ABSTRACT
The draft genomes of Lactobacillus plantarum strains isolated from Asian fermented foods, infant feces, and shrimp intestines were sequenced and compared to those of well-studied strains. Among 28 strains of L. plantarum, variations in the genomic features involved in ecological adaptation were elucidated. The genome sizes ranged from approximately 3.1 to 3.5 Mb, of which about 2,932 to 3,345 protein-coding sequences (CDS) were predicted. The food-derived isolates contained a higher number of carbohydrate metabolism-associated genes than those from infant feces. This observation correlated to their phenotypic carbohydrate metabolic profile, indicating their ability to metabolize the largest range of sugars. Surprisingly, two strains (P14 and P76) isolated from fermented fish utilized inulin. β-Fructosidase, the inulin-degrading enzyme, was detected in the supernatants and cell wall extracts of both strains. No activity was observed in the cytoplasmic fraction, indicating that this key enzyme was either membrane-bound or extracellularly secreted. From genomic mining analysis, a predicted inulin operon of fosRABCDXE, which encodes β-fructosidase and many fructose transporting proteins, was found within the genomes of strains P14 and P76. Moreover, pts1BCA genes, encoding sucrose-specific IIBCA components involved in sucrose transport, were also identified. The proteomic analysis revealed the mechanism and functional characteristic of the fosRABCDXE operon involved in the inulin utilization of L. plantarum. The expression levels of the fos operon and pst genes were upregulated at mid-log phase. FosE and the LPXTG-motif cell wall anchored β-fructosidase were induced to a high abundance when inulin was present as a carbon source.
IMPORTANCE Inulin is a long-chain carbohydrate that may act as a prebiotic, which provides many health benefits to the host by selectively stimulating the growth and activity of beneficial bacteria in the colon. While certain lactobacilli can catabolize inulin, this has not yet been described for Lactobacillus plantarum, and an associated putative inulin operon has not been reported in this species. By using comparative and functional genomics, we showed that two L. plantarum strains utilized inulin and identified functional inulin operons in their genomes. The proteogenomic data revealed that inulin degradation and uptake routes, which related to the fosRABCDXE operon and pstBCA genes, were widely expressed among L. plantarum strains. The present work provides a novel understanding of gene regulation and mechanisms of inulin utilization in probiotic L. plantarum generating opportunities for synbiotic product development.
KEYWORDS: lactic acid bacteria, genomes, prebiotic, fos operon, β-fructosidase
INTRODUCTION
Inulin-type fructans are among the prebiotic substances found in several fruits and vegetables, such as artichokes, chicory, bananas, garlic, and asparagus (1). Inulin chains can be found in many lengths, with a degree of polymerization (DP) varying from 2 to 60, or even more (2). Inulin and its partially hydrolyzed fructooligosaccharides (FOS) are linear d-fructose polymers linked by a β (2-1) glycosidic bond, with a terminal glucose or fructose unit (3). The β (2-1) linkages protect these fructans from digestion in the upper gastrointestinal tract of humans. However, these prebiotics can be hydrolyzed by β-fructosidase-producing bacteria residing in the colon (3). Inulin-type fructans are readily degraded by the majority of bifidobacteria (4). On the other hand, inulin degradation is quite rare among lactobacilli, and is mainly limited to some strains of L. paracasei, L. casei, L. acidophilus, and L. delbrueckii (5–8).
The ability of lactobacilli to metabolize prebiotics, especially long-chain FOS of the GFn and FFn types, has been documented. Several pathways, including the fos (9, 10), msm (8), and pts1BCA (11) operons, are reportedly associated with FOS (fructooligosaccharide) metabolism in Lactobacillus species. In L. plantarum WCSF1, proteins encoded by the pts1BCA genes involved in the sucrose phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) can transport short-chain FOS (scFOS) into the cytosol, which are then further digested by intracellular β-fructofuranosidase, encoded by sacA (11). The fosRABCDXE operon is involved in the FOS utilization pathway of L. paracasei strain 1195 and encodes components of a putative fructose/mannose PEP-dependent PTS and a β-fructosidase precursor (FosE). FosE contains an N-terminal signal peptide sequence and an LPQAG cell wall anchor motif in the C-terminal region, indicating its location outside the cell (9, 10). Another system for FOS utilization that has been genetically characterized in L. acidophilus NCFM is the msm operon. The msm operon encodes an ABC transport system (msmEFGK) and a putative intracellular β-fructosidase (bfrA), which were found to hydrolyze sucrose, inulin-type fructans, and inulin (8).
Although many strains of L. plantarum have been found to exhibit a FOS-degrading ability, none can degrade the long-chain inulin (12). Thus far, only L. plantarum no. 14 has been reported to grow in the presence of inulin with a DP range of 5 to 30 (5). In this study, we aimed to compare the genomic features and metabolic properties of 28 strains of L. plantarum isolated from Asian fermented foods, infant feces, and shrimp intestines. Apart from the insights in the core and pan genomes of the L. plantarum, this approach enabled the identification and characterization of a new inducible operon involved in efficient utilization of inulin, a natural fructose polymer with high industrial relevance that, to the best of our knowledge, has not been described as being degradable by L. plantarum.
RESULTS AND DISCUSSION
The genomic diversity of Lactobacillus spp. has been shown to be unusually high and indicates that L. plantarum and its two subspecies are part of a deeply rooted group (13). It is known that L. plantarum strains are quite versatile, but there have been no comparative next-generation genomics studies with functional analyses that can shed light on novel functions. In this study, the genomic and phenotypic characterizations were determined and compared among 28 strains of L. plantarum, the earlier analyzed L. plantarum strain WCFS1 isolated from the oral cavity, the type strain L. plantarum DSM20246, L. plantarum subsp. plantarum DSM20174, and L. plantarum subsp. argentoratensis DSM16365 (see Table S1 in the supplemental material). The collection of 28 strains included 13 strains newly isolated from fermented rice and fish from Thailand, since comparative hybridization analysis has shown that L. plantarum strains from Asian fermented foods are quite divergent from those found in other habitats (14). Moreover, this collection also contained several L. plantarum isolates from marine fish, shrimp intestine, and other foods, as well as isolates of human intestinal origin, including strains from Asian infants of various ages, isolated under aerobic and anaerobic conditions (see Table S1). All strains were compared for their metabolic and genomic properties under identical circumstances and using the same technology to reveal new functionalities, as described below.
General genomic features of L. plantarum and its predicted core and pan genomes.
All Asian strains of L. plantarum showed variation in their genome size ranging from approximately 3.1 to 3.5 Mb, with an overall GC content of 44.1 to 45.1%. The predicted numbers of protein-coding sequences (CDS) ranged from 2,932 to 3,345 (see Table S1). No plasmids were detected in any of the newly sequenced genomes, as opposed to L. plantarum WCSF1, which harbors three plasmids (15). The total gene pool of the L. plantarum species was calculated based on the genome comparison between all Asian L. plantarum strains with the best quality genome sequence of L. plantarum WCSF1 (16) (see Fig. S1). The pan genome was estimated to include 4,408 orthologous genes, of which 2,280 were common to the core genomes of all analyzed strains. The number of genes shared between the 28 L. plantarum strains and strain WCSF1 ranged from 2,445/2,841 (86.1%) to 2,630/2,841 (92.6%), with a median number of 2,555/2,841 (89.9%). None of the Asian strains shared the complete set of 2,841 genes present in L. plantarum WCSF1.
