz model.
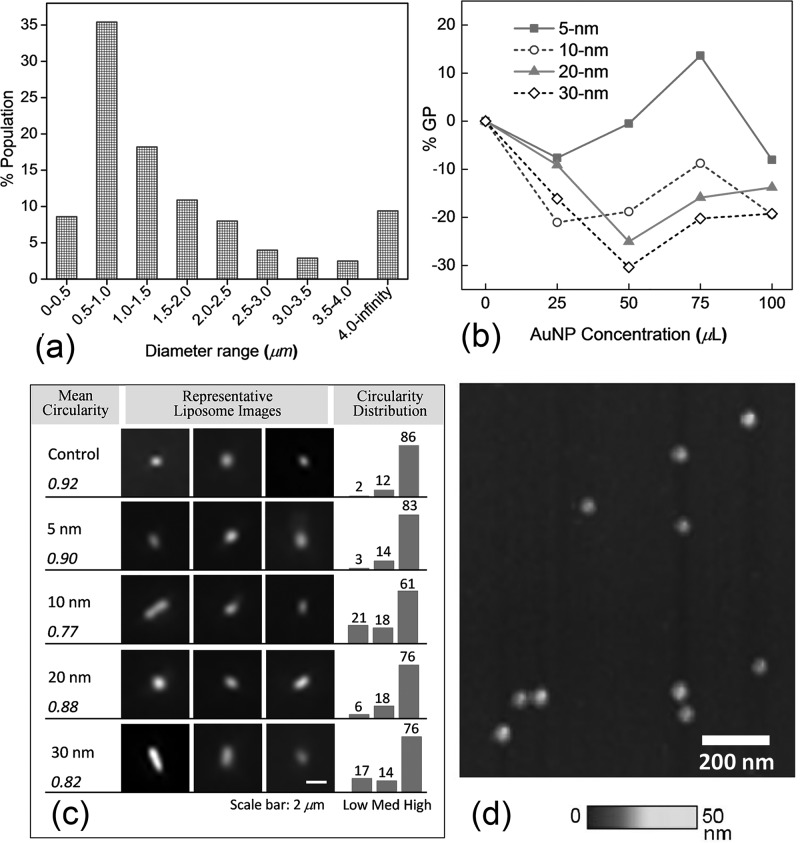
Figure 2(a) shows the size distribution of as-prepared liposomes analyzed from fluorescence images. Approximately 44% of as-prepared liposomes are LUVs (diameter less than 1 μm) and 56% are GUVs (diameter larger than 1 μm).28 Note that the size was determined by fluorescing pixels, usually larger than the actual particle size. Therefore, this result should be taken for the perception of an approximate size range in the experiment. Given the wide size range and varied polydispersity, more accurate size characterization technique such as DLS (dynamic light scattering) is not suitable here as the scattering from submicron liposomes will be overwhelmed by that from larger liposomes.
FIG. 2.
(a) Size distribution of as-prepared liposomes from fluorescence image analysis. (b) Percent changes in generalized polarization (% GP) of control liposomes vs. treatment of various sizes and concentrations of AuNPs. (c) Representative fluorescence images of liposomes under various AuNP treatment (center). The mean values of circularity (left). The histograms of percent populations for low (0–0.5), medium (0.5–0.8), and high (0.8–1) circularities (right). (d) Size uniformity of 20-nm AuNPs imaged by QNM AFM.
