ABSTRACT
While a majority of academic studies concerning acetone, butanol, and ethanol (ABE) production by Clostridium have focused on Clostridium acetobutylicum, other members of this genus have proven to be effective industrial workhorses despite the inability to perform genetic manipulations on many of these strains. To further improve the industrial performance of these strains in areas such as substrate usage, solvent production, and end product versatility, transformation methods and genetic tools are needed to overcome the genetic intractability displayed by these species. In this study, we present the development of a high-efficiency transformation method for the industrial butanol hyperproducer Clostridium saccharoperbutylacetonicum strain N1-4 (HMT) ATCC 27021. Following initial failures, we found that the key to creating a successful transformation method was the identification of three distinct colony morphologies (types S, R, and I), which displayed significant differences in transformability. Working with the readily transformable type I cells (transformation efficiency, 1.1 × 106 CFU/μg DNA), we performed targeted gene deletions in C. saccharoperbutylacetonicum N1-4 using a homologous recombination-mediated allelic exchange method. Using plasmid-based gene overexpression and targeted knockouts of key genes in the native acetone-butanol-ethanol (ABE) metabolic pathway, we successfully implemented rational metabolic engineering strategies, yielding in the best case an engineered strain (Clostridium saccharoperbutylacetonicum strain N1-4/pWIS13) displaying an 18% increase in butanol titers and 30% increase in total ABE titer (0.35 g ABE/g sucrose) in batch fermentations. Additionally, two engineered strains overexpressing aldehyde/alcohol dehydrogenases (encoded by adh11 and adh5) displayed 8.5- and 11.8-fold increases (respectively) in batch ethanol production.
IMPORTANCE This paper presents the first steps toward advanced genetic engineering of the industrial butanol producer Clostridium saccharoperbutylacetonicum strain N1-4 (HMT). In addition to providing an efficient method for introducing foreign DNA into this species, we demonstrate successful rational engineering for increasing solvent production. Examples of future applications of this work include metabolic engineering for improving desirable industrial traits of this species and heterologous gene expression for expanding the end product profile to include high-value fuels and chemicals.
KEYWORDS: ABE fermentation, Clostridium, biofuel, genetics
INTRODUCTION
Given the pressing need for alternative sources of liquid transportation fuels that are both renewable and economically feasible, much attention has surrounded microorganisms capable of converting biomass-derived sugars into suitable replacement fuels (1). Acetone-butanol-ethanol (ABE) fermentation by certain members of the anaerobic bacterial genus Clostridium serves as a promising solution to the need for renewable liquid fuels, either through the direct use of butanol as a “drop-in” fuel (2) or by using all three products as precursors for catalytic conversion to hydrocarbons with molecular weights similar to those found in gasoline, diesel, or aviation fuel (3). In addition to the utility of their end products, solvent-producing Clostridium organisms are valued for their ability to metabolize a variety of carbon sources such as pentoses, hexoses, oligosaccharides, and lignocellulose hydrolysates, permitting significant flexibility in the selection of biological feedstocks (4, 5).
Since the first large-scale implementation of ABE fermentation with Clostridium acetobutylicum during World War I (6), a number of other solventogenic Clostridium species have been discovered and subsequently employed for industrial operation, including C. beijerinckii, C. saccharobutylicum, and C. saccharoperbutylacetonicum (7). Compared to the other major industrial ABE producers, C. saccharoperbutylacetonicum is characterized by its high selectivity toward butanol (as high as 85% of the total solvents produced), low sporulation frequency (a desirable trait for industrial operation), and wide range of metabolizable carbohydrates (8–10). First detailed in 1960 (8), C. saccharoperbutylacetonicum has been the subject of numerous academic studies highlighting the diverse renewable feedstocks that can be utilized by this species, including molasses (11), palm oil (12), cassava (13), sago starch (14), rice bran (15), agricultural waste (16), and lignocellulosic hydrolysate (17–19). Evaluating the species from an engineering perspective, other studies have demonstrated that C. saccharoperbutylacetonicum is amenable to operating in a continuous mode (20–22) as well as incorporating in situ separation strategies such as liquid-liquid extraction (23) and membrane pervaporation (24). Thus, the demonstrated industrial scalability, feedstock flexibility, and downstream processability associated with C. saccharoperbutylacetonicum indicate that this species is highly attractive for use in industrial biofuel production.
Despite the favorable fermentative characteristics of C. saccharoperbutylacetonicum, a major drawback associated with this species is the distinct lack of tools and techniques available for performing genetic manipulations. This is in stark contrast to C. acetobutylicum and C. beijerinckii, for which transformation methods have existed for more than 20 years (25, 26). Building on these methods, rational metabolic engineering of these two species has enabled significant progress in expanding substrate utilization, improving oxygen tolerance, eliminating sporulation, increasing solvent titers/productivities, and enabling the generation of valuable end products beyond ABE (27–31). Without an established transformation method, stable host/vector system, and efficient gene disruption strategy, the types of advances made using rational metabolic engineering in C. acetobutylicum and C. beijerinckii are not possible for C. saccharoperbutylacetonicum without cumbersome and tedious screening of traditional mutagenesis libraries. To our knowledge, only one report from 2007 has detailed a transformation method for any C. saccharoperbutylacetonicum strain (strain N1-4 ATCC 13564) (32). Other than this report and a single follow-up study by the same group in 2008 (33), we were unable to find any reports demonstrating transformation methods or heterologous gene expression for any of the C. saccharoperbutylacetonicum strains [which include strain N1-4 ATCC 13564 and its two derivatives, strain N1-4 (HMT) ATCC 27021 and strain N1-504 ATCC 27022 (9)]. As strain N1-4 ATCC 13564 has long been deaccessioned, we attempted the published transformation method (32) using the publicly available strain N1-4 (HMT) ATCC 27021 (proposed to be the current type strain [9]) and were unable to obtain any transformants. Therefore, we sought to develop a genetic transformation method for C. saccharoperbutylacetonicum strain N1-4 (HMT) ATCC 27021 (hereafter referred to as C. saccharoperbutylacetonicum N1-4), determine if plasmid-based gene overexpression and targeted gene deletion would be possible, and importantly, demonstrate improvements in its fermentation performance using rational metabolic engineering.
Here we report the development of an efficient, robust, and repeatable genetic transformation method for C. saccharoperbutylacetonicum N1-4, along with the first reported targeted gene deletions of any C. saccharoperbutylacetonicum strain. A key finding was the discovery of multiple phenotypic subtypes of C. saccharoperbutylacetonicum N1-4 that displayed dramatic differences in transformability via electroporation. After establishing a repeatable transformation method, we selected 12 genes across the ABE metabolic network for overexpression studies, report batch fermentation data for five strains that displayed altered solvent titers, and report the fermentation performance of three gene deletion strains. These metabolic engineering efforts resulted in several engineered strains with improved ABE production. This study opens the door to future metabolic engineering efforts in C. saccharoperbutylacetonicum N1-4 through gene overexpression and targeted gene deletions, as well as newly developed genome-editing tools recently adapted for use in Clostridium. Moreover, we believe that these advances make C. saccharoperbutylacetonicum N1-4 a competitive candidate for industrial biofuel production.
RESULTS
Initial transformation attempts and identification of morphological subtypes of C. saccharoperbutylacetonicum N1-4.
Many clostridia are notorious for their genetic intractability, often due to their unwillingness to accept foreign DNA. Thus, our first challenge was to develop a protocol capable of overcoming the DNA restriction-modification systems, membrane-associated DNase activity, morphological heterogeneity, and overall low transformation efficiencies displayed by most clostridia (34). Transformation via electroporation was our natural first choice given the high transformation efficiencies compared to conjugation or other DNA transfer methods. Recently, an excellent review (34) of successful DNA transfer procedures for clostridia was published, providing a starting point for establishing appropriate electroporation parameters.
After determining an appropriate liquid and solid growth medium (2× YTG [pH 6.5], consisting of 16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl, 10 g/liter glucose, with an additional 15 g/liter agar for solid medium), and antibiotic for selection (erythromycin [Ery], with a measured MIC of ∼10 μg/ml), we attempted electroporation of mid-log-phase cells (optical density at 600 nm [OD600], ∼ 0.6) using a dam dcm methylated Escherichia coli-Clostridium shuttle vector deemed pWIS_empty, a derivative of the pIMP vector backbone commonly employed for transformations in C. acetobutylicum ATCC 824 (26, 35). Following liquid recovery and incubation on 2× YTG + Ery (20 μg/ml) plates, we observed no transformant colonies. To overcome potential type II DNA endonuclease activity, we performed in vivo methylation of pWIS_empty prior to electroporation using the Bacillus subtilis phage φ3T I methyltransferase (expressed on E. coli plasmid pAN1). This procedure protects plasmid DNA from targeted degradation in C. acetobutylicum ATCC 824 through methylation of 5′-GGCC-3′ and 5′-GCNCG-3′ DNA sequences and was the key to achieving efficient transformations in C. acetobutylicum (25). Still, following DNA methylation and electroporation, no transformant colonies were observed. As electroporation parameters have dramatic effects on transformation efficiency, we tested a wide range of parameters (electroporation voltages, resistances, and buffers) as well as competent cell preparation methods (log-phase versus stationary-phase cells, membrane solubilization, and washing procedures). Again, we did not observe transformants under any of these conditions.
While exploring additional transformation strategies, we happened to make a seemingly unrelated discovery; upon plating on solid 2× YTG medium, three distinct colony morphologies of C. saccharoperbutylacetonicum N1-4 could be observed, which we called regular (R), irregular (I), and sticky (S) (Fig. 1A). While all three morphological subtypes originated from stocks purchased from ATCC (ATCC 27021), one ATCC stock yielded mixtures of R and I variants (no S variants), while an independently purchased ATCC stock yielded only the S morphology, the latter being the subtype used for our initial transformation attempts. The three subtypes were sequenced for 16S rRNA and showed 100% identity agreement with that of the published sequence (NCBI accession number NC_020291.1). Additionally, we confirmed that all three variants harbor the expected 136-kb megaplasmid (NCBI accession number NC_020292.1), which features a number of seemingly unrelated genes as well as many hypothetical proteins (see Fig. S2 in the supplemental material). Thus, the phenotype differences could not be explained by megaplasmid loss, a phenomenon known to lead to strain degeneration (the loss of ability to produce solvents) in C. acetobutylicum ATCC 824 (4).
