ABSTRACT
In pasture-based systems, changes in dairy herd habitat due to seasonality results in the exposure of animals to different environmental niches. These niches contain distinct microbial communities that may be transferred to raw milk, with potentially important food quality and safety implications for milk producers. It is postulated that the extent to which these microorganisms are transferred could be limited by the inclusion of a teat preparation step prior to milking. High-throughput sequencing on a variety of microbial niches on farms was used to study the patterns of microbial movement through the dairy production chain and, in the process, to investigate the impact of seasonal housing and the inclusion/exclusion of a teat preparation regime on the raw milk microbiota from the same herd over two sampling periods, i.e., indoor and outdoor. Beta diversity and network analyses showed that environmental and milk microbiotas separated depending on whether they were sourced from an indoor or outdoor environment. Within these respective habitats, similarities between the milk microbiota and that of teat swab samples and, to a lesser extent, fecal samples were apparent. Indeed, SourceTracker identified the teat surface as the most significant source of contamination, with herd feces being the next most prevalent source of contamination. In milk from cows grazing outdoors, teat prep significantly increased the numbers of total bacteria present. In summary, sequence-based microbiota analysis identified possible sources of raw milk contamination and highlighted the influence of environment and farm management practices on the raw milk microbiota.
IMPORTANCE The composition of the raw milk microbiota is an important consideration from both a spoilage perspective and a food safety perspective and has implications for milk targeted for direct consumption and for downstream processing. Factors that influence contamination have been examined previously, primarily through the use of culture-based techniques. We describe here the extensive application of high-throughput DNA sequencing technologies to study the relationship between the milk production environment and the raw milk microbiota. The results show that the environment in which the herd was kept was the primary driver of the composition of the milk microbiota composition.
KEYWORDS: farm practices, food chain, food safety, metagenomics, microbial source tracking, raw milk microbiota
INTRODUCTION
The impact of the dairy farm environment on the microbial composition of raw milk and raw milk products has been appreciated for some time (1). There are numerous niches that collectively constitute the dairy farm environment, and these harbor a vast array of microbes. The transfer of microbes from the farm environment to raw milk can be influenced be a number of factors, including farmer hygiene, husbandry practices, herd health, and herd housing (2). In turn, the microbial composition of raw milk is critically important to its quality, processability, and safety.
The microbiota composition of dairy farm niches and of raw milk has typically been examined using traditional plate cultivation-based techniques. These culture-based assays are still widely used by industry and target specific phenotypes, e.g., the ability to grow at or survive exposure to particular temperatures (psychrotrophs [3], mesophiles [4], and thermodurics [5]) or the capacity to produce proteases, lipases, or other enzymes (6) or species known to be human pathogens (2). Using these culture-based techniques, Vacheyrou previously examined possible routes of microbial transfer in farms supplying raw milk for Comte style cheese, revealing that the extent to which milk was contaminated varied depending on the type of barns used to house animals (2). However, recent advances in molecular microbiology, and in high-throughput DNA sequencing (HTS) in particular, have allowed for a more in-depth analysis of the flow of microbes through environments (7–12).
Indeed, a study of two artisan cheese-making plants observed that spatial diversification within both plants was indicative of “functional adaptations” by microbial communities colonizing different fomites within each plant. Spatial diversification between plants confirms the phenomenon of a unique production plant (“house”)-associated microbiota, which was postulated to influence the distinct organoleptic properties of products from each facility (11). The facility-specific microbiota developed as a result of the selection pressure introduced by the individual cheese-making processing methods (11). The observation of a niche-specific functional adaptation has also been observed in the microbiota of a winery, with the additional observation that the community was influenced by seasonality (12).
The present proof-of-concept study focuses on the Irish dairy farm system, which is primarily a pasture-based system, in which herds are grazed on pasture for the majority of their lactation curve. However, during the winter months, herds are housed indoors. The transition between environments is an important consideration for dairy producers, since it is accompanied by changes in exposure to microbes from different niches in the environment, as well as dietary changes. Previous, culture-based, efforts to address this question have noted elevated spore counts in bulk tank milk collected from a number of Midwestern American farms during summer months on American farms (13), although elevated numbers of sporeformers can also be an issue when cows are housed indoors if poor quality silage is used (14). Our study also investigates the impact that teat preparation has on the microbiology of raw milk. This farm management practice has been shown to reduce bacterial counts in milk previously (15), but its impact on the raw milk microbiota has not been reported.
Based on the results of the studies highlighted above and in the context of the seasonal milk production system applied in Ireland (all cows calved within a 12-week period), it is reasonable to assume that cattle are exposed to niche-specific microbes when housed indoors during winter months and that these environmental microbes differ significantly from that present when the herd is grazing on pasture during the summer. Such differences would be expected, in turn, to have an impact on the raw milk microbiota. Specifically, we examined the influence that seasonal housing and grazing conditions have on the microbiota of raw cows' milk. We also examined the influence that the farm management practice of teat preparation (prep) has on the raw milk microbiota in both environments. To address these questions, we applied HTS and a Bayesian inference algorithm to examine environmental sources of bacteria, as well as seasonal changes to the raw milk microbiota driven by changes in habitat.
RESULTS
Microbiota alpha and beta diversities of raw milk, teat surface swabs, and environmental samples cluster according to habitat.
Samples were collected from the same herd over two sampling periods. Sampling phases corresponded to when the herd was housed indoors and outdoors on pasture, respectively. Across both sampling phases, milk samples were collected from teat-prepared (prepped) and non-teat-prepped samples. Samples were also classified as either a potential “source” of microorganisms or a “sink” (a sample that is liable to contain bacteria originating from a source). Milk samples both from individual cows and bulk tank milk (BTM) were classified as sinks, and all environmental samples were classified as sources. After sequencing, the alpha and beta diversities of the bacterial populations present were investigated.
Alpha diversity is the diversity determined in each sample, using species richness and evenness to calculate the diversity in each environment. There was no significant difference in alpha diversity between the microbiotas of individual indoor and outdoor milk samples from nonprepped animals. Similarly, there was no significant difference in the alpha diversities of the microbiota of indoor milk sourced from animals who underwent teat prep and those that did not. However, the alpha diversity of the outdoor milk microbiota was significantly higher in OP (prepped outdoors) samples relative to ONP (not prepped outdoors) samples (P = 0.016 [Simpson's diversity index], P = 0.008 [Shannon diversity index]; Table 1). A corresponding analysis of the alpha diversity of the microbiota of the teat surface revealed significantly greater diversity (chao1, Shannon, PD whole tree, and observed species) among OP samples relative to IP (prepped indoors) samples (P = <0.01, 0.026, <0.01, and <0.01, respectively; Table 1). No other significant differences in the alpha diversity of teat microbiota samples were observed.