Genomic clustering and sugar utilization capacity.
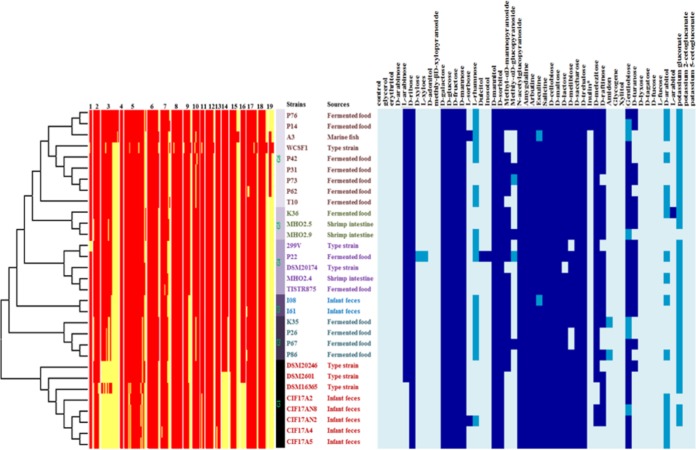
Since the genomic properties of the L. plantarum strains should translate into specific functions, the sugar utilization capacities of all strains were determined and clustered into 6 genomic groups (G1 to G6) based on the presence and absence of genes (Fig. 1). Of note, these genome-based clusters partly correlated to the order seen in the sugar utilization profile. Moreover, the genomic groups also correlated with the origins of the L. plantarum strains, where the strains from infant feces were enriched in G1 and G3, strains from fermented foods were enriched in G2 and G6, and strains from shrimp intestine were enriched in G5 (Fig. 1). The carbohydrate utilization profile further revealed that most L. plantarum strains use a broad range of simple and complex carbohydrates, and all strains grew on a group of 15 sugars, including glucose, galactose, mannose, sucrose, and fructose. The distinct carbohydrate metabolism may reflect their genomic diversity and evolution and adaptation to various environmental niches by gene decay, mutation (17, 18), gain, or loss (19, 20).
FIG 1.
Correlation genome diversity based on carbohydrate gene presence (red) and absence (yellow) (left side) in L. plantarum isolated from various sources by using the Mega 6 program (unweighted pair group method using average linkages [UPGMA] bootstrap value of 1,000, P-distance model) with the phenotypic data (API 50 CH) (right side); dark blue, completely fermented; lighter blue, partially fermented; lightest blue, nonfermented. The numbers at the top of the left figure refer to the PTS system and other transport and metabolic genes involved in sugar metabolism: 1, trehalose biosynthesis; 2, sucrose utilization; 3, fructooligosaccharide (FOS) and raffinose utilization; 4, lactose utilization; 5, maltose and maltodextrin utilization; 6, beta-glucoside metabolism; 7, trehalose uptake and utilization; 8, lactose and galactose uptake and utilization; 9, mannose metabolism; 10, d-tagatose and galactitol utilization; 11, d-gluconate and ketogluconate metabolism; 12, fructose utilization; 13, d-ribose utilization; 14, l-rhamnose utilization; 15, d-sorbitol (d-glucitol) and l-sorbose utilization; 16, l-arabinose utilization; 17, glycerol and glycerol-3-phosphate uptake and utilization; 18, mannitol utilization; and 19, inositol catabolism.
The capability to utilize carbohydrates depends on the presence of a functional transport system and of intracellular metabolic pathways. In this study, we detected genes encoding a PTS system and essential enzymes, which were originally annotated with functions involved in the uptake and metabolism of d-ribose, d-galactose, d-fructose, d-mannose, d-mannitol, d-sorbitol, d-cellobiose, d-maltose, d-lactose, d-saccharose, and d-trehalose (see Fig. S2). Strains isolated from fermented food and shrimp intestine (in G2, G3, G4, G5, and G6) had higher numbers of sugar utilization genes than the infant isolates (in G1). This was also evident from the summary of total genes predicted to be involved in sugar utilization as obtained from the Rapid Annotations using Subsystems Technology (RAST) server (see Fig. S3). Further comparison of genes involved in carbohydrate utilization revealed that L. plantarum isolated from fermented food and shrimp intestine comprised a high variety of genes, while strains isolated from infant feces lost genes involved in the utilization of fructooligosaccharides (FOS), raffinose, l-rhamnose, and l-arabinose (see Fig. S2 and S3). Therefore, the source of isolation displayed a remarkable degree of phenotypic and genotypic variety. There were large differences in carbohydrate availability in the various habitats from which the L. plantarum strains were recovered. The plant-associated niches were abundant in sucrose, trehalose, maltose, cellobiose, raffinose, starch, and inulin, whereas the mammalian intestinal tract was found to contain a variety of diet-derived carbohydrates and substantial quantities of host-derived carbohydrates (21, 22).
Adaptation to early life.
The genomes of the five L. plantarum strains (CIF17A2, CIF17A4, CIF17A5, CIF17AN2, and CIF17AN8) originating from the 5-month-old infant's intestine had slightly lower GC contents and were considerably smaller than those of L. plantarum strains from other sources, including those from older infants and adults (see Table S1). Since infants at this young age are predominantly breast-fed, this implies a specific adaptation of these L. plantarum strains to the mother's milk diet and that the strains in the baby's intestine may be associated with gene loss events, well-known in lactic acid bacteria and often mediated by insertion sequences (13, 23).
The young infant's L. plantarum strains lacked the capacity to utilize arabinose, a typical sugar found only in plants, notably in rice and banana plants that are provided to Thai infants after weaning. This is in line with the observation that the L. plantarum strains from the older and weaned infants (I08 and I61, respectively) and adults (299V) utilize arabinose. In contrast, the L. plantarum strains from the young infants used arabitol, a reduced sugar formed in the human body from arabinose and found in blood, urine, and likely also in the mother's milk. Similarly, the arabinose-utilizing human strains consumed d-turanose, while the young infant's strains did not utilize this plant signaling sugar for growth.
Analysis of inulin-utilizing strains.
Two strains (P14 and P76) showed vigorous growth in the presence of inulin as a sole carbon source, indicating the presence of genes involved in inulin metabolism (Fig. 2). There are some reports of genes encoding enzymes belonging to the β-fructofuranosidase and β-fructosidase family in the genomes of L. plantarum isolates (11, 24). However, the consumption of FOS, inulin-type fructan, or inulin has rarely been reported (11, 25). Hence, we further focused on the conversion of this natural fructose polymer. Two inulin-utilizing strains (P14 and P76) were isolated from a fermented fish product (Pla Pang Dang), which includes sticky rice, salt, and sugar, as well as garlic that may serve as a source of inulin (26).