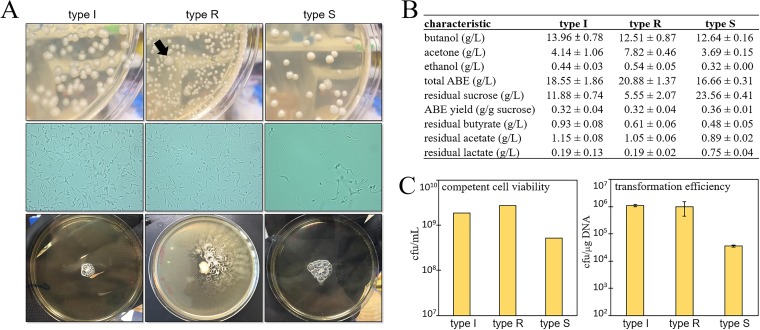
FIG 1.
Comparison of three morphological subtypes of C. saccharoperbutylacetonicum N1-4. (A) Top row, colonies of each subtype cultured on TYA plates. The black arrow on the type R plate indicates an example of the spontaneous appearance of a type I colony. Middle row, bright-field microscopy images of the three subtypes, with samples diluted from liquid 2× YTG cultures (OD600, ∼1.0). Bottom row, motility assays on solid TYA plates. (B) Batch flask fermentation comparison of the three subtypes in PL7S media with samples taken 66 h postinoculation. Fermentations were performed in biological triplicates, for which we report the average. (C) Competent cell viability and transformation efficiency of the three subtypes using the optimized transformation protocol. Electroporations were performed in triplicate.
While type R colonies featured a circular/round form and smooth margin, type I colonies had an irregular form and undulate margin (Fig. 1A). Type S colonies (named for their sticky texture) appeared to have characteristics of both type I and type R, with a slightly irregular form and entire margin, but were convex in elevation (compared to a slightly raised elevation for R and I). Interestingly, the type R phenotype appeared to be unstable; spontaneous conversion to the type I phenotype was observed upon restreaking (Fig. 1A). To probe this phenomenon further, three type R colonies were cultured in separate liquid cultures for 120 h, dilutions were plated onto solid media, and the resulting colony phenotypes (either type R or type I) were enumerated after 48 h of incubation. The results varied widely for the three original cultures; culture A yielded 39% type R colonies, culture B yielded 89% type R colonies, and culture C yielded 100% type R colonies. Thus, the R phenotype was unstable, and the conversion of colonies from type R to type I occurred spontaneously at variable rates. The reverse phenotype conversion (type I to type R) was not detected under any conditions, indicating that the type I morphology was likely stable. Similarly, we did not observe any morphology conversions in the offspring of type S colonies, which appeared to be another stable phenotype.
Additional assays showed high motility for type R cells, low motility for type I and S cells, visible spore formation for all three subtypes, and visibly different cellular morphologies according to light microscopy (Fig. 1A). Slight differences in batch solvent production were observed for the three subtypes, with the highest butanol production by type I (14.0 g/liter) and the highest acetone production by type R (7.8 g/liter) (Fig. 1B). Type S showed lower sucrose utilization, with 23.6 g/liter remaining at the end of the fermentation, potentially related to the lower cell viability observed for this strain, although this subtype displayed a higher ABE yield (0.36 g/g sucrose) than did the other subtypes.
Development of a high-efficiency transformation protocol for C. saccharoperbutylacetonicum N1-4 subtype I.
Two observations indicated that the three C. saccharoperbutylacetonicum N1-4 variants might exhibit different transformation efficiencies. Qualitatively, we observed what appeared to be high extracellular polysaccharide (EPS) production in liquid cultures from type R colonies: after several days of incubation in PL7S medium, culture supernatant remained cloudy and somewhat viscous, and centrifugation resulted in loose pellets (see Fig. S1 in the supplemental material). This was unlike type I and S cultures, which demonstrated very little cloudiness in aged cultures and yielded compact pellets following centrifugation. Second, we observed significantly lower cell viability (in terms of CFU per milliliter) for type S cultures than for I and R types. As both EPS production and cell viability are factors known to impact transformation efficiencies, we expanded our electroporation efforts to all three C. saccharoperbutylacetonicum N1-4 subtypes. Using the same initial electroporation procedure (and dam dcm methylated plasmid) that failed to yield transformants for type S cells, we were able to obtain a substantial number of erythromycin-resistant transformants for type I and R subtypes (transformation efficiencies on the order of 104 CFU/μg DNA). Transformants were confirmed by plasmid purification from liquid cultures, followed by DNA restriction digest gels, which showed bands consistent with the transformed pWIS_empty vector. With improvements in electroporation parameters (by testing voltages between 1.0 and 2.0 kV and resistances between 200 and infinite Ω) and competent cell preparation, we achieved transformation efficiencies of 1.1 × 106 (±9.3 × 104) and 9.8 × 105 (±5.4 × 105) CFU/μg DNA for type I and R cells, respectively (Fig. 1C). Interestingly, we observed an increase of 1 order of magnitude in transformation efficiencies using C. saccharoperbutylacetonicum N1-4 type I by performing competent cell preparation at room temperature (versus 4°C, as is used for preparations of C. acetobutylicum competent cells [25]). Comparing the viabilities of competent cells stocks of the three subtypes, it appeared that the significantly lower transformability of the S subtype was largely due to its decreased viability. Despite this, the optimized transformation method developed using type I cells permitted successful transformations of type S cells, albeit at significantly lower efficiencies. Unexpectedly, the suggested difference in EPS production between type R and I variants did not appear to have a significant impact on transformability.
Development of targeted gene knockout (KO) method for C. saccharoperbutylacetonicum N1-4.
Recent advances in the genetic manipulation of clostridia (primarily C. acetobutylicum ATCC 824 and C. beijerinckii NCIMB 8052) have afforded several methods for targeted gene disruption, including (i) heterologous expression of mobile group II introns with the ClosTron system (36), (ii) allelic exchange via homologous recombination (37, 38), and very recently, (iii) clustered regularly interspaced short palindromic repeat(s) (CRISPR)-Cas9 genome editing (39–41). Given the observed instability and off-target effects associated with mobile group II introns (42) as well as the start-up time required to adapt an efficient CRISPR-Cas9 system for use in C. saccharoperbutylacetonicum N1-4, we chose here to pursue a double-crossover-based allelic exchange (DCAE) method. Given the instability of the subtype R phenotype and the relatively low transformation efficiencies of subtype S cells, we chose to work exclusively with C. saccharoperbutylacetonicum N1-4 subtype I for genetic manipulations.
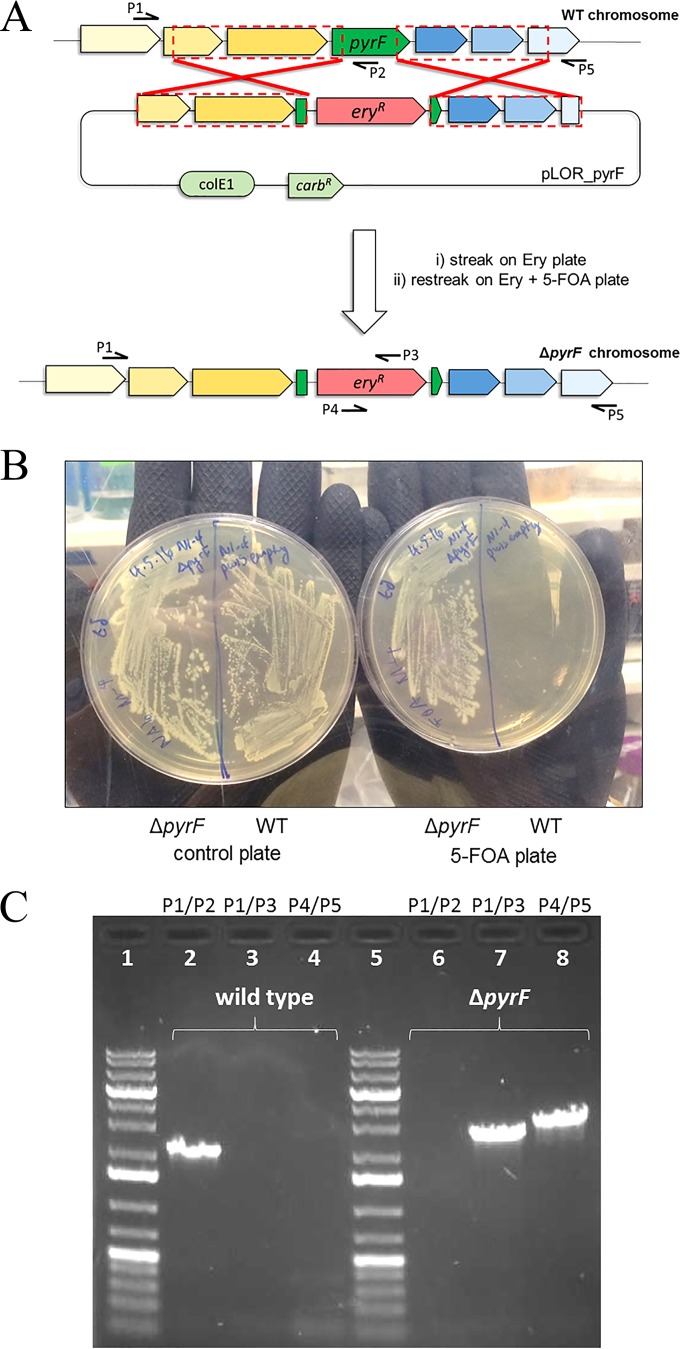
To perform targeted gene KOs, we constructed the plasmid series pLOR, containing an erythromycin resistance-based allelic exchange cassette and the necessary genetic elements for plasmid maintenance in E. coli (ColE1 replicon and ampicillin resistance). Notably, we omitted the Gram-positive replicon RepL (present on the pWIS vector backbone) to avoid the need for plasmid curing. To test this system, we first targeted the pyrF gene from C. saccharoperbutylacetonicum N1-4 (CSPA_RS05335) for deletion, as mutants of pyrF are known to exhibit resistance to the toxic antimetabolite 5-fluoroorotic acid (5-FOA) (37). Additionally, as pyrF is necessary for the biosynthesis of uracil, pyrF mutants should display uracil auxotrophy. For performing this targeted gene KO, we constructed plasmid pLOR_pyrF, containing ∼2-kb regions of homology that flank a majority of the pyrF coding region (Fig. 2A). Following electroporation of type I cells, recovery, and a 72-h incubation on erythromycin-containing plates, we obtained about a dozen transformants. Since pyrF mutants should exhibit resistance to 5-FOA, we cultured one transformant in liquid medium until it reached the late log phase (OD600 of approximately 1.0) and plated dilutions on 2× YTG plates containing 500 mg/liter 5-FOA. Consistent with the expected ΔpyrF phenotype, the transformant was able to grow on the 5-FOA plate, while a wild-type control strain did not display any growth (Fig. 2B). The transformant also displayed the expected uracil auxotrophy as evidenced by its need for uracil supplementation for growth on defined solid media (see Fig. S5 in the supplemental material). This transformant was further evaluated by colony PCR, which confirmed the predicted allelic exchange event (Fig. 2C). Thus, we demonstrated a successful DCAE method for gene KO in C. saccharoperbutylacetonicum N1-4 using a nonreplicating vector.
