ABSTRACT
Candida albicans is among the most common human fungal pathogens, causing a broad range of infections, including life-threatening systemic infections. The cell wall of C. albicans is the interface between the fungus and the innate immune system. The cell wall is composed of an outer layer enriched in mannosylated glycoproteins (mannan) and an inner layer enriched in β-(1,3)-glucan and chitin. Detection of C. albicans by Dectin-1, a C-type signaling lectin specific for β-(1,3)-glucan, is important for the innate immune system to recognize systemic fungal infections. Increased exposure of β-(1,3)-glucan to the immune system occurs when the mannan layer is altered or removed in a process called unmasking. Nanoscale changes to the cell wall during unmasking were explored in live cells with atomic force microscopy (AFM). Two mutants, the cho1Δ/Δ and kre5Δ/Δ mutants, were selected as representatives that exhibit modest and strong unmasking, respectively. Comparisons of the cho1Δ/Δ and kre5Δ/Δ mutants to the wild type reveal morphological changes in their cell walls that correlate with decreases in cell wall elasticity. In addition, AFM tips functionalized with Dectin-1 revealed that the forces of binding of Dectin-1 to all of the strains were similar, but the frequency of binding was highest for the kre5Δ/Δ mutant, decreased for the cho1Δ/Δ mutant, and rare for the wild type. These data show that nanoscale changes in surface topology are correlated with increased Dectin-1 adhesion and decreased cell wall elasticity. AFM, using tips functionalized with immunologically relevant molecules, can map epitopes of the cell wall and increase our understanding of pathogen recognition by the immune system.
KEYWORDS: β-(1,3)-glucan; adhesion force mapping; Candida albicans; Dectin-1; Young's modulus; atomic force microscopy; caspofungin; elasticity; gelatin immobilization; macrophages; indentation force mapping
INTRODUCTION
The polymorphic, commensal yeast Candida albicans is one of the most prevalent fungal pathogens infecting humans. It can infect a broad range of tissues, including skin, mucus membranes, and gastrointestinal and urogenital tracts, and it can also cause life-threatening systemic infections (1, 2). C. albicans presents serious medical challenges, especially for immunocompromised patients, including those with HIV infections, being treated with corticosteroids, undergoing cancer chemotherapy, or having organ transplants. The most serious of these infections are systemic bloodstream infections, and they can have a mortality rate of 40 to 60% (3–5). Although bacteria are the predominant microorganisms, Candida species have emerged as one of the leading causes of hospital-acquired infections and are responsible for 80% of all nosocomial infections caused by fungi (6). Most drugs used to treat systemic fungal infections fall into three classes. The azoles inhibit the synthesis of the essential lipid ergosterol, and the polyene amphotericin B interacts directly with ergosterol to damage the cell membrane (7). The echinocandins inhibit the synthesis of the essential cell wall polymer β-(1,3)-glucan (8). However, antifungal resistance is limiting the effectiveness of both azoles and echinocandins (9–11), and toxicity from treatment with amphotericin B can be a serious problem for patients (12). The development of new antifungal drugs is urgently needed (13–15).
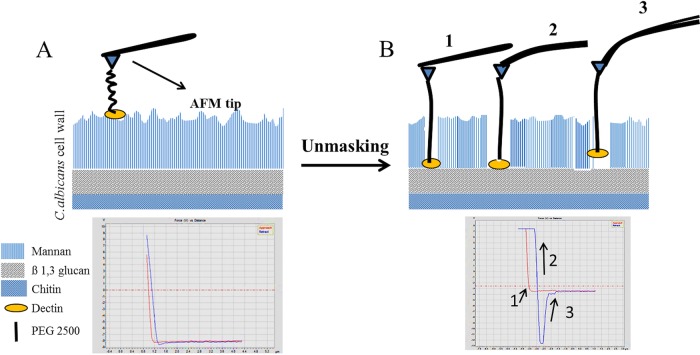
One promising avenue for the development of antifungal therapies is to improve the innate immune system's recognition of the pathogen. A healthy immune system can respond effectively to C. albicans (16), so enhancement of the immune response in immunocompromised patients can potentially be a powerful therapy (17, 18). This requires a comprehensive understanding of the interactions between C. albicans and the human immune system, particularly in the early stages of infection. The C. albicans cell wall consists primarily of polysaccharides and can be divided into three components (Fig. 1A) (19–21). The outer surface layer is enriched in mannose polysaccharides linked to protein to form a mannoprotein barrier (mannan) that acts as a filter for high-molecular-weight materials. The inner layer consists mostly of the glucose polysaccharide β-(1,3)-glucan and a minor amount of β-(1,6)-glucan, which is important for cross-linking other components of the wall. In addition to the glucan polymers, the inner layer contains a small but crucial amount of chitin, a linear β-(1,4)-linked N-acetylglucosamine polymer.
FIG 1.
