ABSTRACT
Many phototrophic flagellates ingest prokaryotes. This mixotrophic trait becomes a critical aspect of the microbial loop in planktonic food webs because of the typical high abundance of these flagellates. Our knowledge of their selective feeding upon different groups of prokaryotes, particularly under field conditions, is still quite limited. In this study, we investigated the feeding behavior of three species (Rhodomonas sp., Cryptomonas ovata, and Dinobryon cylindricum) via their food vacuole content in field populations of a high mountain lake. We used the catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) protocol with probes specific for the domain Archaea and three groups of Eubacteria: Betaproteobacteria, Actinobacteria, and Cytophaga-Flavobacteria of Bacteroidetes. Our results provide field evidence that contrasting selective feeding exists between coexisting mixotrophic flagellates under the same environmental conditions and that some prokaryotic groups may be preferentially impacted by phagotrophic pressure in aquatic microbial food webs. In our study, Archaea were the preferred prey, chiefly in the case of Rhodomonas sp., which rarely fed on any other prokaryotic group. In general, prey selection did not relate to prey size among the grazed groups. However, Actinobacteria, which were clearly avoided, mostly showed a size of <0.5 μm, markedly smaller than cells from the other groups.
IMPORTANCE That mixotrophic flagellates are not randomly feeding in the main prokaryotic groups under field conditions is a pioneer finding in species-specific behavior that paves the way for future studies according to this new paradigm. The particular case that Archaea were preferentially affected in the situation studied shows that phagotrophic pressure cannot be disregarded when considering the distribution of this group in freshwater oligotrophic systems.
KEYWORDS: Archaea, mixotrophic protist, selective feeding
INTRODUCTION
Mixotrophic behavior, the combination of phototrophic and phagotrophic nutritional modes within a single cell, has been increasingly documented in aquatic systems (1, 2). Phagotrophy occurs in a variety of phytoplankton flagellate groups, including Chrysophyceae, dinoflagellates, prymnesiophytes, and cryptophytes, which comprise some picoeukaryotes (3–5). Currently, there is no doubt of the ubiquity of mixotrophy and its significance in the functioning of planktonic systems. Under oligotrophic conditions, phototrophic flagellates can account for up to 80% of total bacterial grazing (4, 6, 7).
Predation by protists is among the primary mortality factors of prokaryotes in planktonic communities and thus is an important selective pressure. It becomes a structuring factor of the abundance, morphology, composition, and activity of bacterial assemblages (8, 9). The impact of protist predation appears to be modulated by the characteristics of the system (e.g., productivity) and predator and prey traits (10). Over the last few decades, efforts have been made to understand the selective feeding behavior of protists. General selection mechanisms have been identified (11). However, the current view is still mainly based on laboratory data, using readily growing species (12). On the other hand, a limited number of field experiments use general grazer groups rather than evaluating predation at the species level (12–14). There is a need to evaluate prey selection under natural conditions comparing flagellate species to determine more specific interactions between microbial predators and prey and assessing the relevance of selective microbial predation in the microbial loop dynamics. The scarcity of studies is due in great part to the difficulties in (i) prokaryote prey identification and (ii) establishing the prey and predator links at the highest possible taxonomic resolution. Prey identification can be addressed by techniques based on DNA fingerprinting (15, 16), which allow ensuing taxonomic changes in prey assemblages (10, 17). Linking prey and predator can be addressed using techniques based on fluorescence in situ hybridization (FISH), which allow detecting targeted prey inside protist food vacuoles (5, 14, 18, 19). These techniques have been performed mostly under experimental conditions (18, 20), and results suggest a high selectivity in the feeding of heterotrophic flagellates and some ciliate species investigated. In contrast, the few in situ measurements showed unclear ingestion patterns for lake flagellates, and even random feeding was proposed for some bacterial groups (14). All in all, there is still limited information on the selective feeding of phagotrophic protists under natural conditions, and more remarkably, there is a huge gap of knowledge about mixotrophic flagellate species.
The catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) protocol (21) is particularly suited to assess the phagotrophy of mixotrophic protists on prokaryotes, since it maintains cell and plastid integrity and allows the visualization of labeled prey against plastid autofluorescence. CARD-FISH can be easily applied to natural assemblages to evaluate in situ prey preference of mixotrophic species and other prokaryotic grazers (14, 19) using the appropriate bacterial and archaeal probes. In the present study, we examined the phagotrophic selectivity of three mixotrophic species under natural conditions. We sampled a deep high-mountain lake in which we expected mixotrophic activity to be enhanced by the ultraoligotrophic conditions. Samples were obtained at different times of day to take into account potential feeding variation and to assess the mean behavior better. The phytoplankton species were identified by the size and shape of the autofluorescent plastid based on a prior taxonomic knowledge of the assemblages, which is an advantage of investigating mixotrophic flagellates at the species level with respect to the heterotrophic ones. The heterotrophic flagellates are usually grouped into operational functional groups (14, 20, 22). In our study, we used fluorescent probes for the domain Archaea and three groups of Eubacteria: Betaproteobacteria, Actinobacteria, and the Cytophaga-Flavobacteria group of Bacteroidetes. These groups dominate aquatic prokaryotes in the Pyrenean lakes and account for more of the 85% of clades present in the lake studied (23).