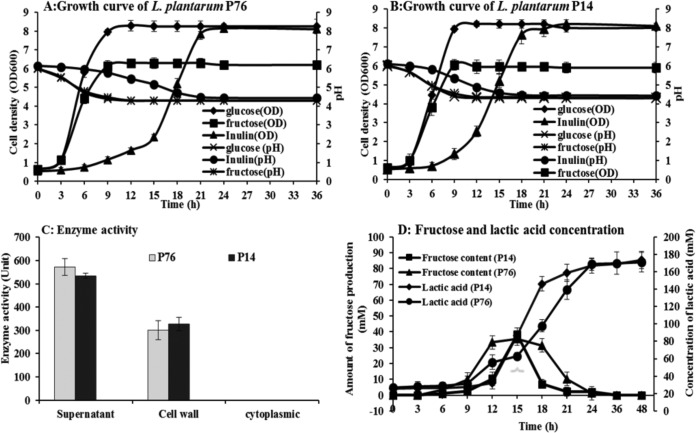
FIG 2.
Growth, β-fructosidase enzyme activity, and products of inulin-grown L. plantarum strains. Growth kinetics on inulin in comparison with other sugars for L. plantarum strains P76 (A) and P14 (B). β-Fructosidase enzyme activities in the various fractions (C). Amounts of lactic acid and fructose produced when grown on inulin (D).
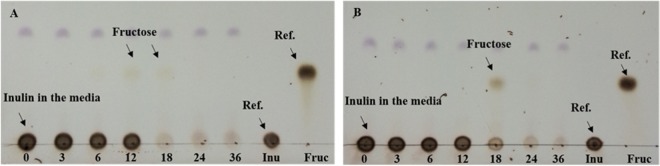
The capacity of L. plantarum strains P14 and P76 to ferment inulin was confirmed by a batch growth experiment in modified MRS-inulin in comparison to growth in the glucose- and fructose-containing media (Fig. 2A and B). Both strains grew faster in glucose (specific growth rates of 0.50 and 0.54 h−1, respectively) than in fructose (specific growth rates of 0.44 and 0.45 h−1, respectively). The stationary phase was reached in approximately 9 h on glucose or fructose compared to 18 to 21 h on inulin. L. plantarum P14 grew faster on inulin than L. plantarum P76. It reached the stationary phase several hours earlier than the P76 strain (after approximately 18 h and 21 h, respectively). In addition, the initial log phases of the P14 and P76 strains were 9 and 15 h, respectively, and their specific growth rates were 0.24 and 0.27 h−1, respectively, at mid-log phase. When grown on inulin, both L. plantarum strains reached optical densities at 600 nm (OD600) of about 8.0, similar that of strains grown on glucose. Lactic acid was the main metabolic end product of the P14 and P76 strains, with concentrations of 166.85 ± 5.25 and 168 ± 7.50 mM, respectively, after incubating for 24 h (Fig. 2D). Inulin utilization analysis showed that strains P14 and P76 consumed 86.28% ± 5.60% and 86.22% ± 4.55% of inulin, respectively, after incubating for 24 h. Thin-layer chromatographic (TLC) analysis of the culture supernatant revealed the presence of fructose after 12-h (P14) and 18-h (P76) incubations (Fig. 3).
FIG 3.
Sugar analysis of culture supernatants from inulin-grown L. plantarum strains. Culture supernatants obtained following the growth of L. plantarum P14 (A) and P76 (B) strains on inulin (time in hours is indicated) were spotted onto thin-layer chromatography silica gel plates and run in an acetic acid-chloroform-water (7:5:1) solvent. Sugars were visualized by spraying the plates with ethanolic 50% sulfuric acid and heating them at 115°C for 5 min. Inulin and fructose were used as a reference (Ref.).
Inulin degradation was confirmed by high-pressure liquid chromatography (HPLC), which showed that 38.38 ± 5.50 and 35.69 ± 4.50 mM fructose was generated by strains P14 and P76 after 6 h and 15 h, respectively, which was then completely metabolized at 24 h (Fig. 2D). This observation reflected the presence of an active β-fructosidase. To further substantiate this, the activity of this particular enzyme was evaluated in three fractions of cell culture using inulin as a substrate, i.e., cell-free supernatant, cell wall extract, and cytoplasmic fractions. The β-fructosidase activity was detected in the cell-free supernatant and cell wall extract fractions, but not in the cytoplasmic one. The highest β-fructosidase enzyme activities from the cell-free supernatants of strains P14 and P76 were observed at 18 h (649.24 ± 47.14 and 572.62 ± 35.92 μmol/ml/min, respectively), and the highest enzyme activities in the cell wall fractions were found at 15 h (367.06 ± 47.58 and 387.70 ± 28.62 μmol/ml/min, respectively) (Fig. 2C). These data provide evidence indicating that the β-fructosidase enzyme was not only secreted extracellularly but was also anchored to the cell wall, like other enzymes possessing an LPXTG anchor motif (27, 28). Results from a previous mutational study of β-fructosidase encoded by the fosE gene provided evidence that the fos operon encodes key components for the utilization of fructooligosaccharide and other, structurally similar carbohydrates in L. paracasei 1195 (9). In addition, the cell wall-anchored FosE was encoded by the last gene of the fosRABCDXE operon, which may encode for the degradation of inulin, as discussed below.
Proteogenomic analysis reveals an inducible inulin operon.
Three pathways, encoded by the fos, msm, and pts1BCA operons, are reportedly involved in FOS and inulin metabolism in Lactobacillus species (8–11). Microarray and other functional analyses revealed that the pts1BCA operon encodes a sucrose PTS transport system involved in metabolizing scFOS in L. plantarum WCSF1 in conjunction with an intracellular β-fructofuranosidase encoded by sacA (11). The pts1BCA gene complex, found in the draft genome sequences of strains P14 and P76, is also predicted to encode a sucrose PTS EIIBCA. It also shares complete identity (100% sequence similarity) with the gene of strain WCSF1. In addition, two other pathways have been described for metabolizing FOS and long-chain carbohydrates (inulin) in Lactobacillus species, but they have not yet been reported in L. plantarum. The fosRABCDXE operons present in the genomes of strains P14 and P76 are predicted to encode components of a fructose/mannose PEP-dependent PTS and a β-fructosidase precursor (the product of the fosE gene; see above) that are involved in the FOS utilization pathway of L. paracasei 1195 (9, 10). The presence of an N-terminal signal peptide sequence and an LPQAG cell wall anchor motif in the C-terminal regions of the deduced FosE precursor amino acid sequences predicts that the enzymes are cell wall associated, indicating that FOS may be extracellularly hydrolyzed, and the subsequent uptake of the hydrolysis products may be mediated by the FosABCDX PTS (9, 10). Another system for FOS utilization was previously described for L. acidophilus NCFM, where genes encoding the ABC transport system (msmEFGK) and a putative intracellular β-fructosidase (bfrA) were located in a multiple sugar metabolism (msm) operon. The bfrA gene product hydrolyzes sucrose, inulin-type fructan, and inulin (8). Therefore, in this study, the fosRABCDXE operon is likely involved in inulin utilization by L. plantarum strains P14 and P76. This operon has not yet been observed in other L. plantarum strains.