The graphs of %GP changes of AuNP-treated liposomes from the control are shown in Figures 2(b). The averages of four independent experiments for each size and concentration combination were plotted in the graphs. Note that a positive %GP change suggests a transition toward gel phase whereas a negative change for a transition toward fluid phase. Negative %GP changes are found for almost all cases except for 5-nm AuNPs at 75 μl, suggesting that AuNPs tend to make as-prepared liposomes more fluidic or disorderly packed. In particular, the most pronounced GP change of −30% occurred with 30-nm AuNPs at 50 μl and the highest positive %GP change of +14% was found with 5-nm AuNPs at 75μl. Given the variability in size distribution of liposomes among different batch of preparations, conclusions can be drawn here that AuNPs from 5 to 30 nm tend to make as-prepared liposomes more fluidic and larger AuNPs (greater than 5 nm) impact the lipid packing more effectively than 5-nm AuNPs. Note that DMPC bilayers feature the lipid-chain melting/disordering at 24 °C; therefore, our experiments conducted at 22 °C may cause an enhanced impact by AuNPs as seen in the data.29,30
The fluidization increases the liposome surface area because the head groups of lipids lose the triangular lattice order and adopt a more disordered arrangement with increased inter-molecular spacing.31 An increased surface area with a given volume should undergo a shape change from spheres to oblongs because the volume-to-surface ratio decreases.32 To test this hypothesis of fluidization-led-to shape changes, the liposome circularity analysis was conducted at AuNP treatment of 100 μl for different sizes. Figure 2(c) shows the representative fluorescence images of Laurdan-labeled liposomes treated with different size AuNPs juxtaposed with the histograms of percent populations for liposomes of low (0–0.5), medium (0.5–0.8), and high (0.8–1) circularities. The liposomes without AuNPs have 86% high-circularity population, the greatest among all analyzed. Rod-like liposomes were found frequently with 10 and 30-nm AuNP treatment, as evidenced by much greater low-circularity population of 21% and 17%, respectively. Consequently, the mean circularities from 10 and 30-nm AuNPs registered the lowest values of 0.77 and 0.82. In Figure 2(b) at 100 μl, 10 and 30-nm AuNPs induced greatest negative %GP changes of −19%, followed by 20-nm AuNPs of −14% and 5-nm of −8%. The mean circularities followed the exact same order of %GP changes with 10, 30, 20, 5-nm AuNPs and control, corresponding to 0.77, 0.82, 0.88, 0.90, and 0.92. The total agreement between %GP changes and mean circularities supports the aforementioned hypothesis that AuNP-induced membrane fluidization leads to less circular liposomes.
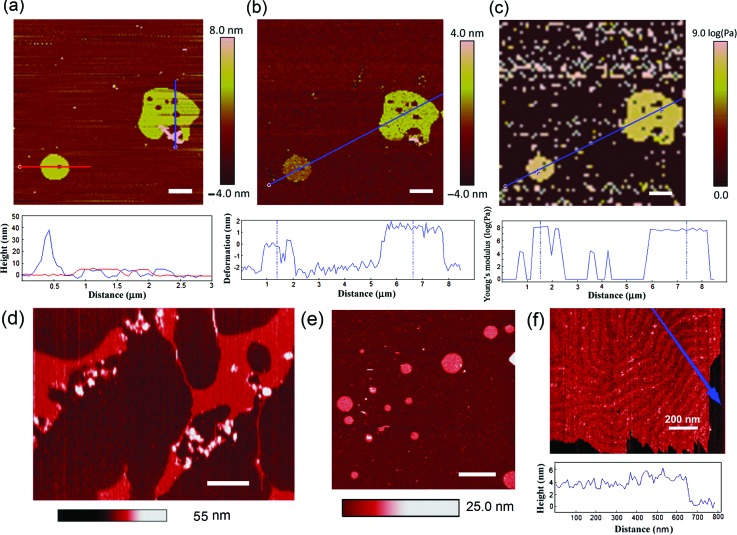
The nonspecific adsorption of charged nanoparticles onto the single-component phospholipid bilayer and the disruption of lipid bilayers have been observed previously.32–36 The AuNPs used in this work were prepared by citrate reduction of chloroauric acid (HAuCl4) and stabilized in 0.1 mM PBS.37 The citrate capped AuNPs carry negative surface charges. They show good size uniformity as confirmed with AFM at 20 nm (Figure 2(d)). Previous studies have shown both experimentally38 and theoretically18 that negatively charged AuNPs can disrupt zwitterionic membranes and form nanoscale “holes” or “pores” on lipid bilayers.39 The formation of pores in supported lipid bilayers was previously observed under AFM for various nanoparticles including peptides, proteins, polymers, AuNPs, and silica nanoparticles.35 In Figure 3(a), the AFM height image of supported DMPC bilayers shows similar AuNP-induced pores. As many as five pores can be seen on the large lipid patch, whereas only one pore is found on the small patch. The diameters of the pores range approximately from 170 to 310 nm. A sizable aggregate of AuNPs in pinkish hue can be found adjoining the lower part of the large lipid patch. These features are confirmed with the height profiles (Figure 3(a)): the tall peak at 40 nm features AuNP aggregates; the single lipid bilayer has an average thickness of 6 nm; the pores level with the substrate. In Figure 3(b), an average of 3.5-nm deformation was generated on the large lipid patch while approximately 2 nm of deformation was generated on the small lipid patch. The greater indentation generated on the large patch than on the small one is likely a result from a higher density of AuNPs on the large patch (Figure 3(a)). This is yet another confirmation that AuNPs make lipid bilayer more fluidic or more elastic. Similar conclusion can be drawn from the Young's modulus image and the section profile in Figure 3(c), where the Young's modulus of the large lipid patch is ∼107.6 Pa versus ∼108.0 Pa for the small patch. In other words, the stiffness of the small lipid patch is 2.5 times that of the large patch. Higher density of AuNPs has softened the large lipid patch. When the concentration of AuNPs is high, multiple ruptures are found on lipid bilayers (Figure 3(d)); when the concentration of AuNPs is low, the lipid bilayers are round and continuous without holes or ruptures (Figure 3(e)). This comparison further confirms the effect of pore formation induced by AuNPs on lipid bilayers. A higher resolution of AFM in Figure 3(f) reveals the ripple phase of a continuous lipid bilayer with few to none AuNP adsorption.38
FIG. 3.