TABLE 1.
Alpha diversity differences between individual milk and teat swab samplesa
| Sample type and parameter | Alpha diversity value (SD) |
P |
||||||
|---|---|---|---|---|---|---|---|---|
| INP | ONP | IP | OP | INP vs ONP | IP vs OP | INP vs IP | ONP vs OP | |
| Milk | ||||||||
| chao1 | 3,139 (1,271) | 2,733 (833) | 3,017 (703) | 3,328 (784) | 0.721 | 0.867 | 0.99 | 0.445 |
| Simpson | 0.98 (0.02) | 0.95 (0.05) | 0.98 (0.02) | 0.98 (0.02) | 0.036 | 0.885 | 0.983 | 0.016 |
| Shannon | 8.25 (1.07) | 7.49 (1.17) | 8.26 (1.07) | 9.02 (0.80) | 0.309 | 0.361 | 1 | 0.008 |
| PD whole tree | 90.3 (29.4) | 70.5 (27.2) | 93.8 (26.1) | 86.3 (23.8) | 0.304 | 0.918 | 0.99 | 0.521 |
| Observed species | 2,914 (1,232) | 2,525 (784) | 2,791 (706) | 3,036 (752) | 0.726 | 0.922 | 0.988 | 0.547 |
| Teat | ||||||||
| chao1 | 3,373 (792) | 4,307 (1,172) | 2,949 (536) | 4,791 (1,219) | 0.187 | <0.001 | 0.742 | 0.699 |
| Simpson | 0.99 (0.01) | 0.99 (0.00) | 0.99 (0.01) | 0.99 (0.00) | 0.99 | 0.716 | 0.962 | 0.997 |
| Shannon | 8.54 (0.67) | 8.84 (0.41) | 8.44 (0.48) | 9.17 (0.67) | 0.695 | 0.026 | 0.977 | 0.612 |
| PD whole tree | 125 (27.0) | 157 (37.7) | 107 (17.8) | 174 (39.9) | 0.156 | <0.001 | 0.589 | 0.665 |
| Observed species | 3,194 (767) | 4,090 (1,119) | 2,725 (500) | 4,526 (1,188) | 0.19 | <0.001 | 0.655 | 0.741 |
INP, no prep indoors; ONP, no prep outdoors; IP, prep indoors; OP, prep outdoors; prep, preparation.
Beta diversity is the diversity between different samples; it provides a measure of dissimilarity between samples. The Bray-Curtis principle coordinate plot of beta diversity (Fig. 1A) depicts all samples from this study, with data points colored by sample origin and shaped according to their designation as a source or sink. In this plot it can be observed that samples (soil, grass, bedding, silage, teat surface indoor, teat surface outdoor, fecal indoor pool, fecal outdoor pool, indoor milk, and outdoor milk [individual and BTM]) form clusters, which in turn are further separated from one another based on habitat (outdoor/indoor). More specifically, there is a clear separation between samples depending on whether they were collected from an indoor or an outdoor environment. Feces, teat, individual milk samples, and BTM samples also separate based on which environment they were sampled from (indoor/outdoor) (Fig. 1A). There are more similarities between samples taken from the same habitat. This includes environmental samples (grass and soil [outdoor] and bedding and silage [indoor]), as seen by the overlaps in the ellipses. Within both habitats, it is apparent that there is an overlap between data points representing the milk sample microbiota and that of teat swab samples, reflecting similarities in their beta diversities (Fig. 1A). Teat prep did not result in further subclusters within the milk or teat samples (see Fig. S1 in the supplemental material). Fecal pool samples from both habitats separate from one another and are located in relatively close proximity to the corresponding milk and teat samples from the same environment (Fig. 1A).
FIG 1.
(A) Bray-Curtis PCoA plot of milk and environmental samples. (B) Bray-Curtis Network plot of milk and environmental samples. SourceSink indicates whether a sample is classified as a potential source of contamination or a sink for contaminating communities. ENV_dif indicated the sample origin.
Network analysis shows relationships between raw milk and environmental samples.
Network plots are a useful graphical tool to illustrate relationships between microbiota data sets. The nodes in this network plot represent samples, and the edges that connect nodes indicate correlations between samples. The network analysis shows relationships that exist between the environmental samples and milk samples (Fig. 1B). Consistent with the beta diversity data, it is particularly notable that, of the environmental microbiota samples, the fecal pools and teat microbiota are most closely related to the microbiota of the milk samples, thereby identifying feces and the teat surface as important sources of contamination. These relationships reflect the habitat (indoor or outdoor) from which the samples were collected. There are more edges linking indoor fecal pool samples with indoor BTM samples than there are linking outdoor fecal pool samples with outdoor BTM samples. Some of the outdoor milk samples are not linked to any of the outdoor sources by edges. This suggests that these niches are not substantial sources of microbial contaminants in these milk samples.
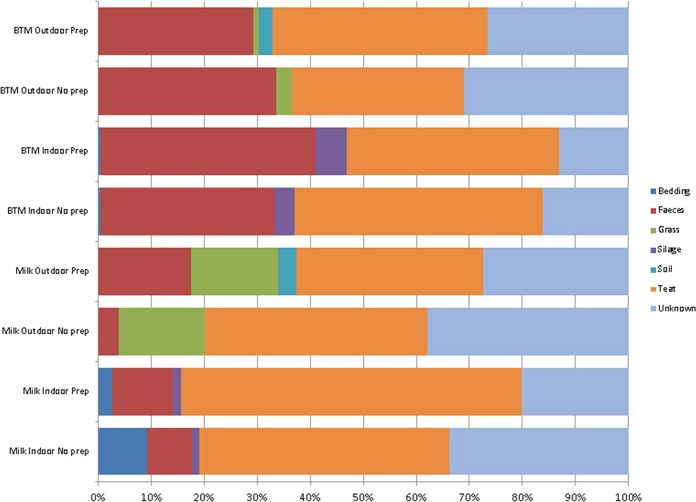
SourceTracker analysis further highlights the contribution of fecal and teat sources to the raw milk microbiota.
