ABSTRACT
The alginate-producing bacterium Pseudomonas fluorescens utilizes the Entner-Doudoroff (ED) and pentose phosphate (PP) pathways to metabolize fructose, since the upper part of its Embden-Meyerhof-Parnas pathway is defective. Our previous study indicated that perturbation of the central carbon metabolism by diminishing glucose-6-phosphate dehydrogenase activity could lead to sugar phosphate stress when P. fluorescens was cultivated on fructose. In the present study, we demonstrate that PFLU2693, annotated as a haloacid dehalogenase-like enzyme, is a new sugar phosphate phosphatase, now designated Spp, which is able to dephosphorylate a range of phosphate substrates, including glucose 6-phosphate and fructose 6-phosphate, in vitro. The effect of spp overexpression on growth and alginate production was investigated using both the wild type and several mutant strains. The results obtained suggested that sugar phosphate accumulation caused diminished growth in some of the mutant strains, since this was partially relieved by spp overexpression. On the other hand, overexpression of spp in fructose-grown alginate-producing strains negatively affected both growth and alginate production. The latter implies that Spp dephosphorylates the sugar phosphates, thus depleting the pool of these important metabolites. Deletion of the spp gene did not affect growth of the wild-type strain on fructose, but the gene could not be deleted in the alginate-producing strain. This indicates that Spp is essential for relieving the cells of sugar phosphate stress in P. fluorescens actively producing alginate.
IMPORTANCE In enteric bacteria, the sugar phosphate phosphatase YigL is known to play an important role in combating stress caused by sugar phosphate accumulation. In this study, we identified a sugar phosphate phosphatase, designated Spp, in Pseudomonas fluorescens. Spp utilizes glucose 6-phosphate, fructose 6-phosphate, and ribose 5-phosphate as substrates, and overexpression of the gene had a positive effect on growth in P. fluorescens mutants experiencing sugar phosphate stress. The gene was localized downstream of gnd and zwf-2, which encode enzymes involved in the pentose phosphate and Entner-Doudoroff pathways. Genes encoding Spp homologues were identified in similar genetic contexts in some bacteria belonging to several phylogenetically diverse families, suggesting similar functions.
KEYWORDS: Pseudomonas fluorescens, alginate biosynthesis, sugar phosphate stress response, phosphatase, PFLU2693, YigL, sugar phosphate phosphatase
INTRODUCTION
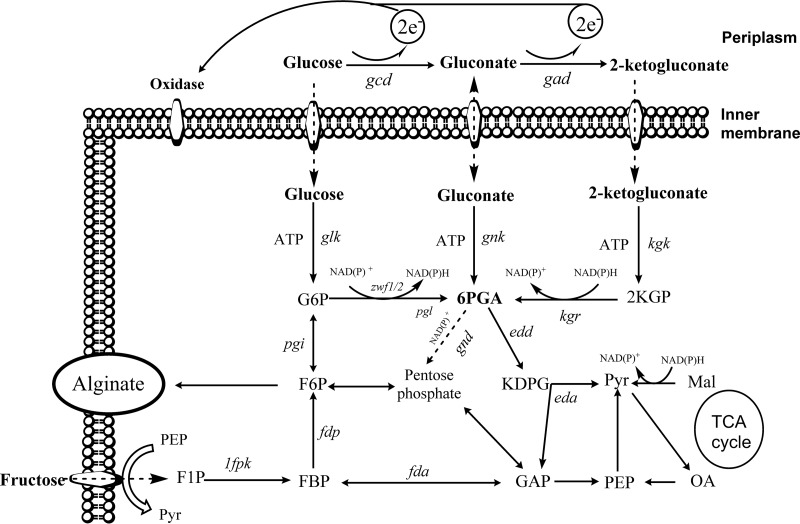
Pseudomonas central carbon metabolism has been well studied during recent decades (1–3). Unlike Escherichia coli, which utilizes the phosphotransferase system type G (PtsG) as the major glucose transporter (4), most pseudomonads import glucose through an active transport system (5). In contrast, fructose transport is mediated by the phosphoenolpyruvate (PEP)-dependent phosphotransferase system in these organisms. Pseudomonas species lack the 6-phosphofructokinase gene required for glycolysis, and glucose is converted to glyceraldehyde-3-phosphate (GAP) and pyruvate (Pyr) via the Entner-Doudoroff (ED) pathway. In addition, several studies have already shown that fructose is mainly metabolized through the ED route by converting fructose-1,6-bisphosphate (FBP) to fructose-6-phosphate (F6P) and then to glucose-6-phosphate (G6P) (1, 6) (Fig. 1).
FIG 1.
Glucose and fructose metabolic pathways in P. fluorescens. Abbreviations: G6P, glucose-6-phosphate; 6PGA, 6-phosphogluconate; 2KGP, 2-keto-6-phosphogluconate; KDPG, 2-keto-3-deoxy-6-phosphogluconate; F6P, fructose-6-phosphate; FBP, fructose-1,6-bisphosphate; F1P, fructose-1-phosphate; GAP, glyceraldehyde-3-phosphate; Pyr, pyruvate; DPGA, 1,3-diphosphoglycerate; 3PGA, 3-phosphoglycerate; 2PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate; OA, oxaloacetate; Mal, malate; Gad, gluconate dehydrogenase; Gcd, glucose dehydrogenase; Kgk, 2-ketogluconate kinase; Gnk, gluconokinase; Glk, glucokinase; Kgr, 2-KGP reductase; Gnd, 6PGA dehydrogenase; Zwf1-2, G6P dehydrogenases; Pgi, phosphoglucose isomerase; Pgl, 6-phosphogluconolaktonase: Edd, 6PGA dehydratase; Eda, KDPG aldolase; 1Fpk, 1-phosphofructokinase; Fdp, fructose-1,6-bisphosphatase; Fda, fructose-1,6-bisphosphate aldolase. Figure abridged from reference 10.
Alginate is a polysaccharide with a wide variety of industrial and medical applications, and it is composed of variable ratios of β-d-mannuronic acid (M) and α-l-guluronic acid (G). Two genera of bacteria, Pseudomonas and Azotobacter, are capable of producing alginate. Alginate biosynthesis uses F6P as a precursor to generate GDP-mannuronic acid, which then is polymerized, transported through the periplasm, and secreted through the AlgE porin in the outer membrane (7, 8).