(a) The AFM height image of supported DMPC bilayers incubated with AuNPs and imaged under 1× PBS at low QNM force of 3.3 nN. The height profiles along two section lines across the big (blue) and small (red) bilayer patches respectively are shown below. (b) The AFM mapping of bilayer deformation under 1× PBS at medium QNM force of 6.6 nN and the deformation profile along the section line. (c) Strain-dependent elastic modulus map and the section profile at low QNM force at 0.33–1.5 nN. (d) AFM topographic images of DMPC bilayer disruption by high concentration AuNPs under 1× PBS. (e) AFM topographic image of the DMPC bilayer with a low concentration of AuNPs. (f) AFM topographic image and height section plot along the blue arrow of a DMPC bilayer with few AuNP(s) under 1× PBS in the “ripple” phase.38 Scale bars: 1 μm or otherwise specified.
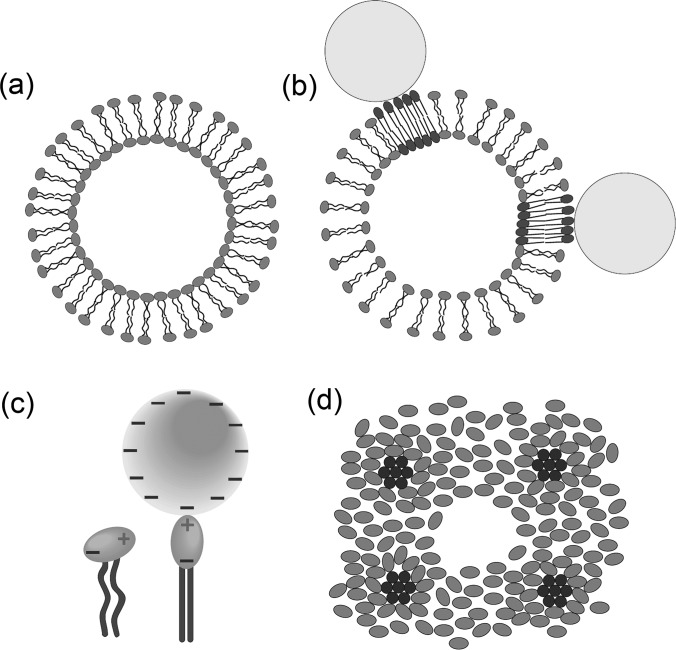
The adsorbed negatively charged AuNPs have been previously reported to stiffen phosphatidylcholine (PC) lipid bilayers at molecular length scale and restructure the bound lipid molecules into a raft-like phase.39 Combining this and the results above, we hypothesize a localized stiffening and long-range fluidization model. First, AuNPs adsorb onto liposome surfaces through attractive Coulomb potential (Figures 4(a) and 4(b)). Upon adsorption, the phosphocholine (PC) head group (Figure 1(a)) switch from a leaning position to a stand-up position due to dipole-charge interaction (Figure 4(c)). The electrostatic forces draw otherwise loosely packed lipid molecules to tightly pack around the AuNP binding sites, thereby forming raft-like domains (Figure 4(b)). As the distance of lipids from adsorbed AuNPs increases, the attractive force decreases. When the lipids are far from the adsorbed AuNPs, the Coulomb attraction is negligible. However, a perturbation arising from stiffened AuNP-adsorbed lipids affects the lipid packing far away. As these lipids have to cover more surface area vacated by AuNP-adsorbed lipids, the lipid-lipid spacing increases, and therefore fluidity increases as observed in %GP and AFM studies. The raft-like domains of AuNP-bound lipids, on the other hand, should increase the GP value. However, this contribution is rather small and only partially cancels out the GP decrease due to the long-range lipid fluidization. Assuming 100% AuNP adsorption (the actual binding is lower), the ratio of DMPC pairs per AuNP (supplementary material) ranges from 4 × 103 (5 nm at 100 μl) to 5 × 106 (30 nm at 25 μl), translating into area of unbound lipid bilayer thousands to millions times that of the raft-like AuNP-adsorbed patches. The actual binding determined by adsorption isotherm will be lower than 100%. Therefore, fluidization dominates gelation, leading to an overall increased fluidity as observed. However, when liposomes are more densely adsorbed with AuNPs, more rigidified lipid domains contribute to GP increase, therefore more likely counteracting the long-range GP decrease. That explains one exception of increased GP observed for 5-nm AuNP at 75 μl, the second highest density of AuNP coverage among all tested. Given the variations in AuNP binding and liposome size distribution from preparation to preparation, it is acceptable that the experiment did not follow exactly the calculation-based trend.