The SourceTracker model assumes that each individual community (milk, soil, grass, feces, teat, bedding, and silage) is a mixture of communities deposited from other known or unknown source environments and, using a Bayesian approach, the model provides an estimate of the proportion of the community originating from each of the different sources. When a community contains a mixture of taxa that do not match any of the potential source environments studied, that portion of the community is assigned to an “unknown” source. The analysis revealed that the teat surface was the most significant contributor of microbes in milk samples regardless of habitat or teat preparation. Teat surface contaminants constitute a higher proportion of total contaminants in indoor milk compared than in outdoor milk, both for individual and for BTM samples. Feces was the next most important source of contaminants and had a greater influence on indoor, than outdoor, milk samples, particularly in BTM samples (Fig. 2).
FIG 2.
SourceTracker results highlight the percentages of inferred sources of contamination in BTM and individual milk samples.
Taxonomic analysis of raw milk, teat surface, and herd fecal microbiota.
Graphs representing the microbiota at the family level in the various sample sets are provided in the supplemental data (see Fig. S2 and S3). The compareGroups function was used in R to compare differences in microbial composition between samples. Operational taxonomic units (OTU) that differ significantly can be found in the supplemental material (see Tables S1 to S3 in the supplemental material). In milk samples from individual animals that did not undergo a teat prep treatment, it was noted that indoor samples contained higher relative proportions of, for example, Eremococcus, Ruminococcus, Prevotella, uncultured Corynebacteriales bacterium, and Ruminococcaceae Incertae Sedis (P = 0.012, 0.012, 0.02, 0.022, and 0.028, respectively) and lower proportions of Pseudomonas, Acinetobacter, Lactococcus, and Tumebacillus (P = 0.003, 0.008, 0.002, and 0.014, respectively) relative to outdoor milk samples. Quantitative PCR (qPCR) analysis to determine total bacterial numbers showed that there was significantly more bacteria in indoor milk samples than the equivalent outdoor milk samples (P = 0.003) (Table 2). When the corresponding milk samples from individual teat-prepped animals were compared, it was noted that 25 genera were present in significantly different proportions in indoor milk samples relative to outdoor-milk samples. Sixteen of these OTU were higher in indoor samples, these include Eremococcus, Alloiococcus, Trichococcus, Prevotella, and Psychrobacter, which were all more abundant in indoor samples (P = 0.001, 0.001, 0.001, 0.02, and 0.019, respectively). Nine OTU were higher in PO samples, including Flavobacterium, Sphingomonas, and Tumebacillus (P = 0.009, 0.014, and 0.021, respectively). There was no significant difference in total bacterial numbers between the indoor and outdoor milk samples from teat prepped cows (P = 0.598) (Table 2).
TABLE 2.
qPCR determinations of total bacteria numbers for individual milk samples and comparisons of total bacterial numbers present in individual milk samples from different conditions
| Analysisa | Total no. of bacteriab | P |
|---|---|---|
| qPCR | ||
| INP | 335,500 | |
| IP | 424,333 | |
| ONP | 49,600 | |
| OP | 416,000 | |
| Comparisons | ||
| INP vs IP | 0.758 | |
| INP vs ONP | 0.003 | |
| IP vs OP | 0.598 | |
| ONP vs OP | 0.004 |
INP, no prep indoors; ONP, no prep outdoors, IP, prep indoors, OP, prep outdoors. prep, preparation.
That is, the number of copies of the 16S rRNA gene.
The taxonomic data also facilitated an analysis of the specific effects of teat prep on the bacterial composition of the milk produced. In indoor milk samples from individual animals, it was noted that proportions of Pseudomonas were higher in samples from cows which had undergone teat prep (P = 0.035), suggesting that, among the indoor teat microbiota, Pseudomonas was relatively less sensitive to the antimicrobial effects of the teat prep in indoor samples. qPCR analysis demonstrated that there was no significant difference in total bacterial numbers because of the teat prep (P = 0.758) (Table 2). Pseudomonas, Lactococcus, and Lactobacillus were among nine genera present in outdoor milk samples that were influenced by teat prep. In the case of the aforementioned genera, proportions were higher in samples when no teat prep was carried out (P = 0.011, 0.025, and 0.03, respectively). There were significantly fewer total bacteria in milk samples from nonprepped animals samples than in samples from prepped animals in the outdoor environment (P = 0.004) (Table 2).
The microbiota composition of the teat swabs was also assessed, and it was established that, in samples where teat prep did not occur, 18 genera differed significantly in their relative abundance between indoor and outdoor samples. Trichococcus, Proteiniphilum, and Eremococcus, as well as Corynebacterium, were more abundant in indoor samples (P = 0.012, 0.021, 0.044, and 0.039, respectively), while a further 11 OTU were present in significantly higher proportions in outdoor samples. In samples where teat preparation was carried out, 60 genera differed significantly between indoor and outdoor samples. Of these, 21 samples, including Eremococcus, Proteiniphilum, Corynebacterium, Psychrobacter Bifidobacterium, Trichococcus, and Prevotella, were significantly higher in indoor samples (P = 0.001, 0.001, 0.002, 0.002, 0.003, 0.004, and 0.005, respectively) and 39 genera, including Stenotrophomonas, Xanthomonas and Rhizobium, (P = 0.001, 0.001, and 0.003, respectively) were significantly more prevalent in outdoor samples. Among the outdoor teat samples, there were no significant differences between prepped and nonprepped samples. Among the corresponding indoor teat samples, the proportions of Variovorax and Devosia were higher in teat samples that were not treated (P = 0.033 and 0.043) (see Table S2 in the supplemental material).
In addition, it is noteworthy from the stacked bar charts (see Fig. S1B and D in the supplemental material) that the composition of individual milk samples differs considerably from that of BTM. More specifically, higher proportions of Micrococcaceae and Flavobacteriaceae were observed in all individual milk sample types, and Prevotella and Rikenellaceae were more prevalent in BTM samples.
Finally, the availability of fecal pool samples from both the indoor and outdoor environment facilitated a comparison of their composition. At the genus level, 15 genera, including Prevotella, Bacteroides, and Treponema, were more prevalent in indoor fecal pool samples (P = 0.001, 0.002, and 0.021), and a further eight genera, including Phocaeicola and Paludibacter, were more prevalent in outdoor fecal pool samples (P = 0.027 and 0.036) (see Table S3 in the supplemental material).
