ABSTRACT
The requirements for bacterial chemotaxis and motility range from dispensable to crucial for host colonization. Even though more than 50% of all sequenced prokaryotic genomes possess at least one chemotaxis signaling system, many of those genomes contain multiple copies of a chemotaxis gene. However, the functions of most of those additional genes are unknown. Most motile bacteria possess at least one CheY response regulator that is typically dedicated to the control of motility and which is usually essential for virulence. Borrelia burgdorferi appears to be notably different, in that it has three cheY genes, and our current studies on cheY2 suggests that it has varied effects on different aspects of the natural infection cycle. Mutants deficient in this protein exhibit normal motility and chemotaxis in vitro but show reduced virulence in mice. Specifically, the cheY2 mutants were severely attenuated in murine infection and dissemination to distant tissues after needle inoculation. Moreover, while ΔcheY2 spirochetes are able to survive normally in the Ixodes ticks, mice fed upon by the ΔcheY2-infected ticks did not develop a persistent infection in the murine host. Our data suggest that CheY2, despite resembling a typical response regulator, functions distinctively from most other chemotaxis CheY proteins. We propose that CheY2 serves as a regulator for a B. burgdorferi virulence determinant that is required for productive infection within vertebrate, but not tick, hosts.
KEYWORDS: Borrelia burgdorferi, Lyme disease, spirochetes, chemotaxis, response regulator, CheY, virulence, chemotaxis/motility, tick-mouse, flagella, pathogenesis
INTRODUCTION
Borrelia burgdorferi is the causative organism of Lyme disease, which is the most common vector-borne illness in the United States and Europe (1, 2). During its natural enzootic cycle, the organism must survive within a vertebrate host (usually a rodent) and a tick vector of an Ixodes species. Spirochete-infected ticks efficiently transmit the bacteria during a blood meal to the vertebrate host, where the organisms disseminate from the initial site of inoculation in the dermis to several distant tissues, such as tibiotarsal joints, heart, and the nervous system, where they persist to produce various clinical manifestations (3–5). B. burgdorferi is a highly motile organism whose motility is provided by flagella that are enclosed by the outer membrane and thus are called periplasmic flagella (6–9). Motility is absolutely required for migration of B. burgdorferi from the skin to distant tissues, establishment of persistent infection in mice, transmission from the tick vector to the murine host, and optimal survival in ticks (6, 7, 10). Chemotaxis is also important for the spirochetal infectious life cycle, as these pathways are involved in directing bacterial motility during the different stages of infection (11–13).
Chemotaxis, which uses a two-component signaling system, has been extensively studied in Escherichia coli and Salmonella enterica serovar Typhimurium; thus, these organisms serve as model organisms (14–17). In these systems, the two-component system is initiated when a membrane-bound protein, called the methyl-accepting chemotaxis protein (MCP), binds a ligand. This action causes signal transduction from the MCP to a histidine kinase, CheA, via a linker protein known as CheW. CheA autophosphorylates, which then transfers its phosphate to CheY. Phosphorylated CheY in turn binds to the flagellar switch proteins FliM and FliN to alter cellular behavior (18). Binding to the flagellar proteins causes a change in the direction of flagellar rotation from the default counterclockwise (CCW) to clockwise (CW) rotation. When peritrichous flagella of E. coli rotate CCW, the bacterial cell runs, whereas CW rotation results in tumbling, which serves to reorient the swimming direction. Although phosphorylated CheY (CheY-P) autodephosphorylates, a phosphatase known as CheZ in E. coli (or CheX/CheC in other bacteria/spirochetes) dephosphorylates the CheY-P, resulting in CW rotations. Thus, the levels of CheY-P determine whether a cell runs or tumbles (19–21).
The chemotaxis signaling system is conserved among prokaryotes (22). The B. burgdorferi genome possesses a sophisticated chemotaxis system with multiple motility- and chemotaxis-related operons (23, 24). Genomic sequencing as well as in vitro functional analyses indicate that B. burgdorferi encodes multiple copies of the chemotaxis genes, including two histidine kinases (CheA1 and CheA2), three response regulatory proteins (CheY1, CheY2, and CheY3), three coupling proteins (CheW1, CheW2, and CheW3), two sets of chemotaxis adaptation proteins, CheB (CheB1 and CheB2) and CheR (CheR1 andCheR2), five MCPs, and one cytoplasmic chemoreceptor (24–29). B. burgdorferi also possesses a CheX phosphatase which we recently reported generates enhanced activity via CheD (12, 30, 31). Moreover, all of the motility and chemotaxis operons of B. burgdorferi are transcribed by the σ70 promoter (23). While the roles of many of those chemotaxis genes have been evaluated in vitro, only three (cheA2, cheD, and cheY3) have currently been investigated in the natural infectious life cycle of B. burgdorferi (11–13).
B. burgdorferi is a long (10 to 20 μm) and thin (0.3 μm) organism that possesses 7 to 11 periplasmic flagella attached to each end of the cell (9, 32). Tracking B. burgdorferi motility reveals three different swim modes: run, flex, and reverse. Runs occur when the periplasmic flagellar motors at one end of the cell rotate in the opposite direction of the motors at the other end (CW rotation in motors at one end of the cell and CCW rotation in motors at the other end). Spirochetes flex when the motors at both ends rotate in the same direction, i.e., both rotating either CW or CCW. Cell reversal occurs in translating cells when the motors at each end reverse their direction of rotation. The spirochete flex is thought to be equivalent to the E. coli or S. Typhimurium tumble (21, 26, 31, 33, 34).
The three B. burgdorferi cheY genes are located in three separate operons. Using functional and phosphorylation analyses, we have previously reported that these response regulatory proteins are phosphorylated by both of the histidine kinases, CheA1 and CheA2 (25, 31). However, only cheY3 was found to be essential for motility and chemotaxis in vitro (25). Specifically, while wild-type (WT) B. burgdorferi cells are observed to run, pause/flex, and then change their swimming direction, cheY3 mutant (ΔcheY3) cells constantly run in one direction without reversing and are subsequently deficient in chemotaxis (25). Recently, cheY3-mediated chemotaxis was found to be crucial for motility in vivo, including dissemination and viability in mice and ticks (13). Moreover, the CheY proteins do not functionally overlap each other (25). Since B. burgdorferi survives primarily within the disparate environments of tick and mammalian hosts in nature, we assume that one or more cheY genes are required for viability in ticks and/or persistence in the murine host, and these different proteins may provide different functions within these hosts. CheY proteins were also reported to be important for host tissue colonization by several species of pathogenic bacteria, such as Helicobacter pylori and Vibrio cholerae (35–39). For our current studies, we intend to delineate the importance of the CheY2 response regulator for dissemination and persistence within both their arthropod and murine hosts. Our findings suggest that CheY2 provides different functions within these two hosts compared to our previous observations with a ΔcheY3 mutant, and we propose a model suggesting how CheY2 could operate in the Lyme disease spirochete.
RESULTS
Construction of two independent cheY2 mutants.
The B. burgdorferi cheY2 gene is located at the end of an operon consisting of cheW2-orf0566-cheA1-cheB2-orf0569-cheY2 genes (Fig. 1A) (26). This operon is transcribed by the σ70 subunit of RNA polymerase (25, 26). We have previously reported that cheY2 was not essential for motility or chemotaxis in vitro (25). However, those mutants were constructed in a high-passage, noninfectious strain and cannot be evaluated for their role in the enzootic life cycle of B. burgdorferi (in mouse or tick-mouse models of Lyme disease) (25). To assess the role of cheY2 in the B. burgdorferi pathogenic cycle, the gene was inactivated singly using a kanamycin or a streptomycin resistance cassette in two different wild-type clones, B31-A3 (A3) and B31-A3-K10 (K10), respectively. The two independent single mutants were confirmed by genotyping using PCR (data not shown) and immunoblotting using B. burgdorferi CheY2-specific polyclonal antiserum (Fig. 1B) (25).