FIG 2.
(A) Diagram of the DCAE method applied to targeted KO of the C. saccharoperbutylacetonicum N1-4 pyrF gene. Dashed red boxes indicate the ∼2-kb regions of homology present in pLOR_pyrF. Red X's represent homologous recombination events taking place between the C. saccharoperbutylacetonicum N1-4 chromosome and pLOR_pryF. P1 to P5 indicate annealing regions for primers used in colony PCR confirmation of successful mutants. (B) Demonstration of a successful pyrF mutant. Liquid 2× YTG cultures (OD600, ∼1.0) of ΔpyrF and wild-type (WT) control strains were streaked and incubated (72 h) on a 2× YTG plate (left) and a 2× YTG–500 mg/liter 5-FOA plate (right). (C) DNA electrophoresis gel displaying results of colony PCRs for the isolated pyrF mutant as well as a wild-type control. PCR bands from lanes 7 and 8 were gel extracted and analyzed by Sanger sequencing to further confirm the desired allelic exchange event.
Manipulation of N1-4 fermentative pathways using rational metabolic engineering.
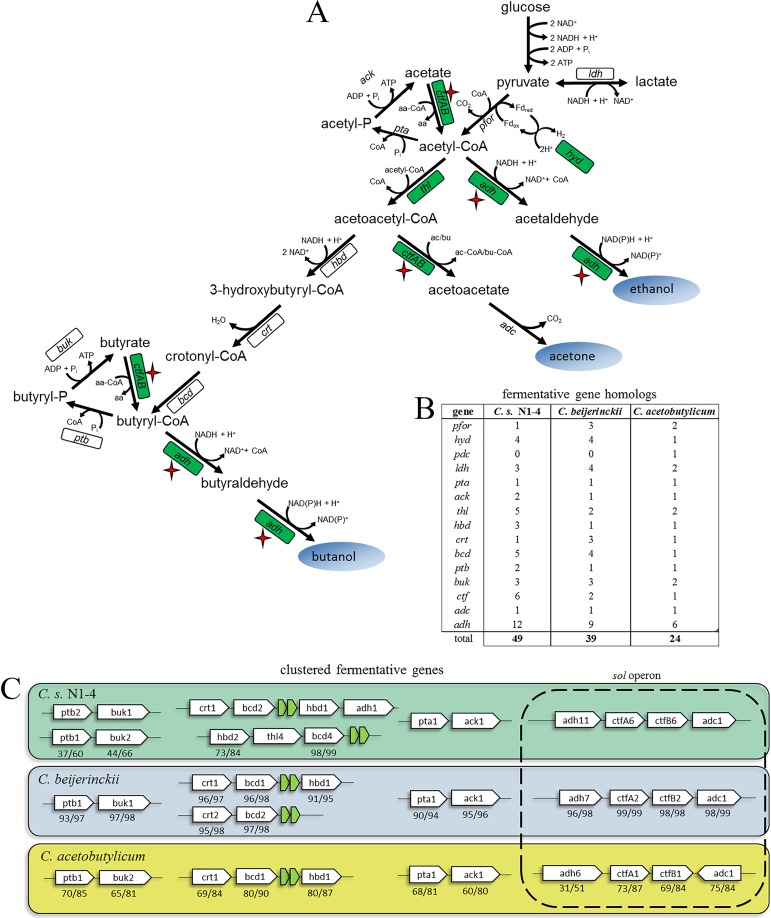
With a viable transformation method, host/vector system for gene expression, and targeted gene KO method, we sought to apply these techniques for rational metabolic engineering of C. saccharoperbutylacetonicum N1-4 fermentative pathways (Fig. 3A). We began by performing a bioinformatic analysis of the putative ABE-related genes present in the recently published C. saccharoperbutylacetonicum N1-4 genome (43, 44) and comparing these to the confirmed ABE pathway genes present in the well-studied C. beijerinckii NCIMB 8052 (a close genetic relative of C. saccharoperbutylacetonicum N1-4 [45]) and the model ABE producer C. acetobutylicum ATCC 824 (Fig. 3C). Interestingly, we found many redundancies in the putative ABE pathway genes present in C. saccharoperbutylacetonicum N1-4 compared to the other two species (Fig. 3B). Despite these differences, there are still clear similarities in the clustering of certain ABE-related genes (for instance, the so-called sol operon of adh-ctfA-ctfB-adc genes necessary for solvent production in C. acetobutylicum) for the three species.
FIG 3.
(A) Putative ABE fermentative pathways for C. saccharoperbutylacetonicum N1-4 based on C. acetobutylicum ABE metabolism. Genes with homologs selected for initial gene overexpression studies are boxed. The subset of these genes with reported batch fermentation data are highlighted in green. A red star indicates that a strain with a deletion in this gene was generated and evaluated by batch fermentation. Enzymes: pfor, pyruvate:ferredoxin oxidoreductase; hyd, hydrogenase; pdc, pyruvate decarboxylase; ldh, lactate dehydrogenase; pta, phosphotransacetylase; ack, acetate kinase; thl, thiolase; hbd, 3-hydroxybutylryl-CoA dehydrogenase; crt, crotonase; bcd, butyryl-CoA dehydrogenase complex; ptb, phosphotransbutyrylase; buk, butyrate kinase; ctf, CoA transferase; adc, acetoacetate decarboxylase; adh, alcohol and/or aldehyde dehydrogenase. Metabolites: aa-CoA, acetoacetyl-CoA; ac-CoA, acetyl-CoA; bu-CoA, butyryl-CoA; aa, acetoacetate; ac, acetate; bu, butyrate. (B) Numbers of fermentative gene homologs in three solventogenic clostridia based on bioinformatic analysis. Further details are presented in Table S2 in the supplemental material. (C) Diagram of clustered fermentative genes for C. saccharoperbutylacetonicum N1-4 (NCBI accession numbers NC_020291.1 and NC_020292.1), C. beijerinckii NCIMB 8052 (NCBI accession number NC_009617.1), and C. acetobutylicum ATCC 824 (NCBI accession numbers NC_003030.1 and NC_001988.2). Further details are presented in Table S2 in the supplemental material. Numbers below genes indicate the corresponding protein percent identity/similarity compared to the primary C. saccharoperbutylacetonicum N1-4 homolog.
Without any prior knowledge on the function of these 49 predicted ABE pathway genes, we selected 12 genes/gene pairs for targeted gene expression studies (Table 2). Highlighted in Fig. 3A, we selected genes that spanned most of the primary ABE fermentative pathways to probe which of these genes had major bearings on the end product profile of batch fermentations. We purposefully included genes from clusters homologous to those found in C. beijerinckii and C. acetobutylicum (including ptb2, buk1, and adh11), in addition to genes outside these conserved clusters (including thl4, adh5, and ctfAB2) to investigate whether these redundant homologs might also be important for ABE fermentation in C. saccharoperbutylacetonicum N1-4. We cloned these 12 genes/gene pairs into the pWIS vector backbone under the control of the C. acetobutylicum crotonase (crt) promoter, which we found to be transcribed constitutively in C. saccharoperbutylacetonicum N1-4. Of these 12 starting vectors, 10 yielded a high number of C. saccharoperbutylacetonicum N1-4 (subtype I) transformants, while two constructs (both expressing bcd genes) did not yield any transformants.
TABLE 2.
List of native C. saccharoperbutylacetonicum N1-4 genes selected for gene overexpression studiesa
| Vector | Locus tag | Gene locus | Gene annotation |
|---|---|---|---|
| pWIS2 | ptb2 | CSPA_RS01245 | Phosphate butyryltransferase |
| buk1 | CSPA_RS01250 | Butyrate kinase | |
| pWIS3 | crt1 | CSPA_RS02130 | 3-Hydroxybutyryl-CoA dehydratase |
| bcd2 | CSPA_RS02135 | Acyl-CoA dehydrogenase | |
| pWIS4 | hbd1 | CSPA_RS02150 | 3-Hydroxybutyryl-CoA dehydrogenase |
| adh1 | CSPA_RS02155 | NADPH-dependent butanol dehydrogenase | |
| pWIS6 | ldh1 | CSPA_RS05375 | l-Lactate dehydrogenase |
| pWIS7 | hbd2 | CSPA_RS10210 | 3-Hydroxybutyryl-CoA dehydrogenase |
| pWIS8 | thl4 | CSPA_RS10215 | Acetyl-CoA acetyltransferase |
| pWIS9 | bcd4 | CSPA_RS10220 | Acyl-CoA dehydrogenase |
| pWIS10 | ctfA2 | CSPA_RS10270 | CoA transferase subunit alpha |
| pWIS10 | ctfB2 | CSPA_RS10275 | CoA transferase subunit beta |
| pWIS11 | adh2 | CSPA_RS10510 | Aldehyde-alcohol dehydrogenase |
| pWIS12 | adh11 | CSPA_RS27680 | Aldehyde dehydrogenase |
| pWIS13 | hyd1 | CSPA_RS11980 | [FeFe] hydrogenase group A |
| pWIS14 | adh5 | CSPA_RS18045 | Aldehyde-alcohol dehydrogenase |
Gene annotations are based on NCBI designations. Genes/vectors used in detailed batch fermentations are highlighted in boldface.