FIG. 4.
The electrostatic forces between lipid head groups and charged AuNPs draw otherwise loosely packed liposome molecules (a) to tightly pack around the binding sites and forming raft-like domains (b). (c) The head group of a DMPC molecule switch from the leaning to the stand-up position under the dipole-charge interaction with a citrate-capped AuNP. (d) A possible top-view of lipid head group arrangement of a pore formed by multiple AuNP adsorptions. Note that the ordered domains (in dark shade) are stressed regions with AuNPs sitting on top but removed for viewing convenience.
Another consequence induced by adsorbed AuNPs is the membrane pore formation, which we consider as a secondary effect of altered lipid packing. We argue that the pores are ruptures resulting from the lateral pressure induced by multiple AuNP adsorptions,16 rather than from the insertion of AuNPs. A simulation has suggested that a hole is transiently formed before AuNP entry and closed after AuNP translocation.40 Also, the insertion of charged AuNPs in this study is not energetically favorable because of the hydrophobic lipidic tails. In addition, the pore sizes found on the AFM image (Figure 3(a)) range from 170 to 310 nm, much larger than individual AuNPs, making penetration-caused pore formation by single AuNPs unlikely but possible if AuNP concentration is high enough.16 Figure 4(d) illustrates a possible top-view lipid head group arrangement of pore formation. Note that the ordered domains are stressed regions induced by bound AuNPs; however, AuNPs are removed for viewing purpose.
In summary, citrate stabilized AuNPs from 5 to 30 nm generally increase the fluidity of as-prepared DMPC liposomes as found in fluorescence shifts of Laurdan and AFM nanomechanical characterization. The increased fluidity leads to an increased surface area, which results in liposome shape changes from circular to less circular, as confirmed in fluorescence images. The localized stress in lipids induced by electrostatically adsorbed AuNPs was hypothesized to cause the dominant long-range effect of fluidization of unbound lipid membranes. A secondary effect of the AuNP induced lateral pressure is the membrane rupture or pore formation, as observed in multi-mode AFM under fluid. What we found in this study supports the supposition in an earlier article that “nanoparticle-induced reconstruction of the phase state offers a new mechanism to modulate stiffness.”33 The rigid patches on cell membranes, or lipid rafts, are traditionally held as liquid-ordered domains stiffened by enrichment of cholesterol and sphingolipids.41 In this study, the adsorption of AuNPs was found to offer an alternative approach of modulating membrane packing: they stiffen the lipids bound to them whereas fluidize lipids far from the binding sites. Understanding the factors that modulate lipid packing is important for the discovery of alternative therapeutic methods for diseases that are linked to membrane integrity such as high blood pressure and cancer metastasis.42
See supplementary material for liposome and AFM sample preparation, AFM force curves, and the calculations of ratio of DMPC pairs per AuNP.