Pseudomonas fluorescens strain Pf201 is a nonpathogenic mutant strain that was developed in our group by random mutagenesis of the wild-type strain NCIMB10525 and studied as a stable alginate producer (9). In our recent study focusing on the role of genes involved in central carbon metabolism of Pf201 on alginate production (10), it was hypothesized that the observed growth deficiencies of the glucose-6-phosphate dehydrogenase (G6PD) (encoded by zwf-1 and zwf-2) mutants of the NCIMB10525 strain grown on fructose resulted from accumulation of hexose phosphates (G6P and F6P) causing sugar phosphate stress. It was shown that the growth defect was less severe in a zwf-1 mutant of an alginate-producing strain, possibly because alginate production may drain away a portion of the excessive sugar phosphates (10). This hypothesis motivated our current study to investigate any possible sugar phosphate stress response in Pf201 further.
A sugar phosphate stress response has been studied in E. coli and Salmonella (11, 12) using the nonmetabolizable glucose analog α-methyl glucoside (αMG), which is taken up efficiently by PtsG and represses cell growth. Recently, Papenfort et al. provided strong evidence that the phosphatase YigL in Salmonella enterica serovar Typhimurium is needed for dephosphorylation of accumulated hexose phosphate prior to efflux by an unidentified efflux pump (11). Sugar phosphate stress is still not well understood, but current evidence indicates that accumulation of sugar phosphates leads to depletion of other carbon metabolites or phosphorus (13–16). Sugar phosphatases might then be used to alleviate this problem. However, this creates a futile cycle of phosphorylation and dephosphorylation, and E. coli utilizes phosphatases as an immediate response, while the more lasting solution is to downregulate sugar uptake and phosphorylation (15).
YigL belongs to the vast superfamily of haloacid dehalogenase (HAD)-like hydrolases. Characterized enzymes belonging to this superfamily function as haloacid dehalogenases, phosphonatases, phosphatases, and β-phosphoglucomutases (17). In spite of very low full-length sequence similarity within the family, three motifs are highly conserved: motif 1 (D-D-[T/V][L/V]) contains an absolutely conserved aspartate contributing to the nucleophile attack on the substrate phosphate group, resulting in formation of a covalent intermediate; motif 2 (S/T) consists of serine or threonine residues binding the substrate phosphoryl group; and finally, motif 3 (K-D) contains a strictly conserved lysine residue followed at some distance by a conserved aspartate (18).
In the present study, we sought to investigate the hypothesized sugar phosphate stress and potential sugar phosphatase involvement in response to such a stress in P. fluorescens. We identified a putative HAD-like hydrolase enzyme in P. fluorescens with some homology to YigL from S. Typhimurium. The enzyme showed sugar phosphatase activity in vitro, and the effect of overexpression and inactivation of its gene on growth and alginate production of P. fluorescens was studied further.
RESULTS
G6PDH mutants accumulate increased amounts of G6P and F6P.
In our earlier study (19), we found that mutant NCIMB10525Δzwf-1Δzwf-2, which does not produce any active G6PD enzymes, hardly grew on fructose. The effect was less severe in the single mutant NCIMB10525Δzwf-1. For alginate-producing strains it was found that strain Pf201ΔalgCΔzwf-1::TnKB60 grew more slowly than its parent, Pf201ΔalgC::TnKB60, but the difference was less pronounced than that between the corresponding non-alginate-producing strains NCIMB10525 and NCIMB10525Δzwf-1. We suggested that the observed growth defect was partly due to accumulation of G6P or F6P and that alginate production would drain away some of these hexose phosphates, thus diminishing the stress (19).
A first step toward validating this hypothesis would be to show that the G6PDH mutants accumulate more hexose phosphates than the wild-type strain. Since the zwf-1 zwf-2 double mutant displayed the most severe growth defect in the wild-type background, an in-frame deletion of zwf-2 was constructed in the alginate-producing strain Pf201ΔalgCΔzwf-1::TnKB60. Since it proved to be impossible to delete zwf-1 from the alginate-producing strain Pf201, the mutants were made in a derivative where algC has been deleted, and then a new copy controlled by the inducible Pm-G5 promoter was delivered by the transposon TnKB60 (10). AlgC is necessary for alginate biosynthesis, and these genetic manipulations provided strains that produce small amounts of alginate in the absence of the inducer m-toluate and large amounts of alginate when the Pm-G5 promoter is induced.
The zwf-2 deletion is an in-frame deletion designed to avoid any polar effects on expression of the downstream genes. Strain Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60 was complemented with transposon TnSM1 containing zwf-2 under the control of the Pm-G5-promoter. As expected, the complemented strain, Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60::TnSM1, grew similarly to Pf201ΔalgCΔzwf-1::TnKB60 (results not shown). TnSM1 has previously been shown to complement the growth defects in strain NCIMB10525Δzwf-1Δzwf-2 (10). This indicates that the observed phenotype is not caused by a polar effect on any genes downstream of zwf-2.
For sugar phosphate quantification, the wild-type strain NCIMB10525, the alginate-producing strain Pf201ΔalgC::TnKB60, and zwf-1 zwf-2 double mutants of these two strains were cultivated for 24 h in DEF3 containing fructose as the carbon source. The cells were harvested and the intracellular levels of G6P and F6P were measured as described in Materials and Methods. The results showed that the wild-type strain contained about twice as much G6P and F6P as the alginate-producing control strain. However, the mutants lacking glucose 6-phosphate dehydrogenase enzymes contained 20 times more of these hexose-phosphates than the control strains (Table 1).
TABLE 1.
Quantification of G6P and F6P in P. fluorescens strainsa
| Strainb | Level [μg/OD600 (SD)] of: |
|
|---|---|---|
| G6P | F6P | |
| NCIMB10525 | 0.027 (0.007) | 5.6 (2.0) |
| NCIMB10525Δzwf-1 Δzwf-2 | 0.55 (0.043) | 93 (2.4) |
| NCIMB10525Δzwf-1 Δzwf-2::TnRH1 | 0.43 (0.031) | 72 (13) |
| Pf201ΔalgC::TnKB60 | 0.005 (0.000) | 1.8 (0.6) |
| Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60 | 0.25 (0.018) | 54 (15) |
| Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60::TnRH1 | 0.172 (0.018) | 39 (4.3) |
The strains were cultivated for 24 h in DEF3 medium containing fructose as the sole carbon source, as well as m-toluate and triclosan.
spp and algC are expressed from Pm-G5 promoters in TnRH1 and TnKB60, respectively.
Identification of a putative hexose phosphate phosphatase gene in P. fluorescens.