DISCUSSION
The objective of this proof-of-concept study was to harness the power of next-generation DNA sequencing technologies to investigate the influence that seasonal housing and teat preparation have on raw milk microbiota from individual cows and in BTM. Furthermore, information potentially revealing the extent to which different microbial niches in the milk production environment influence the microbiota of raw milk was also generated. Although culture-based investigations to study the source of microorganisms in raw milk have primarily focused on BTM in the past, in the present study samples from a small subset of individual animals were also included. While analysis did not reveal differences between the microbiota alpha diversities of indoor and outdoor milk samples, beta diversity analysis highlighted a clear separation between samples from an indoor environment versus those from an outdoor environment. No distinct separation pattern was observed when samples were colored by teat preparation treatment (see Fig. S1 in the supplemental material). Thus, this analysis demonstrates that habitat had a greater impact on the raw milk microbiota than teat preparation.
The SourceTracker algorithm was used as a complementary means of identifying the likely source within the dairy farm environment (soil, silage, bedding, grass, teat, and feces) of bacteria ultimately found in raw milk and, in the process, also reveals the influence of seasonal housing and farm management practices. Regardless of habitat or treatment, teat surface was again identified as the greatest contributor to the raw milk microbiota, followed by feces. This is consistent with a previous (culture-based) study, which proposed that the teat skin was a source of microbial populations in raw milk and that farm management and animal grazing practices influenced the diversity and microbiota of raw milk (16).
The taxonomic results also show that habitat had a much greater influence on the raw milk and teat microbiota than teat prep. For instance, in milk samples from cows that were not subjected to teat prep, Gram-positive and gut-associated genera, such as Ruminococcus, Eremococcus, and Ruminococcaceae Incertae Sedis, were more prevalent in indoor than in outdoor samples, and uncultured Corynebacteriales were more prevalent in indoor than in outdoor samples. Ruminococcus and Ruminococcaceae Incertae Sedis are both gut-associated genera, although, from a dairy perspective, Ruminococcaceae Incertae Sedis has previously been found in continental-type cheese (17), and Ruminococcus has been detected in raw milk (18), and in the present study these organisms were detected in higher proportions in INP (no prep indoors) milk than in ONP (no prep outdoors) milk. While relatively little is known about the uncultured Corynebacteriales, the cultured equivalent contains species known to cause mastitis (19), as well as other species that are found on the surfaces of surface-ripened cheese (20). Similarly, the other genus noted, Eremococcus, has not been well characterized, although a typed strain does exist, having been isolated from the vaginal discharge of a thoroughbred horse (21). The proportions of the Gram-negative genus Prevotella, which is typically gut associated, were also higher in indoor samples, while, for the outdoor samples, the Gram-negative genera Pseudomonas and Acinetobacter, as well as the Gram-positive genus Lactococcus, were among the more dominant organisms. Pseudomonas and Acinetobacter are both dairy spoilage-associated genera (6) that can have a negative impact on dairy product quality. Lactococci are best known for their positive contribution to the production of fermented dairy products but can also be isolated from outdoor environments such as grass (22). These results indicate that indoor milk is more likely to have higher proportions of host/gut-associated microbes than outdoor milk, while, unsurprisingly, outdoor milk is more likely to contain higher proportions of environmental bacteria.
For milk samples from cows that were teat prepped prior to milking, lactic acid bacteria (LAB), such as Eremococcus, Alloiococcus, and Trichococcus spp., as well as Psychrobacter sp., are also found in significantly higher proportions in IP samples. Interestingly, Alloiococcus has not been described in raw milk previously, having instead being associated with human ear infections (23). Trichococcus has been found in raw milk and dairy waste (24), and Psychrobacter has previously been found in teat apexes (25) and in cheese (26). Again, in the corresponding OP milk samples, soil bacteria such as Flavobacterium, Sphingomonas, and Tumebacillus spp., were detected in higher proportions. This finding indicates that outdoor milk is more likely to contain increased proportions of soil-associated microbes, whereas indoor milk is more likely to have higher proportions of host/gut bacteria. The proportions of LAB found in milk appear to be low in comparison to other studies (18); this is perhaps due to the protocol used, which did not incorporate enzymatic lysis.
In teat swab samples, Gram-positive genera such as Corynebacterium, Trichococcus, and Eremococcus and Gram-negative genera such as Proteiniphilum were significantly more prevalent in INP samples than in ONP samples. Proteiniphilum has previously been associated with the feces of dairy cattle (27). A number of soil-type OTU were observed to be significantly elevated in ONP samples relative to INP teat swab samples. This indicates that the transmission of soil-type bacteria to the teat is greater in periods where cows are grazing outdoors, potentially leading to subsequent transmission from the teat to milk. In teat samples that were prepped, Corynebacterium, Eremococcus, and Trichococcus spp. were again more abundant in IP teat samples. Bifidobacterium was also present in greater proportions in these samples. Although Bifidobacterium is typically associated with the gastrointestinal tracts of mammals (27), it may be significant that prep has previously been shown to cause an increase in Actinobacteria proportions on the teat surface (15). With regard to Gram-negative bacteria, Proteiniphilum, Psychrobacter, and Prevotella spp. were all significantly more abundant in IP teat swab samples than in OP samples. In outdoor samples that were teat prepped, many soil-type bacteria, including Rhizobium, Xanthomonas, and Stenotrophomonas, were significantly more prevalent compared to OP samples. Thus, soil-type bacteria, also detected on ONP teat surfaces, persist even when teat prep occurs.