RESULTS
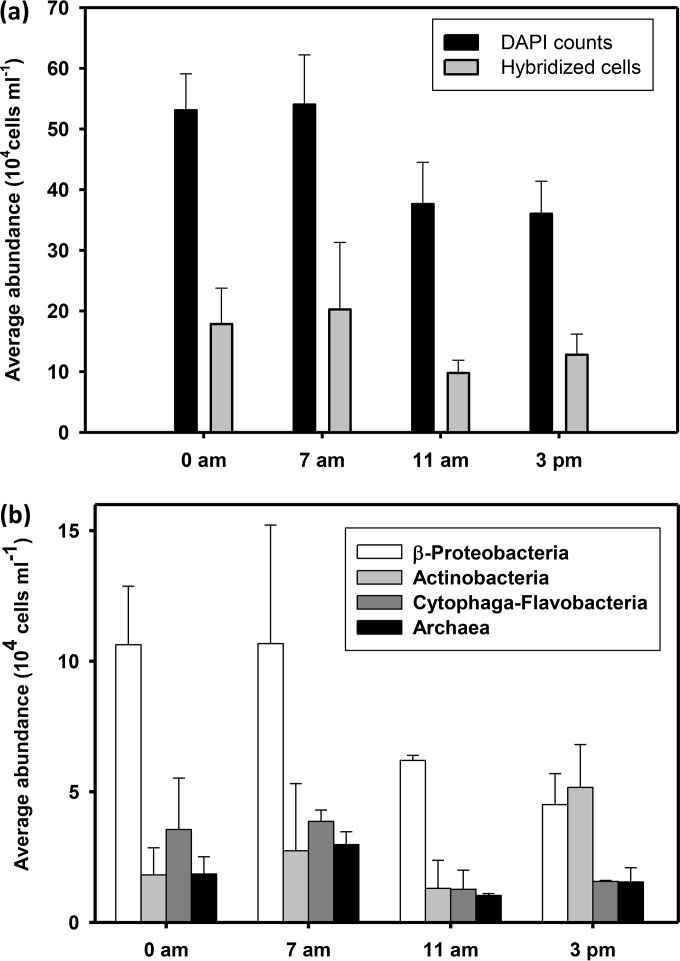
Total 4′,6-diamidino-2-phenylindole (DAPI) counts ranged from 2.8 × 105 to 6.3 × 105 cells ml−1, and 27 to 37% of such total counts were hybridized with the four probes used in this study (Fig. 1a). No significant differences were found in the total amount of hybridized cells between samplings (Fig. 1a) (P value of >0.05 by analysis of variance [ANOVA]). The three groups of Bacteria considered together accounted for 99 to 104% of cells hybridized by probe EUB338, which is generic for the domain Bacteria, indicating that no major Bacteria group was missing from our study.
FIG 1.
(a) Prokaryotic cell abundance (average ± standard deviation [SD] total DAPI counts) and the total amount of cells hybridized by the four probes used in this study (averages ± SD; n = 8) at the four sampling times. (b) Prokaryotic assemblage composition as hybridized cell abundance (averages ± SD; n = 2) of the four targeted groups at the four sampling times.
From the four tested prokaryotic groups, Betaproteobacteria was often the most abundant, ranging from 12.4% ± 3.3% to 20.2% ± 8.8% of total DAPI counts. Only at the last sampling time did the group Actinobacteria show higher abundance, reaching 15.6% ± 2.7% of total DAPI counts (Fig. 1b). Proportions of Cytophaga-Flavobacteria of Bacteroidetes were slightly variable, representing between 4.0% ± 2% and 7.2% ± 4.5% of total DAPI counts, whereas Archaea ranged between 2.9% ± 0.8% and 6% ± 0.4%.
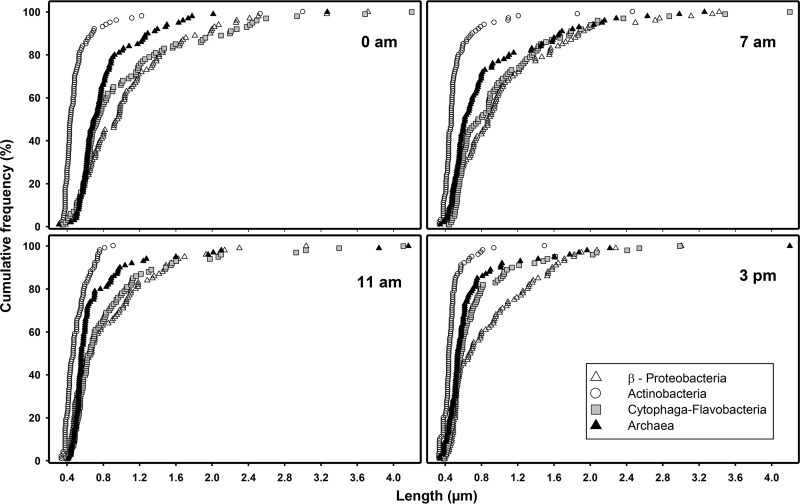
Cell length frequency distributions evidenced that Actinobacteria were always smaller and had more uniform sizes than the others prokaryotic groups (Fig. 2). A temporal tendency can be observed in Archaea and Cytophaga-Flavobacteria distributions: cells were larger at night and progressively declined in size during the day.
FIG 2.
Cell length cumulative distributions for the four targeted prokaryotic groups at the four sampling times.
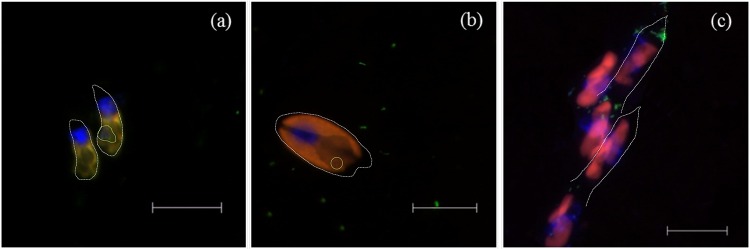
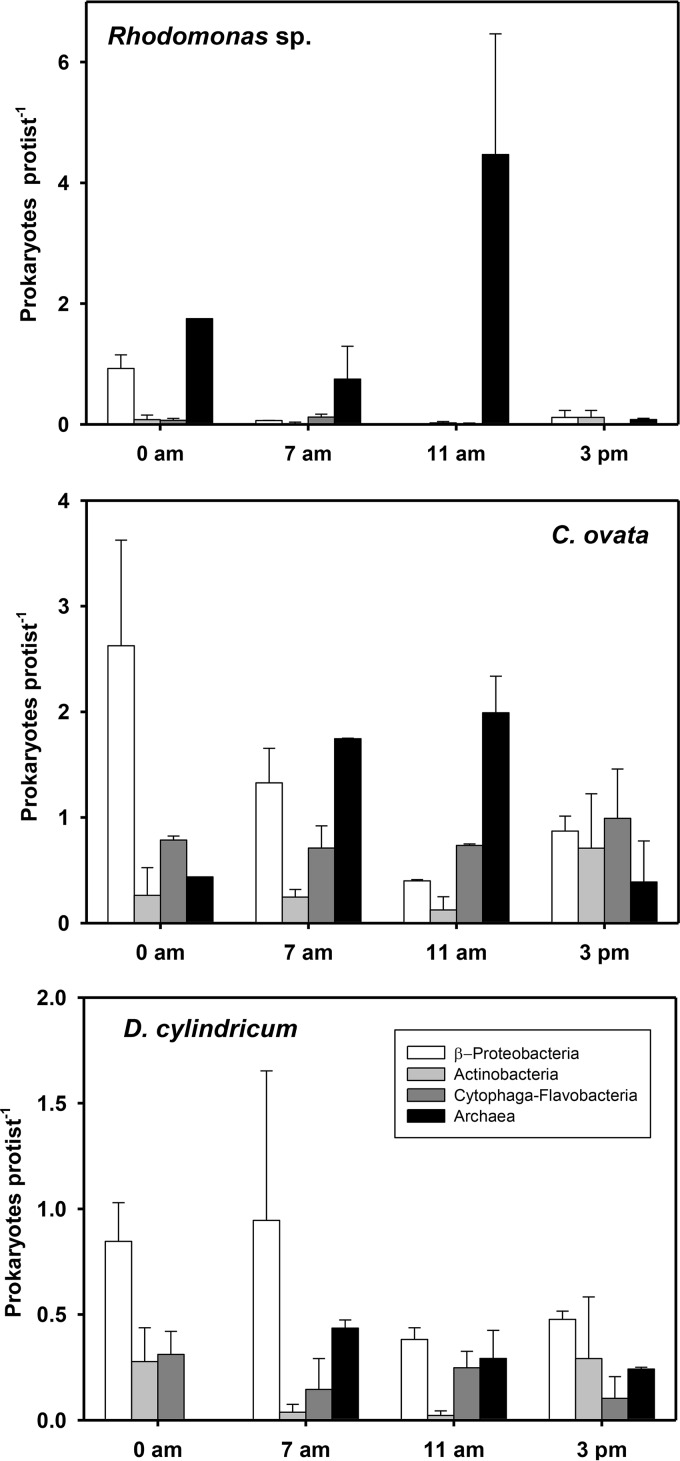
Three mixotrophic species (two species of Cryptophyta [Rhodomonas sp. and Cryptomonas ovata] and one species of Chrysophyceae [Dinobryon cylindricum]) were selected to assess the phagotrophic activity and describe their feeding behavior. These species were easy to recognize by fluorescence microscopy and large enough (Table 1) to accurately quantify the hybridized prokaryote cells inside them (Fig. 3). Their abundance and the percentage of feeding cells observed for each probe are included in Table 1. In some cases, cells actively feeding accounted for more than half of the population (e.g., Rhodomonas sp. feeding on Archaea or C. ovata on Betaproteobacteria), whereas in others