FIG 1.
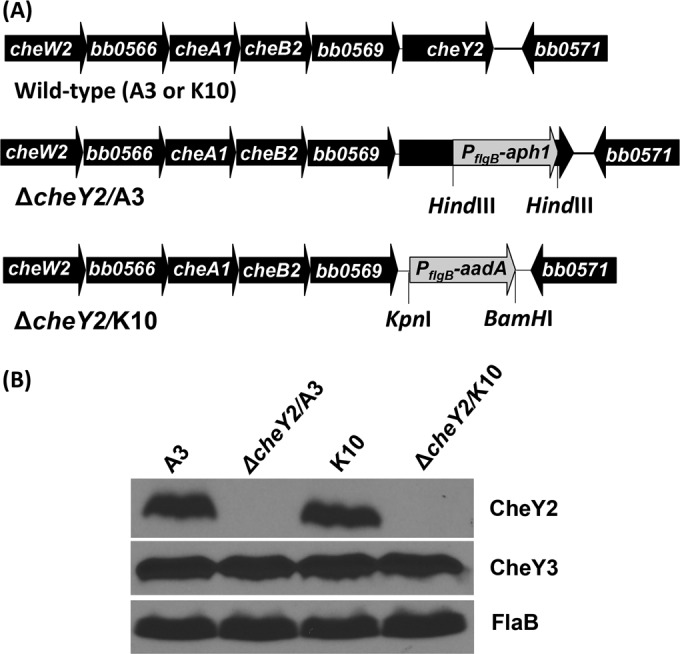
Inactivation and confirmation of two independent ΔcheY2 mutants. (A) The B. burgdorferi cheY2 operon consists of cheW2, bb0566, cheA1, cheB2, bb0569, and cheY2 genes (diagram is not to scale). The cheY2 gene was inactivated by inserting the PflgB-aph1 or PflgB-aadA cassette using allelic exchange mutagenesis in the B31-A3 (A3) or B31-A3-K10 (K10) wild-type background, respectively. (B) Inactivation of the cheY2 gene was confirmed by immunoblotting using whole-cell lysates from the indicated B. burgdorferi cells that were subjected to SDS-PAGE followed by probing with B. burgdorferi CheY2-specific polyclonal antisera. The antiserum reacted with the 10-kDa CheY2 protein in wild-type cells, as expected, and this protein is absent from the ΔcheY2 mutants (both clones). FlaB and CheY3 were used as loading controls.
Because the targeted cheY2 gene is located at the end of the operon, a polar effect on downstream gene expression is unlikely. Moreover, using the same protocol, we have created a cheY2 mutant in the high-passage-number avirulent strain that exhibits no phenotypic alterations (25). Additionally, to exclude the possibility of a secondary alteration elsewhere in the genome, we attempted to complement the mutants using an intact cheY2 gene, as described previously (6, 40). Even though our group has successfully generated numerous B. burgdorferi mutants in the past, our multiple attempts to complement the mutant in cis or in trans were unsuccessful, which is not uncommon due to the challenging nature of B. burgdorferi genetic manipulation. To best address this issue, we constructed two independent cheY2 mutants instead of complementing the mutant, as others have done in the past (7, 41–45). Since a complemented strain was not constructed or analyzed, the observed phenotypes (discussed below) could result from a secondary mutation elsewhere in the genome. Importantly, both mutant cell types showed similar phenotypes, suggesting that the characteristics observed with the current cheY2 mutants (ΔcheY2/A3 and ΔcheY2/K10) appear to be attributed to the cheY2 mutation. Furthermore, linear and circular endogenous plasmids of the mutants were verified by PCR, and all clones retained the plasmids possessed by the parental wild-type cells (data not shown) (40, 46).
In vitro chemotaxis and motility phenotype of ΔcheY2/A3 and ΔcheY2/K10 mutants.
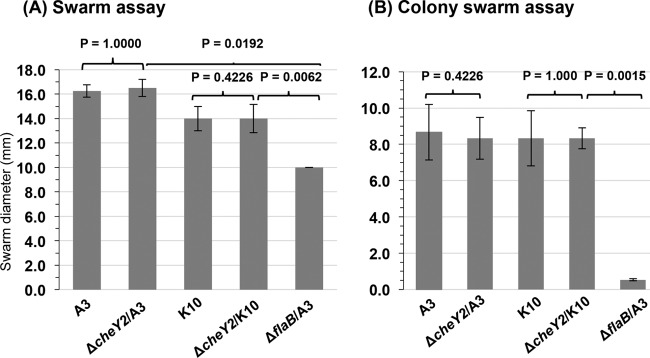
To determine if the two ΔcheY2 strains exhibit any altered motility or chemotaxis phenotype, we assessed both the bacterial swimming rate and swarm plate chemotaxis assays, as described previously (25, 47, 48). Both ΔcheY2 strains exhibit swimming patterns (run-pause/flex-reverse) and motility phenotypes that are indistinguishable from their respective wild-type parental cells (swimming velocity, 7.5 ± 1.1 μm/s versus 7.1 ± 0.9 μm/s by the wild-type clones; data not shown). Furthermore, swarm plate assays indicated that the chemotaxis phenotype of either ΔcheY2 strain was not significantly altered compared to the parental wild-type cells (Fig. 2A). A modified swarm plate assay was also performed to more accurately assess the chemotactic behavior of individual bacteria (12, 13). The swarm plate assay is a group event where millions of bacteria attempt to migrate from the initial site of inoculation (in a semisolid plate) as they respond to chemotactic materials and metabolize neighboring nutrients, resulting in a swarm ring. However, results obtained from such an assay may not accurately determine the chemotactic ability of an individual spirochete. Accordingly, we plated 20 to 50 B. burgdorferi cells in a semisolid plate (the same plates as those used for the swarm plate assays) to determine the chemotactic ability of individual cells by measuring their colony swarm size. Prolonged incubation of those plates produced colony sizes that were not statistically different from those of the parental wild-type cells (P = 0.38 and P = 0.27, respectively) (Fig. 2B). Together, these data are the same as those reported previously, which suggest that a mutation in cheY2 has little to no effect on motility or chemotaxis in vitro (25).
FIG 2.
B. burgdorferi ΔcheY2 mutants are not deficient in chemotaxis in vitro. (A) Swarm plate assays using 106 B. burgdorferi cells from the indicated strains were spotted onto 0.35% soft-agarose plates. Plates were incubated for 6 days, and the diameter of each clone was measured (in millimeters). A nonmotile ΔflaB mutant was used as a control (7). Bars represent means ± standard deviations from 3 independent plates. (B) B. burgdorferi wild-type and mutant cells were plated (10 to 50 cells per plate) separately in 0.35% soft agarose containing BSK growth medium that was diluted 10× with PBS and incubated for 4 weeks. Numbers are indicative of the means ± standard deviations of the means from at least 12 individual colonies (per clone per assay). Statistical analyses were performed using Student's paired t test to determine the P values between the wild type and mutants. A P value of <0.05 between strains was considered significant.
ΔcheY2/A3 and ΔcheY2/K10 mutants are severely attenuated in their abilities to persist or disseminate within C3H/HeN mice.