Using the data generated, it is possible to assess the impact of teat preparation on the milk and teat microbiota composition by comparing data from animals that were or were not subjected to a treatment (during the same season). In milk samples, LAB, such as Lactococcus and Lactobacillus, as well as Pseudomonas, were more prevalent in ONP samples, suggesting that the application teat prep significantly reduced the numbers of these microbes in raw milk. There were no significant differences between OP and ONP teat swab samples. Among indoor teat samples, soil-type Proteobacteria, such as Variovorax and Devosia, were more abundant in INP teats than in IP teats. Variovorax has previously been found in hay (2), and Devosia has previously been found in raw milk (28). It was surprising that teat prep increased the numbers of total bacteria in both indoor and outdoor milk. Alpha diversity was also found to have increased in milk from cows where the teats were prepped prior to milking compared to milk from cows where teat preparation was omitted. It may be that the teat preparation process, including forestripping and drying, weakens the attachment of commensal and contaminating teat canal bacteria and results in their being shed into the milk in greater numbers. This result contrasts with findings from culture-based analysis on the impact of teat prep on raw milk, which showed that this treatment reduced bacterial diversity (15) or counts (29). Further studies will be required to reexamine the influence that teat preparation has on the raw milk microbiota. Another important consideration is that the farm used in this study is a research farm where stringent hygiene practices are upheld. This could perhaps limit the impact that teat preparation has on the raw microbiota.
There were considerable differences observed between the individual milk and BTM microbiotas (see Fig. S1 in the supplemental material). This may be due to microorganisms in the BTM being acquired from the milking machine and pipes. Indeed, this possibility has been highlighted previously (30) but not in the context of DNA-based analysis. Further explorations to definitively establish the basis for these differences is merited.
The availability of fecal microbiota data from multiple samples also facilitated comparative analysis of these samples. It was apparent that the beta diversities of the herd fecal pool microbiota differed significantly between the two sampling periods. From a taxonomic perspective, eight genera were found to be significantly more prevalent in outdoor herd fecal samples, and 15 genera were found to be significantly more prevalent in indoor herd fecal pool samples. Treponema, Prevotella, and Bacteroides spp. were among the gut-associated genera that were more prevalent in indoor samples. Treponema has previously been associated with digital dermatitis in cattle (31) and in the bovine rumen (32). Phocaeicola and Paludibacter have also been positively associated with valerate in the rumen (33) and were more prevalent in outdoor samples. This difference in fecal microbiota may be influenced by habitat, host physiological changes, or dietary changes associated with the differing habitats. It is also possible that the transmission of bacteria from a fecal origin may differ based on habitat due to the differences in the microbiota observed here.
In the present study, high-throughput DNA sequencing facilitated the analysis of the microbiota of raw milk samples in parallel with samples from a dairy farm environment. The results provide a more detailed insight into the composition of these microbial populations while also allowing an examination of the relationship between the microbiota of these environments and of raw milk. This analysis highlights that herd habitat is a significant driver for milk microbiota composition and that teat prep has a much more limited impact on raw milk microbiota. In the process, it became apparent that high-throughput sequencing can be an extremely insightful tool to help elucidate the movement of microbes from the environment into the food chain.
MATERIALS AND METHODS
Treatment and sample collection.
Samples were collected from the same herd of Holstein-Friesian dairy cows (n = 60) from the Moorepark Research Farm (Fermoy, Coounty Cork, Ireland) during February (average days in milk [ADIM] = 140) and May (ADIM = 200) of 2015. The milking parlor and equipment were cleaned after each milking as outlined previously (34). Sampling phases corresponded to when cows were housed indoors (February) and outdoors on pasture (May). During the indoor sampling period (February), cows were fed grass silage within a cubicle house with automatic scraper cleaning of the central passageway. Cubicle beds were fitted with rubber mats with a daily allowance of ground limestone added to the backend of the cubicle. Cows managed in the outdoor sampling period (May) grazed on perennial ryegrass pasture on a 24-h rotational grazing regime. The herd was milked in a 30-unit, 80° side-by-side milking parlor (Dairymaster, Causeway, County Kerry, Ireland). Although most studies incorporating molecular methods focus only on the bulk tank milk (BTM), in this instance, milk from three individual cows was also tested. Three cows with a somatic cell count lower than 100,000 cells/ml were chosen for specific individual sampling before commencement of the study and were used throughout the study. Milk and teat swab samples were collected twice weekly from these three cows throughout the study during the morning milking.
Two premilking teat preparation treatments were applied within each sampling phase. One treatment comprised of washing teats with running water, drawing of foremilk, and an application of a premilking teat disinfectant (Deosan Teat-Foam; Deosan, Johnson Diversey [Ireland], Ltd., Dublin, Ireland), followed at least 30 s later by drying using individual paper towels, prior to attaching the milking cluster (prep). The second treatment involved no teat preparation prior to cluster attachment for milking (non-prep). For both indoor and outdoor sampling periods, the teat treatments applied were as follows: week 1, all animals had teats prepped prior to milking; week 2, animals were not prepped; week 3, teats were prepped prior to milking; and week 4, no teat preparation was carried out. All cows in the herd were subjected to each teat preparation treatment at each day of sampling. Environmental samples (feces, bedding, silage grass, and surface soil) were collected twice a week on days 1 and 3, apart from the teat swab samples, which were collected after the teat preparation treatment was applied and prior to cluster attachment for milking on days 2 and 4. Microbial DNA was extracted from all samples using the Powersoil kit (Mo Bio, Carlsbad, CA). Due to the different sample types, the preprocessing protocol for samples varied. At morning's milking on days 2 and 4 of each sampling week, all four teats from the cows were swabbed using one sterile cotton swab per teat (Sarstedt, Ireland). The swabs were dipped in a solution of 3 ml of NaCl (0.09%) prior to swabbing to improve recovery (35). Swabs were drawn across the teat orifice and up the side of each teat, avoiding contact with the udder hair. The four swabs from each cow were then pooled in an NaCl solution (12 ml) in a sterile 15-ml Falcon tube (Sarstedt) and vortexed for 2 min. This resulted in one teat pool for each cow sampled at each time point. The pool, including liquid and swab heads, was then centrifuged for 5 min at 900 × g to separate the swab heads from the liquid. The supernatant was then removed and transferred to another sterile 15-ml Falcon tube. Each pool was then centrifuged at 5,444 × g for 30 min at 4°C. The supernatant was then carefully removed, and the resulting pellet was dissolved in the lysis solution from the Powersoil microbead tubes.