most individuals showed no prey inside (e.g., Rhodomonas sp. feeding on Actinobacteria or Cytophaga-Flavobacteria). The average food vacuole content of the three flagellate species changed among the different surveys performed (Fig. 4). C. ovata and D. cylindricum presented a similar feeding variation. Rhodomonas vacuole content had an elevated variation with significant differences among surveys (P value of <0.05 by Kruskal-Wallis test) (Fig. 4).
TABLE 1.
Protist average data
| Species | Size (μm) | Abundance (cells ml−1)a | Percentage of active feeding cellsa |
|||
|---|---|---|---|---|---|---|
| Betaproteobacteria | Actinobacteria | Cytophaga-Flavobacteria | Archaea | |||
| Rhodomonas sp. | 11 by 6 | 28 ± 8 | 9.4 ± 12.3 | 3.3 ± 2.6 | 2.9 ± 3.2 | 52.5 ± 47.7 |
| Cryptomonas ovata | 30 by 14 | 16 ± 6 | 50 ± 19.4 | 19.2 ± 14.7 | 44.1 ± 9.9 | 36.6 ± 16 |
| Dinobryon cylindricum | 14 by 6 | 139 ± 47 | 28.5 ± 9 | 7.8 ± 7.3 | 11.5 ± 5.4 | 13.3 ± 10.2 |
Data are averages ± standard deviations.
FIG 3.
Epifluorescence microscope photographs of the three mixotrophic flagellate species studied. (a) Rhodomonas sp. (b) Cryptomonas ovata. (c) Dinobryon cylindricum. Presented is CARD-FISH staining with ARCH915 probe (a and c) and BET42a probe (b), showing chloroplast autofluorescence (red), DAPI-stained nucleus (blue), and targeted prokaryotes (green). Dashed white lines represent cell outlines, and green lines surround food vacuoles. Scale bars, 12 μm (a) and 20 μm (b and c).
FIG 4.
Food vacuole content (average ± SD [n = 2] number of cells hybridized by the four probes) for the three mixotrophic flagellates at the four sampling times.
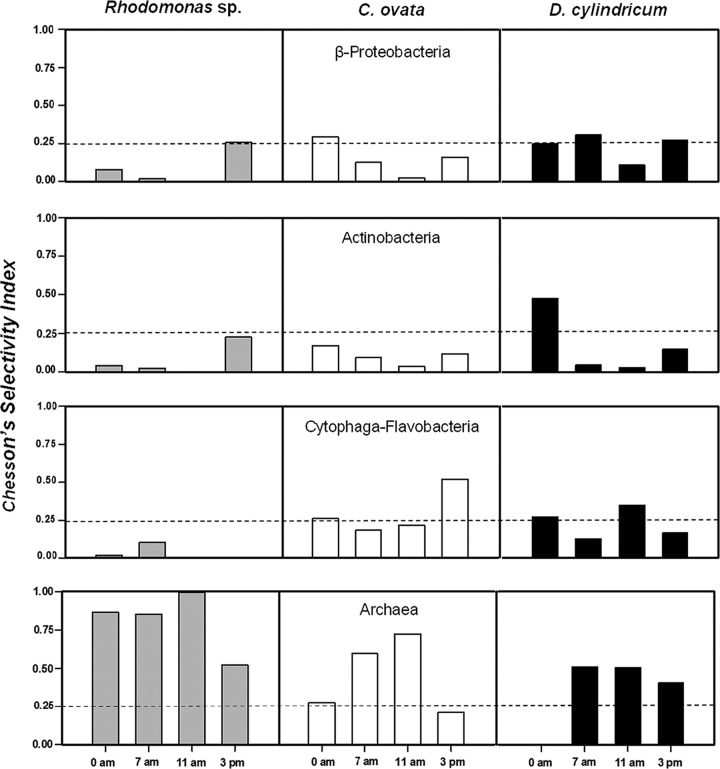
The main ingested prokaryotic group differed among the three protists (Fig. 4), and Chesson's selectivity index (αi) clearly indicates that the three species were not randomly feeding (Fig. 5). Archaea were usually positively selected by the three protists. The preference was extreme in the case of Rhodomonas sp., which hardly grazed on any other prokaryotic group. In contrast, Actinobacteria were always apparently avoided, and only once did D. cylindricum positively select this group. Finally, Betaproteobacteria and Cytophaga-Flavobacteria were only strongly avoided by Rhodomonas and randomly grazed or slightly avoided by C. ovata and D. cylindricum.
FIG 5.
Chesson's selectivity index for the three mixotrophic species (plots distributed vertically) and for each of the targeted prokaryotic groups (plots distributed horizontally) at each sampling time. The horizontal dashed lines indicate the value above which positive selection of a particular prokaryotic group is assumed (0.25 = 1/n, where n is the number of prokaryotic groups).