Since these data supported our hypothesis that P. fluorescens experiences sugar phosphate stress, it seemed plausible that the bacterium encodes an enzyme that could relieve such stress. The amino acid sequence of S. enterica YigL (GenBank accession number AAL22806.1) was used in a BLAST search against the translated P. fluorescens SBW25 genome sequence. The best hit was the translated PFLU2693 (accession number CAY48924.1), displaying 28% identity and 46% similarity over the full length of S. enterica YigL. The PFLU2693 protein possesses all three motifs that are conserved in the HAD-like hydrolase superfamily (not shown) (18). The protein is also the closest P. fluorescens SBW25 relative to two other E. coli proteins, YidA and YbiV, both known to be active on sugar phosphates (20). These proteins all belong to the COF subfamily (IPR000150) of the HAD-like hydrolase superfamily that is supposed to also include sugar phosphate phosphatases. Based on this and the experimental results detailed below, we designated the gene spp, for sugar phosphate phosphatase. Interestingly, spp is located downstream of gnd and zwf-2, both of which encode enzymes involved in the pentose phosphate (PP) pathway (1). The reading frames of gnd and zwf-2 overlap, as do those of zwf-2 and spp, indicating that all three genes belong to one operon.
Spp homologs in other bacterial species.
To assess how widespread this protein is in nature, the amino acid sequence of Spp was compared to that of proteins in the nonredundant protein sequence database (NCBI). The best hits were, as expected, against other strains of the genus Pseudomonas that belong to the gammaproteobacteria. However, nearly all proteins with full-length homology were identified in bacteria of the P. fluorescens and P. syringae groups. No full-length homologs were detected within the P. aeruginosa group, and only P. putida strain MC4-5222 and P. oryzihabitans, of the P. putida group, encoded Spp homologs. The genomic context was checked for the best hit for each species encoding a full-length Spp homolog, and in all cases the putative spp gene was preceded by gnd and zwf genes.
A new BLAST search was then performed excluding all bacteria belonging to the genus Pseudomonas. The 100 best hits all displayed an identity between 39% and 56% over at least 95% of the Spp amino acid sequence (Fig. 2). Interestingly, the bacteria harboring genes encoding Spp homologs belonged to several different genera of Proteobacteria, Cyanobacteria, and Actinobacteria, and the most represented group was the alphaproteobacteria. For all identified alpha- and gammaproteobacteria and cyanobacteria, and some of the betaproteobacteria and actinomycetes, the spp homolog was located in gene clusters together with other genes belonging to the ED or PP pathway. zwf and gnd genes were found in most of these clusters (Fig. 2).
FIG 2.
Gene contexts of Spp (COF) homolog-encoding genes. The 100 proteins most similar to the full-length amino acid sequence of Spp (COF family protein) were identified by a BLAST search (34) against GenBank. The genus Pseudomonas was excluded from the search. The gene contexts from representatives of each genus are displayed using standard gene names. Genes encoding enzymes involved in the ED or PP pathway are shown as colored boxes.
Analyses of the in vitro activity of the Spp protein.
In order to be able to verify the hypothesis that spp encodes a hexose phosphate phosphatase, it was necessary to express and purify the encoded protein. Expression vector pSM12, encoding a C-terminally His-tagged Spp, was constructed as described in Materials and Methods and introduced into E. coli RV308, and the resulting strain was used for protein production. The purified protein was analyzed by SDS-PAGE, and a protein band with the expected mass for Spp (29.9 kDa) was identified (Fig. 3a). This band was also detected on a Western blot using antibodies against the His tag (not shown). The purified protein displayed phosphatase activity when p-nitrophenyl phosphate (pNPP) was used as the substrate (not shown).
FIG 3.

Purification and substrate specificities of Spp. (a) SDS-PAGE of purified Spp displaying a dominant protein band (lane 1) and protein standard (lane 2). The arrows mark the standard protein bands with the approximate sizes 35 and 27 kDa. (b) The phosphatase activity was measured using 0.1 mM ribose 5-phosphate (R5P), F6P, G6P, pyridoxal 5-phosphate (P5P), 6-phosphogluconate (6PG), and AMP as substrates. Samples from the reaction mixture (see Materials and Methods for details) were taken at different time points, and released Pi was quantified using MGR. The values represent the means from three different experiments, and standard deviations are indicated using error bars.
As described above, our hypothesis was that the growth defect of NCIMB10525Δzwf-1Δzwf-2 was caused by the observed accumulation of G6P and/or F6P (10), and we wanted to assess the activity of Spp on those substrates. The purified enzyme was incubated with G6P and F6P separately, and the release of phosphate was monitored by taking samples from the reaction mixtures and terminating the reaction by addition of malachite green reagent (MGR). For F6P, Km was determined to be 0.7 mM, kcat was 9.6 s−1, and the specificity coefficient, kcat/Km, was 13,000 s−1 M−1. The corresponding values for G6P were 1.8 mM, 11.4 s−1, and 6,400 s−1 M−1, respectively, indicating that F6P is a preferred substrate compared to G6P. The R2 value for the lines used to determine Km and Vmax was 0.99 in both cases. These numbers are in a range similar to that found for E. coli HAD phosphatases (20). These results confirmed our hypothesis that spp encodes a sugar phosphate phosphatase.
In E. coli, several phosphatases of the same family have been shown to accept a range of substrates (20). Given that spp is found in the same operon as two genes encoding enzymes in the PP pathway, we tested R5P and 6PG, both of which are metabolites in this pathway. We also tested P5P, which is a derivative of R5P and a good substrate for E. coli YigL (20), and, finally, AMP, which is a fairly unrelated compound. The results show that at a substrate concentration of 0.1 mM, the reaction rate was highest for R5P (Fig. 3b). The initial reaction rate for P5P was only 4% of that of R5P, while the activity on 6PG and AMP was hardly above the noise level (21, 22). Still, the in vitro data indicated that Spp could relieve the cell from stress caused by accumulated F6P and G6P.
The effect of spp overexpression on growth and alginate production.
F6P is utilized for production of the alginate precursor GDP-mannuronic acid (Fig. 1), and therefore alginate production competes with the central carbon metabolism for this vital metabolite (23, 24). As a consequence, alginate-producing P. fluorescens strains usually reach a lower cell density than nonproducers due to F6P drainage for alginate biosynthesis (19, 25). Overproduction of a hexose phosphate phosphatase would also be expected to have a negative effect on growth, since it would introduce a futile cycle and deplete the cell of important metabolites. An F6P phosphatase thus would be expected to compete for F6P with pathways for both central metabolism and alginate biosynthesis (Fig. 1). If Spp has hexose phosphate phosphatase activity in vivo, then overexpression of the gene would be expected to result in weaker growth and lower alginate production.