To substantiate the expression of the fosRABCDXE operon in L. plantarum P14 and P76, these strains were grown on inulin and the proteins produced were analyzed by shotgun proteomics. The results revealed an increased abundance of β-fructosidase, FosE, and other products of this operon (Table 1). β-Fructosidase (FosE) is a key enzyme in inulin degradation and hydrolyzes the termini of nonreducing 2,1- and 2,6-linked beta-d-fructofuranose residues in fructans. In the presence of inulin, 10- and 25-fold increased abundances of FosE were measured in strains P76 and P14, respectively (LBP14_150 and LBP76_3198, respectively) (Table 1). The 2,900-bp and 2,386-bp fosE genes of P14 and P76, respectively, were located next to fosX and coded in the same direction (Fig. 4). β-Fructosidase belongs to glycosyl hydrolase family 32 and contains conserved amino acid residues essential for its catalytic activity (29). However, the Gram-positive anchor was found in the FosE amino acid sequences from strains P14 and P76. The alignment of these amino sequences with those of fructan hydrolases of L. casei ATCC 334 (BfrA), L. paracasei 1195 (FosE), and Bacillus subtilis 168 (SacC) revealed that these enzymes were closely related to those encoded by bfrA and fosE, with sequence identities of 96 and 89.8%, respectively. On the other hand, the sequence shared only moderate sequence identity (38.6%) with levanase of B. subtilis 168. A dendrogram based on the deduced amino acid sequences can be seen in Fig. S4 in the supplemental material. These proteins were similar to each other (100% identity) and to β-fructosidase sequences of L. casei and L. paracasei.
TABLE 1.
Proteome analysis of inulin-grown L. plantarum strains
| Gene | Protein annotation for indicated L. plantarum straina |
Annotation | Fold changeb |
||
|---|---|---|---|---|---|
| P14 | P76 | P14 | P76 | ||
| fosR | LBP14_156 | —c | Transcription regulator related to NorR | 6.31 | — |
| fosA | LBP14_155 | — | PTS system, fructose/mannose-inducible, IIA component | 104.71 | — |
| fosB | LBP14_154 | LBP76_3114 | PTS system, fructose/mannose-inducible, IIB component | 60.26 | 58.88 |
| fosC | LBP14_153 | — | PTS system, fructose/mannose-inducible, IIC component | 537.03 | — |
| fosD | LBP14_152 | LBP76_3112 | PTS system, fructose/mannose-inducible, IID component | 213.80 | 64.57 |
| fosE | LBP14_150 | LBP76_3198 | Sucrose-6-phosphate hydrolase (beta-fructosidase) | 26.92 | 11.75 |
| sacK1 | LBP14_1639 | LBP76_917 | Fructokinase | 165.96 | 245.47 |
| pts1BCA | LBP14_1640 | LBP76_918 | PTS system, sucrose-specific, IIB, IIC, and IIA components | 1258.93 | 72.44 |
| sacC | LBP14_1641 | LBP76_919 | Sucrose-6-phosphate hydrolase (EC 3.2.1.26) | 288.40 | 9.77 |
| fruK | LBP14_1067 | LBP76_2192 | 1-Phosphofructokinase | 21.38 | 20.89 |
| — | LBP14_1804 | LBP76_1082 | Aerobic glycerol-3-phosphate dehydrogenase | 151.36 | 630.96 |
| araT2 | LBP14_2218 | LBP76_1662 | N-Acetyl-l,l-diaminopimelate aminotransferase | 177.83 | 109.65 |
| — | LBP14_1423 | LBP76_705 | Maltose-6-phosphate glucosidase | 54.95 | 380.19 |
| — | LBP14_2003 | LBP76_999 | PTS system, cellobiose-specific, IIC component | 33.88 | 154.88 |
| pts6C | LBP14_1721 | LBP76_1344 | PTS system, cellobiose-specific, IIC component | 20.89 | 22.39 |
| pts33BCA | LBP14_1357 | — | PTS system, beta-glucoside-specific, IIB, IIC, and IIA components | 354.81 | — |
| pbg10 | LBP14_1356 | — | 6-Phospho-beta-glucoside | 125.89 | — |
| — | LBP14_566 | — | LSU ribosomal protein L35p | 630.96 | — |
| dtpT | LBP14_3012 | — | Di-/tripeptide permease (DtpT) | 194.98 | — |
| mvaS | LBP14_1040 | — | Hydroxymethylglutaryl-CoA synthase | 151.36 | — |
| pps | LBP14_909 | — | Phosphoenolpyruvate synthase | 125.89 | — |
| mleR1 | LBP14_2541 | — | Malolactic regulator | 107.15 | — |
| nox5 | — | LBP76_584 | NADH peroxidase (Npx) | — | 269.15 |
| glpK | — | LBP76_1081 | Glycerol kinase | — | 234.42 |
| minC | — | LBP76_2308 | Septum site-determining protein MinC | — | 83.18 |
| — | — | LBP76_427 | ATP/GTP-binding protein | — | 83.18 |
| — | — | LBP76_1793 | ErfK/YbiS/YcfS/YnhG family protein, putative | — | 81.28 |
| elaC | — | LBP76_2187 | RNase Z | — | 72.44 |
| coaA | — | LBP76_1274 | Pantothenate kinase | — | 67.61 |
| ppx3 | — | LBP76_1220 | Exopolyphosphatase | — | 64.57 |
| pheS | — | LBP76_54 | Phenylalanyl-tRNA synthetase alpha chain | — | 60.26 |
| — | — | LBP76_1033 | Hypothetical protein | — | 53.70 |
| dnaI | — | LBP76_19 | Helicase loader (DnaI) | — | 47.86 |
| mecA | — | LBP76_2882 | Negative regulator of genetic competence (MecA) | — | 42.66 |
| pox1 | — | LBP76_1225 | Pyruvate oxidase | — | 39.81 |
| — | — | LBP76_2377 | Fumarate reductase, flavoprotein subunit precursor | — | 39.81 |
| pflA | — | LBP76_489 | Pyruvate formate-lyase activating enzyme | — | 36.31 |
| pps | — | LBP76_365 | Phosphoenolpyruvate synthase | — | 35.48 |
| — | — | LBP76_659 | Galactitol-1-phosphate 5-dehydrogenase | — | 32.36 |
P14 and P76 were grown on inulin and on glucose.
Quantitative proteome analysis was performed on cell extracts. Inulin-induced proteins with a >5-fold change versus glucose are listed.
—, not available.