DISCUSSION
CARD-FISH performance.
Technical capacity for distinguishing the food vacuole content of protists is crucial to study their selective feeding. The CARD-FISH protocol (21) offers this possibility. Critical steps in the procedure are probe selection and the percentage of hybridized cells achieved. The probes used in the present study included the most abundant prokaryotic groups described in the plankton of the Pyrenean high-mountain lakes (24). Previous studies in Lake Redon and in 17 lakes in the same area of the Pyrenees have shown that Betaproteobacteria, Bacteroidetes, and Actinobacteria always account for more than 75% of the total Bacteria clades and more than 85% in Lake Redon (see Fig. S2 from reference 23). The remaining clades were divided into 7 secondary groups and other minority ones. Our results show that no primary Bacteria group was missing from our assessment. The three groups of Bacteria considered accounted for 99 to 104% of cells hybridized by probe EUB338. There is no reason to assume that we were missing any large part of the prokaryotic community with our probes, as long as we considered the three most important groups of Bacteria in addition to Archaea. The same probes were used to characterize Bacteria composition in plankton from alpine lakes of other ranges (25, 26).
The relatively low hybridization compared with DAPI counts (27 to 37%) is due to the efficiency of the procedure. Low efficiency may be due to detection technical issues and to the cell physiological states (27). Cells with low rRNA content are common in samples from oligotrophic environments due to low cell metabolic activity or dormant state. Different assessments provide a small percentage of active cells in the plankton of the Pyrenean lakes, with respiring prokaryotes ranging between 2 and 7%, whereas levels of viable cells were seldom higher than 50% in deep Pyrenean lakes (28). Even though CARD-FISH appears suitable for the detection of prokaryotes with small quantities of rRNA molecules (29), if cells are scarcely active or small, the percentage of hybridized cells compared to DAPI counts declines (27, 30). Therefore, the ratio of CARD-FISH to DAPI counts is often low and highly variable in lake plankton samples (19, 26, 31) and is expected to be particularly low in an ultraoligotrophic lake such as Redon. In a recent phosphorus enrichment experiment performed in this lake, the percentage of hybridized cells (using the same four probes we applied here) increased from 36% of DAPI counts under low-P and lake conditions to nearly 100% in the most productive mesocosm (see the supplemental material). This is clear evidence that the apparent low hybridizing efficiency we obtained is not a probe problem but a constraint resulting from the low level of activity of many cells. In our study, the percentage of hybridization did not significantly change over time (Fig. 1) and thus did not affect our objective to compare the food vacuole content to the plankton assemblage composition.
The ARCH915 probe has been widely used to detect archaeal cells. Occasionally, some biases of the probe-hybridizing members of the phylum Bacteroidetes have been reported (32–34). This doubling hybridization can inflate the Archaea counts. It does not seem to occur in our case for several reasons. No correlation was found between ARCH915 and CF319a counts in water samples or in vacuoles. Indeed, contrasting extreme values between the two groups were found in some protist food vacuoles (Fig. 4). Also, the Archaea proportions in the water samples of our study (3 to 6%) were similar to those in previous studies in Lake Redon and other Pyrenean lakes using 16S rRNA gene tag sequencing, namely, 0 to 8% (35) and 0 to 6% (36).
Selective predation.
The significance of predation on bacterial activity and community structure in natural aquatic systems has been demonstrated (10, 37–40). Consequently, effort is currently placed on understanding the details and dynamics of grazing by protists (11). From laboratory experiments, evidence exists that certain taxa feed selectively (41, 42). Under natural conditions, little is known about flagellate grazing preferences and whether they tend to be specialist or generalist predators. In a recent microcosm study, Glücksman et al. (17) showed that closely related and morphologically similar flagellated species can have different impacts on natural bacterial communities. Beyond phylogeny, some general protist traits, namely, cell size and morphological plasticity, explained variation in prey composition. Accordingly, our results show that two highly related species (i.e., the two cryptophytes), inhabiting the same system and feeding in the same prokaryotic assemblage, markedly differ in their selectivity (Fig. 4 and 5).
The species studied are closer to specialist predators than generalists (Fig. 5). Rhodomonas sp., the smallest species analyzed, showed a more differentiated feeding pattern (Fig. 4). Unfortunately, the comparison of only three species does not have the statistical power to evaluate the relationship between traits and selectivity. In any case, we show that selectivity may be extreme and thus has substantial implications for the dynamics of the prokaryotic assemblages. The three protist species analyzed were the most abundant mixotrophs in the phytoplankton community at the time of the study (Table 1). The selectivity, at least at the high taxonomic levels shown by our results, paves the way for studies focusing on finer prokaryotic taxonomy, using more specific probes and evaluating whether grazing by protists constitutes a differential selective pressure within each of the large prokaryotic groups, with ecological and evolutionary implications.
Cell size and grazing pressure.
Protist prey selection may occur at various feeding steps, namely, capture, prey processing, ingestion, and digestion (11, 43). Prey size is a trait that is easy to measure. There is evidence indicating that flagellates tend to graze on a limited size range of prokaryotic cells, thus removing medium-sized cells and shifting the size distribution of the prey toward larger and smaller cells (44–46). In our case, the positively selected Archaea showed average cell sizes similar to those of Betaproteobacteria and Cytophaga-Flavobacteria, although large cells (i.e., quartile 75% in Fig. 2) were slightly smaller in Archaea than in the other two groups. Very likely, factors other than cell size must explain the strong preference for Archaea. During the sampling day, the cell length declined with time in both Archaea and Cytophaga-Flavobacteria, which may reflect the dynamic effect of grazing throughout the day (Fig. 2), yet other factors may have an influence, such as cell division. In all samples, Actinobacteria were markedly smaller than the members of the other prokaryotic groups (Fig. 2) and were less affected by protist grazing (Fig. 5). This observation agrees with previous studies that indicate lower grazing pressure on Actinobacteria by heterotrophic nanoflagellates (18, 20) and by the mixotrophic Chrysophyceae Ochromonas sp. (47). Indeed, if small size (i.e., <0.5 μm) constitutes a refuge against grazing, Actinobacteria may be negatively selected only in appearance, either because the limiting size might be an evolved defense mechanism (and thus cells tend to be smaller than those in other groups) or, on the contrary, because preference by grazers is so high that the group is permanently confined to the small-size refuge. It has been suggested that cell miniaturization alone is not sufficient to explain grazing avoidance (48) and that other resistance mechanisms, such as wall structures present in Gram-positive Actinobacteria, are involved in determining a limited edibility (49). Research on prokaryotic grazing is still in its infancy, but our results indicate that random grazing cannot be the paradigm.