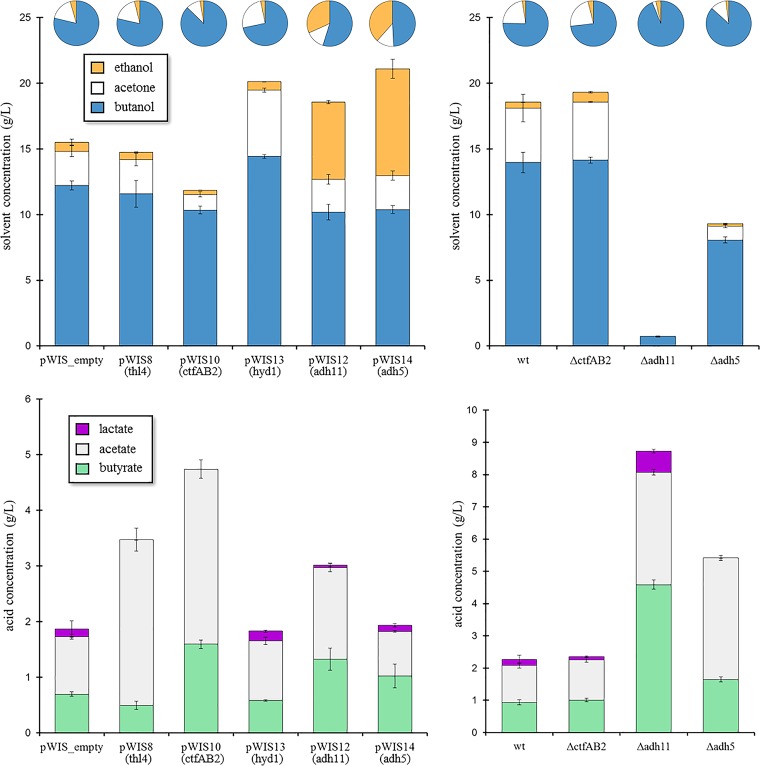
To evaluate the 10 generated C. saccharoperbutylacetonicum N1-4 overexpression strains, we performed preliminary batch fermentations in an anaerobic chamber with PL7S growth medium, a derivative of the commonly used clostridial growth medium (CGM; see Materials and Methods). We selected sucrose (70 g/liter) as the carbon source, given that the feedstock sugarcane juice (primarily sucrose) is an attractive low-cost industrial carbon source (46), in addition to the fact that C. saccharoperbutylacetonicum N1-4 has long been known to readily ferment sucrose-based feedstocks (11). Of the 10 strains, 5 appeared to show significant deviations in their end product profiles relative to a control strain and were used as the basis for detailed batch fermentation analysis (see Table 2). Fermentations of C. saccharoperbutylacetonicum N1-4/pWIS8, C. saccharoperbutylacetonicum N1-4/pWIS10, C. saccharoperbutylacetonicum N1-4/pWIS12, C. saccharoperbutylacetonicum N1-4/pWIS13, and C. saccharoperbutylacetonicum N1-4/pWIS14, and the control strain, C. saccharoperbutylacetonicum N1-4/pWIS_empty, revealed significant differences in both ABE titers and ratios among the six strains (Fig. 4). While C. saccharoperbutylacetonicum N1-4/pWIS10 (overexpressing the coenzyme A [CoA] transferase genes ctfA2 and ctfB2) exhibited decreased ABE production (with a 15% decrease in butanol and a >50% decrease in acetone and ethanol titers), C. saccharoperbutylacetonicum N1-4/pWIS13 (overexpressing the hydrogenase gene hyd1) showed significantly increased butanol and acetone titers (18% and 93% increases, respectively). Additionally, while C. saccharoperbutylacetonicum N1-4/pWIS12 and C. saccharoperbutylacetonicum N1-4/pWIS14 (overexpressing the alcohol/aldehyde dehydrogenase genes adh11 and adh5, respectively) showed slight decreases in butanol titers, substantial increases in ethanol production for both of these strains (8.5- and 11.8-fold-higher ethanol titers, respectively) made for higher overall ABE titers. In particular, C. saccharoperbutylacetonicum N1-4/pWIS14 produced the highest total ABE titer (21.1 g/liter) and ABE yield (0.39 g/g sucrose) observed in this study (see Table S3 in the supplemental material). Residual acid (butyrate and acetate) levels were consistent with the solvent production for the six strains, as relatively high overall acid accumulation was observed for the strains with decreased ABE production (C. saccharoperbutylacetonicum N1-4/pWIS8 and C. saccharoperbutylacetonicum N1-4/pWIS10).
FIG 4.
Batch solvent (top row) and acid (bottom row) production for engineered strains of C. saccharoperbutylacetonicum N1-4. Top left and bottom left plots display results for gene overexpression strains (with pWIS_empty as the control strain), while top right and bottom right plots display results for gene deletion strains (with vectorless wild-type [wt] type I as the control strain). Genes overexpressed in pWIS-harboring strains are identified in parentheses below their respective strain names. Fermentations were performed under anaerobic conditions in PL7S media (70 ml). Endpoint samples were taken 66 h after inoculation. Distribution of solvent end products are represented with pie charts for each strain. Further details are provided in Table S3 in the supplemental material.
Based on the above observations, we attempted to create C. saccharoperbutylacetonicum N1-4 mutants targeting the genes that hampered ABE production when overexpressed: thl4 (expressed on C. saccharoperbutylacetonicum N1-4/pWIS8) and ctfA2 and ctfB2 (expressed on C. saccharoperbutylacetonicum N1-4/pWIS10), to explore if any beneficial effects could be observed by deleting these genes. Additionally, we wanted to determine if either adh5 or adh11 encoded an ethanol-specific dehydrogenase (rather than a bifunctional ethanol/butanol dehydrogenase); therefore, strains with deletions of these genes were also desired. Following the procedure that we employed for deleting the pyrF gene, we created KO vectors targeting the four genetic loci (pLOR_thl, pLOR_ctfAB2, pLOR_adh5, and pLOR_adh11) and performed transformations in C. saccharoperbutylacetonicum N1-4 (subtype I). After 72 h of incubation on 2× YTG solid medium, 4 to 12 colonies appeared on each of the transformation plates. The desired double-crossover mutants were isolated at a frequency of roughly one double-crossover KO for every eight screened colonies except for thl4, for which we were able to isolate only single-crossover mutants despite repeated efforts and extensive subcultures.
While batch fermentation of the ΔctfAB2 strain showed improvements relative to the C. saccharoperbutylacetonicum N1-4/pWIS_empty strain, comparison to the vectorless type I wild type showed comparable batch fermentation performance (Fig. 4). Thus, deletion of ctfAB2 did not appear to significantly affect ABE production, while overexpression proved harmful to solvent production. Interestingly, deletion of either adh5 or adh11 resulted in significantly decreased ABE production. In particular, deletion of adh5 led to decreases in titers of butanol, acetone, and ethanol of 42%, 75%, and 58%, respectively (compared to the vectorless wild type); while deletion of adh11 abolished acetone and ethanol production and decreased the butanol titer to less than 1 g/liter. High acid accumulation in both Δadh5 and Δadh11 mutants confirmed the inability of these strains to effectively reduce CoA-bound intermediates to the typical alcohol products.
DISCUSSION
Since the discovery of an efficient gene transfer method over 20 years ago, C. acetobutylicum ATCC 824 has served as host for the vast majority of genetic engineering studies performed on solventogenic clostridia. This has been due largely to the well-studied physiology, fermentative metabolism, and industrial performance of this organism, dating back to the early 20th century. In the last 10 years, long-awaited progress in genetic tools for use in clostridia has afforded advanced genetic manipulations of C. acetobutylicum. Despite this, advanced genetic tools are of no use to genetically intractable organisms, as is the case for most known Clostridium species. Recent achievements in developing efficient transformation methods for historically intractable Clostridium species (e.g., C. pasteurianum, C. cellulovorans, and C. ljungdahlii) (47–49) have permitted novel genetic engineering efforts for these species. These accomplishments encourage the development of transformation methods and genetic tools for historically intractable solventogenic strains, particularly those with a history of strong industrial performance.
In this study, we present the development and application of a high-efficiency transformation method for the industrial butanol hyperproducer C. saccharoperbutylacetonicum strain N1-4 (HMT) ATCC 27021. Key to the development of this transformation method, we identified three distinct colony morphologies (types R, I, and S), which displayed significant differences in transformability. Presently, it is unclear whether these distinct colony morphologies arise from genetic or epigenetic factors. The high frequency and unpredictability of type R to type I conversions suggest that epigenetic influences may be at play, although it is unclear which factors might stimulate this conversion. As type I and type S colonies were stable phenotypes under all conditions that we evaluated, mutations at one or more genetic loci could be responsible for the observed differences between these two subtypes. Comparing early descriptions of C. saccharoperbutylacetonicum colony morphologies (summarized in reference 9) to those that we observed, either type R or type S colonies seem to be consistent with these records, which reported domed, white, smooth colonies with entire/undulated margins. Although we did not find previous reports of plate motility assays being performed, our results demonstrated that type R and type S colonies could easily be distinguished using this assay (Fig. 1A). Differences in ABE production between the subtypes raised further questions as to the genetic or epigenetic factors which distinguish these subtypes. For the purposes of this study, we chose to perform all strain engineering using type I cells, given their stable phenotype and high transformation efficiencies, despite the higher total ABE production offered by subtype R (Fig. 1B). However, factors other than ABE titers are important in evaluating the industrial utility of different strains—certainly the production of EPS-like material from liquid cultures of type R colonies could present challenges for industrial applications.