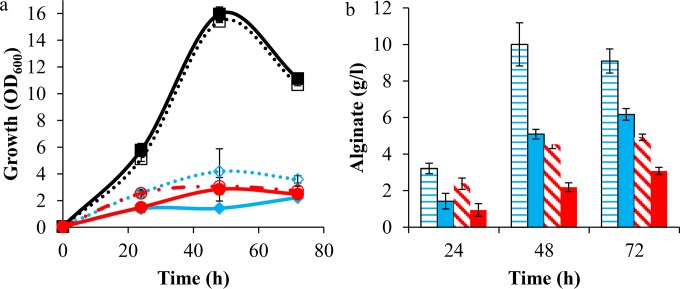
spp was cloned on a transposon vector where its expression is controlled by the inducible Pm-G5 promoter; the transposon was designated TnRH1. The transposon vector was transferred to strain NCIMB10525 and its alginate-producing derivatives Pf201 and Pf201ΔalgC::TnKB60. The resulting strains were cultivated in DEF3 medium containing fructose as a carbon source, where growth and, when applicable, alginate production were measured (Fig. 4). Both growth and alginate production in Pf201 and Pf201ΔalgC::TnKB60 were negatively affected by overproduction of Spp. Similar results were obtained for several independent transposon insertion strains (data not shown). These results show that Spp expressed from transposon TnRH1 caused the phenotype expected for hexose phosphate phosphatase overexpression in an alginate producer. However, overexpression of spp did not affect growth of the wild-type strain NCIMB10525 (Fig. 4).
FIG 4.
Effect of Spp overproduction from TnRH1 on growth (a) and alginate production (b) in P. fluorescens. Strains included NCIMB10525 (□ in panel a; alginate deficient), NCIMB10525::TnRH1 (■ in panel a; alginate deficient), Pf201 (♢ in panel a; horizontal stripes in panel b), Pf201::TnRH1 (◆ in panel a; solid blue bar in panel b), Pf201ΔalgC::TnKB60 (○ in panel a; diagonal stripes in panel b), and Pf201ΔalgC::TnKB60::TnRH1 (● in panel a; solid red bar in panel b). The growth values represent the means from at least two independent biological replicates, and standard deviations are indicated using error bars.
Inactivation of spp does not influence growth of wild-type P. fluorescens.
The nonmetabolizable glucose analog α-methyl-glucoside (α-MG) is taken up by a glucose transporter in E. coli and Salmonella and then converted to α-MG-6-phosphate (α-MG6P) (4). This compound accumulates in the cytoplasm and can hamper bacterial growth (26). Papenfort et al. showed that loss of YigL in E. coli grown on α-MG plates resulted in growth deficiency because of the inability to dephosphorylate α-MG6P and promote sugar efflux, while this mutant was able to survive on glucose (11). In order to investigate how essential Spp is for P. fluorescens growth, we decided to inactivate the corresponding gene. A plasmid containing a mutated version of spp with an in-frame deletion, as well as some sequences flanking this gene, was constructed and used to create strain NCIMB10525Δspp by double homologous recombination, leading to an in-frame deletion of an internal part of spp. When the recombinant strain was cultivated in DEF3 with fructose, it grew similarly to the wild-type strain (data not shown). Therefore, the Spp phosphatase is not essential for growth of the non-alginate-producing strain on fructose. However, after several attempts, we did not succeed in inactivating spp in either of the two alginate-producing strains, Pf201 and Pf201ΔalgC::TnKB60.
The effect of spp overexpression on G6PD mutants grown on fructose.
If the decreased growth rates observed in the G6PDH mutants result from hexose phosphate stress, overexpression of spp would be expected to ease the problem. Accordingly, the transposon vector pRH1, encoding Spp, was transferred to NCIMB10525Δzwf-1, NCIMB10525Δzwf-1Δzwf-2, Pf201ΔalgCΔzwf-1::TnKB60, Pf201ΔalgCΔzwf-1, and Δzwf-2::TnKB60, and recombinant strains containing TnRH1 were isolated. These strains were then cultivated in DEF3 medium containing fructose and m-toluate (to induce spp expression), as were the corresponding strains without TnRH1.
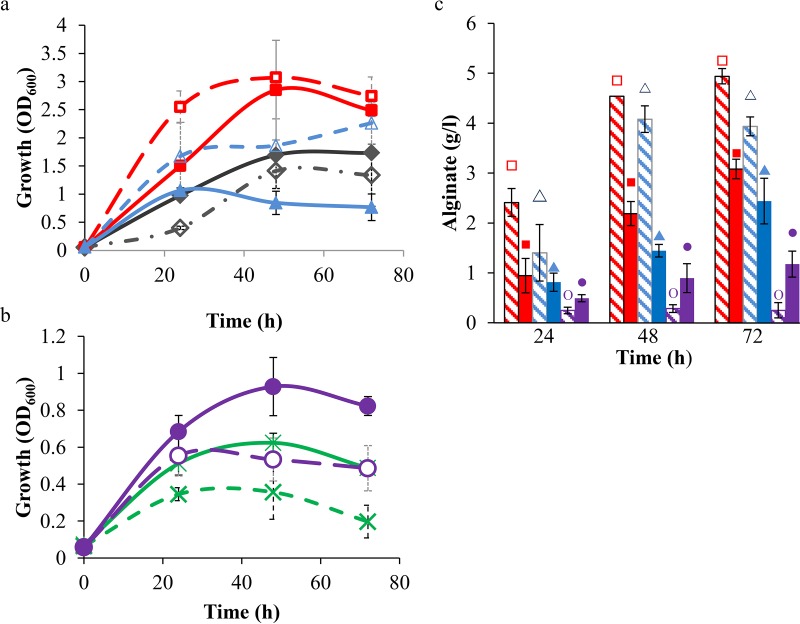
Overexpression of spp from TnRH1 in strain Pf201ΔalgCΔzwf-1::TnKB60 yielded results similar to those for Pf201ΔalgC::TnKB60; both growth and alginate production were negatively affected (Fig. 5a and c). However, the long lag phase observed for NCIMB10525Δzwf-1 grown on fructose was not observed when strain NCIMB10525Δzwf-1::TnRH1 was monitored (Fig. 5a), indicating a positive effect of additional Spp in the non-alginate-producing zwf-1 mutant.
FIG 5.
Effect of Spp overproduction from TnRH1 on growth and alginate production of P. fluorescens glucose 6-phosphate dehydrogenase mutants cultivated on fructose. (a and b) Growth profiles. The Δzwf-1Δzwf-2 double mutants were plotted in panel b using a different scale due to poorer growth. (c) Alginate production of the Pf201 derivatives. Symbols and colors: NCIMB10525Δzwf-1 (black, ◇ and ◆), NCIMB10525Δzwf-1Δzwf-2 (green, ×), Pf201ΔalgC::TnKB60 (red, □ and ■), Pf201ΔalgCΔzwf-1::TnKB60 (blue, △ and ▲), Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60 (purple, ○ and ●). Strains without TnRH1 are displayed with open symbols and hatched lines and bars, and the corresponding strains containing TnRH1 are displayed using filled symbols and bars and solid lines. The presented data are averages from three biological replicates, and standard deviations are indicated using error bars.