To determine if the ΔcheY2 strains are able to establish an infection in the mammalian host, we intradermally inoculated 5 × 106 in vitro-grown B. burgdorferi wild-type and ΔcheY2 mutant bacteria in separate groups of mice (n = 4). Immediately before the injections, we verified by PCR that the endogenous B. burgdorferi plasmids are retained in the wild-type clones and their respective isogenic mutants (not shown). Four weeks postinoculation, the mice were euthanized and bacterial persistence was determined by the growth of the spirochetes from the isolated mouse tissues (ear skin, ankle joints, and urinary bladder) incubated in Barbour-Stoenner-Kelly II (BSK-II) growth medium. While the wild-type spirochetes were detected from all mouse tissues assessed, mutant B. burgdorferi (both clones) was detected in only a single bladder tissue out of 12 total tissue samples from four mice (Table 1). To validate the bacterial outgrowth analysis, DNA was isolated from each of those mouse tissues except the bladder tissue, followed by PCR to detect B. burgdorferi flaB DNA. Our PCR data indicate that DNAs from all of the mouse tissue samples were able to detect wild-type bacterial genomes (both clones), whereas none of the tissue samples were positive for the mutant genomes (both clones) (Table 1 and data not shown). These results suggest that CheY2 is crucial for establishing a disseminated infection in the murine host.
TABLE 1.
ΔcheY2 spirochetes are severely attenuated in murine infection via regrowth analysesa
| B. burgdorferi clone | Dose per mouse | No. of mouse tissues colonized/no. analyzed | No. of mice infected/no. analyzed |
|---|---|---|---|
| WT (A3) | 5 × 106 | 11/12 | 4/4 |
| ΔcheY2/A3 | 5 × 106 | 01/12 | 1/4b |
| WT (K10) | 5 × 106 | 10/12 | 4/4 |
| ΔcheY2/K10 | 5 × 106 | 01/12 | 1/4b |
C3H/HeN mice were injected intradermally/subcutaneously using the indicated in vitro-grown spirochete clones. Mice were euthanized 4 weeks post injection, and infectivity was determined by reisolation of B. burgdorferi from the tissue samples (ear, joint, and bladder from each mouse). Doses shown are the actual numbers of spirochetes injected in each mouse.
ΔcheY2/A3 or ΔcheY2/K10 mutant was detected in only one bladder tissue of a mouse.
To better delineate whether the attenuated ΔcheY2 strains are able to survive at the injection site skin tissue of the mice and/or disseminate from the skin to the distant tissues or whether the bacteria were efficiently cleared by the immune responses, groups of mice were inoculated with wild-type and mutant B. burgdorferi and subsequently euthanized at various times (24 h and 1 and 2 weeks postinjection). Tissue samples from each mouse were processed to recover live spirochetes from the culture as well as for bacterial genome quantification by quantitative PCR (qPCR). As shown in Table 2, the wild-type and ΔcheY2 cells were reisolated from all injection site skin tissues when the mice were euthanized 24 h postinjection. When the mice were euthanized 1 week postinjection, wild-type cells were recovered from the injection site as well as from the distant tissues (ear or bladder), suggesting bacterial dissemination. Alternatively, ΔcheY2 cells were not recovered from the injection site of any mouse tissues assessed at ≥1 week postinfection and were only recovered from a single distant ear skin tissue site out of 12 tissues processed from four mice (Table 2). Moreover, when the mice were euthanized 2 weeks postinjection, wild-type spirochetes were found to be disseminated to the distant tissues in all four mice. However, the mutant spirochetes were not able to be reisolated from any mouse tissues (Table 2).
TABLE 2.
B. burgdorferi ΔcheY2 mutant is attenuated for persistent infection and dissemination within mice via regrowth analysesa
| B. burgdorferi clone and euthanization time postinjection | No. cultures positive/no. tested |
No. of mice infected/no. tested | |||
|---|---|---|---|---|---|
| Injection site skin | Distant skin | Bladder | Joint | ||
| WT (A3) | |||||
| 24 h | 4/4 | 0/4 | 0/4 | 0/4 | 4/4 |
| 1 week | 2/4 | 2/4 | 1/4 | 0/4 | 3/4 |
| 2 week | 0/4 | 4/4 | 4/4 | 4/4 | 4/4 |
| ΔcheY2/A3 | |||||
| 24 h | 3/4b | 0/4 | 0/4 | 0/4 | 3/4 |
| 1 week | 1/4 | 0/4 | 0/4 | 0/4 | 1/4 |
| 2 week | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
C3H/HeN mice were injected intradermally/subcutaneously (into ear skin for 24 h postinjection or dorsal skin for 1 to 2 weeks postinjection) using the indicated in vitro-grown spirochete clones. Approximately 5 × 106 spirochetes were inoculated per mouse as verified by CFU. Mice were euthanized at the indicated time points, and infectivity was determined by reisolation of B. burgdorferi from the tissue samples.
An injection site skin tissue could have been processed mistakenly.
To determine if mutant spirochetes were disseminated to the distant tissues but were unable to be accurately detected by regrowth assays and/or were cleared/reduced by the host immune responses, parallel mouse tissue samples were processed for DNA isolation followed by B. burgdorferi genome quantitation using qPCR (Fig. 3). At 24 h postinjection, the numbers of ΔcheY2 genomes in the skin injection site were more than those detected for wild-type bacteria, suggesting that our inoculum of ΔcheY2 mutant was actually greater than that of wild-type bacteria. However, by 1 week postinjection the numbers of ΔcheY2 genomes at the injection site were significantly reduced compared to the original inoculum at 24 h and were over 100-fold less than those of the wild-type bacterial genomes at 1 week postinjection at the inoculation site (P = 0.006), suggesting that only wild-type B. burgdorferi multiplied. Additionally, wild-type bacteria were detected at distant skin sites at 1 week postinjection, whereas no ΔcheY2 genomes were detected at the distant site at this time, again suggesting that this mutant is unable to efficiently disseminate from the inoculation site (Fig. 3). Finally, at 2 weeks postinjection, ΔcheY2 genomes were barely detected in the skin injection site and were never detected in any distant tissue assessed, whereas wild-type genomes were detected in both the local inoculation site and all distant tissues assessed (Fig. 3). Overall, these qPCR data match fairly well with the bacterial reisolation shown in Table 2, although the qPCR appears to be more sensitive in detecting the ΔcheY2 bacteria, potentially due to a decreased ability of these mutants to migrate out of the collected tissues for the tube-based regrowth assays. Importantly, these data suggest that the ΔcheY2 bacteria are attenuated both in their ability to establish a persistent infection at the skin inoculation site as well as their ability to disseminate to distant target tissues and/or evade immune clearance at those sites.
FIG 3.
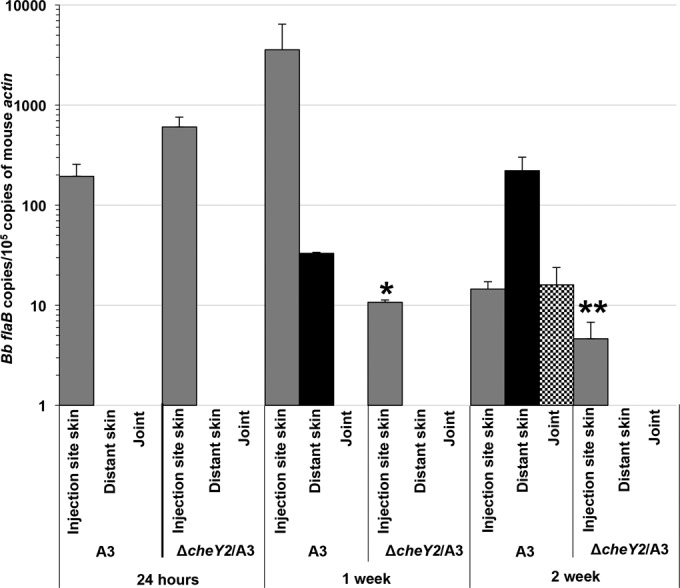
Relative number of B. burgdorferi genomes in mouse tissues as determined by qPCR. Groups of C3H/HeN mice (n = 4 per clone per time point) were injected with 5 × 106 wild-type or ΔcheY2 mutant bacteria via the intradermal dorsal or ear skin route (24 h). Mice were euthanized at the indicated time points. Mouse tissues were processed for DNA purification followed by qPCR. The number of B. burgdorferi flaB copies was normalized to the number of mouse β-actin copies. Each bar represents means of data from four mouse tissues ± standard deviations of the means. Bb, B. burgdorferi. *, P < 0.006; **, P < 0.01. A bar is not shown for some tissues because bacterial genome was not detected.