Milk samples from the selected three cows were collected within sterilized sampling bottles using the Weighall milk meter on days 2 and 4 of each sampling week (Dairymaster). Then, 60 ml of individual milk was used for each extraction. BTM samples representing the complete herd were collected after the morning milking on days 2 and 4. These were collected using 30-ml sterile Blue Dippa sample tubes (Ocon Chemicals). A 60-ml portion of the BTM was used for each extraction. For both individual milk and BTM, milk was aseptically transferred to 15-ml Falcon tubes (Sarstedt), and centrifuged at 5,444 × g for 30 min at 4°C. The fat layer was carefully removed, and the supernatant was decanted. The resulting pellets were then washed using sterile phosphate-buffered saline (PBS) and centrifuged at 14,000 × g for 1 min. The four pellets for each individual milk and BTM sample were then pooled to give four samples (three individual milk samples and one BTM sample). Cell pellets were then dissolved in the lysis solution from the microbead tubes from the Powersoil kit.
For fecal pool samples, a pool of the herd's fecal samples was created at each day of sampling. Two fecal pools were collected for each week of sampling on days 1 and 3. To make this pool, equivalent amounts of fecal material were collected from five random cow pats, and the pool was then homogenized for 2 min by vortexing at full speed. DNA was extracted from 250 mg of this fecal pool.
Surface soil samples were collected on days 1 and 3 from the paddock from which the herd were grazing. These samples were collected, taking care to avoid collecting feces or grass using a disposable spatula (VWR, Ireland), 250 mg of surface soil was used for the soil sample extractions. For bedding, silage and grass samples, 20 g of material was aseptically collected using sterile forceps (VWR) and scissors (for grass samples) (both from Medguard, Meath, Ireland) and stored in stomacher bags. For bedding samples, 4 g of bedding material was collected from five cubicles from which the herd had been occupying to create a 20-g bedding sample; two bedding samples were collected for each week of the indoor sampling period. For silage samples, 20 g of silage was collected from where the herd was feeding; two silage samples were collected on each week of the indoor sampling period. For the grass samples, 20 g of grass was aseptically collected from the paddock in which the herd had been grazing when outdoors; two grass samples were collected for each week of the outdoor sampling period. Then, 180 ml of sterile PBS was added to each stomacher bag, and the samples were homogenized in a stomacher. The resultant mixture was then aliquoted into 50-ml Falcon tubes and centrifuged at 900 × g for 5 min to remove solids. After this, the supernatant was filtered through 0.45-μm-pore size nitrocellulose filter membrane (Merck Millipore). After filtration, the membrane was aseptically cut into microbead tubes (Powersoil kit) using a sterile scissors and forceps.
The sample numbers collected included surface soil (n = 8), feces (n = 16, 8) indoor pools and 8 outdoor pools), silage (n = 8), and bedding (n = 8), as well as teat swabs (n = 48, of which 40 subsequently yielded amplicons (10 indoor no prep [INP], 11 indoor prep [IP], 11 outdoor prep [OP], and 8 outdoor no prep [ONP]), individual milk samples (n = 48, of which 47 subsequently yielded amplicons [12 INP, 12 IP, 11 OP, and 12 ONP]), bulk tank milk (BTM; n = 14, 4 INP, 3 IP, 3 ONP, and 4 OP), and grass (n = 8).
After the samples had been preprocessed and lysis solution was added, C1 solution lysis solution (preheated to 60°C) was added to all samples, followed by incubation for 10 min at 60°C with vortexing every 2 min for 30 s. After this incubation, the samples were mechanically lysed at full speed for 10 min using a TissueLyser (Qiagen) and then processed according to the Powersoil kit protocol. DNA was quantified and quality checked by gel electrophoresis and spectrophotometry using a NanoDrop 1000 instrument (Thermo Fisher Scientific, Inc.).
16S rRNA amplicon sequencing.
The V3-V4 variable region of the 16S rRNA gene was amplified from the 149 DNA extracts using the 16S metagenomic sequencing library protocol (Illumina). PCRs were completed on the template DNA. Initially, the DNA was amplified with primers specific to the V3-V4 region of the 16S rRNA gene which also incorporates the Illumina overhang adaptor (forward primer, 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; reverse primer, 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC) (36). Each PCR mixture contained DNA template (ca. 10 to 12 ng), 5 μl of forward primer (1 μM), 5 μl of reverse primer (1 μM), 12.5 μl of 2× Kapa HiFi Hotstart ready mix (Anachem, Dublin, Ireland), and PCR-grade water to a final volume of 25 μl. For the environmental samples (surface soil, fecal, silage, swabs, bedding, and grass), PCR amplification was carried out as follows. The lid was heated to 110°C. Samples were then subjected to 95°C for 3 min; followed by 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; followed in turn by 72°C for 5 min; and finally held at 4°C. For the milk samples, the same cycling parameters were used, except 32 cycles were used instead of 25 cycles. PCR products were visualized using gel electrophoresis (1× TAE buffer, 1.5% agarose, 100 V) and cleaned using AMPure XP magnetic beads (Labplan, Dublin, Ireland). After this, a PCR was performed on the purified DNA (5 μl) to index each of the samples, allowing the samples to be pooled for sequencing on three flow cells and subsequently demultiplexed for analysis. The samples were indexed randomly to prevent any run bias in analysis. Two Nextera XT indexing primers (Illumina, Sweden) were used per sample. Each PCR mixture contained 5 μl of index 1 primer (N7xx), 5 μl of index 2 primer (S5xx), 25 μl of 2× Kapa HiFi Hotstart ready mix, and 10 μl of PCR-grade water. PCRs were performed as described above, with eight amplification cycles. The PCR products were visualized using gel electrophoresis and subsequently cleaned (as described above). The samples were quantified using Qubit (Bio-Sciences, Dublin, Ireland), along with the broad-range DNA quantification assay kit (BioSciences), and the samples were then pooled in an equimolar fashion. The pooled sample was run on an Agilent Bioanalyzer for quality analysis prior to sequencing. The sample pool (4 nM) was denatured with 0.2 N NaOH, diluted to 4 pM, and combined with 10% (vol/vol) denatured 4 pM PhiX, prepared according to Illumina guidelines. The samples were sequenced on the MiSeq sequencing platform in the Teagasc sequencing facility, using a 2×250 cycle V3 kit, according to standard Illumina sequencing protocols. Reads were deposited in the European Nucleotide Archive database under accession number PRJEB16770 (secondary accession number ERP018630).
Bioinformatic and statistical analysis.