Schematic of AFM ligand-receptor interactions during unmasking of β-(1,3)-glucan. In these cartoons, the AFM tip has been functionalized by attaching Dectin-1 to the cantilever tip. Dectin-1 can access the β-(1,3)-glucan layer if the mannan layer is disturbed. (A) In untreated wild-type cells, the wall consists of two main layers, with mannan in the outer layer and β-(1,3)-glucan and chitin in the inner layer. As the cantilever, with Dectin-1 attached, approaches the cell wall surface of wild-type cells, it rarely interacts with β-(1,3)-glucan due to the overlying mannan layer. Therefore, the cantilever approaches the surface of the cell, touches the surface, and withdraws without making contact with the β-(1,3)-glucan layer. (B) In cells where β-(1,3)-glucan is more exposed through the mannan layer, the cantilever, with Dectin-1 attached, contacts the β-(1,3)-glucan (1). As the cantilever begins to withdraw, strain appears on the cantilever, and the cantilever bends (2). As the cantilever continues to be withdrawn, the applied force builds until the adhesion is ruptured, and the cantilever snaps off the surface (3).
Macrophages recognize Candida through fungus-specific pathogen-associated molecular patterns (PAMPs) such as β-(1,3)-glucan. The Dectin-1 receptor on macrophages specifically recognizes and binds β-(1,3)-glucan and is part of the mechanism by which these first-responder cells detect fungi (23). Increased exposure of β-(1,3)-glucan to the immune system can occur when the mannan layer in the cell wall is altered or removed by mutations or antifungal drugs such as caspofungin. This “unmasking” can lead to an increase in Dectin-1 binding and thereby can improve macrophage recognition (Fig. 1) (24, 25). It is therefore possible that the development of approaches that increase unmasking in vivo might enhance immune responses and facilitate adjunctive therapies.
An understanding of unmasking at the nanoscale level is needed to elucidate this process, and this is at an early stage. Recently, Lin et al. examined the β-(1,3)-glucan distribution during caspofungin-mediated unmasking using direct stochastic optical reconstruction microscopy (dSTORM). This study revealed that when fluorescently labeled soluble Dectin-1 binding probes were used to detect unmasked β-glucan, single- and multiple-point exposure sites were observed (26). However, the size and frequency of multiple-point sites increased during caspofungin-induced unmasking, and the frequency of single-point sites also increased. This nanoscale analysis was performed at a ∼20-nm precision. The dSTORM approach yielded a nanoscale description of unmasking points and revealed patterns of β-glucan exposure. However, these studies do not show the distribution of the rest of the cell wall polymers relative to β-glucan.
Complementary studies of the effects of caspofungin on cell wall ultrastructure have been performed by using atomic force microscopy (AFM) and reveal the three-dimensional topology of the wall along with effects on the physical properties of the wall such as elasticity (27–29). By pressing the cantilever tip against the surface of the cell, differences in both surface elasticity (30–33) and indentation (34, 35) can be measured. Previous studies reported that wild-type C. albicans cells treated with caspofungin have decreased surface elasticity (Young's modulus) compared to that of untreated cells (29). However, other work has shown that surface elasticity is increased compared to that of wild-type cells following treatment with caspofungin (36). Thus, there is some controversy regarding how caspofungin impacts cell wall elasticity. An open question is whether other conditions that cause unmasking, such as cell wall mutations, will cause effects similar to those of caspofungin or differ in some respects.
In addition to measuring changes in surface topology and elasticity, AFM can also spatially identify and measure molecular adhesion between a probe attached to the tip of the cantilever and a cell surface (37–41). Previous studies employing Saccharomyces cerevisiae cells expressing the C. albicans adhesion protein Als5p attached to the AFM cantilever measured adhesion events on hydrophobic and fibronectin-coated substrates (42). In a similar experiment, antibodies to the adhesion protein Als5p attached to the cantilever tip were used to bind to an Als5p adhesion protein on the surface of C. albicans and initiate force-induced nanodomains over the entire cell surface (43). In addition, multiple lectins and antibodies were used on functionalized AFM tips to examine the flexibility of outer cell wall mannan polymers and inner cell wall glucan and chitin polymers in C. albicans, Candida glabrata, and S. cerevisiae as well as to compare adhesins and mannans in C. albicans yeast cells and hyphae (44, 45). Furthermore, functionalized AFM tips have been used to characterize the manner in which macrophages bind to C. albicans cells. Binding was shown to depend on mannose binding lectins from macrophages, although the precise lectins and their contributions were not identified, and binding is impacted by the flexible nature of the macrophage membrane (46).
In this study, we employ several mutants with various levels of unmasking, along with AFM, to interrogate the impact of β-glucan exposure on nanoscale topology and cell wall elasticity, as well as the distribution of β-glucan exposure, as seen with a Dectin-1-functionalized AFM probe. We also measure the forces of binding of Dectin-1 to masked versus unmasked β-(1,3)-glucan. We examine the cell wall topography in the wild type and in two mutants, the cho1Δ/Δ and kre5Δ/Δ mutants, that exhibit unmasking. The cho1Δ/Δ strain is a phosphatidylserine (PS) synthase mutant that exhibits increased chitin levels and a modest level of β-(1,3)-glucan unmasking that occurs in a punctate pattern (25, 47). The kre5Δ/Δ mutant exhibits stronger defects in cell wall composition than the cho1Δ/Δ mutant, including increased chitin and β-(1,3)-glucan and decreased β-(1, 6)-glucan and mannan levels and stronger unmasking (24, 48, 49). We also compared these mutants to wild-type cells treated with caspofungin such that they were unmasked, and they exhibited increased β-(1,3)-glucan exposure.