Using a DCAE method for generating KOs permitted, to our knowledge, the first reported examples of targeted gene deletions in C. saccharoperbutylacetonicum. Furthermore, high transformation efficiencies (∼1 × 106 CFU/μg DNA) permitted the use of a nonreplicating KO vector (pLOR), avoiding the need for plasmid curing. While we were able to generate four mutant strains using this technique, a counterselection system for the elimination of single-crossover mutants is highly desirable for future gene deletions. It is unknown if our inability to generate a few of the desired KOs was due to allelic exchange inefficiency or the essential nature of the targets. Unfortunately, we were unable to apply a recently published counterselection method based on a C. perfringens lactose-inducible promoter (38), as we observed transcription of this promoter with or without lactose addition in C. saccharoperbutylacetonicum N1-4 (data not shown). We are currently working to employ a pyrF-based selection system (working with the ΔpyrF strain developed in this study), as we have since identified additional antibiotic markers effective in C. saccharoperbutylacetonicum N1-4. Counterselection is particularly important for DCAE gene deletions in C. saccharoperbutylacetonicum N1-4, which seemed to have a relatively low double-crossover rate. Low homologous recombination frequencies could also be addressed by including a resolvase protein (RecU) on the pLOR backbone, as this has proven to be an effective method of improving recombination efficiencies in other Clostridium species (50).
To select targets for rational metabolic engineering in C. saccharoperbutylacetonicum N1-4, we performed bioinformatic comparisons to published C. acetobutylicum and C. beijerinckii genomes (Fig. 3B and C). These revealed similarities in fermentative gene clustering (most notably, the sol operon), but also a large number of redundant ABE-related genes in the C. saccharoperbutylacetonicum N1-4 genome. Curiously, while some redundant genes are highly homologous (e.g., bcd2 and bcd4 have 98% identical and 99% similar protein sequences), others showed significant divergence (e.g., buk1 and buk2 have only 37% identical and 60% similar protein sequences). The consequences of these gene redundancies in the context of ABE fermentation are currently unclear; further work is required to determine which homologs are actually expressed over the course of a fermentation, evaluate whether any synergy is achieved in expressing genes with redundant function, and assess the impact of deleting “unnecessary” homologs. Toward addressing these questions, we emphasized selecting multiple gene homologs across the ABE metabolic network for our plasmid-based overexpression studies.
The reason for the slightly decreased batch performance of the thl4 overexpression strain (C. saccharoperbutylacetonicum N1-4/pWIS8) is somewhat unclear. The acetate accumulation observed (3.0 g/liter) was unexpected, as thiolase is predicted to direct carbon flux away from the acetyl-CoA/acetyl-P/acetate cycle (Fig. 3A). One possible explanation is that thl4 is relatively inefficient compared to the four other predicted copies of the thiolase gene in the N1-4 genome. If valid, thl4 overexpression may cause nonproductive substrate (acetyl-CoA) titration, placing additional stress on upstream pathways to increase acetyl-CoA pools. The pWIS10 strain (overexpressing ctfAB2) demonstrated a similar behavior in batch fermentation (reduced solvent production and increased acid accumulation), although the effect was more pronounced in this strain. A similar substrate titration effect due to a relatively inefficient CtfAB2 (acting on acetoacetyl-CoA) could be a possible explanation for this behavior. Thus, we postulate that CtfAB2 is not the primary CoA transferase in C. saccharoperbutylacetonicum N1-4, also supported by the retained ability of the ΔctfAB2 strain to reassimilate acetate and butyrate. This example demonstrates the strategy of strain improvement through deletion of potentially inefficient redundant genes, although in this case, ctfAB2 deletion did not significantly affect the fermentation profile. Deletion of the five other putative CoA transferase genes would serve to explore this strategy further.
The batch fermentation performance of strains C. saccharoperbutylacetonicum N1-4/pWIS12 and C. saccharoperbutylacetonicum N1-4/pWIS14 (expressing adh11 and adh5, respectively) is consistent with the predicted aldehyde dehydrogenase activity for these proteins, as total alcohol production increased (primarily ethanol) for these strains. E. coli heterologous expression and in vitro analyses of Adh11 (referred to as “Bld” in these studies) have previously been performed for applications in synthetic 1,3-butanediol (51) and 1,4-butanediol (52) pathways, demonstrating the ability of Adh11 to reduce butyryl-CoA derivatives to their corresponding aldehydes. As evidenced by the increased ethanol production observed for adh11 and adh5 overexpression strains, Adh11 and Adh5 appear to also have significant activity toward acetyl-CoA as a substrate. Under this assumption, ethanol (rather than butanol) production may have increased due to increased metabolic flux toward acetaldehyde from acetyl-CoA, outweighing the flux directed from acetyl-CoA toward butyryl-CoA (and eventually butanol). Therefore, protein engineering of Adh11/Adh5 to improve the substrate specificity for butyryl-CoA (and decrease the substrate specificity for acetyl-CoA) could be an effective strategy to improve butanol titers in C. saccharoperbutylacetonicum N1-4. From our results alone, we are unable to confirm the bifunctional aldehyde/alcohol dehydrogenase activity predicted for Adh5, as the C. saccharoperbutylacetonicum N1-4 genome is predicted to encode several other alcohol dehydrogenases (Fig. 3B). Deletion of adh5 and adh11 revealed significant disruptions in solvent production, indicating that multiple dehydrogenases are likely involved in alcohol production in C. saccharoperbutylacetonicum N1-4. Efforts to characterize the roles of different aldehyde/alcohol hydrogenases in C. acetobutylicum (most recently described in references 53 and 54) have taken a similar reverse genetics approach and have been successful in identifying butanol-specific dehydrogenases useful for improving butanol selectivities. This approach could easily be extended to C. saccharoperbutylacetonicum N1-4 by evaluating mutants/overexpression strains of all 12 predicted adh genes (see Table S2 in the supplemental material).
The improved solvent (particularly acetone) production by the pWIS13 strain (overexpressing the hydrogenase gene hyd1) was unexpected given that increased hydrogen production has previously been shown to result in decreased butanol production for C. acetobutylicum (55). In C. acetobutylicum, reduced ferredoxin competitively donates reducing equivalents to either HydA or ferredoxin:NAD(P)H oxidoreductase, leading to increased NAD(P)H production (and thus, butanol production) for strains with reduced hydrogenase activity (56). Quantification of the hydrogen evolution of the hyd1 overexpression strain versus a control strain could aid in explaining our results, as well as performing gene overexpression/deletion studies for the other three predicted hydrogenase genes in C. saccharoperbutylacetonicum N1-4 (Table S2).
In summary, we have demonstrated a methodology for highly efficient transformations, genetic machinery for plasmid-based gene overexpression, targeted gene KOs through allelic exchange using a nonreplicating vector, and successful rational metabolic engineering approaches for use in the industrial ABE producer Clostridium saccharoperbutylacetonicum strain N1-4 (HMT) ATCC 27021. This study serves as a foundation for future metabolic engineering efforts of this prominent solvent producer to not only improve fermentation performance metrics (e.g., solvent titers, productivities, and yields, oxygen tolerance, substrate specificity, cell viability), but also to serve as a platform for the production of other high-value products (e.g., higher alcohols and petrochemical precursors). Additionally, this study opens the door for advanced genetic manipulations of this organism using recent achievements in adapting CRISPR/Cas9 systems for use in clostridia (40, 41). Before this is possible, future studies are required to identify additional promoters, antibiotic markers, and counterselection methods able to function in C. saccharoperbutylacetonicum N1-4. With these tools and techniques in hand, we believe that future engineered strains of C. saccharoperbutylacetonicum N1-4 will be better able to meet the stringent demands required for an economically viable biofuel production process.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1 and Table 2.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Bacterial strains | ||
| E. coli XL1-Blue | Cloning strain | Stratagene/Agilentb |
| C. saccharoperbutylacetonicum strains | ||
| N1-4 (HMT) | Wild-type strain | ATCC 27021 |
| N1-4 ΔpyrF | ΔpyrF::Eryr | This study |
| N1-4 ΔctfAB2 | ΔctfAB2::Eryr | This study |
| N1-4 Δadh5 | Δadh5::Eryr | This study |
| N1-4 Δadh11 | Δadh11::Eryr | This study |
| Plasmids | ||
| pIMP1 (= pWIS_empty) | E. coli-Clostridium shuttle vector; Eryr; repL; Ampr; colE1 | 26, 35 |
| pWIS_2 through pWIS_14 | pWIS_empty with C. acetobutylicum Pcrt and Thbd; strain N1-4 gene/gene pair to be overexpressed | This study |
| pKO_mazF | Thr Eryr; repL; colE1; bgaR and PbgaL upstream of mazF | 38 |
| pLOR_empty | Pcrt upstream of Eryr; Ampr; colE1 | This study |
| pLOR_pyrF | pLOR_empty with Eryr flanked by ∼2-kb pyrF upstream and downstream homologous regions | This study |
| pLOR_ctfAB2 | pLOR_empty with Eryr flanked by ∼2-kb ctfA2-ctfB2 upstream and downstream homologous regions | This study |
| pLOR_adh5 | pLOR_empty with Eryr flanked by ∼2-kb adh5 upstream and downstream homologous regions | This study |
| pLOR_adh11 | pLOR_empty with Eryr flanked by ∼2-kb adh11 upstream and downstream homologous regions | This study |
Eryr, erythromycin resistance; repL, pIM13 Gram-positive origin of replication; Ampr, ampicillin resistance; colE1, E. coli replication origin; Pcrt, crotonase promoter; Thbd, 3-hydroxybutyryl-CoA dehydrogenase terminator; Thr, thiamphenicol resistance; PbgaL, C. perfringens bgaL promoter; bgaR, C. perfringens divergent regulator of PbgaL; mazF, toxin gene.
Agilent Technologies, Santa Clara, CA.
Culture media and growth conditions.