Acknowledgments
This work was made possible by the Delaware INBRE program, supported by a grant from the National Institute of General Medical Sciences—NIGMS (P20 GM103446) from the National Institutes of Health and the state of Delaware. This work was also supported by grants from the CREST Program (No. 1242067) of National Science Foundation, and the Intramural Research Program of the National Institutes of Health, including National Institute of Biomedical Imaging and Bioengineering (ZIA EB000015-09 to A.J.). Q.L. wishes to thank OSCAR Imaging Facility at Delaware State University for access to ISS-K2 spectrofluorometer and fluorescence microscopes.
References
- 1. Monopoli M. P., Åberg C., Salvati A., and Dawson K. A., Nat. Nanotechnol. , 779 (2012). 10.1038/nnano.2012.207 [DOI] [PubMed] [Google Scholar]
- 2. Lai Z. W., Yan Y., Caruso F., and Nice E. C., ACS Nano , 10438 (2012). 10.1021/nn3052499 [DOI] [PubMed] [Google Scholar]
- 3. Daniel M.-C. and Astruc D., Chem. Rev. , 293 (2004). 10.1021/cr030698+ [DOI] [PubMed] [Google Scholar]
- 4. Connor E., Mwamuka J., Gole A., Murphy C., and Wyatt M., Small , 325 (2005). 10.1002/smll.200400093 [DOI] [PubMed] [Google Scholar]
- 5. Bhattacharya R. and Mukherjee P., Adv. Drug Delivry Rev. , 1289 (2008). 10.1016/j.addr.2008.03.013 [DOI] [PubMed] [Google Scholar]
- 6. Dykman L. and Khlebtsov N., Chem. Soc. Rev. , 2256 (2012). 10.1039/C1CS15166E [DOI] [PubMed] [Google Scholar]
- 7. Tiwari P. M., Vig K., Dennis V. A., and Singh S. R., Nanomaterials , 31 (2011). 10.3390/nano1010031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh P., Han G., De M., Kim C. K., and Rotello V. M., Adv. Drug Delivery Rev. , 1307 (2008). 10.1016/j.addr.2008.03.016 [DOI] [PubMed] [Google Scholar]
- 9. Duncan B., Kim C., and Rotello V. M., J. Controlled Release , 122 (2010). 10.1016/j.jconrel.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Papasani M. R., Wang G., and Hill R. A., Nanomed. Nanotechnol. Biol. Med. , 804 (2012). 10.1016/j.nano.2012.01.008 [DOI] [Google Scholar]
- 11. Mieszawska A. J., Mulder W. J. M., Fayad Z. A., and Cormode D. P., Mol. Pharm. , 831–847 (2013). 10.1021/mp3005885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chuang Y.-C., Li J.-C., Chen S.-H., Liu T.-Y., Kuo C.-H., Huang W.-T., and Lin C.-S., Biomaterials , 6087 (2010). 10.1016/j.biomaterials.2010.04.026 [DOI] [PubMed] [Google Scholar]
- 13. Wang S., Chen K.-J., Wu T.-H., Wang H., Lin W.-Y., Ohashi M., Chiou P.-Y., and Tseng H.-R., Angew. Chem. Int. Ed. , 3777 (2010). 10.1002/anie.201000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee K., Lee H., Bae K. H., and Park T. G., Biomaterials , 6530 (2010). 10.1016/j.biomaterials.2010.04.046 [DOI] [PubMed] [Google Scholar]
- 15. Pike L. J., J. Lipid Res. , 655 (2003). 10.1194/jlr.R200021-JLR200 [DOI] [PubMed] [Google Scholar]
- 16. Heimburg T., Thermal Biophysics of Membranes ( Wiley-VCH, Weinheim, 2007), Chap. 12, p. 261. [Google Scholar]
- 17. Maiorano G., Sabella S., Sorce B., Brunetti V., Malvindi M. A., Cingolani R., and Pompa P. P., ACS Nano , 7481 (2010). 10.1021/nn101557e [DOI] [PubMed] [Google Scholar]
- 18. Lin J., Zhang H., Chen Z., and Zheng Y., ACS Nano , 5421 (2010). 10.1021/nn1010792 [DOI] [PubMed] [Google Scholar]
- 19. Tatur S., Maccarini M., Barker R., Nelson A., and Fragneto G., Langmuir , 6606 (2013). 10.1021/la401074y [DOI] [PubMed] [Google Scholar]