RESULTS
Exposure of β-(1,3)-glucan on the cell surface of C. albicans.
In order to further characterize the nanoscale effects of unmasking on the cell wall architecture, we chose to analyze two mutants that exhibit differential defects in cell wall structure and microscopic unmasking patterns. First, we chose the kre5Δ/Δ mutant, because it exhibits extensive defects in cell wall composition, including increased chitin and β-(1,3)-glucan and decreased mannan and β-(1,6)-glucan levels, and appears to have β-(1,3)-glucan unmasking over the whole cell (24, 48, 49). Second, for contrast, we chose the cho1Δ/Δ PS synthase mutant, which also exhibits β-(1,3)-glucan exposure, but only in punctate regions around yeast-form cells (25). Additionally, this mutant does not have gross alterations in all cell wall polymers, like the kre5Δ/Δ mutant. Only its cell wall chitin level is increased, whereas the levels of other cell wall polymers, β-(1,3)-glucan, β-(1,6) glucan, and mannan, are unchanged (data not shown) (47). Both mutants exhibit increased recognition by macrophages, as evidenced by greater binding with anti-β-(1,3)-glucan antibodies and soluble Dectin-1 (sDectin-1) as well as increased elicitation of tumor necrosis factor alpha (TNF-α) from macrophages (24, 25).
We first directly compared the patterns of unmasking of the wild-type (SC5314), kre5Δ/Δ, and cho1Δ/Δ strains using secondary immunofluorescence with a β-(1,3)-glucan-specific primary antibody (Fig. 2). As expected, the kre5Δ/Δ mutant exhibits whole-cell unmasking (Fig. 2A) that is quantitatively greater than that of the cho1Δ/Δ mutant (Fig. 2B). The cho1Δ/Δ mutant exhibits punctate regions of unmasking (Fig. 2A), and this is significantly greater than the unmasking quantified for the wild type, but significantly less than that for the kre5Δ/Δ mutant (Fig. 2B).
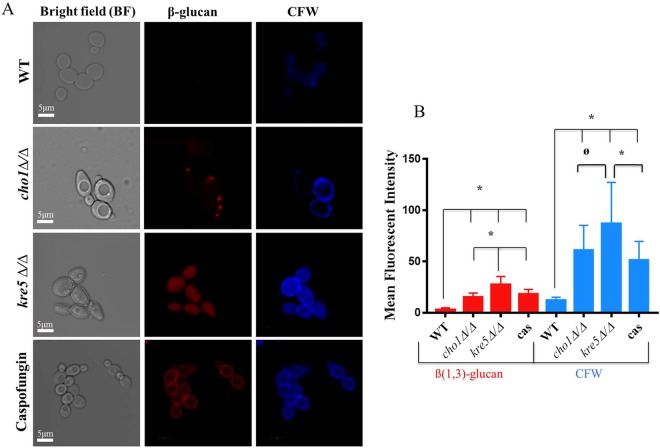
FIG 2.
β-(1,3)-Glucan exposure and chitin levels are increased in cho1Δ/Δ, kre5Δ/Δ, and caspofungin-treated wild-type (WT) cells compared to those in untreated wild-type cells. (A) Cells were stained with anti-β-(1,3)-glucan primary antibodies, Cy3-labeled secondary antibodies, and calcofluor white (CFW). Three biological replicates were carried out for all experiments. (B) Graph of the relative mean fluorescent intensity of each strain, as quantified by Image J. These data reveal that kre5Δ/Δ mutant cells, caspofungin (cas)-treated cells, and cho1Δ/Δ mutant cells have greater exposure of β-(1,3)-glucan and higher levels of chitin than the wild type (*, P < 0.0001; ø, P < 0.0026).
In addition to examining β-(1,3)-glucan exposure, we also examined the distribution and levels of chitin by staining with calcofluor white (CFW). Both the cho1Δ/Δ and kre5Δ/Δ mutants exhibited higher levels of chitin than those in wild-type cells (Fig. 2A). In wild-type cells, CFW stains mainly the bud neck and bud scar, while the mutants show major increases in chitin levels throughout the cell wall, and kre5Δ/Δ cell staining is more pronounced than cho1Δ/Δ cell staining, based on both visual inspection (Fig. 2A) and quantification with ImageJ (Fig. 2B). Wild-type cells treated with caspofungin exhibited increased β-(1,3)-glucan exposure and increased chitin levels that were distributed over the whole cells, more similar to the kre5Δ/Δ mutant but of lower intensity (Fig. 2A and B).
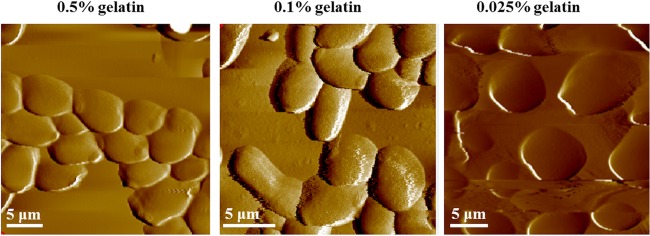
C. albicans cells were immobilized and imaged in liquid on mica coated with different percentages of gelatin.
In order to begin examining the ultrastructure of the C. albicans cell wall, we used atomic force microscopy (AFM), as it gives high-resolution images of the cell wall in a liquid environment without damaging the sample. This allows real-time imaging of metabolically active samples (50–53). First, we needed to immobilize the cells on a suitable surface. This was achieved by using gelatin-coated mica surfaces. This positively charged surface was developed for imaging bacterial cells in a liquid environment (52–54) and was extended here for mounting C. albicans cells. In Fig. 3, the cells were immobilized on the surface of a mica sheet covered with various percentages of gelatin (0.5 to 0.025%) and imaged in water. The cells are dispersed uniformly, making it easier to detect cells and image them by AFM. A lower percentage of gelatin (0.025%) was used, as it seemed to reduce the level of aggregation, which was higher with higher percentages of gelatin (i.e., 0.1 to 0.5%) (51).