It is noteworthy that the negative effect of zwf-1 deletion on growth was less pronounced for the alginate-producing strain, which grew better than strain NCIMB10525Δzwf-1. The same was seen for the zwf-1 zwf-2 double mutants (Fig. 5b). However, no double mutant reached an optical density at 600 nm (OD600) above 1, not even when spp was overexpressed. Expression of spp from TnRH1 further resulted in significantly higher alginate production in Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60::TnRH1 than in strain Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60 (Fig. 5c). When G6P and F6P were measured in the zwf-1 zwf-2 double mutants, the strains overexpressing spp were shown to contain somewhat less of these hexose phosphates than the corresponding strains without TnRH1 (Table 1). Still, the amount was far above that determined for the control strains. The obtained results suggest that accumulation of hexose phosphates is one of the reasons for the observed growth defects in the G6PDH mutant strains, since overexpression of spp from transposon TnRH1 had a positive effect in the strains assumed to suffer from such stress.
DISCUSSION
In a previous study (19), we found that the non-alginate-producing P. fluorescens zwf-1 zwf-2 double mutant grown on fructose exhibited a severe growth defect. In the zwf-1 single mutant, the growth defect was less pronounced, likely because the PP pathway still operates in the normal direction and provides part of the metabolites needed for growth (Fig. 1). We postulated that one of the reasons for this growth impairment is the result of G6P accumulation, based on the observation that alginate-producing zwf-1 mutants grew relatively better. Quantification of G6P and F6P has proven that these mutants do accumulate far more G6P and F6P than the wild-type strain (Table 1). We have further shown that a previously uncharacterized YigL-like protein encoded by PFLU2693 is able to dephosphorylate R5P, G6P, and F6P in vitro (Fig. 3). This protein was designated Spp.
The growth profile of the wild-type-based spp deletion mutant grown on fructose showed no difference compared to that of the wild-type strain (data not shown). However, we were not able to inactivate spp in alginate-producing strains. Alginate-producing cells have a higher fructose uptake rate, and the metabolite flow toward F6P is increased (6, 24). It is feasible that we were unable to delete spp from the alginate-producing strains because the increased flow to F6P might cause temporary sugar phosphate imbalances that are detrimental to the cells. Furthermore, spp overexpression in the alginate-producing strain negatively affected growth and alginate production, while it did not influence the growth profile of the non-alginate-producing strain (Fig. 4). This might be explained using the observation that compared to the wild-type strain, the alginate-producing strain Pf201 grows more slowly (Fig. 4a). spp overexpression creates a futile cycle of phosphorylation-dephosphorylation that consumes ATP, and for the alginate-producing strain where the situation already may be close to critical, this leads to further reduction of growth. The situation is different in the wild type, which does not produce alginate and probably has an energy surplus (24). Therefore, this strain can cope with overexpression of spp, and an ATP drain due to the futile cycle does not have any drastic effects on growth.
Moreover, alginate production may lower the pH of the medium, leading to a stress response, and this makes the cell more vulnerable to additional stress (10). Similarly, none of the E. coli HADs, including YigL, were essential for growth when the cells experienced no sugar phosphate stress (11, 20). In contrast, E. coli Δpgi ΔYigL was not able to grow on glucose due to accumulation of G6P and lack of the phosphatase YigL for aiding the stress response (11).
It would be expected that for mutants experiencing enhanced sugar phosphate levels, alginate production may alleviate some of the stress, and both Pf201ΔalgCΔzwf-1::TnKB60 and Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60 mutants grew somewhat better than their non-alginate-producing counterparts (Fig. 5a and b). Overexpression of spp resulted in diminished growth and alginate production for Pf201ΔalgCΔzwf-1::TnKB60, while it was positive for the other tested glucose dehydrogenase mutants. These data indicate that as long as the alginate production and the native-level expression of Spp are sufficient to maintain an acceptable concentration of sugar phosphates, overproduction of Spp negatively influences both growth and alginate production, because it dephosphorylates G6P and/or F6P and makes them unavailable for growth and alginate production.
spp is located in the same operon as two genes, gnd and zwf-2, involved in the PP pathway (Fig. 1). We found that genes encoding putative sugar phosphate phosphatases with significant homology to Spp are located in gene clusters encoding proteins involved in the PP or ED pathway in many other bacteria. This indicates that utilization of these pathways in some environments poses a risk of sugar phosphate imbalance. Still, Spp homologs sharing more than 40% identity are found in only a limited number of bacteria, even among those preferring the ED pathway to the standard Embden-Meyerhof-Parnas pathway. This suggests either that sugar phosphate stress is not a problem in these strains or that they encode sugar phosphate phosphatases that are less similar to Spp.
Given that the spp homologs are found together with genes encoding enzymes involved in the PP pathway, it is interesting that Spp was able to degrade all three tested substrates that are components of this cycle. Recently, it was demonstrated that sugar phosphatases can function as metabolite repair enzymes by removing rare, but toxic, side products synthesized by core metabolic enzymes (21, 22). It is not unlikely that Spp in P. fluorescens has other main substrates than the ones tested here.
YigL has been shown to be important for combating sugar phosphate stress in enteric bacteria (11). Our data suggest that the Spp phosphatase of P. fluorescens, which shares some homology with YigL, plays an important role in sugar phosphate balancing when P. fluorescens experiences F6P/G6P accumulation. This further implicates that sugar phosphate stress is a factor that should be considered when the carbon metabolism of a bacterium is engineered to obtain high-level production of a desired compound.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and constructed plasmids used in this study are described in Table 2. The E. coli strains S17.1 and S17.1 (λpir) were used as hosts for cloning and replication of suicide plasmids, while E. coli RV308 was used for protein production. E. coli strains were routinely grown in L broth (LB) (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl). P. fluorescens strains (wild-type NCIMB10525 and derivatives) were cultivated at 225 rpm at 30°C in LB or in DEF3 minimal medium supplemented with 20 g/liter of fructose (10). When required, antibiotics were added at the following concentrations: triclosan (Tric), 25 μg/ml; apramycin (Am), 50 μg/ml; ampicillin (Ap), 200 μg/ml; kanamycin (Km), 50 μg/ml; tetracycline (Tc), 15 μg/ml. To protect produced alginate from degradation by lyases, protease enzymes (0.15 ml/liter of alkalase [2.4 liters] and neutrase [0.5 liters]) were added to alginate production media. Alginate production was quantified using alginate lyases as described earlier (30). All alginate assays were performed in triplicates.