Clostridium saccharoperbutylacetonicum strain N1-4 (HMT) ATCC 27021 was cultured at 34°C in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) containing an atmosphere of 97% nitrogen and 3% hydrogen. For transformation procedures, strains were grown in 2× YTG medium (16 g/liter tryptone, 10 g/liter yeast extract, 5 g/liter NaCl, 10 g/liter glucose with an additional 15 g/liter agar for solid media) with the pH adjusted to 6.5. For comparing colony morphologies, solid TYA medium was used (6 g/liter tryptone, 2 g/liter yeast extract, 3 g/liter ammonium acetate, 0.2 g/liter MgSO4, 0.01 g/liter FeSO4, 40 g/liter glucose, 0.5 g KH2PO4, and 15 g/liter agar) with the pH adjusted to 6.5. For growing overnight cultures and preparing glycerol stocks, PL7G media, a derivative of clostridial growth medium (CGM) (57) was used (30 g/liter glucose, 5 g/liter yeast extract, 2.67 g/liter ammonium sulfate, 1 g/liter NaCl, 0.75 g/liter monobasic sodium phosphate, 0.75 g/liter dibasic sodium phosphate, 0.5 g/liter cysteine-HCl, 0.7 g/liter magnesium sulfate heptahydrate, 20 mg/liter manganese sulfate heptahydrate, and 20 mg/liter iron sulfate heptahydrate, with the initial pH set to 6.3).
Escherichia coli XL1-Blue (Agilent Technologies, Santa Clara, CA) was grown aerobically in Luria-Bertani (LB) medium at 37°C. Clostridium and E. coli strains were maintained as 20% (vol/vol) glycerol stocks stored at −80°C.
For appropriate Clostridium strains, culture medium was supplemented with erythromycin (40 μg/ml). Antibiotic was omitted in flask fermentation cultures to avoid negative effects on the fermentation cycle. Carbenicillin (100 μg/ml) was added to E. coli culture medium as needed. 5-FOA was purchased as a 100-mg/ml solution (dimethyl sulfoxide [DMSO]) from Zymo Research (Irvine, CA); 500 mg/liter 5-FOA was added to growth media as indicated.
Flask fermentations.
For performing flask fermentations, PL7S medium was used, which is identical to PL7G medium with the following exceptions: 70 g/liter sucrose was used in place of glucose, and 300 mg/liter adenine was added. Additionally, Antifoam 204 (10 μl antifoam/70 ml culture; Sigma, St. Louis, MO) and solid CaCO3 (6 g/liter, for pH control) were added to flask fermentation cultures just prior to inoculation. All overnight cultures, subcultures, and fermentation cultures of Clostridium were cultured anaerobically at 34°C without agitation. Initial overnight cultures were started from freshly streaked (<5 days old) individual colonies on 2× YTG plates. To perform flask fermentations, a mid-/late-exponential-phase overnight culture (OD600, 0.6 to 1.0; PL7G medium) was used to inoculate a subculture in PL7S medium (pH 6.3) with a 10% inoculum. At mid/late exponential phase (OD600, 0.6 to 1.0), 7 ml of subculture was used to inoculate 63 ml of fresh PL7S medium (supplemented with antifoam and CaCO3) in a 125-ml glass shake flask with foil caps. All flask fermentations were performed as biological triplicates. Endpoint samples for high-performance liquid chromatography (HPLC) analysis (1 ml) were taken 66 h postinoculation.
Analytical procedures.
Cell concentrations were monitored via absorbance using a spectrophotometer to determine the OD600. Sucrose, acetone, butanol, ethanol, acetate, butyrate, and lactate concentrations were measured using a Shimadzu Prominence UFLC system with refractive index and diode array detection (Shimadzu America, Inc., Columbia, MD). Prior to analysis, samples of culture supernatant were filtered with 0.22 μm polyvinylidene difluoride (PVDF) syringe filters. Compounds were separated with a Bio-Rad Aminex HPX-87H column (300 mm by 7.8 mm) and detected by measurement of refractive index (sucrose, butanol, ethanol, acetate, lactate) or UV absorbance (acetone, 265 nm; butyrate, 208 nm). The following operating conditions were used: mobile phase, 0.01 N sulfuric acid; flow rate, 0.7 ml/min; run time, 35 min; column temperature, 35°C.
Bright-field microscopy.
Bright-field microscopy of the three subtypes of C. saccharoperbutylacetonicum N1-4 was performed with a Leica DM5000 B fluorescence microscope (Wetzlar, Germany) fitted with a Leica DFC490 camera. Samples of liquid culture (OD600, ∼1.0) started from colonies of the three subtypes were diluted 100-fold in 0.85% NaCl. Samples were observed using a 40× objective lens.
Motility assays.
Glycerol stocks of subtypes R, I, and S were inoculated into liquid medium (2× YTG). After overnight incubation (OD600, ∼0.6), the cultures were plated on solid TYA and incubated anaerobically for 48 h. Single colonies of each subtype (confirmed visually) were picked with pipette tips and stabbed into the center of fresh soft agar (5 g/liter agar instead of the normal 15 g/liter) TYA plates. Plates were incubated anaerobically for 48 h at 34°C before imaging.
Plasmid construction.
Oligonucleotides were synthesized by IDT (Coralville, IA). Table S1 in the supplemental material lists all relevant oligonucleotide sequences used. Vector pWIS_empty was identical to the received pIMP_empty (25, 35). All derivatives of pWIS_empty included the C. acetobutylicum ATCC 824 crotonase (crt) promoter with a synthetic ribosome binding site (RBS), followed by the coding region for the selected gene(s) to be overexpressed, and finally, the C. acetobutylicum ATCC 824 hydroxybutyryl-CoA dehydrogenase (encoded by hbd) rho-independent terminator. This expression cassette was inserted downstream of the Gram-positive replicon RepL-coding sequence on the pWIS_empty backbone using Gibson assembly (58). All native C. saccharoperbutylacetonicum N1-4 genes used for gene overexpression studies were PCR amplified from purified C. saccharoperbutylacetonicum N1-4 genomic DNA. DNA primers used to amplify these genes contained 27-bp overhangs to permit Gibson assembly with the crt promoter region (forward primer) and hbd terminator region (reverse primer). The pWIS vector backbone was amplified as two regions (pWIS_bk A, 3.0 kb, primers pWIS_bk_F and pWIS_bk_R2; pWIS_bk B, 2.0 kb, primers pWIS_bk_F2 and pWIS_bk_R), which overlap at a 24-bp sequence. Gibson assembly for pWIS series vectors included pWIS_bk A, pWIS_bk B, and the amplified native N1-4 gene containing 27-bp 5′ and 3′ overhangs (Table S1). After transformation into E. coli XL1-Blue, putative transformants were cultured and underwent plasmid Miniprep purification (Qiagen North America, Germantown, MD). Purified plasmid DNA was screened by restriction digest testing, followed by Sanger sequencing.
The pLOR vector series for performing targeted gene deletions in C. saccharoperbutylacetonicum N1-4 was derived from elements of pIMP1 and pKO_mazF. A 677-bp region containing the E. coli origin of replication ColE1 was amplified from pIMP1 using primers pLOR_P1_F & pLOR_P1_R. A 1,233-bp region containing the ampicillin resistance gene (Ampr) was amplified from pIMP1 using primers pLOR_P2_F and pLOR_P2_Ro. An 858-bp region containing the erythromycin resistance (Eryr) gene driven by the C. acetobutylicum thiolase (thl) promoter was amplified from pIMP1 using primers pLOR_P3_Fo and pLOR_P3_R. A 44-bp region containing a spacer sequence present in pKO_mazF was included upstream of Pthl-Eryr by addition of an overlapping sequence to the 5′ end of pLOR_P3_F. A 287-bp region containing a terminator sequence was amplified from pKO_mazF to follow the Pthl-Eryr cassette using primers pLOR_P4_Fo and pLOR_P4_R. These four PCR products underwent Gibson assembly along with PCR-amplified upstream homologous region (UHR) and downstream homologous region (DHR) (both approximately 2.0 kb) designed to target the pyrF gene (using primers UHR_pyrF_F/UHR_pyrF_R, and DHR_pyrF_F/DHR_pyrF_R to amplify the pyrF UHR and DHR, respectively); 24-bp overhang sequences were included on primers of neighboring PCR products to permit assembly. Following transformation, plasmid purification, confirmation by test digestion, and sequencing, pLOR_pyrF served as the template for all other pLOR assemblies. For assembly of other pLOR constructs, appropriate UHR and DHR PCR products were assembled with the ColE1/AmpR region from pLOR_pyrF (with primers pLOR_P2_F and pLOR_P1_R), and the Pthl-Eryr cassette (with primers pLOR_P3_Fs and pLOR_P4_R) (to be flanked by the UHR and DHR regions); 24-bp overhang sequences were included on all primers for the UHR and DHR PCRs to permit annealing to the pLOR vector backbone elements. As above, all vectors were confirmed by test digestion and sequencing.
Plasmid and genomic DNA isolation in C. saccharoperbutylacetonicum N1-4.
For plasmid DNA isolation in C. saccharoperbutylacetonicum N1-4, 5 ml of stationary-phase liquid cultures (OD600 > 2.0) underwent plasmid Miniprep purification according to the manufacturer's instructions (Qiagen North America, Germantown, MD). Genomic DNA isolation of C. saccharoperbutylacetonicum N1-4 was performed using a modified alkaline lysis method. Stationary-phase culture (10 ml) was prepared as for plasmid purification and centrifuged (room temperature, 3,500 × g, 15 min). The pellet was resuspended in 5 ml SET buffer (75 mM NaCl, 25 mM EDTA pH 8.0, 20 mM Tris-HCl [pH 7.5]), and lysozyme was added to a final concentration of 2 mg/ml. The mixture was incubated for 60 min at 37°C with gentle mixing every 15 min, and lysis was induced by addition of 660 μl of 1 M NaOH solution containing 10% (wt/vol) SDS. The resulting solution was mixed by inversion, proteinase K was added to a final concentration of 0.5 mg/ml, and the mixture was incubated for 1 h at 55°C. Phenol-chloroform (equal volume, 1:1; room temperature) was added, and the solution was mixed by inversion for 5 min. After centrifugation (room temperature, 3,500 × g, 10 min), the aqueous phase was removed by pipetting, and 3 M sodium acetate (10% by volume in the resulting mixture) was added. After mixing by inversion, 2 volumes of ethanol were added to precipitate genomic DNA, and the DNA was harvested with a glass hook and washed with 70% ethanol. After 15 min of drying, the precipitated DNA was resuspended in low Tris-EDTA (TE) buffer (10 mM Tris, 0.1 mM EDTA [pH 8.0]) and incubated at 50°C until dissolved. This procedure resulted in 1 ml of approximately 1 μg/μl genomic DNA.