FIG 3.
Immobilization of C. albicans cells onto mica surfaces coated with a solution containing different percentages of gelatin. Mica sheets were first coated with gelatin (0.5 to 0.025%) and allowed to dry overnight. Next, 100 μl (∼1 × 104 cells/ml) of Candida albicans cells in sodium acetate buffer was pipetted onto the gelatin-coated mica sheet. After 5 min of incubation, the surfaces were washed with water and placed into the AFM instrument for imaging in distilled water.
Cell wall unmasking correlates with increased cell surface roughness.
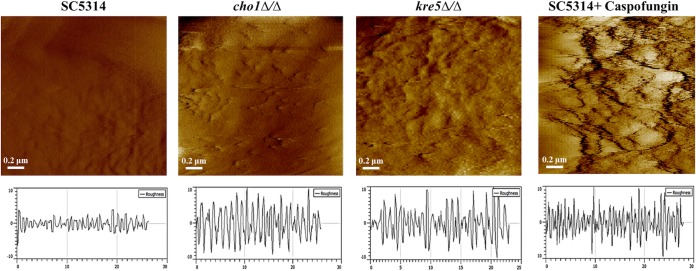
Once immobilization was achieved, we examined how the nanoscale topology of unmasked mutants differs from that of the wild type using AFM in the deflection mode in water (Fig. 4). The cell walls of both mutants appear rougher than that of the wild type (SC5314), which appears smooth. The CAF2 strain, which is closely related to the background strain of the kre5Δ/Δ mutant (48), was also examined by AFM and is similar to SC5314, so all studies reported herein discuss SC5314 as the reference wild-type strain. Of the two mutants, the kre5Δ/Δ mutant has the roughest appearance, followed by the cho1Δ/Δ mutant, which has a rough appearance in specific regions. Roughness can be quantified (Fig. 4, black trace below each image) and expressed as the root mean square (RMS) surface roughness, which was 34.35 ± 5.81 nm for the kre5Δ/Δ strain, 29.67 ± 3.21 nm for the cho1Δ/Δ strain, and 8.21 ± 2.028 nm for the wild type. The increased rough areas in cho1Δ/Δ mutant cells are more sporadic, which may correlate with the punctate unmasking in this mutant (Fig. 2). The rough appearance in these two mutants was similar to that found in wild-type cells that were treated with caspofungin (Fig. 4). The RMS surface roughness of caspofungin-treated wild-type cells was 39.5 ± 6.91 nm.
FIG 4.
Topographic 2- by 2-μm images of the cell wall surface of C. albicans wild-type cells with or without caspofungin treatment and cho1Δ/Δ and kre5Δ/Δ mutant cells. Cells were mounted onto gelatin-coated mica and imaged in water in contact mode with cantilevers having spring constants of 0.01 or 0.03 N/m by using a 5500 PicoPlus AFM instrument. PicoPlus AFM software was used to measure surface roughness, which is shown in the plot below each corresponding image. The plot for each strain was the average from measurements made for 30 cells.
The cell walls of the kre5Δ/Δ and cho1Δ/Δ mutants exhibit decreased elasticity.
The increased roughness of the cell wall was hypothesized to correlate with changes in cell wall elasticity. To test this, force volume maps, across a 2-μm2 area on the top of the cell, were recorded to assess cell wall elasticity. Each map contains an array of 32-by-32 points, with each point being an average of 3 force curves.
