Abstract
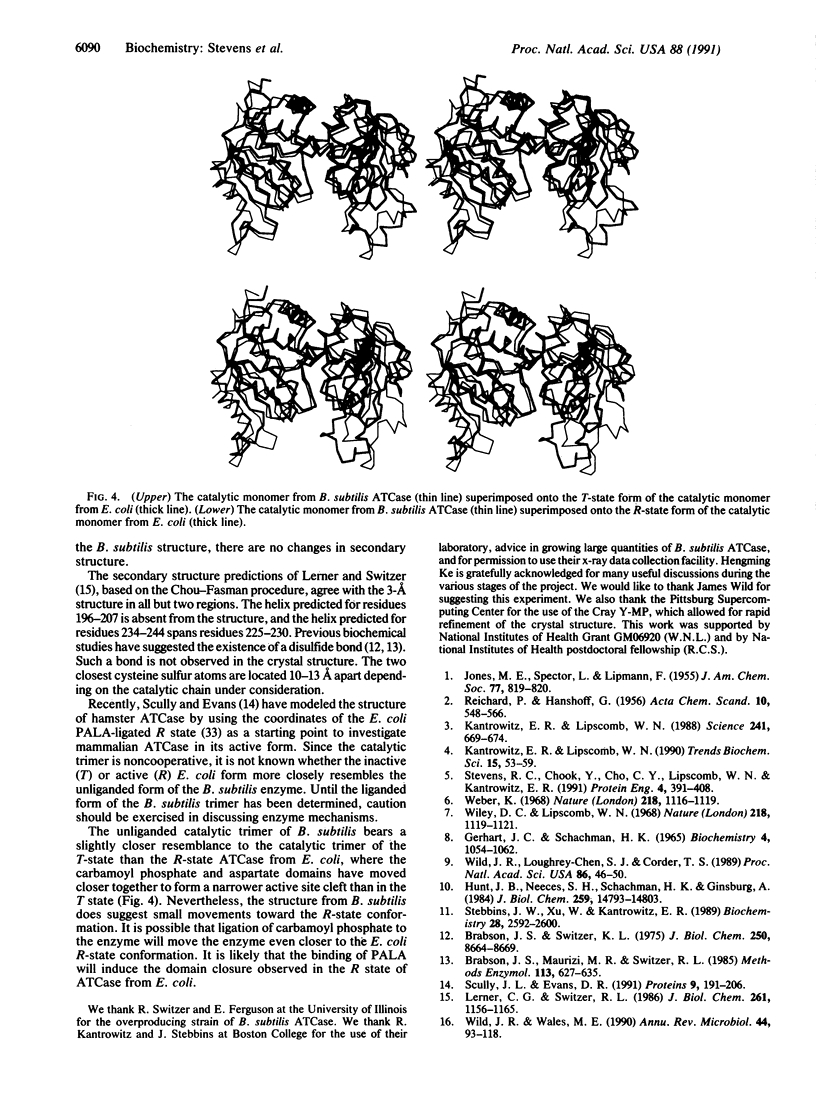
The three-dimensional structure of Bacillus subtilis aspartate transcarbamoylase (ATCase; aspartate carbamoyltransferase; carbamoyl-phosphate:L-aspartate carbamoyltransferase, EC 2.1.3.2) has been solved by the molecular replacement method at 3.0 A resolution and refined to a crystallographic R factor of 0.19. The enzyme crystallizes in the space group C2 with unit cell dimensions a = 258.5, b = 153.2, and c = 51.9 A and beta = 97.7 degrees. The asymmetric unit is composed of three monomers related by noncrystallographic threefold symmetry. A total of 295 of 304 amino acid residues have been built into the monomer. The last 9 residues in the C terminus were not included in the final model. Each monomer consists of 34% alpha-helix and 18% beta-strand. Three solvent-exposed loop regions (residues 69-84, 178-191, and 212-229) are not well defined in terms of electron density. The catalytic trimer of ATCase from B. subtilis shows great similarity to the catalytic trimer in Escherichia coli ATCase, which was used in constructing the model for molecular replacement. The unliganded trimer from B. subtilis, which is not cooperative, resembles the T (inactive) state slightly more than the R (active)-state form of the E. coli trimer. However, certain regions in the B. subtilis trimer exhibit shifts toward the E. coli R-state conformation.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brabson J. S., Maurizi M. R., Switzer R. L. Aspartate transcarbamylase from Bacillus subtilis. Methods Enzymol. 1985;113:627–635. doi: 10.1016/s0076-6879(85)13084-3. [DOI] [PubMed] [Google Scholar]
- Brabson J. S., Switzer R. L. Purification and properties of Bacillus subtilis aspartate transcarbamylase. J Biol Chem. 1975 Nov 25;250(22):8664–8669. [PubMed] [Google Scholar]
- Brünger A. T., Krukowski A., Erickson J. W. Slow-cooling protocols for crystallographic refinement by simulated annealing. Acta Crystallogr A. 1990 Jul 1;46(Pt 7):585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- Gerhart J. C., Schachman H. K. Distinct subunits for the regulation and catalytic activity of aspartate transcarbamylase. Biochemistry. 1965 Jun;4(6):1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- Hamlin R. Multiwire area X-ray diffractometers. Methods Enzymol. 1985;114:416–452. doi: 10.1016/0076-6879(85)14029-2. [DOI] [PubMed] [Google Scholar]
- Howard A. J., Nielsen C., Xuong N. H. Software for a diffractometer with multiwire area detector. Methods Enzymol. 1985;114:452–472. doi: 10.1016/0076-6879(85)14030-9. [DOI] [PubMed] [Google Scholar]
- Hunt J. B., Neece S. H., Schachman H. K., Ginsburg A. Mercurial-promoted Zn2+ release from Escherichia coli aspartate transcarbamoylase. J Biol Chem. 1984 Dec 10;259(23):14793–14803. [PubMed] [Google Scholar]