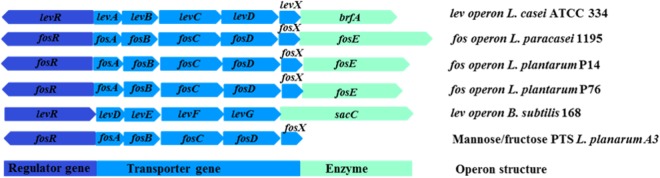
FIG 4.
Comparative gene organization of the L. plantarum inulin utilization operon. The predicted inulin degradation genes of L. plantarum strains P76 and P14 are shown in comparison with those from the fos operon of L. paracasei 1195 and the lev operon of L. casei ATCC 334. The fructose/mannose PTS system of L. plantarum A3 is also depicted. Dark blue, regulator gene; blue, PTS genes involved in sugar transport; light green, beta-fructosidase gene.
The proteomics analysis of strains P14 and P76 also provided evidence for the inducible expression of the sugar PTS genes in the presence of inulin (Table 1). Increases of 60- and 540-fold were observed for the FosABCD transporter in strains P14 and P76, respectively. FosE and FosABCDX are encoded by the fosRABCDXE operon, which was previously documented in a strain of L. paracasei (9, 10). The identified putative FosABCD transporter located in the genomes of L. plantarum P14 and P76 comprises genes encoding a transcriptional regulator (FosR) and the EIIA, IIB, IIC, IID, and EII components of a fructose/mannose-specific PTS. The structural organizations and gene sequences of these operons were extremely similar to those of the putative fos operon of L. paracasei 1195 and the putative levanase operon of L. casei ATCC 334 (Fig. 4). The products of fosR genes (LBP14_156 and LBP76_3116) exhibited 69% and 47% amino acid sequence identity with the fosR transcriptional regulator of L. paracasei 1195 (GenBank accession no. ABD57313.1) and a putative levanase operon transcriptional regulator in L. casei ATCC 334 (GenBank accession no. ABJ69406.1), respectively. The structural genes consist of four genes (FosABCD) that encode the EIIABCD components of the fructose/mannose PTS. FosA and FosB are presumably responsible for phosphorylating substrates, while FosC and FosD are likely specific for fructose and/or mannose (30). The nucleotide sequences of FosB, FosC, and FosD shared 85.6, 91.5, and 69.1% identities, respectively, with the structural genes of L. paracasei 1195 and L. casei ATCC 334, whereas FosA is similar (49% identity) to L. paracasei 1195 and L. casei ATCC 334. The fosX gene encodes a 109-residue putative EII component that shares 69% identity with a hypothetical protein in L. paracasei 1195 (GenBank accession no. ABD57318.1) and L. casei ATCC 334 (GenBank accession no. ABJ69401.1).
In L. acidophilus and Bifidobacterium breve, ABC transporters were observed to be involved in FOS uptake (8, 31), but these systems were not found in the inulin-utilizing P14 and P76 strains of L. plantarum described here. However, these strains were found to contain an additional sucrose PTS, encoded by pts1BCA, which was upregulated during inulin fermentation in strains P14 and P76, resulting in 1,259-fold (LBP14_1640) and 72-fold (LBP76_918) increased abundances, respectively. Moreover, expression of sucrose-6-phosphate hydrolase (sacC) was increased 288-fold (LBP14_1641) and 9.7-fold (LBP76_919) while expression of fructokinase (sacK1) was increased 165- and 245-fold in strains P14 and P76, respectively, when grown on inulin (Table 1). The upregulated sucrose-6-phosphate hydrolase in strains P14 and P76 revealed a 99% similarity to beta-fructofuranosidase in other L. plantarum strains (accession no. AGE_37932.1 and WP_02400235.1), which hydrolyzed sucrose and scFOS. In addition, pts33BCA may be involved in metabolizing glucose residues present as terminal sugars of certain types of inulin (32, 33). The proposed pathway of inulin utilization is depicted in Fig. 5.
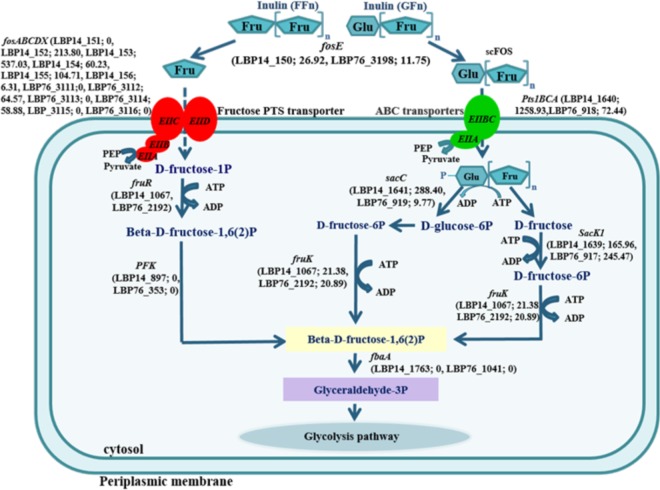
FIG 5.
Proposed model for inulin degradation by L. plantarum strains P14 and P76 based on the proteogenomic analysis. The locus tags and fold induction of protein based on proteomic data are indicated for L. plantarum strains P76 and P14. fosE, beta-fructofuranosidase; fosABCDX, the putative fos operon; sacK1, fructokinase; fruR, 1-phosphofructokinase; PFK, 6-phosphofructokinase; fbaA, fructose-bisphosphate aldolase class II.
Conclusion.
Twenty-eight Asian isolates of L. plantarum were investigated at the genomic and phenotypic levels. Obviously, genome size correlated with the isolation sites. The strains isolated from foods typically harbored more genes than those isolated from humans. Although information related to their niches might parallel their ecological fitness and adaptation, each strain harbors sets of strain-specific genes. To further illustrate the association between genotype and phenotype, we reported and examined genes involved in carbohydrate utilization among L. plantarum strains in this study. Specifically, strains P14 and P76 harbored the operon fosRABCDXE and pts1BCA in their genomes, enabling these two strains to hydrolyze inulin. We further demonstrated the expression and the functionality of the inulin metabolic pathway using a proteomic approach.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Twenty-eight strains of L. plantarum were isolated and selected from various sources, including infant feces, marine fish, shrimp intestines, fermented fish, and fermented rice noodle products from Thailand (see Table S1 in the supplemental material). All strains were classified as L. plantarum according to their 16S rRNA nucleotide sequences and sugar utilization profiles (using the API 50 CH kit; see below for further details). Additionally, these strains were differentiated from the closest species, L. pentosus and L. paraplantarum, based on the nucleotide sequences of recA and dnaK genes. Seven infant strains (CIF17A2, CIF17A4, CIF17A5, CIF17AN2, CIF17AN8, I08, and I61) (34, 35) as well as the marine fish and shrimp intestine strains (A3, MHO2.4, MHO2.5, and MHO2.9) (36, 37) were previously described. The 13 fermented-food isolates were cultured from Thai fermented rice noodles and fish essentially as described previously (38) (kindly provided by A. Nuylert and K. Kanjanasmith). L. plantarum TISTR 875 (39) was isolated from a Thai fermented food (pickled cabbage) donated by the Thailand Institute of Scientific and Technological Research (Bangkok, Thailand). The L. plantarum type strain DSM20246, L. plantarum subsp. plantarum DSM20174, and L. plantarum subsp. argentoratensis DSM16365 and DSM2601 (previously described as L. arizonensis; see reference 40) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ, Germany), L. plantarum strain 299V was isolated from a fermented ProViva drink produced with strain 299v (41), and L. plantarum strain WCSF1 was obtained from the Laboratory of Microbiology at Wageningen University (The Netherlands). All L. plantarum strains/isolates were cultivated in MRS broth at 37°C for 24 h.