A number of 250-bp paired-end reads were assembled using FLASH (fast length adjustment of short reads to improve genome assemblies) (37). Further processing of paired-end reads, including quality filtering based on a quality score of >25 and the removal of mismatched barcodes and sequences below length thresholds, was completed using QIIME (38). A total of 32,766,563 reads were generated after filtering, with an average of 219,909 per sample. Denoising, chimera detection, and clustering into OTU (97% identity) were performed using USEARCH v7 (64-bit) (39). OTU were aligned using PyNAST (python nearest alignment space termination; a flexible tool for aligning sequences to a template alignment), and taxonomy was assigned using BLAST against the SILVA SSURef database release 111. Samples were then rarefied to an even depth of sequences per sample. Alpha diversity was generated in QIIME, and the compareGroups function (40) was then was then used to determine any statistically significant differences (P < 0.05) and generate standard deviations between samples based on conditions using analysis of variance. Beta diversity was calculated in R, using Phyloseq (41) and Bray-Curtis distances. Principal coordinate analysis (PCoA) plots were visualized using ggplot2 (42). Confidence ellipses were generated using stat_ellipse in the ggplot2 package (42). Network analysis was also carried out using phyloseq and ggplot2. The SourceTracker algorithm (9) was also used to investigate possible sources of environmental contamination in milk from both sampling periods. SourceTracker analysis was carried out at a depth of 13,500, with 100 burn-ins and 10 restarts. The compareGroups function was used in R to compare differences in microbial composition between individual milk, teat swab, and fecal pool samples; the Kruskal-Wallis test was applied in this instance with Benjamini-Hochberg corrections (43) to highlight any statistically significant differences (P < 0.05 after correction).
qPCR.
qPCR was carried out on individual milk samples to determine the total bacteria levels in each sample using the 16S rRNA gene. qPCR was carried out as described previously (44), except the equivalent volume of Kappa SYBR Fast (Roche Diagnostics) was used instead of SYBR green for the present study. Samples, negative controls (where template DNA was replaced with PCR-grade water), and standards were evaluated in triplicate (technical replicates).
Accession number(s).
Reads were deposited in the European Nucleotide Archive database under accession number PRJEB16770 (secondary accession number ERP018630).
Supplementary Material
ACKNOWLEDGMENTS
We thank John Paul Murphy and all the staff at the Animal and Grassland Research and Innovation Centre Dairy Unit, Teagasc, Moorepark, Fermoy. We also thank Fiona Fouhy and Fiona Crispie of the Food Bioscience Department in Teagasc Moorepark.
This study was funded through a Teagasc Walsh Fellowship (2013030) to C.J.D. and internal Teagasc funding (RMIS6364) to P.D.C.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02694-16.
REFERENCES
- 1.Sevi A, Albenzio M, Muscio A, Casamassima D, Centoducati P. 2003. Effects of litter management on airborne particulates in sheep houses and on the yield and quality of ewe milk. Livestock Production Sci 81:1–12. doi: 10.1016/S0301-6226(02)00228-2. [DOI] [Google Scholar]
- 2.Vacheyrou M, Normand A-C, Guyot P, Cassagne C, Piarroux R, Bouton Y. 2011. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int J Food Microbiol 146:253–262. doi: 10.1016/j.ijfoodmicro.2011.02.033. [DOI] [PubMed] [Google Scholar]
- 3.Vithanage NR, Dissanayake M, Bolge G, Palombo EA, Yeager TR, Datta N. 2016. Biodiversity of culturable psychrotrophic microbiota in raw milk attributable to refrigeration conditions, seasonality and their spoilage potential. Int Dairy J 57:80–90. doi: 10.1016/j.idairyj.2016.02.042. [DOI] [Google Scholar]
- 4.Mhone TA, Matope G, Saidi PT. 2011. Aerobic bacterial, coliform, Escherichia coli and Staphylococcus aureus counts of raw and processed milk from selected smallholder dairy farms of Zimbabwe. Int J Food Microbiol 151:223–228. doi: 10.1016/j.ijfoodmicro.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 5.Doyle CJ, Gleeson D, Jordan K, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2015. Anaerobic sporeformers and their significance with respect to milk and dairy products. Int J Food Microbiol 197:77–87. doi: 10.1016/j.ijfoodmicro.2014.12.022. [DOI] [PubMed] [Google Scholar]
- 6.Hantsis-Zacharov E, Halpern M. 2007. Culturable psychrotrophic bacterial communities in raw milk and their proteolytic and lipolytic traits. Appl Environ Microbiol 73:7162–7168. doi: 10.1128/AEM.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kembel SW, Meadow JF, O'Connor TK, Mhuireach G, Northcutt D, Kline J, Moriyama M, Brown G, Bohannan BJ, Green JL. 2014. Architectural design drives the biogeography of indoor bacterial communities. PLoS One 9:e87093. doi: 10.1371/journal.pone.0087093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. 2011. Microbial biogeography of public restroom surfaces. PLoS One 6:e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, Bushman FD, Knight R, Kelley ST. 2011. Bayesian community-wide culture-independent microbial source tracking. Nat Methods 8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokulich NA, Bergsveinson J, Ziola B, Mills DA. 2015. Mapping microbial ecosystems and spoilage-gene flow in breweries highlights patterns of contamination and resistance. eLife 4:e04634. doi: 10.7554/eLife.04634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bokulich NA, Mills DA. 2013. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl Environ Microbiol 79:5214–5223. doi: 10.1128/AEM.00934-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bokulich NA, Ohta M, Richardson PM, Mills DA. 2013. Monitoring seasonal changes in winery-resident microbiota. PLoS One 8:e66437. doi: 10.1371/journal.pone.0066437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buehner KP, Anand S, Garcia A. 2014. Prevalence of thermoduric bacteria and spores on 10 midwest dairy farms. J Dairy Sci 97:6777–6784. doi: 10.3168/jds.2014-8342. [DOI] [PubMed] [Google Scholar]
- 14.Gleeson D, O'Connell A, Jordan K. 2013. Review of potential sources and control of thermoduric bacteria in bulk-tank milk. Irish J Agric Food Res 52:217–227. [Google Scholar]