Preference for Archaea.
In the last decade, several studies have shown the broad distribution and abundance of Archaea in aquatic ecosystems (50–52) and their potential relevance in the sulfur, nitrogen, and carbon cycles (33, 53–55). However, their specific ecological interactions within the microbial communities remain largely unknown. There was no evidence that Archaea behaved fundamentally differently from Bacteria on predatory interactions (56). Our results indicate that this is not always the case. The three mixotrophic flagellate species studied here selected Archaea positively. The amount of such a prokaryotic group inside the protist food vacuoles was always larger than that expected by random feeding (Fig. 5). In Rhodomonas sp., the preference for Archaea was extreme. It might be thought that the Archaea dominance in the vacuoles responds to a stable symbiosis. However, this appears unlikely. The average Archaea densities within the vacuoles and the number of Rhodomonas cells without any Archaea inside markedly changed between the four sampling times (Fig. 4 and Table 1). This pattern favors the view of selective predation and relatively quick digestion rather than a stable symbiosis.
The differences among the three protist species suggest that the strength of grazing on Archaea can highly depend on the protist present. Nevertheless, the three species show a preference for Archaea, which raises questions on the importance of grazing in determining the low proportion of this group in many planktonic microbial assemblages.
The reason for high positive selectivity on Archaea remains highly speculative. It could be related to a poor development of resistance mechanisms in this domain, or it could be related to unknown stoichiometric features of the Archaea; it has been shown that bacteria with a low C-to-P ratio may be ingested at higher rates by flagellates (57, 58). The reason also could be the chemical composition of some particular group of the Archaea within them, as they differ in cell wall structure. It could even be some chemical cues released to water that could enhance encounter rates between prey and predator (59). Archaea studies in Lake Redon indicated that most of the 16S rRNA gene sequences matched Thaumarchaeota (close to 90%), and only a few Euryarchaeota were detected. MG 1.1 dominated most of the water column in summer. This clone has >97% identity with Nitrosoarchaeum limnia, an ammonium-oxidizing archaeon (35). However, as new primers are applied, new groups of Euryarchaeota are found in the lake (60). In any case, the high positive selectivity on Archaea observed in the mixotrophs of Lake Redon paves the way toward new ecological and physiological studies on Archaea. Protist grazing is a factor that cannot be ignored for understanding the distribution and abundance of Archaea in planktonic microbial assemblages.
MATERIALS AND METHODS
Sampling site.
Lake Redon is an ultraoligotrophic high mountain lake located at 2,240 m above sea level in the central Pyrenees. It has a surface area of 24 ha and maximum and mean depths of 73 and 32 m, respectively. The lake is dimictic, well oxygenated throughout the water column, and usually covered by ice and snow 6 months a year (61). The productivity patterns and seasonal changes in the water column are typical for high mountain lakes (62, 63). This lake has been widely studied in the last 30 years. A general description of its physical, chemical, and temporal changes can be found in references 64 and 65, and a detailed description of the microbial plankton composition is in reference 66.
Sampling.
On 9 August 2004, during the summer stratification period, integrated samples (0 to 60 m) of the water column were taken at the deepest point of the lake at midnight (12 a.m.), dawn (7 a.m.), morning (11 a.m.), and afternoon (3 p.m.). The four sampling times aimed to estimate the consistency of potential selective behaviors and roughly approximate the time scale of digestion in case the number of ingested prey markedly fluctuated. The goal was not to investigate daily patterns with only one sampling day. The water samples were screened through a 40-μm mesh net to remove large zooplankton and subsequently divided into two subsamples. One of the subsamples was fixed with 0.5% (vol/vol) alkaline Lugol's solution followed by 2% buffered (pH 7) 0.2-μm-pore-size filtered formaldehyde, and several drops of 3% sodium thiosulfate were used to decolor Lugol's fixation by following the method of Medina-Sánchez et al. (21). After 1 h of fixation at room temperature, 24 aliquots of each sample (12 of 90 ml each for protists and 12 of 10 ml each for prokaryotes) were gently filtered (<100 mm Hg) onto respective 25-mm-diameter polycarbonate Millipore membrane filters (type RTTP, 1-μm pore size for protists; type GTTP, 0.2-μm pore size for prokaryotes). Filters were then rinsed twice with Milli-Q water, allowed to air dry, and stored at −20°C until further processing. The second subsample was preserved with 0.5% (vol/vol) alkaline Lugol's solution to identify algal species (67).
Phagotrophic activity.
Three different horseradish peroxidase (HRP)-labeled probes for the main Bacteria groups found in the plankton of the Pyrenean lakes (23, 24, 68) and one for the domain Archaea were used to hybridize the filters (Table 2). The probe EUB338, which targets most Bacteria, was used only to evaluate the hybridization yield of the other probes. Two replicate filters were processed for each probe and sample. Alexa 488-labeled tyramide was used for signal amplification, and filters were counterstained with DAPI (1 μg ml−1 final concentration) and mounted on glass slides by using Citifluor (Citifluor Ltd., United Kingdom). Slides were stored at −20°C in the dark until subsequent counting.
TABLE 2.
Oligonucleotide probes used in this study
| Probe name | Specificity | Sequence (5′–3′) | % FAa | Reference |
|---|---|---|---|---|
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 55 | 69 |
| ARCH915 | Archaea | GTGCTCCCCCGCCAATTCCT | 40 | 21 |
| CF319a | Cytophaga-Flavobacteria of Bacteroidetes | TGGTCCGTGTCTCAGTAC | 55 | 70 |
| HGC69a | Actinobacteria | TATAGTTACCACCGCCGT | 30 | 71 |
| BET42a | Betaproteobacteria | GCCTTCCCACTTCGTTT | 55 | 71 |
Formamide (FA) concentration in the hybridization buffer.
Slides were examined with a Zeiss Axio Imager epifluorescence microscope at ×1,000 magnification for bacterial groups, Archaea, and smaller protists (<10 μm) and at ×400 magnification for larger protists (>10 μm). The microscope was equipped with an X-Cite 120 light, appropriate filter sets for DAPI (Zeiss filter set 01; BP365/12 FT396 LP397) and Alexa-Fluor 488 (Zeiss filter set 09; BP450-490 FT510 LP515, o24 DBP485/20 DFT500/600 BP515-540 + LP610), a coupled camera (AxioCam MRm), and PC-based image analysis software (AxioVision 4.8). For prokaryotes, a minimum of 500 cells was counted to establish the total abundance (DAPI counts) and the number of hybridized cells for each specific probe to estimate their abundance and percentage of hybridization. Also, cell area and perimeter were measured by image analysis software in at least 100 cells of each filter. A characteristic length was calculated using the square root of the cell area.