ΔcheY2 mutant spirochetes are unable to infect mice by tick bite.
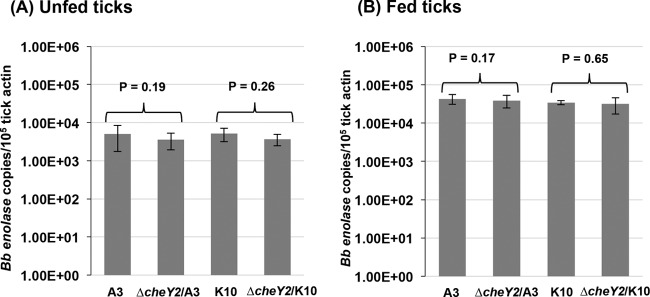
The B. burgdorferi natural enzootic cycle requires that the bacteria establish persistent infection within both tick and vertebrate hosts. Because the ΔcheY2 mutants were severely attenuated in mice by needle inoculation, it is impractical to assess their ability to infect naive ticks by natural acquisition (i.e., via feeding on infected mice, followed by determination of spirochete transmission [tick to mouse]) (6, 7, 48). To address the possibility that the ΔcheY2 strain can establish infection in naive mice via tick bite, Ixodes scapularis nymphs were artificially infected by immersion with either the wild type or isogenic ΔcheY2 mutants before being encapsulated and allowed to feed on naive C3H/HeN mice (15 nymphs per mouse, 3 mice per strain per assay). Seven days after ticks dropped off the mice, ticks were surface sterilized and then squashed individually to isolate genomic DNA to determine spirochete densities using enolase gene-specific qPCR, as described previously (Fig. 4) (6). The ΔcheY2 strain displayed densities similar to those of wild-type bacteria in ticks both before (i.e., unfed) and after (i.e., fed) feeding on mice (P = 0.19 and P = 0.26, respectively, in unfed ticks; P = 0.17 and P = 0.65, respectively, in fed ticks), suggesting they persist within fed and unfed ticks similar to wild-type bacteria. To determine if infected ticks transmit the ΔcheY2 spirochetes in mice and establish persistent infection, mouse tissues were collected either 48 h or 2 weeks after tick repletion. Bacterial reisolation from both the tick-feeding site and distant tissues indicate that no assessed tissues were positive for the ΔcheY2 strain, whereas all tissues from mice fed on by the wild-type infected nymphs demonstrated bacterial growth (Table 3). qPCR analyses performed on mouse tissues from parallel studies produced results similar to those of the regrowth studies (not shown). Together, these results suggest that ΔcheY2 mutants are not able to establish an infection in mice via tick bite, even though the mutants survived normally in nymphal ticks. Finally, these studies observed no differences between mutants generated on both genetic clones (ΔcheY2/A3 and ΔcheY2/K10) in all in vitro or in vivo assays.
FIG 4.
Viability of ΔcheY2 mutants in unfed and fed nymphs. Naive nymphs were artificially infected as described in Materials and Methods. A subset of immersed nymphs was allowed to feed on separate naive C3H/HeN mice. Seven days after feeding, fed (B) and unfed (A) ticks were processed for PCR analysis to determine spirochete-positive ticks, and subsequently qPCR was performed to determine the number of spirochete genomes using enolase gene-specific primers. Results shown are means ± standard deviations of the means from at least five spirochete-positive ticks per clone per assay. Representative data from two independent studies are shown here. A P value of <0.05 between strains is considered significant.
TABLE 3.
ΔcheY2 spirochetes were not able to infect naive C3H/HeN mice by tick bitea
| Nymphs infected with B. burgdorferi clone | No. of mice infected by tick bite/no. challenged |
|---|---|
| WT (A3) | 5/6 |
| ΔcheY2/A3 | 0/6 |
| WT (K10) | 5/6 |
| ΔcheY2/K10 | 0/6 |
Nymphal ticks were infected with the indicated B. burgdorferi strains by immersion in liquid cultures and were subsequently allowed to feed on naive mice. Fifteen infected encapsulated nymphs per mouse were allowed to feed to repletion. Two weeks (or 48 h; data not shown) after feeding, mice were euthanized to determine bacterial growth from the tissues.
DISCUSSION
Chemotaxis signal transduction systems typically govern the rotation of flagellar motors, which allows a bacterium to respond and survive in a challenging environment. Even though more than 50% of all sequenced prokaryotic genomes contain at least one chemotaxis system, many of those genomes possess multiple sets of chemotaxis genes, and at least one of them is dedicated to controlling motility (22, 49–51). In most bacterial species that possess multiple sets of chemotaxis genes, very little is known about what role these additional chemotaxis genes perform. However, there are a few instances in which multiple chemotaxis-like signaling systems appear to have very different roles in the same species. For example, Myxococcus xanthus possesses eight sets of chemotaxis-like genes. Three of them control the frequency of gliding motility reversals, exopolysaccharide (EPS) synthesis, and entry into the myxospore developmental pathway (52–54). These and other recent reports suggest that some chemotaxis-like signaling pathways control other cellular nonchemotactic processes. However, in each of those cases it is not clear whether the effect on transcription is direct or indirect or if those chemotaxis-like proteins possess a DNA binding domain (52, 54–58). Importantly, most typical CheY chemotaxis response regulatory proteins lack the DNA-binding domain; thus, these proteins generally do not modulate gene expression. These CheY proteins instead bind to a flagellar protein (FliM/FliN) to alter cellular swimming behavior (22, 59, 60). Based on this model, similar cheY2 mutant cells would be expected to exhibit altered motility and deficient chemotaxis phenotypes similar to that observed in many bacterial cheY mutants, including the B. burgdorferi cheY3 mutants (13, 25, 50, 56). However, the cheY2 mutants produced in our study exhibited in vitro motility and chemotaxis phenotypes that are indistinguishable from the wild-type parental spirochetes (Fig. 2). This lack of an altered motility/chemotaxis phenotype is not without precedent. The Vibrio cholerae genome was reported to possess five CheY proteins, and only two of them (CheY3 and CheY4) are required for motility/chemotaxis in vitro; the functions of the other CheY proteins are unknown (36, 51). Previously, any cheY mutant that was observed to exhibit an altered motility/chemotaxis phenotype was also found to have deficiencies in virulence; examples include a B. burgdorferi cheY3 mutant and V. cholerae cheY3 and cheY4 mutants (13, 35, 36, 51). Conversely, when a cheY mutation does not alter the motility/chemotaxis phenotype in vitro, those cheY mutants exhibit wild-type abilities in colonizing animal hosts (35, 36, 51). Surprisingly, our observations with B. burgdorferi cheY2 mutants appear to be an exception, as these ΔcheY2 cells appeared normal in vitro but were significantly attenuated in murine infection (Tables 1 and 2 and Fig. 3).