- 20. Van Lehn R. C., Atukorale P. U., Carney R. P., Yang Y.-S., Stellacci F., Irvine D. J., and Alexander-Katz A., Nano Lett. , 4060 (2013). 10.1021/nl401365n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mhashal A. R. and Roy S., PLoS One , e114152 (2014). 10.1371/journal.pone.0114152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sezgin E., Sadowski T., and Simons K., Langmuir , 8160 (2014). 10.1021/la501226v [DOI] [PubMed] [Google Scholar]
- 23. Sanchez S. A., Tricerri M. A., and Gratton E., Proc. Natl. Acad. Sci. , 7314–7319 (2012). 10.1073/pnas.1118288109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu W., So P., French T., and Gratton E., Biophys. J. , 626 (1996). 10.1016/S0006-3495(96)79646-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Parasassi T., Gratton E., Yu W., Wilson P., and Levi M., Biophys. J. , 2413 (1997). 10.1016/S0006-3495(97)78887-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tricerri M. A., Toledo J. D., Sanchez S. A., Hazlett T. L., Gratton E., Jonas A., and Garda H. A., J. Lipid Res. , 669 (2005). 10.1194/jlr.M400340-JLR200 [DOI] [PubMed] [Google Scholar]
- 27. Liao H.-S., Lin J., Liu Y., Huang P., Jin A., and Chen X., Nanoscale , 14814 (2016). 10.1039/C6NR04672J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laouini A., Jaafar-Maalej C., Limayem-Blouza I., Sfar S., Charcosset C., and Fessi H., J. Colloid Sci. Biotechnol. , 147 (2012). 10.1166/jcsb.2012.1020 [DOI] [Google Scholar]
- 29. Akabori K. and Nagle J. F., Soft Matter , 918–926 (2015). 10.1039/C4SM02335H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Charrier A. and Thibaudau F., Biophys. J. , 1094–1101 (2005). 10.1529/biophysj.105.062463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janiak M. J., Small D. M., and Shipley G. G., J. Biol. Chem. , 6068 (1979), see http://www.jbc.org/content/254/13/6068.abstract. [PubMed] [Google Scholar]
- 32. Lipowsky R., Curr. Opin. Struct. Biol. , 531 (1995). 10.1016/0959-440X(95)80040-9 [DOI] [PubMed] [Google Scholar]
- 33. Wang B., Zhang L., Bae S. C., and Granick S., Proc. Natl. Acad. Sci. , 18171 (2008). 10.1073/pnas.0807296105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Leroueil P. R., Hong S., Mecke A., James J., Baker R., Orr B. G., and Holl M. M. B., Acc. Chem. Res. , 335 (2007). 10.1021/ar600012y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leroueil P. R., Berry S. A., Duthie K., Han G., Rotello V. M., McNerny D. Q., Baker J. R., Orr B. G., and Holl M. M. B., Nano Lett. , 420 (2008). 10.1021/nl0722929 [DOI] [PubMed] [Google Scholar]
- 36. Moghadam B. Y., Hou W.-C., Corredor C., Westerhoff P., and Posner J. D., Langmuir , 16318 (2012). 10.1021/la302654s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Toma H. E., Zamarion V. M., Toma S. H., and Araki K., J. Braz. Chem. Soc. , 1158 (2010). 10.1590/S0103-50532010000700003 [DOI] [Google Scholar]
- 38. Needham D. and Evans E., Biochemistry , 8261–8269 (1988). 10.1021/bi00421a041 [DOI] [PubMed] [Google Scholar]
- 39. Montis C., Maiolo D., Alessandri I., Bergese P., and Berti D., Nanoscale , 6452 (2014). 10.1039/c4nr00838c [DOI] [PubMed] [Google Scholar]
- 40. Lin J. and Alexander-Katz A., ACS Nano , 10799 (2013). 10.1021/nn4040553 [DOI] [PubMed] [Google Scholar]
- 41. Simons K. and Ikonen E., Nature , 569 (1997). 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- 42. Hanahan D. and Weinberg R., Cell , 646 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for liposome and AFM sample preparation, AFM force curves, and the calculations of ratio of DMPC pairs per AuNP.