Protists were identified based on their plastid size and shape, as observed by chlorophyll autofluorescence under blue excitation. The identification was facilitated by parallel observation of Lugol's fixed subsample under an inverted light microscope (×600 and ×1,000). A minimum of 100 individuals of the most abundant protist species was assessed for each, and the number of hybridized prokaryotic cells inside was counted.
Data analysis.
Protist species prey selectivity was analyzed according to Chesson's α index (72), as follows:
where ri and pi are the mean percentages of each prokaryotic group (i) inside the protist assessed and in the lake water assemblage, respectively; n is the number of prokaryotic groups distinguished (four in our case). When αi equals 1/n, unselective feeding occurs; when αi is less than 1/n, negative selection occurs, i.e., less of the prokaryotic group i occurs inside the protist than expected from random feeding; when αi is greater than 1/n, positive selection occurs, i.e., more individuals of the prokaryotic group i were ingested by the protist than expected from random feeding. One-way and two-way ANOVAs were performed to compare prokaryotic assemblages among samplings. To test changes on protist ingestion, Kruskal-Wallis nonparametric tests were used (STATISTICA 7.1; StatSoft, Inc.).
Supplementary Material
ACKNOWLEDGMENTS
We thank Juan-Manuel Medina-Sánchez and Lluis Camarero for laboratory and field assistance, respectively.
This research was supported by the Spanish Ministry of Science project ECOFOS (CGL2007-64177/BOS) with a predoctoral scholarship to M.A.B., Spanish Ministry of the Environment project EGALA (OAPN-124/2010), and a Generalitat de Catalunya research grant (SGR 2014-1249).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02317-16.
REFERENCES
- 1.Stoecker DK, Johnson MD, de Vargas C, Not F. 2009. Acquired phototrophy in aquatic protists. Aquat Microb Ecol 57:1–11. doi: 10.3354/ame01340. [DOI] [Google Scholar]
- 2.Sanders RW. 2011. Alternative nutritional strategies in protists: symposium introduction and a review of freshwater protists that combine photosynthesis and heterotrophy. J Eukaryot Microbiol 58:181–184. doi: 10.1111/j.1550-7408.2011.00543.x. [DOI] [PubMed] [Google Scholar]
- 3.Zubkov MV, Tarran GA. 2008. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature 455:224–226. doi: 10.1038/nature07236. [DOI] [PubMed] [Google Scholar]
- 4.Hartmann M, Grob C, Tarran GA, Martin AP, Burkill PH, Scanlan DJ, Zubkov MV. 2012. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc Natl Acad Sci U S A 109:5756–5760. doi: 10.1073/pnas.1118179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann M, Zubkov MV, Scanlan DJ, Lepere C. 2013. In situ interactions between photosynthetic picoeukaryotes and bacterioplankton in the Atlantic Ocean: evidence for mixotrophy. Environ Microbiol Rep 5:835–840. doi: 10.1111/1758-2229.12084. [DOI] [PubMed] [Google Scholar]
- 6.Sanders RW, Gast RJ. 2012. Bacterivory by phototrophic picoplankton and nanoplankton in Arctic waters. FEMS Microbiol Ecol 82:242–253. doi: 10.1111/j.1574-6941.2011.01253.x. [DOI] [PubMed] [Google Scholar]
- 7.Unrein F, Gasol JM, Not F, Forn I, Massana R. 2014. Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J 8:164–176. doi: 10.1038/ismej.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrba J, Nedoma J, Kohout L, Kopáček J, Nedbalová L, Ráčková P, Šcaronimek K. 2003. Massive occurrence of heterotrophic filaments in acidified lakes: seasonal dynamics and composition. FEMS Microbiol Ecol 46:281–294. doi: 10.1016/S0168-6496(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 9.Jürgens K, Pernthaler J, Schalla S, Amann R. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol 65:1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corno G, Jürgens K. 2008. Structural and functional patterns of bacterial communities in response to protist predation along an experimental productivity gradient. Environ Microbiol 10:2857–2871. doi: 10.1111/j.1462-2920.2008.01713.x. [DOI] [PubMed] [Google Scholar]
- 11.Montagnes DJS, Barbosa AB, Boenigk J, Davidson K, Jürgens K, Macek M, Parry JD, Roberts EC. 2008. Selective feeding behaviour of key free-living protists: avenues for continued study. Aquat Microb Ecol 53:83–98. doi: 10.3354/ame01229. [DOI] [Google Scholar]
- 12.Šimek K, Horňák K, Mašin M, Christaki U, Nedoma J, Weinbauer MG, Dolan JR. 2003. Comparing the effects of resource enrichment and grazing on a bacterioplankton community of a meso-eutrophic reservoir. Aquat Microb Ecol 31:123–135. doi: 10.3354/ame031123. [DOI] [Google Scholar]
- 13.Salcher MM, Pernthaler J, Psenner R, Posch T. 2005. Succession of bacterial grazing defense mechanisms against protistan predators in an experimental microbial community. Aquat Microb Ecol 38:215–229. doi: 10.3354/ame038215. [DOI] [Google Scholar]
- 14.Gerea M, Queimaliños C, Schiaffino MR, Izaguirre I, Forn I, Massana R, Unrein F. 2013. In situ prey selection of mixotrophic and heterotrophic flagellates in Antarctic oligotrophic lakes: an analysis of the digestive vacuole content. J Plankton Res 35:201–212. doi: 10.1093/plankt/fbs085. [DOI] [Google Scholar]
- 15.Welsh J, McClelland M. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caetano-Anolles G, Brant B. 1991. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Nat Biotechnol 9:553–557. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- 17.Glücksman E, Bell T, Griffiths RI, Bass D. 2010. Closely related protist strains have different grazing impacts on natural bacterial communities. Environ Microbiol 12:3105–3113. doi: 10.1111/j.1462-2920.2010.02283.x. [DOI] [PubMed] [Google Scholar]
- 18.Jezbera J, Horňak K, Šimek K. 2005. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microbiol Ecol 52:351–363. doi: 10.1016/j.femsec.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Bautista-Reyes F, Macek M. 2012. Ciliate food vacuole content and bacterial community composition in the warm-monomictic crater Lake Alchichica, México. FEMS Microbiol Ecol 79:85–97. [DOI] [PubMed] [Google Scholar]