- 15.Verdier-Metz I, Michel V, Delbes C, Montel M-C. 2009. Do milking practices influence the bacterial diversity of raw milk? Food Microbiol 26:305–310. doi: 10.1016/j.fm.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Verdier-Metz I, Gagne G, Bornes S, Monsallier F, Veisseire P, Delbès-Paus C, Montel M-C. 2012. Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl Environ Microbiol 78:326–333. doi: 10.1128/AEM.06229-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Sullivan DJ, Cotter PD, O'Sullivan O, Giblin L, McSweeney PL, Sheehan JJ. 2015. Temporal and spatial differences in microbial composition during the manufacture of a continental-type cheese. Appl Environ Microbiol 81:2525–2533. doi: 10.1128/AEM.04054-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quigley L, McCarthy R, O'Sullivan O, Beresford TP, Fitzgerald GF, Ross RP, Stanton C, Cotter PD. 2013. The microbial content of raw and pasteurized cow's milk as determined by molecular approaches. J Dairy Sci 96:4928–4937. doi: 10.3168/jds.2013-6688. [DOI] [PubMed] [Google Scholar]
- 19.Hogan J, Smith K, Todhunter D, Schoenberger P. 1988. Rate of environmental mastitis in quarters infected with Corynebacterium bovis and Staphylococcus species. J Dairy Sci 71:2520–2525. doi: 10.3168/jds.S0022-0302(88)79840-9. [DOI] [PubMed] [Google Scholar]
- 20.Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM. 2001. Recent advances in cheese microbiology. Int Dairy J 11:259–274. doi: 10.1016/S0958-6946(01)00056-5. [DOI] [Google Scholar]
- 21.Collins MD, Jovita MR, Lawson PA, Falsen E, Foster G. 1999. Characterization of a novel Gram-positive, catalase-negative coccus from horses: description of Eremococcus coleocola gen. nov. sp. nov. Int J Syst Evol Microbiol 49:1381–1385. [DOI] [PubMed] [Google Scholar]
- 22.Alemayehu D, Hannon JA, McAuliffe O, Ross RP. 2014. Characterization of plant-derived lactococci on the basis of their volatile compounds profile when grown in milk. Int J Food Microbiol 172:57–61. doi: 10.1016/j.ijfoodmicro.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Aguirre M, Collins M. 1992. Phylogenetic analysis of Alloiococcus otitis gen. nov., sp. nov., an organism from human middle ear fluid. Int J Syst Evol Microbiol 42:79–83. [DOI] [PubMed] [Google Scholar]
- 24.Rasolofo EA, St-Gelais D, LaPointe G, Roy D. 2010. Molecular analysis of bacterial population structure and dynamics during cold storage of untreated and treated milk. Int J Food Microbiol 138:108–118. doi: 10.1016/j.ijfoodmicro.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Braem G, De Vliegher S, Verbist B, Heyndrickx M, Leroy F, De Vuyst L. 2012. Culture-independent exploration of the teat apex microbiota of dairy cows reveals a wide bacterial species diversity. Vet Microbiol 157:383–390. doi: 10.1016/j.vetmic.2011.12.031. [DOI] [PubMed] [Google Scholar]
- 26.Quigley L, O'Sullivan O, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2012. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl Environ Microbiol 78:5717–5723. doi: 10.1128/AEM.00918-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim M, Wells JE. 2016. A meta-analysis of bacterial diversity in the feces of cattle. Curr Microbiol 72:145–151. doi: 10.1007/s00284-015-0931-6. [DOI] [PubMed] [Google Scholar]
- 28.Baur C, Krewinkel M, Kranz B, von Neubeck M, Wenning M, Scherer S, Stoeckel M, Hinrichs J, Stressler T, Fischer L. 2015. Quantification of the proteolytic and lipolytic activity of microorganisms isolated from raw milk. Int Dairy J 49:23–29. doi: 10.1016/j.idairyj.2015.04.005. [DOI] [Google Scholar]
- 29.McKinnon CH, Rowlands GJ, Bramley AJ. 1990. The effect of udder preparation before milking and contamination from the milking plant on bacterial numbers in bulk milk of eight dairy herds. J Dairy Res 57:307–318. doi: 10.1017/S0022029900026959. [DOI] [PubMed] [Google Scholar]
- 30.Quigley L, O'Sullivan O, Stanton C, Beresford TP, Ross RP, Fitzgerald GF, Cotter PD. 2013. The complex microbiota of raw milk. FEMS Microbiol Rev 37:664–698. doi: 10.1111/1574-6976.12030. [DOI] [PubMed] [Google Scholar]
- 31.Trott DJ, Moeller MR, Zuerner RL, Goff JP, Waters WR, Alt DP, Walker RL, Wannemuehler MJ. 2003. Characterization of Treponema phagedenis-like spirochetes isolated from papillomatous digital dermatitis lesions in dairy cattle. J Clin Microbiol 41:2522–2529. doi: 10.1128/JCM.41.6.2522-2529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bekele AZ, Koike S, Kobayashi Y. 2011. Phylogenetic diversity and dietary association of rumen Treponema revealed using group-specific 16S rRNA gene-based analysis. FEMS Microbiol Lett 316:51–60. doi: 10.1111/j.1574-6968.2010.02191.x. [DOI] [PubMed] [Google Scholar]
- 33.Mao S, Zhang R, Wang D, Zhu W. 2012. The diversity of the fecal bacterial community and its relationship with the concentration of volatile fatty acids in the feces during subacute rumen acidosis in dairy cows. BMC Vet Res 8:1–13. doi: 10.1186/1746-6148-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell A, Ruegg P, Jordan K, O'Brien B, Gleeson D. 2016. The effect of storage temperature and duration on the microbial quality of bulk tank milk. J Dairy Sci 99:3367–3374. doi: 10.3168/jds.2015-10495. [DOI] [PubMed] [Google Scholar]
- 35.Landers TF, Hoet A, Wittum TE. 2010. Swab type, moistening, and preenrichment for Staphylococcus aureus on environmental surfaces. J Clin Microbiol 48:2235–2236. doi: 10.1128/JCM.01958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fouhy F, Deane J, Rea MC, O'Sullivan Ó Ross RP, O'Callaghan G, Plant BJ, Stanton C. 2015. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One 10:e0119355. doi: 10.1371/journal.pone.0119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magoč T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 40.Subirana IV, Sanz J, Lucas H, Peñafiel G, Giménez J. 2013. compareGroups: descriptive analysis by groups: R package version 2.0. https://cran.r-project.org/web/packages/compareGroups/index.html. [Google Scholar]
- 41.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickham H, Chang W, Wickham MH. 2013. ggplot2. http://ggplot2.org/. [Google Scholar]
- 43.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- 44.Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 56:5811–5820. doi: 10.1128/AAC.00789-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.