Sugar fermentation and other metabolic assays.
Carbohydrate fermentation profiles were determined using the API 50 CH kit (bioMérieux, France) according to the manufacturer's instructions. The inulin-utilizing strains (P14 and P76) were cultured in modified MRS media supplemented with 1% inulin (Sigma, USA) as a main carbon source and incubated at 37°C for 48 h (glucose and fructose were used as controls). Bacterial growth was determined every 3 h for up to 48 h by measuring the optical density at 600 nm, and the pH of the cell-free supernatants was also monitored.
Analysis of sugar and other inulin-degraded metabolites.
To determine the concentrations of glucose and fructose residues in the bacterial cultures, cells were removed by centrifugation at 9,500 × g for 10 min at 4°C. The supernatants were transferred to HPLC vials. Twenty microliters of these supernatants was injected by an autosampler into the HPLC system (Spectra system AS3000) equipped with a Metacarb 67H column (300 mm × 6.5 mm). The sugar molecules were eluted with 0.01 N H2SO4 at a flow rate of 0.8 ml/min. Standard curves for glucose and fructose (0, 2.5, 5, 10, and 20 mM) were prepared. Thin-layer chromatography (TLC) was used to monitor the extent of inulin degradation by spotting the supernatants onto a silica gel plate (Merck, Germany) using 1% inulin and fructose as a control. The plates were developed in a solvent mixture of acetic acid-chloroform-water (7:5:1) as a mobile phase. The sugar spots were visualized by spraying the plates with ethanolic 50% sulfuric acid (ethanol:sulfuric acid, ratio 1:1) and heating them at 115°C for 5 min. In addition, a general phenol-sulfuric acid assay was performed to determine the total carbohydrate contents after incubating for 24 h (42).
β-Fructosidase assays.
Enzyme activity was investigated as previously described (8) with some modifications. Briefly, cells were harvested by centrifugation at 9,500 × g for 10 min at 4°C, and cell pellets were washed twice in 0.1 M potassium phosphate buffer (pH 6.6) and resuspended in 1 ml of the same buffer. The cell suspension was homogenized with a Branson sonicator equipped with a 3-mm tip using six pulses of 30 s with a 1-min incubation on ice in between each pulse. Cell lysates were transferred into new microtubes and cell walls were separated from the cytoplasmic fraction by centrifuging at 9,500 × g for 10 min at 4°C, essentially as described previously (9). The cell wall fraction was then resuspended in 1 ml 0.1 M potassium phosphate buffer (pH 6.6).
Ten microliters of each fraction (culture supernatant, cell wall fraction, or cytoplasmic extract) was added to 190 μl of a 1% (wt/vol) inulin solution. The reaction mixtures were incubated at 37°C for 3 h and heat inactivated at 95°C for 5 min. The β-fructosidase activity was measured by determining the amount of inulin released as fructose, and fructose residue concentrations were determined using a fructose assay kit (EnzyChrom; BioAssay Systems, USA) according to the manufacturer's instructions. All experiments were performed in duplicate. One unit of enzyme activity was defined as the enzyme necessary to release 1 μmol of fructose per min at the specified conditions.
Protein extraction and proteomic analysis.
Bacterial proteins were extracted and then fractionated using SDS-PAGE. Bacterial cells at mid-log phase grown (separately twice) in the presence and absence of inulin were centrifuged and washed three times with phosphate-buffered saline (PBS; pH 6.8). The pellet was resuspended in 0.5 ml 100 mM Tris-HCl (pH 7.5) with 4% SDS and 0.1 M dithiothreitol and transferred into 2-ml low-binding microcentrifuge tubes (Eppendorf, The Netherlands). The cell pellets were lysed using a sonicator as described above. Proteins were denatured by heating at 95°C for 20 min, followed by centrifugation at 9,500 × g for 10 min. Protein concentrations were determined as described previously (43). The protein concentrations were equalized, and each of the samples containing 25 μg of protein mixed with 5 μl of loading buffer (250 mM Tris-HCl [pH 6.8], 0.5 M dithiothreitol, 10% SDS, 0.02% bromophenol blue, and 30% glycerol) was subjected to SDS-PAGE using a 10% acrylamide gel (Miniprotein III system; Bio-Rad, The Netherlands). Gels were stained with Coomassie brilliant blue R250.
Protein extractions, separations, and tryptic digestions were performed as described previously (44). The trypsin-digested peptides were analyzed by nano-liquid chromatography coupled with mass spectrometry (nLC-MS/MS) using a Proxeon nLC and a LTQ-Orbitrap mass spectrometer (44, 45), and all spectral data obtained were analyzed with MaxQuant v.1.3.0.5 (45, 46) using a contaminants database and a protein database generated from L. plantarum P14 and L. plantarum P76 after genomic sequencing and proteomic data were obtained (as described below).
The “label-free quantification” (LFQ) and “match between runs” (set to 2 min) options were enabled. Deamidated peptides were used for protein quantification, and all other quantification settings were kept at the default. Filtering and further bioinformatic analysis of the MaxQuant/Andromeda workflow output and the analysis of the abundances of the identified proteins were performed with the Perseus 1.3.0.4 module (47). Peptides and proteins with a false discovery rate (FDR) of less than 1% were accepted, as were proteins with at least 2 identified peptides, of which at least one was unique and at least one was unmodified. Reversed hits were deleted from the MaxQuant result table. Zero LFQ intensity values were replaced by a value of 15,848 (just below the lowest value measured) to permit sensible ratio calculations. Relative protein quantitation of inulin-grown samples to glucose-grown controls was calculated as the ratio of the average inulin-grown LFQ intensities to the average glucose-grown LFQ intensities.
Genome sequencing and annotation.