TABLE 2.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | Cloning host | Life Technologies |
| S17.1 | RP4 2-Tc::Mu-km::Tn7 pro res mod+ | 27 |
| S17.1 (λpir) | λpir recA thi pro hsdR-M+ RP4 2-Tc::Mu-km::Tn7 TpRSMR | 28 |
| RV308 | Expression host | ATCC 31608 |
| NCIMB10525 | Nonmucoid wild type; Tricr | NCIMB |
| NCIMB10525Δzwf-1 | zwf-1 in-frame deletion mutant of NCIMB10525; Tricr | 10 |
| NCIMB10525Δzwf-1 Δzwf-2 | zwf-2 in-frame deletion mutant of NCIMB10525Δzwf-1; Tricr | 19 |
| NCIMB10525Δspp | spp in-frame deletion mutant of NCIMB10525; Tricr | This study |
| NCIMB10525Δzwf-1::TnRH1 | Derivative of NCIMB10525Δzwf-1 in which spp was overexpressed by inserting the transposon from pRH1; Amr, Tricr | This study |
| NCIMB10525Δzwf-1Δzwf-2::TnRH1 | Derivative of NCIMB10525Δzwf-1Δzwf-2 in which spp was overexpressed by inserting the transposon from pRH1; Amr, Tricr | This study |
| Pf201 | Mucoid mutant derived from NCIMB10525 | 9 |
| Pf201ΔalgC::TnKB60 | Pf201ΔalgC with transposon insertion from pKB60 (containing algC controlled by Pm-G5); Kmr, Tricr | 29 |
| Pf201ΔalgCΔzwf-1::TnKB60 | zwf-1 in-frame deletion mutant of Pf201ΔalgC::TnKB60; Kmr, Tricr | 10 |
| Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60 | zwf-2 in-frame deletion mutant of Pf201ΔalgCΔzwf-1::TnKB60; Kmr, Tricr | This study |
| Pf201::TnRH1 | Derivative of Pf201 in which spp was overexpressed by inserting the transposon from pRH1; Amr, Tricr | This study |
| Pf201ΔalgCΔzwf-1Δzwf-2::TnKB60::TnRH1 | Derivative of Pf201ΔalgC Δzwf-1Δzwf-2::TnKB60 in which spp was overexpressed by inserting the transposon from pRH1; Amr, Tricr | This study |
| Pf201ΔalgCΔzwf-1::TnKB60::TnRH1 | Derivative of Pf201ΔalgCΔzwf-1::TnKB60 in which spp was overexpressed by inserting the transposon from pRH1; Amr, Tricr | This study |
| Plasmids | ||
| pMG48 | RK2-based gene replacement vector; Apr, Tcr | 30 |
| pYQ1 | Transposon delivery vector resistant to apramycin; Amr | 10 |
| pMM8 | Suicide gene replacement vector containing Δzwf-2; Apr, Tcr | 10 |
| pSM10 | Derivative of pMG48 in which 2.1-kb DNA fragment containing spp with an in-frame deletion was inserted; Apr, Tcr | This study |
| pSM1 | Derivative of pYQ1 where zwf-2 is controlled by Pm-G5; Amr | 10 |
| pRH1 | Derivative of pYQ1 in which a 0.917-kb NdeI-NotI PCR DNA fragment encoding spp was inserted so that spp is controlled by Pm-G5; Amr | This study |
| pMV3 | RK2-based inducible expression vector; Apr | 31 |
| pSM11 | Derivative of NdeI-NotI-restricted pMV3 where a 0.917-kb NdeI-NotI DNA fragment from pRH1containing spp was inserted; Apr | This study |
| pSM12 | Derivative of pSM11 where a His tag coding sequence was fused to the 3′ end of the spp gene; Apr | This study |
The expression level from a Pm promoter is dependent on the amount of inducer, allowing for a fine-tuning of the expression level (32). When the Pm-G5 promoter was used to express genes in P. fluorescens, the promoter usually was induced by adding 0.25 mM m-toluate. However, in an initial experiment, strain NCIMB10525Δzwf-1Δzwf-2::TnRH1 was cultivated using different concentrations of m-toluate, and 0.05 mM m-toluate was found to give the best growth improvement result (data not shown). It is probable that too-high expression levels are detrimental to the cell (see Results and Discussion). This inducer concentration was subsequently used in all growth experiments involving TnRH1. Conjugation between E. coli and P. fluorescens strains was accomplished on L agar (L broth containing 15 g/liter agar), and selection of transconjugants and homologous recombination procedures was performed as described earlier (30).
Quantification of G6P and F6P in P. fluorescens.
Single colonies from freshly inoculated plates were transferred to LB containing triclosan and cultivated for 18 h. Two hundred microliters of these cells was transferred to 20 ml DEF3 medium containing fructose, triclosan, and 0.05 mM m-toluate in 125-ml flat-bottom flasks and cultivated for 24 h. The OD600 was recorded, and a culture volume corresponding to 3 OD units was centrifuged to harvest the cells. For alginate-producing cells, the cultures were diluted five times with 0.2 M NaCl in order to obtain alginate-free cell pellets. The cells were resuspended in 100 μl H2O, and 50 μl cold 25% HClO4 was added immediately. The cells were then subjected to three freeze-thaw cycles in liquid nitrogen before cell debris were removed by centrifugation at 15,600 × g for 5 min at 4°C. The pH was adjusted with 40 μl 3 M K2CO3. If needed, more K2CO3 was added to obtain a final pH between 7 and 8. Precipitated KClO4 was removed by centrifugation at 5,000 × g for 5 min at 4°C. The supernatant was incubated at −20°C for 15 min and centrifuged again for 2 min at 9,200 × g and 4°C. Finally, the supernatant was filtered using an Amicon Ultra-0.5 centrifugal filter unit (Millipore). The supernatant was aliquoted and stored at −20°C. G6P and F6P were quantified using the high-sensitivity glucose-6-phosphate assay kit and the fructose-6-phosphate assay kit, respectively (both from Sigma-Aldrich), using the recommended procedure. Between 0.25 and 15 μl supernatant was applied in the assays in order to keep the measurements within the linear range of the assay. Three separate colonies were inoculated for each strain, while the assays were performed in duplicate. Standard deviation is given as the deviation found between the biological replicates.
Construction of plasmids and strains.
All routine DNA standard techniques and cloning methods were performed as detailed previously (29). All amplified DNA fragments were confirmed by DNA sequencing.