Interestingly, nymphal ticks colonized with ΔcheY2 bacteria were not able to establish persistent infection in naive mice, even though the densities of mutant spirochetes were similar to wild-type levels in both fed and unfed ticks that were infected by immersion, suggesting that the CheY2 defect was only apparent either during the transmission from the tick host into mouse tissues and/or when these bacteria were within murine host tissues (Table 3 and Fig. 4). This persistence phenotype within the tick is different from that recently observed by a nonmotile ΔmotB strain possessing an intact chemotaxis system, in that the ΔmotB bacteria maintained wild-type numbers in the unfed tick but were rapidly reduced in the tick after taking a blood meal (6). It was speculated that the nonmotile ΔmotB bacteria were unable to generate certain interactions within the tick environment that allows protection against bactericidal factors present in the ingested blood meal (10). Similarly, a ΔcheY3 spirochete that could achieve normal swimming speeds but was unable to reverse directions both in vitro and in vivo was also shown to possess normal numbers in unfed ticks, but the numbers were reduced in fed ticks (13). B. burgdorferi cheY2 expression is reported to be elevated in fed ticks compared to a mammalian-host adapted condition, indicating this pathway is active subsequent to tick feeding (61). Thus, the chemotaxis/motility properties guided by CheY2 should be sufficient to allow wild-type spirochetes to either migrate to a desired environment within the tick and/or to resist clearance by immune mediators present within the blood materials acquired during feeding. This is also supported by studies demonstrating that even B. burgdorferi strains with limited motility can survive efficiently in fed ticks (our unpublished observations). This finding for host-specific motility/chemotaxis deficiencies is unprecedented, and this paradigm may provide a model system for better understanding the unique virulence mechanisms required to persist within vertebrate versus arthropod hosts.
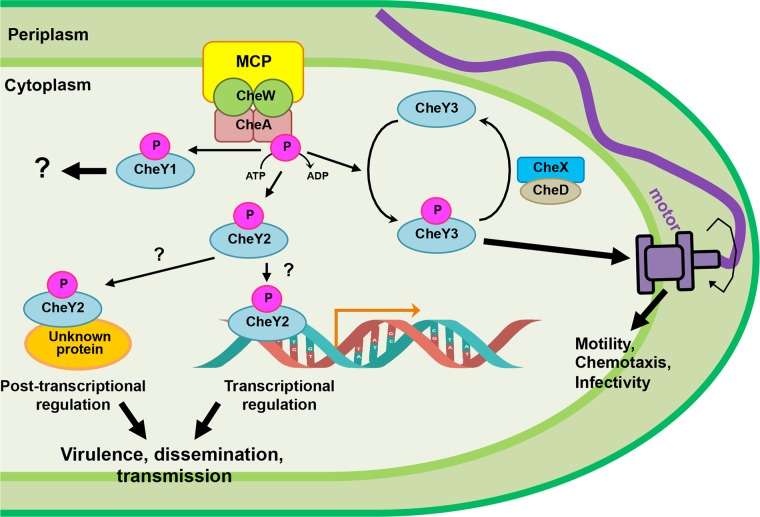
It is currently unclear as to why ΔcheY2 spirochetes show defects in their abilities to disseminate and establish infection in mouse tissues either by tick bite or needle inoculation. Since ΔcheY2 bacteria demonstrate normal motility/chemotaxis in vitro, they appear to be capable of traversing the tight junctions of the tick midgut epithelial cells that is required for transmission of B. burgdorferi from the tick midgut into murine skin, making it less likely that the deficiencies observed in murine infections were due to a defect in transmission, although this is certainly possible. B. burgdorferi cheY2 expression is elevated in fed ticks despite the fact that the operon is transcribed by the σ70 promoter, suggesting this pathway is active subsequent to feeding (61). These findings also suggest that CheY2 possesses some function(s) that is atypical for a chemotaxis response regulator, even though this response regulatory protein contains all of the conserved amino acid residues of CheY and is being phosphorylated by the histidine kinases (25, 26). This would not be unprecedented, as chemotaxis-like signaling systems have been described to perform nonchemotactic cellular functions in several species of bacteria (52–56). We could surmise that CheY2 performs a role other than that of the typical chemotaxis signaling system (Fig. 5), such as binding to an operon promoter or a protein to modulate some virulence determinants in B. burgdorferi (e.g., CheY exerts transcriptional or posttranscriptional regulation). Consequently, a mutation in cheY2 would mediate altered expression of those putative virulence-associated genes, resulting in attenuated mouse infection or dissemination (Fig. 5). Alternatively, since B. burgdorferi is found only within vertebrate or tick hosts during their natural enzootic cycles, the in vitro assay systems used to observe motility/chemotaxis phenotypes likely do not accurately reflect the in vivo environment in either ticks or vertebrate hosts. As such, we realize that the cheY2 mutant may exhibit the altered motility/chemotaxis phenotype only within its native hosts. Ideally, generation of a fluorescently labeled ΔcheY2 clone will allow us to delineate any apparent defects within murine tissues using our intravital microscopy models, as we recently reported for the ΔmotB and ΔcheY3 mutants (6, 13). Regardless, a more detailed analysis of these CheY2-mediated signaling pathways will be required to unravel the complex regulation this distinctive organism performs during its enzootic infection cycle.
FIG 5.
Working model of CheY2 functions in B. burgdorferi enzootic cycle. A simplistic chemotaxis signaling pathway of B. burgdorferi is depicted. Based on our findings, we propose that after phosphorylation by the histidine kinase CheA, CheY3-P binds to the flagellar switch proteins to alter bacterial swimming behavior. Consequently, cheY3-mediated chemotaxis is found to affect dissemination and viability of B. burgdorferi in mice and ticks (13). Dephosphorylation of CheY3-P is mediated by CheX-CheD (12). Our current data suggest that CheY2 does not affect motility or chemotaxis despite having all domains/conserved amino acid residues seen in a classical CheY protein (25). Because the mice are not being infected or partially infected by needle inoculation or tick bite, we propose that CheY2, after being phosphorylated by CheA, acts as a virulence determinant in B. burgdorferi. Instead of controlling motility, this CheY2-P may act as a transcriptional or posttranscriptional regulator to modulate B. burgdorferi gene expression or a protein's function/activity. Alternatively, its elevated expression in fed ticks may alter the bacterial or host gene functions in order for the spirochete to transmit from the vector to the murine host. The function of cheY1 in the enzootic cycle is unknown. As described in the text, B. burgdorferi possesses five MCPs, three CheW and two CheA proteins.
MATERIALS AND METHODS
Ethics statement.
East Carolina University is accredited by the International Association for Assessment and Accreditation of Laboratory Animal Care. Protocols for tick and animal experimentations were approved by the East Carolina University Animal Care and Use Committee.
Bacterial strains and growth conditions.
Low-passage, virulent B. burgdorferi strains B31-A3 and B31-A3-K10 (a kind gift from R. Rego and P. Rosa, Rocky Mountain Laboratories, NIH) were used as wild-type (WT) clones in this study (62, 63). B31-A3-K10 is a derivative of B31-A3 in which the bbe002 gene located in linear plasmid 25 (lp25) was inactivated using a PflaB-aph1 (Kanr) cassette to increase transformation frequency (63). The genome of the parent strain B31 is known to contain 12 linear and 9 circular plasmids, for a total of 21 plasmids, in addition to a 960-kbp linear chromosome (24, 64). The WT clones used in this study retain all endogenous plasmids except circular plasmid 9 (cp9). Construction of the cheY2 mutants in B. burgdorferi strains B31-A3 (referred to as A3) and B31-A3-K10 (K10) are described below. B. burgdorferi cells were grown in liquid Barbour-Stoenner-Kelly II (BSK-II) medium, and cells were plated using plating BSK (P-BSK), which was prepared using 0.5% agarose. Cells were grown at 35°C in a 2.5% CO2 incubator as previously described (65). Escherichia coli strains were cultivated in Luria-Bertani broth (1% tryptone, 1% NaCl, 0.5% yeast extract). When required, culture and plating medium were supplemented with appropriate antibiotics at the following concentrations: 200 μg/ml kanamycin, 100 μg/ml streptomycin or spectinomycin, or 100 μg/ml ampicillin.