For sequencing, genomic DNA from all L. plantarum strains (see Table S1) was extracted from 1.5-ml volume overnight cultures using a commercial kit (MasterPure; Epicentre, Madison, USA). Bacterial genomic DNA was then sequenced on a paired-end (R1 = 326 bp, R2 = 286 bp) Illumina MiSeq run. Adapter sequences and low-quality bases (Q <30) at the ends of reads were trimmed out using cutadapt (version 1.6) (48). Overlapping paired-end reads were merged and extended as single reads by FLASh (Fast Length Adjustment of Short reads) (version 1.2.6) (49). De novo genome assemblies were performed from the extended paired-end and orphan reads using SPAdes assembler (v. 3.1.1) (50). DNA sequencing was performed at the DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki. Gene prediction and annotation were conducted using the RAST server (51). The presence of plasmid sequences was evaluated by using PlasmidFinder version 1.3 (52). The pan and core genomes were evaluated using eggNOG v4.0 (53). The signal peptide prediction was performed using SignalP 4.1 (54), and the transmembrane helices were predicted by the TMHMM server (V.2.0) (55).
Accession number(s).
Genome sequences of the 28 strains were determined, analyzed, and deposited into the GenBank database. The new whole-genome shotgun sequences reported here are deposited at DDBJ/EMBL/GenBank under the accession numbers LEAV00000000 (for L. plantarum 299v), LEAW00000000 (for L. plantarum TISTR875), LEAX00000000 (for L. plantarum DSM 20246), LEAY00000000 (for L. plantarum DSM 2601), LEAZ00000000 (for L. plantarum CIF17AN8), LEBA00000000 (for L. plantarum CIF17AN2), LEBB00000000 (for L. plantarum CIF17A5), LEBC00000000 (for L. plantarum CIF17A4), LEBD00000000 (for L. plantarum CIF17A2), LEBE00000000 (for L. plantarum I08), LEBF00000000 (for L. plantarum A3), LEBG00000000 (for L. plantarum MHO2.9), LEBH00000000 (for L. plantarum MHO2.5), LEBI00000000 (for L. plantarum MHO2.4), LEBJ00000000 (for L. plantarum P86), LEBK00000000 (for L. plantarum P76), LEBL00000000 (for L. plantarum P73), LEBM00000000 (for L. plantarum P67), LEBN00000000 (for L. plantarum P62), LEBO00000000 (for L. plantarum P42), LEBP00000000 (for L. plantarum P31), LEBQ00000000 (for L. plantarum P26), LEBR00000000 (for L. plantarum P22), LEBS00000000 (for L. plantarum T10), LEBT00000000 (for L. plantarum K35), LEBU00000000 (for L. plantarum I61), LEBV00000000 (for L. plantarum P14), and LEBW00000000 (for L. plantarum K36) (see Fig. S1 in the supplemental material). Sequences were compared with those of the previously reported strains WCFS1 (14, 56), L. plantarum subsp. plantarum DSM20174, and L. plantarum subsp. argentoratensis DSM16365 (13).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by grants from the Strategic Scholarships for Frontier Research Network (specific for the southern region), the Office of the Higher Education Commission for the Thai doctoral degree program, the Graduate School of Prince of Songkla University, and the Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands. The research was also partly funded by grants 137389, 141140, and 1272870 from the Academy of Finland, as well as the Spinoza and Gravitation Grant (024.002.002) from the Netherlands Organization for Scientific Research.
We acknowledge the excellent support provided by the Institute of Biotechnology (University of Helsinki, Helsinki, Finland).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02402-16.
REFERENCES
- 1.Van Loo J, Coussement P, De Leenheer L, Hoebregs H, Smits G. 1995. On the presence of inulin and oligofructose as natural ingredients in the Western diet. Crit Rev Food Sci Nutr 35:1–13. doi: 10.1080/10408399509527714. [DOI] [PubMed] [Google Scholar]
- 2.Roberfroid MB. 2005. Introducing inulin-type fructans. Br J Nutr 93:S13–S25. doi: 10.1079/BJN20041350. [DOI] [PubMed] [Google Scholar]
- 3.Roberfroid MB, Delzenne NM. 1998. Dietary fructans. Annu Rev Nutr 18:117–143. doi: 10.1146/annurev.nutr.18.1.117. [DOI] [PubMed] [Google Scholar]
- 4.Roberfroid MB, Van Loo JAE, Gibson GR. 1998. The bifidogenic nature of chicory inulin and its hydrolysis products. J Nutr 128:11–19. [DOI] [PubMed] [Google Scholar]
- 5.Takemura N, Hagio M, Ishizuka S, Ito H, Morita T, Sonoyama K. 2010. Inulin prolongs survival of intragastrically administered Lactobacillus plantarum no. 14 in the gut of mice fed a high-fat diet. J Nutr 140:1963–1969. doi: 10.3945/jn.110.128082. [DOI] [PubMed] [Google Scholar]
- 6.Tsujikawa Y, Nomoto R, Osawa R. 2013. Comparison of degradation patterns of inulin-type fructans in strains of Lactobacillus delbrueckii and Lactobacillus paracasei. Biosci Microbiota Food Health 32:157–165. doi: 10.12938/bmfh.32.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velikova PV, Blagoeva GI, Gotcheva VG, Petrova PM. 2014. Novel Bulgarian Lactobacillus strains ferment prebiotic carbohydrates. J Biosci Biotechnol 2014:55–60. [Google Scholar]
- 8.Barrangou R, Alterman E, Hutkins R, Cano R, Klaenhammer TR. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc Natl Acad Sci U S A 100:8957–8962. doi: 10.1073/pnas.1332765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goh YJ, Zhang C, Benson AK, Schlegel V, Lee JH, Hutkins RW. 2006. Identification of putative operon involved in fructooligosaccharides utilization by Lactobacillus paracasei. Appl Environ Microbiol 72:7518–7530. doi: 10.1128/AEM.00877-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goh YJ, Lee JH, Hutkins RW. 2007. Functional analysis of the fructooligosaccharide utilization operon in Lactobacillus paracasei 1195. Appl Environ Microbiol 73:5716–5724. doi: 10.1128/AEM.00805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saulnier DMA, Molenaar D, de Vos WM, Gibson GR, Kolida S. 2007. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl Environ Microbiol 73:1753–1765. doi: 10.1128/AEM.01151-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cebeci A, Gurakan C. 2003. Properties of potential probiotic Lactobacillus plantarum strains. Food Microbiol 20:511–518. doi: 10.1016/S0740-0020(02)00174-0. [DOI] [Google Scholar]
- 13.Sun Z, Harris HM, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Ross RP, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O'Toole PW. 2015. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siezen R, Tzeneva VA, Castioni A, Wels M, Phan HTK, Rademaker JLW, Starrenburg MJC, Kleerebezem M, Molenaar D, van Hylckama Vlieg JET. 2010. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12:758–773. doi: 10.1111/j.1462-2920.2009.02119.x. [DOI] [PubMed] [Google Scholar]