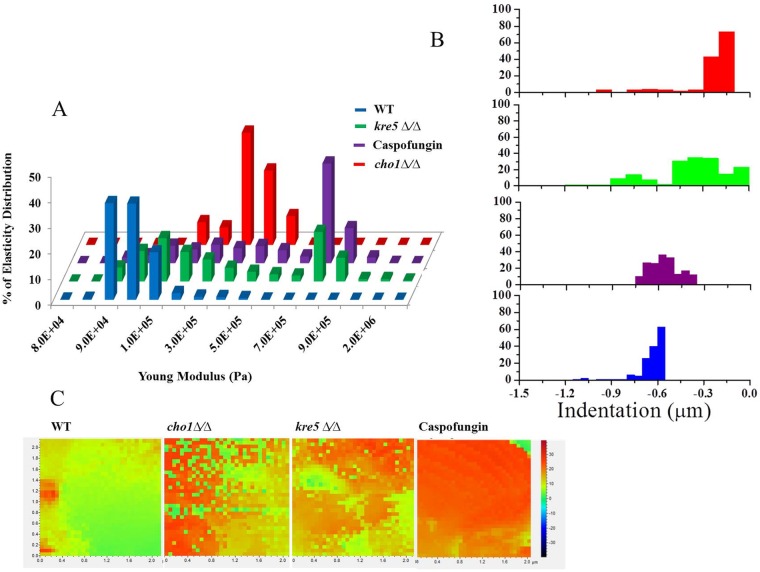
These data were then converted to elasticity and indentation measurements by PicoPlus AFM software to determine Young's modulus for all three strains. Young's modulus measures the ability of the material to undergo stress and strain. Lower Young's modulus values indicate increased elasticity. As a control, caspofungin-treated cells were also tested because this drug inhibits the synthesis of cell wall β-(1,3)-glucan and causes unmasking of β-(1,3)-glucan and increased roughness (Fig. 4) (27, 29). The results of these experiments are shown in Fig. 5A, where the x axis records the Young's modulus and the y axis displays the elasticity distribution. The wild type has a Young's modulus value of ∼9 × 104 Pa, whereas strains affected by cho1Δ/Δ or kre5Δ/Δ mutations or caspofungin (wild type) have Young's moduli that increase to ∼5 × 105 Pa, ∼8 × 105 Pa, or ∼8.2 × 105 Pa, respectively (Fig. 5A). Therefore, the cell walls of these strains are less elastic than those of untreated wild-type cells.
FIG 5.
Elasticity of cho1Δ/Δ, kre5Δ/Δ, and caspofungin-treated cells decreases compared to that of untreated wild-type cells. Force volume maps of the surface of wild-type cells, cho1Δ/Δ and kre5Δ/Δ mutant cells, along with caspofungin-treated wild-type C. albicans cells were taken by scanning a 2-μm2 area on the top of a cell and recording 32-by-32 points, with each point being the average of 3 force curves. These data were converted into elasticity and indentation maps by using PicoPlus AFM software. (A) Percent distributions of Young's modulus values corresponding to the elasticity maps of the wild-type, cho1Δ/Δ, and kre5Δ/Δ strains along with caspofungin-treated cells. (B) Representative histogram of the indentation values for the different strains and caspofungin-treated cells. (C) Heat map for the different strains and caspofungin-treated cells. The color map indicates the modulus of elasticity (in pascals) using force curves generated at each point on the cell surface. The more intense the red color, the less elastic (stiffer) the surface.
Figure 5C shows a heat map of the 32-by-32 point display of the 2-μm2 cell surface, where the transition from green to orange-red corresponds to an increase in Young's modulus. It is evident from this that Young's modulus for the wild type is lower than that for either of the two mutants or the wild type treated with caspofungin. Thus, these cell wall stresses result in decreased elasticity (increased stiffness).
A property closely related to Young's modulus is indentation, which measures the depth that the AFM tip can press into the cell wall. Cells with larger indentations exhibit greater elasticity. Like Young's modulus, indentation measurements indicated that wild-type cells are more elastic than cells of the two mutants or caspofungin-treated cells (Fig. 5B). Nanoindentation measurements were performed on a 2-μm2 area by recording force curves on 32-by-32 point displays for 10 different cells (n = 10). Statistical evaluation of the results by using PicoView 1.20 software shows that the indentation depth for the wild-type cell surface is larger (∼7 × 102 nm ± 34.16 nm) than the indentation depths for the surfaces of kre5Δ/Δ mutant (∼3 × 102 nm ± 31.02 nm), cho1Δ/Δ mutant (∼2 × 102 nm ± 33.04 nm), and caspofungin-treated (∼6 × 102 nm ± 72.61) cells (Fig. 5B). Thus, all the conditions tested here led to decreased elasticity compared to that of the wild type. Compared to the other conditions tested, the kre5Δ/Δ mutant exhibited a broader distribution of elasticity, suggesting that it has a more uneven and perhaps a more disturbed surface.
The unmasked mutants show nanoscale increases in Dectin-1 binding.
As indicated above, AFM images and measurements of elasticity and indentation show that the kre5Δ/Δ and cho1Δ/Δ mutants have disorganized cell walls that lead to distinguishable physical properties. We wanted to analyze the nanoscale spatial organization of the immunologically relevant PAMP β-(1,3)-glucan in the cell walls of unmasked mutant cells. Therefore, AFM tips were functionalized with sDectin-1–Fc (the design of this experiment is represented in Fig. 1). This C-type signaling lectin is specific for β-(1,3)-glucan, and therefore, adhesion force measurements using Dectin-1-functionalized AFM tips can reveal the presence and nanoscale distribution of exposed β-(1,3)-glucan in wild-type, mutant, and caspofungin-treated cells.