Construction of cheY2 mutants.
Construction of the cheY2 inactivation plasmids, electroporation, and plating conditions were described previously (7, 8). To construct a cheY2 mutant in the B31-A3 background, the cheY2 gene (gene locus number bb0570) and flanking DNA was first amplified by PCR from the chromosomal DNA of B. burgdorferi strain B31-A3 using primers CheY2-KO-F (TCTGCTAGGTTTCAAAATAT) and CheY2-KO-R (TGGACTTACCCT TTACATAG), and the product obtained was cloned into plasmid pGEM-T Easy (Promega Inc.). The cheY2 gene was inactivated using a kanamycin resistance cassette (PflgB-aph1), which was inserted at the HindIII sites located within cheY2 (66). DNA containing cheY2-PflgB-aph1 was linearized by restriction digestion to remove the ampicillin marker of the vector and electroporated into competent B31-A3 cells to obtain mutants. Kanamycin-resistant transformants were screened by PCR for proper recombination of the cheY2 inactivation cassette in the genome. Western blot analysis was used to confirm the inactivation of cheY2 using B. burgdorferi CheY2-specific antisera as described below (25).
To construct the cheY2 mutant in B31-A3-K10, the 5′ (968 bp)- and 3′ (919 bp)-flanking regions of the cheY2 gene were amplified by PCR from chromosomal DNA of B. burgdorferi strain B31-A3-K10 using primers CheY2mut-F (CGGATACATCAAAAGTTATAGTAAAAGATG) and CheY2mut.KpnI-R (GGTACCTAATTTTCTCCTAAAACCCT) as well as CheY2mut.BamHI-F (GGATCCGTTTATATTTGCAATTAATTTGT) and CheY2mut-R (GGTGGAGGAAGAGTTGCAAG). A spectinomycin-streptomycin resistance cassette fused with B. burgdorferi flgB promoter (PflgB-aadA) was similarly PCR amplified from a pKFSS1 shuttle vector using primers PflgBStrep.KpnI-F (GGTACCTACCCGAGCTTCA) and PflgBStrep.BamHI-R (GGATCCAAGCTTGACGTCATTA) (67). These three pieces of DNA fragments were individually cloned into plasmid pGEM-T Easy (Promega Inc.). The pieces were cloned together using restriction digestion. First, the 5′-flanking regions of cheY2 and PflgB-aadA were cloned together using KpnI and SacI restriction digestion, yielding plasmid Teasy::5′Y2-PflgB-aadA. The 3′-flaking region of the cheY2 fragment then was cloned into Teasy::5′-cheY2-PflgB-aadA using BamHI and SacI restriction digestion, yielding plasmid Teasy::cheY2_KO-PflgB-aadA. Competent B31-A3-K10 cells were electroporated with cheY2_KO_PflgB-aadA DNA as described above (8, 65). The transformants were selected with streptomycin. The streptomycin-resistant transformants were confirmed by PCR and Western blotting as described above.
SDS-PAGE and Western blot analysis.
SDS-PAGE and immunoblotting with an enhanced chemiluminescent detection method were carried out as reported previously (GE Health Inc.) (6). Protein concentrations of cell lysates were determined with a Bio-Rad protein assay kit using bovine serum albumin as a standard. A volume of cell lysate equivalent to 10 μg of protein was subjected to SDS-PAGE and immunoblotting using B. burgdorferi CheY2-, CheY3-, and FlaB-specific antibodies (25).
Microscopy and computer-assisted motion analysis.
Live B. burgdorferi cells were observed under a Zeiss Axio Imager M1 dark-field microscope connected to an AxioCam digital camera. Exponentially growing cells (2 × 107 to 3 × 107 cells/ml) were mixed with 0.5% 400-mesh methylcellulose (Sigma-Aldrich Co.) and video recorded at room temperature (23°C). Cell swimming behavior and velocity were determined using AxioVision software (47). Results are expressed as velocity (mean distance, in micrometers, traveled by a given strain per second). At least 10 cells from each strain were analyzed (25, 47).
Chemotaxis assays.
Swarm plate chemotaxis assays were performed using our established protocol described previously (65). Approximately 106 cells in a 5-μl volume were spotted onto a 0.35% agarose plate containing plating BSK medium diluted 1:10 in Dulbecco's phosphate-buffered saline. Since B. burgdorferi is a slow-growing organism (8- to 12-h generation time), plates were incubated for 6 days at 35°C in a 2.5% CO2 humidified incubator (6, 7). At least three independent assays were performed. The chemotactic swarming ability of individual bacterial colonies of each strain was determined by plating no more than 50 cells into a 95-mm by 15-mm petri dish containing semisolid P-BSK diluted as described above (12, 13). Plates were incubated for 4 weeks, at which time colony diameters were measured. At least 12 colony diameters were measured for each strain in each assay. The paired Student t test was used to compare wild-type and mutant cell swarm diameters.
Mouse infection studies using needle-injected B. burgdorferi.
Six- to seven-week-old C3H/HeN mice (Charles River Laboratories) were used for infection studies as previously described (41, 48, 68–70). In order to determine the infectious ability of the spirochetes, separate groups of mice were injected intradermally/subcutaneously in ear or dorsal/back skin tissue with in vitro-grown WT A3, WT K10, ΔcheY2/A3, or ΔcheY2/K10 strain at the indicated doses. Spirochetal numbers were determined using a Petroff-Hausser chamber and verified via CFU by plating. Mice were euthanized 24 h, 1 week, 2 weeks, or 4 weeks after injection, and then multiple tissues were harvested aseptically from each mouse. The injection site skin tissue was cut into two equal portions. One part of the skin, as well as a joint, and ear tissue from each mouse (n = 4) were processed for DNA isolation followed by qPCR to determine bacterial density using B. burgdorferi flaB gene-specific primers (6, 7, 40). DNA levels were assessed by amplification of the actin gene for the mouse and flaB gene for B. burgdorferi. Copy numbers for mouse and B. burgdorferi genomes were evaluated by extrapolating to standard curves devised by Bio-Rad CFX Manager 3.1. Final B. burgdorferi numbers were calculated by normalizing B. burgdorferi genomes to 105 mouse genomes. The results are expressed as means ± standard deviations from four tissues collected from four mice. Statistical analyses were performed using Student's paired t test to calculate the significance of the normalized values between wild-type and mutant samples. A P value of 0.05 between samples was considered significant. The other half of the injection site skin, the second joint tissue, and the bladder from each mouse were placed in BSK-II broth for up to 35 days to allow bacterial growth, which is the direct determination of the ability of spirochetes to infect mice and disseminate throughout the body. The presence of spirochetes in the growth medium was determined by dark-field microscopy and is referred to as regrowth, outgrowth, or reisolation.
Assessment of spirochete transmission to mice by encapsulated nymphs.