- 15.van Kranenburg R, Golic N, Bongers R, Leer RJ, de Vos WM, Siezen RJ, Kleerebezem M. 2005. Functional analysis of three plasmids from Lactobacillus plantarum. Appl Environ Microbiol 71:1223–1230. doi: 10.1128/AEM.71.3.1223-1230.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siezen RJ, Francke C, Renckens B, Boekhorst J, Wels M, Kleerebezem M, van Hijum SA. 2012. Complete resequencing and reannotation of the Lactobacillus plantarum WCFS1 genome. J Bacteriol 194:195–196. doi: 10.1128/JB.06275-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 18.Feldgarden M, Byrd N, Cohan FM. 2003. Gradual evolution in bacteria: evidence from Bacillus systematics. Microbiology 49:3565–3573. doi: 10.1099/mic.0.26457-0. [DOI] [PubMed] [Google Scholar]
- 19.McLysaght A, Baldi PF, Gaut BS. 2003. Extensive gene gain associated with adaptive evolution of poxviruses. Proc Natl Acad Sci U S A 100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie JM, Raoult D. 2001. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science 293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 21.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N, Shakhova V, Grigoriev I, Lou Y, Rohksar D, Lucas S, Huang K, Goodstein DM, Hawkins T, Plengvidhya V, Welker D, Hughes J, Goh Y, Benson A, Baldwin K, Lee JH, Diaz-Muniz I, Dosti B, Smeianov V, Wechter W, Barabote R, Lorca G, Altermann E, Barrangou R, Ganesan B, Xie Y, Rawsthorne H, Tamir D, Parker C, Breidt F, Broadbent J, Hutkins R, O'Sullivan D, Steele J, Unlu G, Saier M, Klaenhammer T, Richardson P, Kozyavkin S, Weimer B, Mills D. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Chen C, Ai L, Zhou F, Zhou Z, Wang L, Zhang H, Chen W, Guo B. 2011. Complete genome sequence of the probiotic Lactobacillus plantarum ST-III. J Bacteriol 193:313–314. doi: 10.1128/JB.01159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takagi R, Tsujikawa Y, Nomoto R, Osawa R. 2014. Comparison of the growth of Lactobacillus delbrueckii, L. paracesei and L. plantarum on inulin in co-culture systems. Biosci Microbiota Food Health 33:139–146. doi: 10.12938/bmfh.33.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rittiplang J, Laopaiboon P, Vichitpan K, Danvirutai P. 2007. Lactic acid bacteria from indigenous Loogpang samples of northeastern Thailand and their lactic acid production ability. Thai J Biotechnol 2007:44–48. [Google Scholar]
- 27.Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J Bacteriol 187:4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leenhouts K, Buist G, Kok J. 1999. Anchoring of proteins to lactic acid bacteria. Antonie Van Leeuwenhoek 76:367–376. doi: 10.1023/A:1002095802571. [DOI] [PubMed] [Google Scholar]
- 29.Pons T, Olmea O, Chinea G, Beldarrain A, Marquez G, Acosta N, Rodriguez L, Valencia A. 1998. Structural model for family 32 of glycosyl-hydrolase enzymes. Proteins 33:383–395. [DOI] [PubMed] [Google Scholar]
- 30.Kotrba P, Inui M, Yukawa H. 2001. Bacterial phosphotransferase system (PTS) in carbohydrate uptake and control of carbon metabolism. J Biosci Bioeng 92:502–517. doi: 10.1016/S1389-1723(01)80308-X. [DOI] [PubMed] [Google Scholar]
- 31.Ryan SM, Fitzgerald GF, van Sinderen D. 2005. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl Environ Microbiol 71:3475–3482. doi: 10.1128/AEM.71.7.3475-3482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly G. 2008. Inulin-type prebiotics—a review: part 1. Altern Med Rev 13:315–329. [PubMed] [Google Scholar]
- 33.Michlmayr H, Kneifel W. 2014. β-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS Microbiol Lett 352:1–10. doi: 10.1111/1574-6968.12348. [DOI] [PubMed] [Google Scholar]
- 34.Uraipan S, Hongpattarakere T. 2015. Antagonistic characteristics against food-borne pathogenic bacteria of lactic acid bacteria and bifidobacteria isolated from feces of healthy Thai infant. Jundishapur J Microbiol 8:e18264. doi: 10.5812/jjm.8(5)2015.18264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanjan P, Hongpattarakere T. 2016. Antibacterial metabolites secreted under glucose-limited environment of the mimicked proximal colon model by lactobacilli abundant in infant feces. Appl Microbiol Biotechnol 100:7651–7664. doi: 10.1007/s00253-016-7606-5. [DOI] [PubMed] [Google Scholar]
- 36.Hongpattarakere T, Cherntong N, Wochienchot S, Kolida S. 2012. In vitro prebiotic evaluation of exopolysaccharides produced by marine isolated lactic acid bacteria. Carbohydr Polym 87:846–852. doi: 10.1016/j.carbpol.2011.08.085. [DOI] [PubMed] [Google Scholar]
- 37.Kongnum K, Hongpattarakere T. 2012. Effect of Lactobacillus plantarum isolated from digestive tract of wild shrimp on growth and survival of white shrimp (Litopenaeus vannamei) challenged with Vibrio harveyi. Fish Shellfish Immunol 32:170–177. doi: 10.1016/j.fsi.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez E, Gonzalez B, Gaya P, Nunez M, Medina M. 2000. Diversity of bacteriocins produced by lactic acid bacteria isolated from raw milk. Int Dairy J 10:7–15. doi: 10.1016/S0958-6946(00)00017-0. [DOI] [Google Scholar]
- 39.Hongpattarakere T, Rattanaubon P, Buntin N. 2013. Improvement of freeze-dried Lactobacillus plantarum survival using water extracts and crude fibers from food crops. Food Bioproc Technol 6:1885–1896. doi: 10.1007/s11947-012-1018-z. [DOI] [Google Scholar]
- 40.Swezey JL, Nakamura LK, Abbott TP, Peterson RE. 2000. Lactobacillus arizonensis sp. nov., isolated from jojoba meal. Int J Syst Evol Microbiol 50:1803–1809. doi: 10.1099/00207713-50-5-1803. [DOI] [PubMed] [Google Scholar]
- 41.Adlerberth I, Ahrne S, Johansson ML, Molin G, Hanson LA, Wold AE. 1996. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol 62:2244–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric methods for determination of sugars and related substances. Anal Chem 28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- 43.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 44.Oosterkamp MJ, Boeren S, Plugge CM, Schaap PJ, Stams AJM. 2013. Metabolic response of Alicycliphilus denitrificans strain BC toward electron acceptor variation. Proteomics 13:2886–2894. doi: 10.1002/pmic.201200571. [DOI] [PubMed] [Google Scholar]
- 45.Lu J, Boeren S, de Vries SC, van Valenberg HJF, Vervoort J, Hettinga K. 2011. Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J Proteomics 75:34–43. doi: 10.1016/j.jprot.2011.07.031. [DOI] [PubMed] [Google Scholar]
- 46.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 47.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 48.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 49.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 57:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powell S, Forslund K, Szklarczyk D, Trachana K, Roth A, Huerta-Cepas J, Gabaldón T, Rattei T, Creevey C, Kuhn M, Jensen LJ, von Mering C, Bork P. 2014. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res 42:D231–D239. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petersen TN, Brunak S, Heijne GV, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 55.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 56.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci U S A 100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.