Determination of substrate specificities of Spp necessitated production of a His-tagged version of the protein in E. coli. The gene was first cloned into the protein expression vector pMV3, generating pSM11. In order to remove the gene stop codon and sequence gap placed before His tag in this plasmid, a 0.916-kb PCR fragment of pSM11 containing the His tag and some downstream regions was obtained. This was then fused to an 8.2-kb SbfI-AflII-restricted DNA fragment from pSM11 using the Gibson assembly kit (New England BioLabs). The resulting plasmid, pSM12, was confirmed by DNA sequencing and transformed into E. coli RV308.
In order to inactivate spp, 456 nucleotides were removed in-frame from the spp gene by using two PCR primer pairs (F1/R1 and F2/R2) and the P. fluorescens genome as the template. Primer F2 and R1 contained complementary sequences; therefore, the two generated PCR products could be annealed and used as the template for the third PCR by using F1 and R2 primers. The final PCR product (2.18 kb) was first cloned into pCR-Blunt II-TOPO vector (Zero Blunt TOPO PCR cloning kit; Invitrogen) according to the manufacturer's instructions and then subcloned into the gene replacement vector pMG48. The new plasmid was designated pSM10 and utilized to inactivate spp by homologous recombination as described earlier (30).
In this study, we used the inducible Pm promoter and Pm promoter derivatives to control gene expression.
Heterologous expression and purification of Spp-His.
E. coli RV308 (pSM12) was cultivated in 100 ml LB, and production of Spp-His was induced by addition of m-toluate (0.25 mM) after 2 h. The cells were harvested after 8 h by centrifugation at 13,000 × g for 10 min. The pellet was resuspended in 10 ml binding buffer (HEPES) (50 mM, pH 7.4), NaCl (0.5 M), and imidazole (30 mM) and disrupted by sonication. Cell debris was removed by centrifugation at 30,000 × g for 30 min, followed by filtration of the supernatant using a 0.22-μm filter. The enzyme was then purified on a HisTrap HP column (GE Healthcare Life Sciences) utilizing a 0.03 to 0.5 M imidazole gradient (in a buffer that otherwise was the same as that for binding) for elution. The enzyme was eluted at approximately 0.1 M imidazole. An aliquot of 0.9 μg of the purified Spp protein was electrophoretically separated on SDS-polyacrylamide gels according to Laemmli (33) and then subjected to Western blot analysis as described previously (19).
Measurements of Spp phosphatase activity.
To investigate the phosphatase activity of Spp, general phosphatase activity and substrate specificity assays were carried out according to Kuznetsova et al. (17). In the general assay, p-nitrophenyl phosphate (pNPP) was used as a substrate. Ten microliters of the purified Spp protein fraction was mixed with a 90-μl reaction mixture (4 mM pNPP, 50 mM HEPES K [pH 7.5], and initially 5 mM MgSO4 and 0.5 mM MnCl2) and incubated for 10 min at room temperature. The optimal Mg2+ concentration was subsequently found to be 2.5 mM, while addition of Mn2+ decreased the reaction rate both without and in combination with Mg2+. Addition of 50 mM NaCl also decreased activity.
Substrate specificity was assayed using the malachite green phosphate assay kit (Sigma-Aldrich) using glucose 6-phosphate (G6P), fructose 6-phosphate (F6P), ribose 5-phosphate (R5P), pyridoxal 5-phosphate (P5P), 6-phospho gluconate (6PG), and AMP as substrates. Purified Spp protein was added to the reaction mixture (0.1 mM substrate, 50 mM HEPES K [pH 7.5], 2.5 mM MgCl2) and incubated at room temperature. At set time points, samples (80 μl) were transferred to 96-well plates containing 20 μl malachite green reagent (MGR). The intervals between each time point varied from 15 s to 3 min depending on the specific activity on the various substrates. OD620 was measured after 30 min and compared to the measurements of phosphate standards belonging to the kit. For determination of the kinetic constants for F6P and G6P, substrate concentrations were varied from 0.05 to 2 mM. All enzyme reactions were performed in triplicates.
Bioinformatics.
The P. fluorescens SBW25 complete genome is found in GenBank (www.ncbi.nlm.nih.gov) (accession no. NC_012660.1). Protein BLAST searches were accomplished using BLASTP 2.2.30 on the nonredundant protein database in GenBank.
ACKNOWLEDGMENT
We thank Kjell Domaas Josefsen at SINTEF Materials and Chemistry for help in designing the intracellular metabolite extraction method.
REFERENCES
- 1.Lessie TG, Phibbs PV Jr. 1984. Alternative pathways of carbohydrate utilization in pseudomonads. Annu Rev Microbiol 38:1–12. doi: 10.1146/annurev.mi.38.100184.002043. [DOI] [PubMed] [Google Scholar]
- 2.Lessmann D, Schimz KL, Kurz G. 1975. D-glucose-6-phosphate dehydrogenase (Entner-Doudoroff enzyme) from Pseudomonas fluorescens. Purification, properties and regulation. Eur J Biochem 59:545–559. [DOI] [PubMed] [Google Scholar]
- 3.Chavarria M, Kleijn RJ, Sauer U, Pflüger-Grau K, de Lorenzo V. 2012. Regulatory tasks of the phosphoenolpyruvate-phosphotransferase system of Pseudomonas putida in central carbon metabolism. mBio 3:e00028-12. doi: 10.1128/mBio.00028-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jahreis K, Pimentel-Schmitt EF, Bruckner R, Titgemeyer F. 2008. Ins and outs of glucose transport systems in eubacteria. FEMS Microbiol Rev 32:891–907. doi: 10.1111/j.1574-6976.2008.00125.x. [DOI] [PubMed] [Google Scholar]
- 5.Midgley M, Dawes EA. 1973. The regulation of transport of glucose and methyl alpha-glucoside in Pseudomonas aeruginosa. Biochem J 132:141–154. doi: 10.1042/bj1320141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lien SK, Niedenführ S, Sletta H, Nöh K, Bruheim P. 2015. Fluxome study of Pseudomonas fluorescens reveals major reorganisation of carbon flux through central metabolic pathways in response to inactivation of the anti-sigma factor MucA. BMC Syst Biol 9:6. doi: 10.1186/s12918-015-0148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay ID, Ur Rehman Z, Moradali MF, Wang Y, Rehm BH. 2013. Microbial alginate production, modification and its applications. Microb Biotechnol 6:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rehman ZU, Wang Y, Moradali MF, Hay ID, Rehm BH. 2013. Insights into the assembly of the alginate biosynthesis machinery in Pseudomonas aeruginosa. Appl Environ Microbiol 79:3264–3272. doi: 10.1128/AEM.00460-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gimmestad M, Sletta H, Karunakaran P, Bakkevig K, Ertesvåg H, Ellingsen TE, Skjåk-Bræk G, Valla S. 2002. New mutant strains of Pseudomonas fluorescens and variants thereof, methods of their production, and uses thereof in alginate production. European patent WO2004/011628.