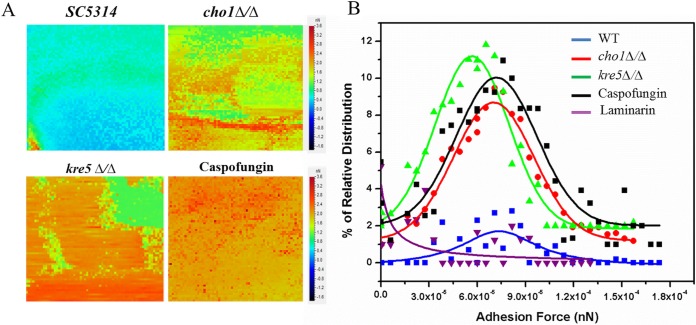
By using the sDectin-1–Fc-functionalized AFM tip to measure the nanoscale distribution of adhesion forces, an adhesion force volume map was generated for a 32- by 32-pixel array of a 2- by 2-μm sample area on the surface of C. albicans. Wild-type cells with or without caspofungin treatment and cho1Δ/Δ and kre5Δ/Δ mutant cells were measured in this manner. As a control, we measured adhesion between the gelatin surface and the functionalized tip, and there was no interaction, indicating that gelatin did not interfere with this experiment. The heat map clearly shows a progression from wild-type cells (blue-green), showing the lowest level of adhesion, to cho1Δ/Δ mutant cells, showing intermediate adhesion (mixture of green, yellow, orange, and red), to kre5Δ/Δ mutant and caspofungin-treated wild-type cells (mostly orange-red), showing the most adhesion (Fig. 6A).
FIG 6.
Adhesion force volume maps using Dectin-1-coated cantilevers recorded on wild-type (SC5314), cho1Δ/Δ, kre5Δ/Δ, and caspofungin-treated wild-type cells. (A) The heat map force curves recorded on the surfaces of cells of the wild-type strain show low-frequency adhesion to the surface of the cell. Force curves collected on kre5Δ/Δ, cho1Δ/Δ, and caspofungin-treated cell surfaces indicate higher-frequency adhesion between the cell surface and the tip. Increased adhesion is indicated by the progression from blue and green to orange and red. (B) Corresponding histograms of the force curves between a Dectin-1-functionalized tip and β-(1,3)-glucan on the surface of wild-type, kre5Δ/Δ, cho1Δ/Δ, and caspofungin-treated cells. The histogram data are derived from 4,096 force curves for each of the surfaces tested. In order to show that the interaction of Dectin-1 on the tip is specific for β-(1,3)-glucan on the surface of the mutants (kre5Δ/Δ or cho1Δ/Δ), soluble β-(1,3)-glucan (laminarin) was injected into the medium. This treatment blocks the Dectin-1 bound to the tip and prevents tip adhesion to the cell surface (purple line).
A histogram of the adhesion force frequency versus rupture strength was generated for cells of the different C. albicans strains and caspofungin-treated wild-type cells. Rupture strength, in this case, is the force required to break adhesion between the Dectin-1-functionalized tips and β-(1,3)-glucan in the cell wall of the fungus (Fig. 6B). There was some interaction between the Dectin-1-coated cantilever tip and the surface of wild-type C. albicans cells, but it was less than that observed for the mutants. It is likely that wild-type cells possess some gaps in the mannan layer or that the surface coatings are dynamically organized, which leads to infrequent exposure of β-(1,3)-glucan and subsequent interaction with the Dectin-1-coated tip. For the wild type, the peak adhesion frequency was ∼1%, while the peak adhesion frequency for the cho1Δ/Δ and kre5Δ/Δ mutants increased to ∼8% and ∼11%, respectively. The peak adhesion frequency for caspofungin-treated cells was ∼10%. These values indicate that the frequency with which the tip interacts with the β-(1,3)-glucan surface is due to increased exposure of the β-(1,3)-glucan layer.
Peak adhesion rupture strengths of wild-type, cho1Δ/Δ mutant, and caspofungin-treated cells are all about the same, which indicates that the same target molecule, β-(1,3)-glucan, is being recognized, but the frequency is higher during unmasking. However, for the kre5Δ/Δ mutant, the peak adhesion rupture strength is lower. The kre5Δ/Δ mutant has diminished β-(1,6)-glucan synthesis (48); β-(1,6)-glucan serves as a cross-linker for the β-(1,3)-glucan layer, and lower cross-linking may affect the structure of the β-(1,3)-glucan layer and subsequently the force required to rupture adhesion.
In control experiments to show that the binding of the functionalized tip to the cell surfaces is dependent on sDectin-1–Fc, soluble β-(1,3)-glucan (laminarin) was added to the imaging buffer. The solid purple line in Fig. 6B shows that the sDectin-1–Fc-coated tip is effectively blocked by laminarin, so no interaction between the tip and the cell surface occurs under these conditions. Thus, the force curves are due only to Dectin-1-dependent adhesion to β-(1,3)-glucan on the cell surface.
DISCUSSION
This study presents a nanoscale map of β-(1,3)-glucan epitope exposure and cell wall morphology in mutants that exhibit unmasking, using AFM. Features common to both mutants, in addition to the increased β-(1,3)-glucan exposure (Fig. 2), are increased overall surface roughness (Fig. 4) and decreased elasticity (Fig. 5). It was of interest to notice that the pattern of β-(1,3)-glucan exposure in the cho1Δ/Δ mutant, which was punctate in appearance, correlated with intermittent regions of surface roughness (Fig. 4). Furthermore, the kre5Δ/Δ mutant, which has global β-(1,3)-glucan exposure, had a more uniform increase in surface roughness.