Transmission of spirochetes from Ixodes scapularis ticks to C3H/HeN mice was assessed using artificially infected nymphs as described previously (6, 48). Naive nymphal ticks were experimentally infected by immersion with exponential-phase (2 to 3 × 107 cells/ml) B. burgdorferi clones, and then we washed the immersed ticks with sterile distilled H2O to remove the surface-attached spirochetes. The ticks were then kept in a humidified chamber for approximately 24 h before allowing them to feed on naive mice (7, 48, 71). Mice were anesthetized, and 15 nymphs were confined to a capsule affixed to the shaved back of a naive C3H/HeN mouse (n = 3 per strain per assay). The ticks were allowed to feed to repletion (3 to 5 days) and then collected from the capsules (6, 72). Fed and unfed (immersed) ticks were surface sterilized using 3% H2O2 and 70% ethanol, individually crushed on day 7 postrepletion to isolate genomic DNA using a DNeasy blood and tissue kit according to the manufacturer's instructions (Qiagen Inc.). The DNA from each tick was utilized for PCR to determine spirochete-positive ticks using B. burgdorferi flaB gene-specific primers. Subsequently, each sample of spirochete-positive tick (n = 5) DNA was used to determine bacterial densities by qPCR using B. burgdorferi enolase gene-specific primers as described previously (40, 73, 74). Numbers of the copies of the B. burgdorferi enolase gene per tick were extrapolated from a standard curve generated using a known amount of plasmid DNA containing the enolase gene as the template and normalized to the tick actin gene. The results are expressed as means ± standard deviations from at least 5 sets of spirochete-positive tick data per clone per assay. Statistical analyses were performed using Student's paired t test to calculate the significance of the normalized values between wild-type and mutant samples. A P value of 0.05 between samples was considered significant.
Tick-fed mice were euthanized at 48 h or 2 weeks postrepletion. A section of skin comprising the tick-feeding site was excised, rinsed in 70% isopropanol, and cut into equal portions. Parts of the tick-bite site skin, ear, bladder, and joint tissues were cultured separately in BSK-II medium for up to 35 days to determine bacterial outgrowth, and the remaining tissues were processed for PCR to detect B. burgdorferi DNA using enolase gene-specific primers (6, 7, 40).
ACKNOWLEDGMENTS
We thank P. Rosa and R. Rego for sharing reagents and E. Novak for help.
This research was supported by grants from the National Institute of Allergy and Infectious Diseases (1R21AI113014) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (1R01AR060834).
REFERENCES
- 1.Mead PS. 2015. Epidemiology of Lyme disease. Infect Dis Clin North Am 29:187–210. doi: 10.1016/j.idc.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Kuehn BM. 2013. CDC estimates 300,000 US cases of Lyme disease annually. JAMA 310:1110. doi: 10.1001/jama.2013.278331. [DOI] [PubMed] [Google Scholar]
- 3.Groshong AM, Blevins JS. 2014. Insights into the biology of Borrelia burgdorferi gained through the application of molecular genetics. Adv Appl Microbiol 86:41–143. doi: 10.1016/B978-0-12-800262-9.00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat Rev Microbiol 10:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisson D, Drecktrah D, Eggers CH, Samuels DS. 2012. Genetics of Borrelia burgdorferi. Annu Rev Genetics 416:513–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sultan SZ, Sekar P, Zhao X, Manne A, Liu J, Wooten RM, Motaleb MA. 2015. Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infect Immun 83:1765–1777. doi: 10.1128/IAI.03097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sultan SZ, Manne A, Stewart PE, Bestor A, Rosa PA, Charon NW, Motaleb MA. 2013. Motility is crucial for the infectious life cycle of Borrelia burgdorferi. Infect Immun 81:2012–2021. doi: 10.1128/IAI.01228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci U S A 97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudryashev M, Cyrklaff M, Baumeister W, Simon MM, Wallich R, Frischknecht F. 2009. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol Microbiol 71:1415–1434. doi: 10.1111/j.1365-2958.2009.06613.x. [DOI] [PubMed] [Google Scholar]
- 10.Motaleb MA, Liu J, Wooten RM. 2015. Spirochetal motility and chemotaxis in the natural enzootic cycle and development of Lyme disease. Curr Opin Microbiol 28:106–113. doi: 10.1016/j.mib.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sze CW, Zhang K, Kariu T, Pal U, Li C. 2012. Borrelia burgdorferi needs chemotaxis to establish infection in mammals and to accomplish its enzootic cycle. Infect Immun 80:2485–2492. doi: 10.1128/IAI.00145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon KH, Hobbs G, Motaleb MA. 2016. Borrelia burgdorferi CheD promotes various functions in chemotaxis and pathogenic life cycle of the spirochete. Infect Immun 84:1743–1752. doi: 10.1128/IAI.01347-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novak EA, Sekar P, Xu H, Moon KH, Manne A, Wooten RM, Motaleb MA. 20 May 2016. The Borrelia burgdorferi CheY3 response regulator is essential for chemotaxis and completion of its natural infection cycle. Cell Microbiol doi: 10.1111/cmi.12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 16.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 17.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol 13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar MK, Paul K, Blair D. 2010. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc Natl Acad Sci U S A 107:9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silversmith RE, Guanga GP, Betts L, Chu C, Zhao R, Bourret RB. 2003. CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89. J Bacteriol 185:1495–1502. doi: 10.1128/JB.185.5.1495-1502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao R, Collins EJ, Bourret RB, Silversmith RE. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol 9:570–575. [DOI] [PubMed] [Google Scholar]
- 21.Boesch KC, Silversmith RE, Bourret RB. 2000. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. J Bacteriol 182:3544–3552. doi: 10.1128/JB.182.12.3544-3552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charon NW, Cockburn A, Li C, Liu J, Miller KA, Miller MR, Motaleb MA, Wolgemuth CW. 2012. The unique paradigm of spirochete motility and chemotaxis. Annu Rev Microbiol 66:349–370. doi: 10.1146/annurev-micro-092611-150145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 25.Motaleb MA, Sultan SZ, Miller MR, Li C, Charon NW. 2011. CheY3 of Borrelia burgdorferi is the key response regulator essential for chemotaxis and forms a long-lived phosphorylated intermediate. J Bacteriol 193:3332–3341. doi: 10.1128/JB.00362-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC, Charon NW. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc Natl Acad Sci U S A 99:6169–6174. doi: 10.1073/pnas.092010499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang K, Liu J, Tu Y, Xu H, Charon NW, Li C. 2012. Two CheW coupling proteins are essential in a chemosensory pathway of Borrelia burgdorferi. Mol Microbiol 85:782–794. doi: 10.1111/j.1365-2958.2012.08139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H, Raddi G, Liu J, Charon NW, Li C. 2011. Chemoreceptors and flagellar motors are subterminally located in close proximity at the two cell poles in spirochetes. J Bacteriol 193:2652–2656. doi: 10.1128/JB.01530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang K, Liu J, Charon NW, Li C. 2015. Hypothetical protein BB0569 is essential for chemotaxis of the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol 198:664–672. doi: 10.1128/JB.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pazy Y, Motaleb MA, Guarnieri MT, Charon NW, Zhao R, Silversmith RE. 2010. Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate. Proc Natl Acad Sci U S A 107:1924–1929. doi: 10.1073/pnas.0911185107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Motaleb MA, Miller MR, Li C, Bakker RG, Goldstein SF, Silversmith RE, Bourret RB, Charon NW. 2005. CheX is a phosphorylated CheY phosphatase essential for Borrelia burgdorferi chemotaxis. J Bacteriol 187:7963–7969. doi: 10.1128/JB.187.23.7963-7969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charon NW, Goldstein SF. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu Rev Genetics 36:47–73. doi: 10.1146/annurev.genet.36.041602.134359. [DOI] [PubMed] [Google Scholar]