Transformation procedure for C. saccharoperbutylacetonicum N1-4.
To prepare competent cell stocks of C. saccharoperbutylacetonicum N1-4, anaerobic overnight cultures (10 ml PL7G, 34°C) were started from 20% (vol/vol) glycerol stocks stored at −80°C. After reaching an OD600 of ∼0.6, overnight cultures were subcultured in 60 ml liquid 2 × YTG (10% inoculum) and incubated for 3 to 5 h until reaching an OD600 of ∼0.6. The subcultures were then centrifuged (room temperature, 3,500 × g, 15 min), the supernatant was removed, and the pellet was resuspended in 10 ml room temperature electroporation buffer (EPB) (270 mM sucrose, 5 mM NaH2PO4 [pH 7.4]). Resuspended cells were centrifuged again (room temperature, 3,500 × g, 10 min), and the pellets were resuspended in 3 ml room temperature EPB, yielding the concentrated (20×) competent cells. The competent cell stock was aliquoted into prechilled 4-mm electroporation cuvettes (500-μl aliquots), 2 μg plasmid DNA was added to each cuvette, and the mixtures were incubated in ice for 30 min. Exponential decay electroporations were performed using a Bio-Rad Gene Pulser Xcell (Hercules, CA) with parameters as follows: voltage, 2.0 kV; resistance, 200 Ω; capacitance, 25 μF. Following electroporation (yielding time constants of ∼ 4.0 ms), cells were immediately resuspended in 10 ml liquid 2× YTG and were allowed to recover at 34°C for 4 h. Recovery cultures were then centrifuged (room temperature, 3,500 × g, 15 min), the supernatant was removed, and pellets were resuspended in 500 μl fresh 2× YTG. The mixture (100 μl) was spread on solid 2× YTG + Ery40 plates and incubated at 34°C for 3 to 4 days. Potential transformant colonies were grown in overnight liquid cultures (2× YTG + Ery40) and replated on solid 2× YTG + Ery40 before undergoing verification.
Targeted gene KO procedure and verification.
The same electroporation method was used to introduce KO vectors (pLOR series) into C. saccharoperbutylacetonicum N1-4, except that 5 μg of plasmid DNA was used. Initial transformant colonies (4–6) were grown in overnight liquid cultures (2× YTG + Ery40) and replated on solid 2× YTG + Ery40. Twelve colonies (2 or 3 per restreaked transformant) were screened via colony PCR using three primer sets (in three separate reactions) (Fig. 2). Two sets were designed to amplify regions spanning from inside the erythromycin resistance gene to outside the regions of homology. Both bands should be present for double-crossover KO strains. The third primer set was designed to amplify a region from inside the wild-type copy of the target gene to outside the regions of homology. This third set was used to rule out contamination of the wild type, as this band should be present only for wild-type strains. After colony PCR screening of the 12 colonies along with one wild-type control (three reactions each), approximately 1 or 2 transformant colonies showed the desired band patterns and were considered valid double-crossover mutants. Sanger sequencing of the colony PCR products associated with the KO strains was also performed to confirm the desired deletion event. If no colonies showed the desired band pattern, colonies that displayed PCR products for all three primer sets (indicating a mixed genotype) were twice restreaked on solid 2× YTG + Ery40. Twelve of the resultant colonies were again screened with the same colony PCR primer sets, and approximately 2 or 3 of these colonies were validated as successful KO strains.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Papoutsakis Lab (University of Delaware) for donating plasmids pIMP1 and pKO_mazF. We acknowledge Mitchell Thompson for assisting in the development of the electroporation protocol and for his insightful discussions and suggestions.
This work was funded by the Energy Biosciences Institute.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02942-16.
REFERENCES
- 1.Antoni D, Zverlov VV, Schwarz WH. 2007. Biofuels from microbes. Appl Microbiol Biotechnol 77:1–17. doi: 10.1007/s00253-007-1163-x. [DOI] [PubMed] [Google Scholar]
- 2.Xue C, Zhao X-Q, Liu C-G, Chen L-J, Bai F-W. 2013. Prospective and development of butanol as an advanced biofuel. Biotechnol Adv 31:1575–1584. doi: 10.1016/j.biotechadv.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Anbarasan P, Baer ZC, Sreekumar S, Gross E, Binder JB, Blanch HW, Clark DS, Toste FD. 2012. Integration of chemical catalysis with extractive fermentation to produce fuels. Nature 491:235–239. doi: 10.1038/nature11594. [DOI] [PubMed] [Google Scholar]
- 4.Lütke-Eversloh T, Bahl H. 2011. Metabolic engineering of Clostridium acetobutylicum: recent advances to improve butanol production. Curr Opin Biotechnol 22:634–647. doi: 10.1016/j.copbio.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Tracy BP, Jones SW, Fast AG, Indurthi DC, Papoutsakis ET. 2012. Clostridia: the importance of their exceptional substrate and metabolite diversity for biofuel and biorefinery applications. Curr Opin Biotechnol 23:364–381. doi: 10.1016/j.copbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Jones DT, Woods DR. 1986. Acetone-butanol fermentation revisited. Microbiol Rev 50:484–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green EM. 2011. Fermentative production of butanol—the industrial perspective. Curr Opin Biotechnol 22:337–343. doi: 10.1016/j.copbio.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Motoyoshi H. July 1960. Process for producing butanol by fermentation. US patent 2,945,786 A.
- 9.Keis S, Shaheen R, Jones DT. 2001. Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int J Syst Evol Microbiol 51:2095–2103. doi: 10.1099/00207713-51-6-2095. [DOI] [PubMed] [Google Scholar]
- 10.Shaheen R, Shirley M, Jones DT. 2000. Comparative fermentation studies of industrial strains belonging to four species of solvent-producing clostridia. J Mol Microbiol Biotechnol 2:115–124. [PubMed] [Google Scholar]
- 11.Hongo M, Murata A. 1965. Bacteriophages of Clostridium saccharoperbutylacetonicum. Agric Biol Chem 29:1135–1145. [Google Scholar]
- 12.Ishizaki A, Michiwaki S, Crabbe E, Kobayashi G, Sonomoto K, Yoshino S. 1999. Extractive acetone-butanol-ethanol fermentation using methylated crude palm oil as extractant in batch culture of Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564). J Biosci Bioeng 87:352–356. doi: 10.1016/S1389-1723(99)80044-9. [DOI] [PubMed] [Google Scholar]
- 13.Thang VH, Kanda K, Kobayashi G. 2010. Production of acetone-butanol-ethanol (ABE) in direct fermentation of cassava by Clostridium saccharoperbutylacetonicum N1-4. Appl Biochem Biotechnol 161:157–170. doi: 10.1007/s12010-009-8770-1. [DOI] [PubMed] [Google Scholar]
- 14.Al-Shorgani NKN, Kalil MS, Yusoff WMW. 2012. Fermentation of sago starch to biobutanol in a batch culture using Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564). Ann Microbiol 62:1059–1070. doi: 10.1007/s13213-011-0347-x. [DOI] [Google Scholar]
- 15.Al-Shorgani NK, Kalil MS, Yusoff WM. 2012. Biobutanol production from rice bran and de-oiled rice bran by Clostridium saccharoperbutylacetonicum N1-4. Bioprocess Biosyst Eng 35:817–826. doi: 10.1007/s00449-011-0664-2. [DOI] [PubMed] [Google Scholar]
- 16.Al-Shorgani NKN, Tibin E-M, Ali E, Hamid AA, Yusoff WMW, Kalil MS. 2014. Biohydrogen production from agroindustrial wastes via Clostridium saccharoperbutylacetonicum N1-4 (ATCC 13564). Clean Technol Environ Policy 16:11–21. doi: 10.1007/s10098-013-0586-6. [DOI] [Google Scholar]
- 17.Chin C, Hwang W, Lee H. 1991. Isolation and characterization of mutants of Clostridium saccharoperbutylacetonicum fermenting bagasse hydrolyzate. J Ferment Bioeng 72:249–253. doi: 10.1016/0922-338X(91)90157-C. [DOI] [Google Scholar]
- 18.Zheng J, Tashiro Y, Wang Q, Sakai K, Sonomoto K. 2015. Feasibility of acetone-butanol-ethanol fermentation from eucalyptus hydrolysate without nutrients supplementation. Appl Energy 140:113–119. doi: 10.1016/j.apenergy.2014.11.037. [DOI] [Google Scholar]
- 19.Noguchi T, Tashiro Y, Yoshida T, Zheng J, Sakai K, Sonomoto K. 2013. Efficient butanol production without carbon catabolite repression from mixed sugars with Clostridium saccharoperbutylacetonicum N1-4. J Biosci Bioeng 116:716–721. doi: 10.1016/j.jbiosc.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 20.Tashiro Y, Takeda K, Kobayashi G, Sonomoto K. 2005. High production of acetone-butanol-ethanol with high cell density culture by cell-recycling and bleeding. J Biotechnol 120:197–206. doi: 10.1016/j.jbiotec.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 21.Richter H, Qureshi N, Heger S, Dien B, Cotta MA, Angenent LT. 2012. Prolonged conversion of n-butyrate to n-butanol with Clostridium saccharoperbutylacetonicum in a two-stage continuous culture with in-situ product removal. Biotechnol Bioeng 109:913–921. doi: 10.1002/bit.24380. [DOI] [PubMed] [Google Scholar]
- 22.Zheng J, Tashiro Y, Yoshida T, Gao M, Wang Q, Sonomoto K. 2013. Continuous butanol fermentation from xylose with high cell density by cell recycling system. Bioresour Technol 129:360–365. doi: 10.1016/j.biortech.2012.11.066. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka S, Tashiro Y, Kobayashi G, Ikegami T, Negishi H, Sakaki K. 2012. Membrane-assisted extractive butanol fermentation by Clostridium saccharoperbutylacetonicum N1-4 with 1-dodecanol as the extractant. Bioresour Technol 116:448–452. doi: 10.1016/j.biortech.2012.03.096. [DOI] [PubMed] [Google Scholar]