Furthermore, the appearance of these rough places on the wall is similar to those of rough places generated by caspofungin in our work (Fig. 4) and those described previously by El-Kirat-Chatel and Dufrene (27) and Formosa et al. (29). In addition, we also observed that our mutants exhibited decreased elasticity, which again is similar to what was observed for caspofungin treatment by Formosa et al. (29). However, this finding contrasts with what was observed by El-Kirat-Chatel and Dufrene (27). We do not fully understand the reasons for these contradictory results. We suspect that in the study by El-Kirat-Chatel and Dufrene (27), the deformation of the cell that they observed indicates that the cell wall was being more heavily damaged than in our study, and as a result, elasticity was decreased because of a loss of structural integrity. In our case, we were attempting to use sublethal doses of caspofungin and observed a loss of elasticity. We think that our results showing a decreased elasticity of the wall reveal what happens during sublethal treatment that causes increased chitin synthesis and unmasking. We were ostensibly using the same concentrations of caspofungin as those used by El-Kirat-Chatel and Dufrene (27), so the precise reasons for such a difference remain unclear.
Previous work (55) indicated that a loss of chitin leads to increased elasticity and that an increased chitin level has the opposite effect. Both mutants analyzed in this study also show increased chitin levels (Fig. 2) (47–49). It is of interest to determine if the enhanced surface roughness observed in this study is caused by increased chitin levels. One can envision a model in which increased chitin synthesis deforms the overall structure of the wall, creating bulges that result in increases in surface roughness. This leads to other questions. Do these bulges influence unmasking and/or how well mutants are detected by the immune system? Observations of unmasking using dSTORM revealed clusters of unmasking foci (26). Do the regions of clustered β-(1,3)-glucan exposure revealed by dSTORM correlate with peaks of roughness in the cell wall observed by AFM? How often does surface roughness correlate with unmasking, and does this impact the exposure of epitopes to the immune system in unique ways? Again, this will be better addressed by expanding this analysis to study additional mutants and strains.
In addition to these topological observations of the wall, a functionalized AFM tip was used to measure the specific binding force between Dectin-1 and β-(1,3)-glucan residues on the surface of living C. albicans mutant and wild-type cells. The binding forces between the wild type and the mutants were very similar, but the frequencies of binding of the functionalized tips to kre5Δ/Δ and cho1Δ/Δ mutant cells and wild-type cells treated with caspofungin were higher than that for wild-type cells (Fig. 6). This indicates that unmasking leads to greater frequencies of interactions rather than increased binding strength. This also demonstrates a novel approach to mapping surface epitopes by using immunologically relevant lectins.
Finally, in this study, we used gelatin-coated mica to immobilize cells, allowing a versatile approach to immobilization that can be expanded to other strains, morphologies, and functionalizations of the tip. In combination with other work on imaging and immobilization of cells (28, 51, 56), our approach to analyzing interactions between host lectins and pathogens using functionalized tips provides another method to address nanoscale interactions in fungal immunology. Exposure of β-(1,3)-glucan plays an important role in the detection of C. albicans by macrophage cells, and therapies to enhance exposure could potentially aid in the control and elimination of C. albicans infections. In addition, future efforts to identify site-specific molecular tags that, when attached to the AFM cantilever, interact with the surface of C. albicans cells with sufficient binding strength and frequency should be pursued. These tags conjugated with antifungal agents would offer an additional approach for treating C. albicans infections.
MATERIALS AND METHODS
Strains, growth media, and chemicals.
C. albicans SC5314 (57), the cho1Δ/Δ mutant (47), and the kre5Δ/Δ mutant (a green fluorescent protein [GFP]-expressing version of the kre5Δ/Δ mutant carrying the ENO1::PENO1-EGFP-NATr plasmid, derived from KAH3 [48, 58]) were used throughout this study. For all experiments, strains were grown overnight at 30°C in YPD (1% yeast extract, 2% peptone, and 2% dextrose) medium under aerobic conditions, and the next day, the cells were diluted into fresh YPD medium at an optical density at 600 nm (OD600) of 0.1 and grown for 3 h at 30°C. In some experiments, wild-type cells were also treated with 50 ng/ml caspofungin (Merck, Kenilworth, NJ, USA) for 3 h before imaging by AFM. Three biological replicates were carried out for all experiments. The following media and chemicals were obtained from Sigma-Aldrich (St. Louis, MO): aminopropyltriethoxy silane (APTES), trimethylamine, acid–polyethylene glycol 2500 (PEG 2500)–N-hydroxysuccinimide (NHS), EDC [1-ethyl-3-(dimethylaminopropyl)carbodiimide], and sulfo-NHS (N-hydroxysulfosuccinimide). The following media and chemicals were obtained from Thermo Fisher Scientific (Waltham, MA): MES [2-(N-morpholino)ethanesulfonic acid], YPD, and bovine serum albumin (BSA). Calcofluor white (CFW) was obtained from Fluka Analytical (catalog number 18909), sDectin-1–Fc was produced by Robert Wheeler's laboratory at the University of Maine, β-(1,3)-glucan antibody was obtained from Biosupplies Australia Pty. Ltd., and goat anti-mouse antibody-Cy3 or horseradish peroxidase (HRP) was obtained from Jackson ImmunoResearch Laboratories (West Grove, PA, USA).
Fluorescence microscopy to visualize β-(1,3)-glucan and chitin.