- 34.Fosnaugh K, Greenberg EP. 1988. Motility and chemotaxis of Spirochaeta aurantia: computer-assisted motion analysis. J Bacteriol 170:1678–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butler SM, Camilli A. 2005. Going against the grain: chemotaxis and infection in Vibrio cholerae. Nat Rev Microbiol 3:611–620. doi: 10.1038/nrmicro1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler SM, Camilli A. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc Natl Acad Sci U S A 101:5018–5023. doi: 10.1073/pnas.0308052101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGee DJ, Langford ML, Watson EL, Carter JE, Chen YT, Ottemann KM. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun 73:1820–1827. doi: 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect Immun 73:803–811. doi: 10.1128/IAI.73.2.803-811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the Borrelia burgdorferi cyclic-di-GMP binding protein PlzA reveals a role in motility and virulence. Infect Immun 79:1815–1825. doi: 10.1128/IAI.00075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart PE, Bestor A, Cullen JN, Rosa PA. 2008. A tightly regulated surface protein of Borrelia burgdorferi is not essential to the mouse-tick infectious cycle. Infect Immun 76:1970–1978. doi: 10.1128/IAI.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf J, Schwartz I. 2011. Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog 7:e1002102. doi: 10.1371/journal.ppat.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. 2009. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol 71:1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dresser AR, Hardy PO, Chaconas G. 2009. Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog 5:e1000680. doi: 10.1371/journal.ppat.1000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller CL, Karna SL, Seshu J. 2013. Borrelia host adaptation regulator (BadR) regulates rpoS to modulate host adaptation and virulence factors in Borrelia burgdorferi. Mol Microbiol 88:105–124. doi: 10.1111/mmi.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purser JE, Norris SJ. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc Natl Acad Sci U S A 97:13865–13870. doi: 10.1073/pnas.97.25.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motaleb MA, Pitzer JE, Sultan SZ, Liu J. 2011. A novel gene inactivation system reveals an altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol 193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sultan SZ, Pitzer JE, Miller MR, Motaleb MA. 2010. Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol 77:128–142. doi: 10.1111/j.1365-2958.2010.07191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szurmant H, Ordal GW. 2004. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev 68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hyakutake A, Homma M, Austin MJ, Boin MA, Hase CC, Kawagishi I. 2005. Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J Bacteriol 187:8403–8410. doi: 10.1128/JB.187.24.8403-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandyopadhaya A, Chaudhuri K. 2009. Differential modulation of NF-kappaB-mediated pro-inflammatory response in human intestinal epithelial cells by cheY homologues of Vibrio cholerae. Innate Immun 15:131–142. doi: 10.1177/1753425908100454. [DOI] [PubMed] [Google Scholar]
- 52.Kirby JR, Zusman DR. 2003. Chemosensory regulation of developmental gene expression in Myxococcus xanthus. Proc Natl Acad Sci U S A 100:2008–2013. doi: 10.1073/pnas.0330944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McBride MJ, Weinberg RA, Zusman DR. 1989. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthus show sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci U S A 86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Z, Ma X, Tong L, Kaplan HB, Shimkets LJ, Shi W. 2000. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol 182:5793–5798. doi: 10.1128/JB.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berleman JE, Bauer CE. 2005. A che-like signal transduction cascade involved in controlling flagella biosynthesis in Rhodospirillum centenum. Mol Microbiol 55:1390–1402. doi: 10.1111/j.1365-2958.2005.04489.x. [DOI] [PubMed] [Google Scholar]
- 56.Berleman JE, Bauer CE. 2005. Involvement of a Che-like signal transduction cascade in regulating cyst cell development in Rhodospirillum centenum. Mol Microbiol 56:1457–1466. doi: 10.1111/j.1365-2958.2005.04646.x. [DOI] [PubMed] [Google Scholar]
- 57.Black WP, Yang Z. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J Bacteriol 186:1001–1008. doi: 10.1128/JB.186.4.1001-1008.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SH, Butler SM, Camilli A. 2001. Selection for in vivo regulators of bacterial virulence. Proc Natl Acad Sci U S A 98:6889–6894. doi: 10.1073/pnas.111581598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kofoid EC, Parkinson JS. 1988. Transmitter and receiver modules in bacterial signaling proteins. Proc Natl Acad Sci U S A 85:4981–4985. doi: 10.1073/pnas.85.14.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol 422:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iyer R, Caimano MJ, Luthra A, Axline D Jr, Corona A, Iacobas DA, Radolf JD, Schwartz I. 2015. Stage-specific global alterations in the transcriptomes of Lyme disease spirochetes during tick feeding and following mammalian host adaptation. Mol Microbiol 95:509–538. doi: 10.1111/mmi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elias AF, Stewart PE, Grimm D, Caimano MJ, Eggers CH, Tilly K, Bono JL, Akins DR, Radolf JD, Schwan TG, Rosa P. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect Immun 70:2139–2150. doi: 10.1128/IAI.70.4.2139-2150.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rego RO, Bestor A, Rosa PA. 2011. Defining the plasmid-encoded restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol 193:1161–1171. doi: 10.1128/JB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol 35:490–516. [DOI] [PubMed] [Google Scholar]
- 65.Motaleb MA, Miller MR, Bakker RG, Li C, Charon NW. 2007. Isolation and characterization of chemotaxis mutants of the Lyme disease Spirochete Borrelia burgdorferi using allelic exchange mutagenesis, flow cytometry, and cell tracking. Methods Enzymol 422:421–437. doi: 10.1016/S0076-6879(06)22021-4. [DOI] [PubMed] [Google Scholar]
- 66.Bono JL, Elias AF, Kupko JJ III, Stevenson B, Tilly K, Rosa P. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol 182:2445–2452. doi: 10.1128/JB.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frank KL, Bundle SF, Kresge ME, Eggers CH, Samuels DS. 2003. aadA confers streptomycin resistance in Borrelia burgdorferi. J Bacteriol 185:6723–6727. doi: 10.1128/JB.185.22.6723-6727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jewett MW, Lawrence K, Bestor AC, Tilly K, Grimm D, Shaw P, VanRaden M, Gherardini F, Rosa PA. 2007. The critical role of the linear plasmid lp36 in the infectious cycle of Borrelia burgdorferi. Mol Microbiol 64:1358–1374. doi: 10.1111/j.1365-2958.2007.05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karna SL, Prabhu RG, Lin YH, Miller CL, Seshu J. 2013. Contributions of environmental signals and conserved residues to the functions of carbon storage regulator A of Borrelia burgdorferi. Infect Immun 81:2972–2985. doi: 10.1128/IAI.00494-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seemanapalli SV, Xu Q, McShan K, Liang FT. 2010. Outer surface protein C is a dissemination-facilitating factor of Borrelia burgdorferi during mammalian infection. PLoS One 5:e15830. doi: 10.1371/journal.pone.0015830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mulay VB, Caimano MJ, Iyer R, Dunham-Ems S, Liveris D, Petzke MM, Schwartz I, Radolf JD. 2009. Borrelia burgdorferi bba74 is expressed exclusively during tick feeding and is regulated by both arthropod- and mammalian host-specific signals. J Bacteriol 191:2783–2794. doi: 10.1128/JB.01802-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patton TG, Dietrich G, Dolan MC, Piesman J, Carroll JA, Gilmore RD Jr. 2011. Functional analysis of the Borrelia burgdorferi bba64 gene product in murine infection via tick infestation. PLoS One 6:e19536. doi: 10.1371/journal.pone.0019536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J Exp Med 199:641–648. doi: 10.1084/jem.20031960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang X, Yang X, Kumar M, Pal U. 2009. BB0323 function is essential for Borrelia burgdorferi virulence and persistence through tick-rodent transmission cycle. J Infect Dis 200:1318–1330. doi: 10.1086/605846. [DOI] [PMC free article] [PubMed] [Google Scholar]