- 24.Ikegami T, Negishi H, Nakayama S, Kobayashi G, Sakaki K. 2014. Pervaporative concentration of biobutanol from ABE fermentation broths by Clostridium saccharoperbutylacetonicum using silicone rubber-coated silicalite-1 membranes. Sep Purif Technol 132:206–212. doi: 10.1016/j.seppur.2014.05.030. [DOI] [Google Scholar]
- 25.Mermelstein LD, Papoutsakis ET. 1993. In vivo methylation in Escherichia coli by the Bacillus subtilis phage φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 59:1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SY, Bennett GN, Papoutsakis ET. 1992. Construction of Escherichia coli-Clostridium acetobutylicum shuttle vectors and transformation of Clostridium acetobutylicum strains. Biotechnol Lett 14:427–432. doi: 10.1007/BF01021259. [DOI] [Google Scholar]
- 27.Lütke-Eversloh T. 2014. Application of new metabolic engineering tools for Clostridium acetobutylicum. Appl Microbiol Biotechnol 98:5823–5837. doi: 10.1007/s00253-014-5785-5. [DOI] [PubMed] [Google Scholar]
- 28.Hillmann F, Döring C, Riebe O, Ehrenreich A, Fischer R-J, Bahl H. 2009. The role of PerR in O2-affected gene expression of Clostridium acetobutylicum. J Bacteriol 191:6082–6093. doi: 10.1128/JB.00351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao H, Li Z, Jiang Y, Yang Y, Jiang W, Gu Y, Yang S. 2012. Metabolic engineering of D-xylose pathway in Clostridium beijerinckii to optimize solvent production from xylose mother liquid. Metab Eng 14:569–578. doi: 10.1016/j.ymben.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Li X, Milne CB, Janssen H, Lin W, Phan G, Hu H, Jin Y-S, Price ND, Blaschek HP. 2013. Development of a gene knockout system using mobile group II introns (Targetron) and genetic disruption of acid production pathways in Clostridium beijerinckii. Appl Environ Microbiol 79:5853–5863. doi: 10.1128/AEM.00971-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wischral D, Zhang J, Cheng C, Lin M, De Souza LMG, Pessoa FLP, Pereira N Jr, Yang S-T. 2016. Production of 1,3-propanediol by Clostridium beijerinckii DSM 791 from crude glycerol and corn steep liquor: Process optimization and metabolic engineering. Bioresour Technol 212:100–110. doi: 10.1016/j.biortech.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama S, Irie R, Kosaka T, Matsuura K, Yoshino S, Furukawa K. 2007. New host-vector system in solvent-producing Clostridium saccharoperbutylacetonicum strain N1-4. J Gen Appl Microbiol 53:53–56. doi: 10.2323/jgam.53.53. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama S, Kosaka T, Hirakawa H, Matsuura K, Yoshino S, Furukawa K. 2008. Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1-4. Appl Microbiol Biotechnol 78:483–493. doi: 10.1007/s00253-007-1323-z. [DOI] [PubMed] [Google Scholar]
- 34.Pyne ME, Bruder M, Moo-Young M, Chung DA, Chou CP. 2014. Technical guide for genetic advancement of underdeveloped and intractable Clostridium. Biotechnol Adv 32:623–641. doi: 10.1016/j.biotechadv.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Walter KA, Mermelstein LD, Papoutsakis ET. 1994. Host-plasmid interactions in recombinant strains of Clostridium acetobutylicum ATCC 824. FEMS Microbiol Lett 123:335–341. doi: 10.1111/j.1574-6968.1994.tb07245.x. [DOI] [Google Scholar]
- 36.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods 70:452–464. doi: 10.1016/j.mimet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR, Hogsett DA, Caiazza NC. 2010. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol 76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Hinai MA, Fast AG, Papoutsakis ET. 2012. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl Environ Microbiol 78:8112–8121. doi: 10.1128/AEM.02214-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Nostrand JDV, He Z, Zhou J. 2015. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. Appl Environ Microbiol 81:4423–4431. doi: 10.1128/AEM.00873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang Z-T, Seo S-O, Lynn P, Lu T, Jin Y-S, Blaschek HP. 2016. Bacterial genome editing with CRISPR-Cas9: deletion, integration, single nucleotide modification, and desirable “clean” mutant selection in Clostridium beijerinckii as an example. ACS Synth Biol 5:721–732. doi: 10.1021/acssynbio.6b00060. [DOI] [PubMed] [Google Scholar]
- 41.Li Q, Chen J, Minton NP, Zhang Y, Wen Z, Liu J, Yang H, Zeng Z, Ren X, Yang J, Gu Y, Jiang W, Jiang Y, Yang S. 2016. CRISPR-based genome editing and expression control systems in Clostridium acetobutylicum and Clostridium beijerinckii. Biotechnol J 11:961–972. doi: 10.1002/biot.201600053. [DOI] [PubMed] [Google Scholar]
- 42.Steiner E, Dago AE, Young DI, Heap JT, Minton NP, Hoch JA, Young M. 2011. Multiple orphan histidine kinases interact directly with Spo0A to control the initiation of endospore formation in Clostridium acetobutylicum. Mol Microbiol 80:641–654. doi: 10.1111/j.1365-2958.2011.07608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Cerro C, Felpeto-Santero C, Rojas A, Tortajada M, Ramón D, García JL. 2013. Genome sequence of the butanol hyperproducer Clostridium saccharoperbutylacetonicum N1-4. Genome Announc 1(2):e00070-13. doi: 10.1128/genomeA.00070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poehlein A, Krabben P, Dürre P, Daniel R. 2014. Complete genome sequence of the solvent producer Clostridium saccharoperbutylacetonicum strain DSM 14923. Genome Announc 2:e01056-14. doi: 10.1128/genomeA.01056-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keis S, Bennett CF, Ward VK, Jones DT. 1995. Taxonomy and phylogeny of industrial solvent-producing clostridia. Int J Syst Evol Microbiol 45:693–705. [DOI] [PubMed] [Google Scholar]
- 46.Mariano AP, Dias MOS, Junqueira TL, Cunha MP, Bonomi A, Filho RM. 2013. Butanol production in a first-generation Brazilian sugarcane biorefinery: technical aspects and economics of greenfield projects. Bioresour Technol 135:316–323. doi: 10.1016/j.biortech.2012.09.109. [DOI] [PubMed] [Google Scholar]
- 47.Pyne ME, Moo-Young M, Chung DA, Chou CP. 2013. Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnol Biofuels 6:50. doi: 10.1186/1754-6834-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang X, Xu M, Yang S-T. 2015. Metabolic and process engineering of Clostridium cellulovorans for biofuel production from cellulose. Metab Eng 32:39–48. doi: 10.1016/j.ymben.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 49.Leang C, Ueki T, Nevin KP, Lovley DR. 2013. A genetic system for Clostridium ljungdahlii: a chassis for autotrophic production of biocommodities and a model homoacetogen. Appl Environ Microbiol 79:1102–1109. doi: 10.1128/AEM.02891-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tracy BP, Jones SW, Papoutsakis ET. 2011. Inactivation of σE and σG in Clostridium acetobutylicum illuminates their roles in clostridial-cell-form biogenesis, granulose synthesis, solventogenesis, and spore morphogenesis. J Bacteriol 193:1414–1426. doi: 10.1128/JB.01380-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kataoka N, Vangnai AS, Ueda H, Tajima T, Nakashimada Y, Kato J. 2014. Enhancement of (R)-1,3-butanediol production by engineered Escherichia coli using a bioreactor system with strict regulation of overall oxygen transfer coefficient and pH. Biosci Biotechnol Biochem 78:695–700. doi: 10.1080/09168451.2014.891933. [DOI] [PubMed] [Google Scholar]
- 52.Hwang HJ, Park JH, Kim JH, Kong MK, Kim JW, Park JW, Cho KM, Lee PC. 2014. Engineering of a butyraldehyde dehydrogenase of Clostridium saccharoperbutylacetonicum to fit an engineered 1,4-butanediol pathway in Escherichia coli. Biotechnol Bioeng 111:1374–1384. doi: 10.1002/bit.25196. [DOI] [PubMed] [Google Scholar]
- 53.Yoo M, Croux C, Meynial-Salles I, Soucaille P. 2016. Elucidation of the roles of adhE1 and adhE2 in the primary metabolism of Clostridium acetobutylicum by combining in-frame gene deletion and a quantitative system-scale approach. Biotechnol Biofuels 9:92. doi: 10.1186/s13068-016-0507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai Z, Dong H, Zhang Y, Li Y. 2016. Elucidating the contributions of multiple aldehyde/alcohol dehydrogenases to butanol and ethanol production in Clostridium acetobutylicum. Sci Rep 6:28189. doi: 10.1038/srep28189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim BH, Bellows P, Datta R, Zeikus JG. 1984. Control of carbon and electron flow in Clostridium acetobutylicum fermentations: utilization of carbon monoxide to inhibit hydrogen production and to enhance butanol yields. Appl Environ Microbiol 48:764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasconcelos I, Girbal L, Soucaille P. 1994. Regulation of carbon and electron flow in Clostridium acetobutylicum grown in chemostat culture at neutral pH on mixtures of glucose and glycerol. J Bacteriol 176:1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green EM, Boynton ZL, Harris LM, Rudolph FB, Papoutsakis ET, Bennett GN. 1996. Genetic manipulation of acid formation pathways by gene inactivation in Clostridium acetobutylicum ATCC 824. Microbiology 142:2079–2086. doi: 10.1099/13500872-142-8-2079. [DOI] [PubMed] [Google Scholar]
- 58.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.