- 20.Jezbera J, Horňák K, Šimek K. 2006. Prey selectivity of bacterivorous protists in different size fractions of reservoir water amended with nutrients. Environ Microbiol 8:1330–1339. doi: 10.1111/j.1462-2920.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- 21.Medina-Sánchez JM, Felip M, Casamayor EO. 2005. Catalyzed reported deposition-fluorescence in situ hybridization protocol to evaluate phagotrophy in mixotrophic protists. Appl Environ Microbiol 71:7321–7326. doi: 10.1128/AEM.71.11.7321-7326.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Šimek K, Weinbauer MG, Hornák K, Jezbera J, Nedoma J, Dolan JR. 2007. Grazer and virus-induced mortality of bacterioplankton accelerates development of Flectobacillus populations in a freshwater community. Environ Microbiol 9:789–800. doi: 10.1111/j.1462-2920.2006.01201.x. [DOI] [PubMed] [Google Scholar]
- 23.Barberan A, Casamayor EO. 2014. A phylogenetic perspective on species diversity, beta-diversity and biogeography for the microbial world. Mol Ecol 23:5868–5876. doi: 10.1111/mec.12971. [DOI] [PubMed] [Google Scholar]
- 24.Hervas A, Casamayor EO. 2009. High similarity between bacterioneuston and airborne bacterial community compositions in a high mountain lake area. FEMS Microbiol Ecol 67:219–228. doi: 10.1111/j.1574-6941.2008.00617.x. [DOI] [PubMed] [Google Scholar]
- 25.Pérez MT, Sommaruga R. 2006. Differential effect of algal- and soil-derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnol Oceanogr 51:2527–2537. doi: 10.4319/lo.2006.51.6.2527. [DOI] [Google Scholar]
- 26.Salcher MM, Pernthaler J, Posch T. 2010. Spatiotemporal distribution and activity patterns of bacteria from three phylogenetic groups in an oligomesotrophic lake. Limnol Oceanogr 55:846–856. doi: 10.4319/lo.2010.55.2.0846. [DOI] [Google Scholar]
- 27.Bouvier T, del Giorgio PA. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol Ecol 44:3–15. doi: 10.1016/S0168-6496(02)00461-0. [DOI] [PubMed] [Google Scholar]
- 28.Sarmento H, Casamayor EO, Auguet J-C, Vila-Costa M, Felip M, Camarero L, Gasol JM. 2015. Microbial food web components, bulk metabolism, and single-cell physiology of piconeuston in surface microlayers of high-altitude lakes. Front Microbiol 6:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshino T, Yilmaz LS, Noguera DR, Daims H, Wagner M. 2008. Quantification of target molecules needed to detect microorganisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl Environ Microbiol 74:5068–5077. doi: 10.1128/AEM.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posch T, Franzoi J, Prader M, Salcher MM. 2009. New image analysis tool to study biomass and morphotypes of three major bacterioplankton groups in an alpine lake. Aquatic Microb Ecol 54:113–126. doi: 10.3354/ame01269. [DOI] [Google Scholar]
- 31.Auguet JC, Casamayor EO. 2008. A hotspot for cold crenarchaeota in the neuston of high mountain lakes. Environ Microbiol 10:1080–1086. doi: 10.1111/j.1462-2920.2007.01498.x. [DOI] [PubMed] [Google Scholar]
- 32.Pernthaler A, Preston CM, Pernthaler J, DeLong EF, Amann R. 2002. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl Environ Microbiol 68:661–667. doi: 10.1128/AEM.68.2.661-667.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llirós M, Alonso-Sáez L, Gich F, Plasencia A, Auguet O, Casamayor EO, Borrego CM. 2011. Active bacteria and archaea cells fixing bicarbonate in the dark along the water column of a stratified eutrophic lagoon. FEMS Microbiol Ecol 77:370–384. doi: 10.1111/j.1574-6941.2011.01117.x. [DOI] [PubMed] [Google Scholar]
- 34.Plasencia A, Gich F, Fillol M, Borrego CM. 2013. Phylogenetic characterization and quantification of ammonia-oxidizing archaea and bacteria from Lake Kivu in a long-term microcosm incubation. Int Microbiol 16:177–189. [DOI] [PubMed] [Google Scholar]
- 35.Auguet JC, Triado-Margarit X, Nomokonova N, Camarero L, Casamayor EO. 2012. Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake. ISME J 6:1786–1797. doi: 10.1038/ismej.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortiz-Alvarez R, Casamayor EO. 2016. High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environ Microbiol Rep 8:210–217. doi: 10.1111/1758-2229.12370. [DOI] [PubMed] [Google Scholar]
- 37.Gasol JM, Pedrós-Alió C, Vaqué D. 2002. Regulation of bacterial assemblages in oligotrophic plankton systems: results from experimental and empirical approaches. Antonie Van Leeuwenhoek 81:435–452. doi: 10.1023/A:1020578418898. [DOI] [PubMed] [Google Scholar]
- 38.Grossart HP, Jezbera J, Horňák K, Hutalle KML, Buck U, Šimek K. 2008. Top-down and bottom-up induced shifts in bacterial abundance, production and community composition in an experimentally divided humic lake. Environ Microbiol 10:635–652. doi: 10.1111/j.1462-2920.2007.01487.x. [DOI] [PubMed] [Google Scholar]
- 39.Bell T, Bonsall MB, Buckling A, Whiteley AS, Goodall T, Griffiths RI. 2010. Protists have divergent effects on bacterial diversity along a productivity gradient. Biol Lett 6:639–642. doi: 10.1098/rsbl.2010.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sintes E, del Giorgio PA. 2014. Feedbacks between protistan single-cell activity and bacterial physiological structure reinforce the predator/prey link in microbial foodwebs. Front Microbiol 5:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boenigk J, Matz C, Jürgens K, Arndt H. 2002. Food concentration-dependent regulation of food selectivity of interception-feeding bacterivorous nanoflagellates. Aquat Microb Ecol 27:195–202. doi: 10.3354/ame027195. [DOI] [Google Scholar]
- 42.Zwirglmaier K, Spence E, Zubkov MV, Scanlan DJ, Mann NH. 2009. Differential grazing of two heterotrophic nanoflagellates on marine Synechococcus strains. Environ Microbiol 11:1767–1776. doi: 10.1111/j.1462-2920.2009.01902.x. [DOI] [PubMed] [Google Scholar]
- 43.Roberts EC, Wootton EC, Davidson K, Jeong HJ, Lowe CD, Montagnes DJ. 2011. Feeding in the dinoflagellate Oxyrrhis marina: linking behaviour with mechanisms. J Plankton Res 33:603–614. doi: 10.1093/plankt/fbq118. [DOI] [Google Scholar]