- Jones T. A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamoylase: the molecular basis for a concerted allosteric transition. Trends Biochem Sci. 1990 Feb;15(2):53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- Kantrowitz E. R., Lipscomb W. N. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988 Aug 5;241(4866):669–674. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Quigley G., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Unit cell transormations in yeast phenylalanine transfer RNA crystals. J Mol Biol. 1973 Apr 5;75(2):429–432. doi: 10.1016/0022-2836(73)90032-6. [DOI] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. 2.5 A structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-L-aspartate. J Mol Biol. 1987 Feb 5;193(3):527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Monaco H. L., Crawford J. L., Lipscomb W. N. Three-dimensional structures of aspartate carbamoyltransferase from Escherichia coli and of its complex with cytidine triphosphate. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5276–5280. doi: 10.1073/pnas.75.11.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully J. L., Evans D. R. Comparative modeling of mammalian aspartate transcarbamylase. Proteins. 1991;9(3):191–206. doi: 10.1002/prot.340090305. [DOI] [PubMed] [Google Scholar]
- Stebbins J. W., Xu W., Kantrowitz E. R. Three residues involved in binding and catalysis in the carbamyl phosphate binding site of Escherichia coli aspartate transcarbamylase. Biochemistry. 1989 Mar 21;28(6):2592–2600. doi: 10.1021/bi00432a037. [DOI] [PubMed] [Google Scholar]
- Stevens R. C., Chook Y. M., Cho C. Y., Lipscomb W. N., Kantrowitz E. R. Escherichia coli aspartate carbamoyltransferase: the probing of crystal structure analysis via site-specific mutagenesis. Protein Eng. 1991 Apr;4(4):391–408. doi: 10.1093/protein/4.4.391. [DOI] [PubMed] [Google Scholar]
- Stevens R. C., Gouaux J. E., Lipscomb W. N. Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-A resolution. Biochemistry. 1990 Aug 21;29(33):7691–7701. doi: 10.1021/bi00485a019. [DOI] [PubMed] [Google Scholar]
- Weber K. New structural model of E. coli aspartate transcarbamylase and the amino-acid sequence of the regulatory polypeptide chain. Nature. 1968 Jun 22;218(5147):1116–1119. doi: 10.1038/2181116a0. [DOI] [PubMed] [Google Scholar]
- Wild J. R., Loughrey-Chen S. J., Corder T. S. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1989 Jan;86(1):46–50. doi: 10.1073/pnas.86.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. C., Lipscomb W. N. Crystallographic determination of symmetry of aspartate transcarbamylase. Nature. 1968 Jun 22;218(5147):1119–1121. doi: 10.1038/2181119a0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]