- 10.Maleki S, Mærk M, Valla S, Ertesvåg H. 2015. Mutational analyses of glucose dehydrogenase and glucose-6-phosphate dehydrogenase genes in Pseudomonas fluorescens reveal their effects on growth and alginate production. Appl Environ Microbiol 81:3349–3356. doi: 10.1128/AEM.03653-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. 2013. Small RNA-mediated activation of sugar phosphatase mRNA regulates glucose homeostasis. Cell 153:426–437. doi: 10.1016/j.cell.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rice JB, Vanderpool CK. 2011. The small RNA SgrS controls sugar phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 39:3806–3819. doi: 10.1093/nar/gkq1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godinho LM, de Sa-Nogueira I. 2011. Characterization and regulation of a bacterial sugar phosphatase of the haloalkanoate dehalogenase superfamily, AraL, from Bacillus subtilis. FEBS J 278:2511–2524. doi: 10.1111/j.1742-4658.2011.08177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J, Chassy BM. 1983. Intracellular hexose-6-phosphate:phosphohydrolase from Streptococcus lactis: purification, properties, and function. J Bacteriol 156:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards GR, Patel MV, Lloyd CR, Vanderpool CK. 2013. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J Bacteriol 195:4816–4825. doi: 10.1128/JB.00705-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richards GR, Vanderpool CK. 2012. Induction of the Pho regulon suppresses the growth defect of an Escherichia coli sgrS mutant, connecting phosphate metabolism to the glucose-phosphate stress response. J Bacteriol 194:2520–2530. doi: 10.1128/JB.00009-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuznetsova E, Proudfoot M, Sanders SA, Reinking J, Savchenko A, Arrowsmith CH, Edwards AM, Yakunin AF. 2005. Enzyme genomics: application of general enzymatic screens to discover new enzymes. FEMS Microbiol Rev 29:263–279. doi: 10.1016/j.femsre.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Calderone V, Forleo C, Benvenuti M, Thaller MC, Rossolini GM, Mangani S. 2006. A structure-based proposal for the catalytic mechanism of the bacterial acid phosphatase AphA belonging to the DDDD superfamily of phosphohydrolases. J Mol Biol 355:708–721. doi: 10.1016/j.jmb.2005.10.068. [DOI] [PubMed] [Google Scholar]
- 19.Maleki S, Almaas E, Zotchev SB, Valla S, Ertesvåg H. 2016. Alginate biosynthesis factories in Pseudomonas fluorescens: localization and correlation with alginate production level. Appl Environ Microbiol 82:2027–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. 2006. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem 281:36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 21.Collard F, Baldin F, Gerin I, Bolsee J, Noel G, Graff J, Veiga-da-Cunha M, Stroobant V, Vertommen D, Houddane A, Rider MH, Linster CL, Van Schaftingen E, Bommer GT. 2016. A conserved phosphatase destroys toxic glycolytic side products in mammals and yeast. Nat Chem Biol 12:601–607. doi: 10.1038/nchembio.2104. [DOI] [PubMed] [Google Scholar]
- 22.Huang L, Khusnutdinova A, Nocek B, Brown G, Xu X, Cui H, Petit P, Flick R, Zallot R, Balmant K, Ziemak MJ, Shanklin J, de Crecy-Lagard V, Fiehn O, Gregory JF III, Joachimiak A, Savchenko A, Yakunin AF, Hanson AD. 2016. A family of metal-dependent phosphatases implicated in metabolite damage-control. Nat Chem Biol 12:621–627. doi: 10.1038/nchembio.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daddaoua A, Krell T, Ramos JL. 2009. Regulation of glucose metabolism in Pseudomonas: the phosphorylative branch and Entner-Doudoroff enzymes are regulated by a repressor containing a sugar isomerase domain. J Biol Chem 284:21360–21368. doi: 10.1074/jbc.M109.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lien SK, Sletta H, Ellingsen TE, Valla S, Correa E, Goodacre R, Vernstad K, Borgos SEF, Bruheim P. 2013. Investigating alginate production and carbon utilization in Pseudomonas fluorescens SBW25 using mass spectrometry-based metabolic profiling. Metabolomics 9:403–417. doi: 10.1007/s11306-012-0454-0. [DOI] [Google Scholar]
- 25.Borgos SEF, Bordel S, Sletta H, Ertesvåg H, Jakobsen Ø, Bruheim P, Ellingsen TE, Nielsen J, Valla S. 2013. Mapping global effects of the transcription factor MucA in Pseudomonas fluorescens through genome-scale metabolic modeling. BMC Syst Biol 7:19. doi: 10.1186/1752-0509-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pikis A, Hess S, Arnold I, Erni B, Thompson J. 2006. Genetic requirements for growth of Escherichia coli K12 on methyl-alpha-D-glucopyranoside and the five alpha-D-glucosyl-D-fructose isomers of sucrose. J Biol Chem 281:17900–17908. doi: 10.1074/jbc.M601183200. [DOI] [PubMed] [Google Scholar]
- 27.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 28.de Lorenzo V, Cases I, Herrero M, Timmis KN. 1993. Early and late response of TOL promoters to pathway inducers: identification of postexponential promoters in Pseudomonas putida with lacZ-tet bicistronic reporters. J Bacteriol 175:6902–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bakkevig K, Sletta H, Gimmestad M, Aune R, Ertesvåg H, Degnes K, Christensen BE, Ellingsen TE, Valla S. 2005. Role of the Pseudomonas fluorescens alginate lyase (AlgL) in clearing the periplasm of alginates not exported to the extracellular environment. J Bacteriol 187:8375–8384. doi: 10.1128/JB.187.24.8375-8384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gimmestad M, Sletta H, Ertesvåg H, Bakkevig K, Jain S, Suh S-J, Skjåk-Bræk G, Ellingsen TE, Ohman DE, Valla S. 2003. The Pseudomonas fluorescens AlgG protein, but not its mannuronan C5-epimerase activity, is needed for alginate polymer formation. J Bacteriol 185:3515–3523. doi: 10.1128/JB.185.12.3515-3523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakke I, Berg L, Aune TE, Brautaset T, Sletta H, Tøndervik A, Valla S. 2009. Random mutagenesis of the Pm promoter as a powerful strategy for improvement of recombinant-gene expression. Appl Environ Microbiol 75:2002–2011. doi: 10.1128/AEM.02315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winther-Larsen HC, Blatny JM, Valand B, Brautaset T, Valla S. 2000. Pm promoter expression mutants and their use in broad-host-range RK2 plasmid vectors. Metab Eng 2:92–103. doi: 10.1006/mben.1999.0143. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]