- 44.Hahn MW, Höfle MG. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol 35:113–121. doi: 10.1111/j.1574-6941.2001.tb00794.x. [DOI] [PubMed] [Google Scholar]
- 45.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 46.Jürgens K, Matz C. 2002. Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81:413–434. doi: 10.1023/A:1020505204959. [DOI] [PubMed] [Google Scholar]
- 47.Pernthaler J, Posch T, Šimek J, Vrba J, Pernthaler A, Glöckner FO, Nubel U, Psenner R, Amann R. 2001. Predator-specific enrichment of Actinobacteria from a cosmopolitan freshwater clade in mixed continuous culture. Appl Environ Microbiol 67:2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anderson R, Kjelleberg S, McDougald D, Jürgens K. 2011. Species-specific patterns in the vulnerability of carbon-starved bacteria to protist grazing. Aquat Microb Ecol 64:105–116. [Google Scholar]
- 49.Tarao M, Jezbera J, Hahn M. 2009. Involvement of cell surface structures in size-independent grazing resistance of freshwater Actinobacteria. Appl Environ Microbiol 75:4720–4726. doi: 10.1128/AEM.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeLong EF, Pace NR. 2001. Environmental diversity of Bacteria and Archaea. Syst Biol 50:470–478. doi: 10.1080/10635150118513. [DOI] [PubMed] [Google Scholar]
- 51.Robertson CE, Harris JK, Spear JR, Pace NR. 2005. Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol 8:638–642. doi: 10.1016/j.mib.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Auguet J-C, Barberan A, Casamayor EO. 2010. Global ecological patterns in uncultured Archaea. ISME J 4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- 53.Nicol GW, Schleper C. 2006. Ammonia-oxidising Crenarchaeota: important players in the nitrogen cycle? Trends Microbiol 14:207–212. doi: 10.1016/j.tim.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 54.Berg IA, Kockelkorn D, Ramos-Vera WH, Say RF, Zarzycki J, Hügler M, Alber BE, Fuchs G. 2010. Autotrophic carbon fixation in archaea. Nat Rev Microbiol 8:447–460. doi: 10.1038/nrmicro2365. [DOI] [PubMed] [Google Scholar]
- 55.Auguet J-C, Nomokonova N, Camarero L, Casamayor E. 2011. Seasonal changes of freshwater ammonia-oxidizing archaeal assemblages and nitrogen species in oligotrophic alpine lakes. Appl Environ Microbiol 77:1937–1945. doi: 10.1128/AEM.01213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jürgens K. 2007. Predation on bacteria and bacterial resistance mechanisms: comparative aspects among different predator groups in aquatic systems. Microbiol Monogr 4:57–92. doi: 10.1007/7171_053. [DOI] [Google Scholar]
- 57.John EH, Davidson K. 2001. Prey selectivity and the influence of prey carbon: nitrogen ratio on microflagellate grazing. J Exp Mar Biol Ecol 260:93–111. doi: 10.1016/S0022-0981(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 58.Shannon S, Chrzanowski T, Grover J. 2007. Prey food quality affects flagellate ingestion rates. Microb Ecol 53:66–73. doi: 10.1007/s00248-006-9140-y. [DOI] [PubMed] [Google Scholar]
- 59.Roberts EC, Legrand C, Steinke M, Wootton EC. 2011. Mechanisms underlying chemical interactions between predatory planktonic protists and their prey. J Plankton Res 33:833–841. doi: 10.1093/plankt/fbr005. [DOI] [Google Scholar]
- 60.Restrepo-Ortiz CX, Casamayor EO. 2013. Environmental distribution of two widespread uncultured freshwater Euryarchaeota clades unveiled by specific primers and quantitative PCR. Environ Microbiol Rep 5:861–867. doi: 10.1111/1758-2229.12088. [DOI] [PubMed] [Google Scholar]
- 61.Catalan J. 1989. The winter cover of a high-mountain Mediterranean lake (Estany Redó, Pyrenees). Water Resour Res 25:519–527. [Google Scholar]
- 62.Catalan J, Ventura M, Brancelj A, Granados I, Thies H, Nickus U, Korhola A, Lotter AF, Barbieri A, Stuchlík E, Lien L, Bitušík P, Buchaca T, Camarero L, Goudsmit GH, Kopáćek J, Lemcke G, Livingstone DM, Müller B, Rautio M, Šiško M, Sorvari S, Šporka F, Strunecký O, Toro M. 2002. Seasonal ecosystem variability in remote mountain lakes: implications for detecting climatic signals in sediment records. J Paleolimnol 28:25–46. doi: 10.1023/A:1020315817235. [DOI] [Google Scholar]
- 63.Catalan J, Camarero L, Felip M, Pla S, Ventura M, Buchaca T, Bartumeus F, de Mendoza G, Miró A, Casamayor E, Medina-Sánchez JM, Bacardit M, Maddi A, Bartrons M, de Quijano DD. 2006. High mountain lakes: extreme habitats and witnesses of environmental changes. Limnetica 25:551–584. [Google Scholar]
- 64.Catalan J. 1988. Physical properties of the environment relevant to the pelagic ecosystem of a deep high-mountain lake (Estany Redo, Central Pyrenees). Oecologia Aquatica 9:89–123. [Google Scholar]
- 65.Catalan J. 1992. Evolution of dissolved and particulate matter during the ice-covered period in a deep, high-mountain lake. Can J Fish Aquat Sci 49:945–955. doi: 10.1139/f92-105. [DOI] [Google Scholar]
- 66.Felip M, Camarero L, Catalan J. 1999. Temporal changes of microbial assemblages in the ice and snow cover of a high mountain lake. Limnol Oceanogr 44:973–987. doi: 10.4319/lo.1999.44.4.0973. [DOI] [Google Scholar]
- 67.Sournia A. 1978. Phytoplankton manual. UNESCO, Paris, France. [Google Scholar]
- 68.Llorens-Marès T, Auguet JC, Casamayor EO. 2012. Winter to spring changes in the slush bacterial community composition of a high-mountain lake (Lake Redon, Pyrenees). Environ Microbiol Rep 4:50–56. [DOI] [PubMed] [Google Scholar]
- 69.Pernthaler A, Pernthaler J, Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 71.Warnecke F, Sommaruga R, Sekar R, Hofer J, Pernthaler J. 2005. Abundances, identity, and growth sate of Actinobacteria in mountain lakes of different UV transparency. Appl Environ Microbiol 71:5551–5559. doi: 10.1128/AEM.71.9.5551-5559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chesson J. 1983. The estimation and analysis of preference and its relationship to foraging models. Ecology 64:1297–1304. doi: 10.2307/1937838. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.