We measured the distribution of chitin and β-(1,3)-glucan on the cell surface using confocal microscopy. We used CFW for chitin staining, and we used anti-β-(1,3)-glucan primary antibody and a goat anti-mouse-Cy3 secondary antibody for staining of β-(1,3)-glucan exposed on the cell wall (25). For each experiment, the staining protocol was identical across all samples, and all samples were imaged under identical conditions and on the same day. Three biological replicates were carried out for all experiments. For fluorescence imaging of β-glucan and chitin, C. albicans strains were grown overnight in YPD medium at 30°C. The next day, cells were inoculated at an OD600 of 0.1 into fresh YPD medium and grown for 3 more hours to reach log phase. Cells were washed three times with PBS (phosphate-buffered saline) and blocked for 30 min with PBS plus 3% BSA, after which they were incubated on ice with anti-β-(1,3)-glucan antibody at a 1:600 dilution for 90 min. After rigorous washing in PBS to remove unbound primary antibody, cells were incubated with a secondary goat anti-mouse antibody conjugated to Cy3 in PBS plus 5% BSA at room temperature for 20 min. Cells were washed in PBS again, and the pellet was resuspended in 500 μl of 0.01 mg/ml calcofluor white. The tubes were covered with aluminum foil and incubated on a rocker at room temperature for 5 min. The cells were washed three times with 1 ml water and resuspended in 100 μl water prior to observation under a confocal microscope. For an antibody negative control, only the primary or secondary antibody was added. Finally, the fluorescence intensities were calculated for ∼32 individual cells/condition by using ImageJ software (NIH [http://rsb.info.nih.gov/ij]).
Preparation of cells for AFM.
C. albicans cells were grown overnight in YPD medium at 30°C with shaking at 250 rpm. The next day, the cells were diluted into fresh YPD medium and incubated at 30°C with shaking at 250 rpm for 3 more hours. Cells in 1.5-ml aliquots were harvested in a microcentrifuge at 5,000 rpm, resuspended, and washed 3 times with 10 ml sodium acetate buffer (29), and 100 μl of cells in buffer (∼1 × 104 cells/ml) was pipetted onto a freshly cleaved mica wafer coated with gelatin, incubated for 5 min, rinsed in a stream of water, and mounted in the AFM wet cell prior to imaging in water (54). The mica surfaces that we used were freshly cleaved by applying clear adhesive tape to each surface and removing multiple layers until the surface appeared to be completely flat. A 0.5% gelatin solution was prepared by dissolving 0.5 g of gelatin (porcine gelatin, catalog number G6144; Sigma-Aldrich) in 100 ml of water that had been heated to boiling. Gelatin concentrations from 0.5% down to 0.025%, in water, worked well, so cells adhered to the surface and did not move during scanning. Subsequently, the cleaved mica was dipped into a gelatin solution that was heated to 60°C and withdrawn in a single motion. The mica was then placed edge down onto filter paper with the upper edge leaning against a surface. After drying overnight, these gelatin surfaces were ready to use for at least a month.
AFM imaging and force measurements.
C. albicans cells grown in YPD medium and suspended in sodium acetate buffer were centrifuged at 5,000 rpm in a microcentrifuge and rinsed 2 times in water. After deposition onto a gelatin-coated mica surface and incubation for 5 min, the surface was rinsed with water and placed into the AFM wet cell. The cells were imaged in water by using a 5500 PicoPlus AFM instrument (Keysight Technologies Inc., Santa Rosa, CA). The instrument was operated, using the PicoView 1.20.2 system, in the contact mode for imaging using MLCT probes (Bruker AFM Probes, Camarillo, CA) with either the C or D cantilever with spring constants of 0.01 or 0.03 N/m, respectively. A tip radius of 10 nm and a Poisson ratio of 0.5 were placed into PicoView software and used to calculate cell elasticity and indentation. The applied force was kept in a range of 3 to 5 nN for both imaging and force spectroscopy. The same cantilever was used for obtaining force volume maps on the wild type and both mutants (34). The force volume map was obtained over a 2- by 2-μm scan of the uppermost area of the cell surface, with an average of six 32-by-32 force volume maps being collected for each sample. Each of the 32-by-32 points of the force volume map is the average of 3 force curves. The data presented are averages of data from three experiments. Images were taken at a line scan speed of 1 line per s with either 256 or 512 pixels per line.
Measurements of elasticity (30–33) and indentation (34, 35, 59, 60) were performed on wild-type (SC5314; CIA4), cho1Δ/Δ, and kre5Δ/Δ cells and on wild-type cells treated with caspofungin, as follows. Prior to measurement of the elasticity of the cell surface, a force curve was generated on a mica sheet covered with gelatin and used as the reference point. After imaging of a sample, force curves were generated for each approach, and the slope of each curve was compared to the reference point to calculate force distance curves and elasticity. The force distance curves were transformed into force indentation by subtracting the cantilever deflection on a mica sheet. The same cantilever was used for obtaining force volume maps on the wild type and both mutants (34, 61). This occurred over a 2- by 2-μm force volume scan of the uppermost area of the cell surface with an average of six 32-by-32 points taken and with an average of 3 force curves per point collected for each sample. The data presented are averages of data from three experiments.
Adhesion force between Dectin-1 bound to the cantilever and β-(1,3)-glucan on the cell surface.
For adhesion experiments (37–41), AFM tips were functionalized with sDectin-1–Fc to act as a binding probe for interacting with the β-(1,3)-glucan layer on the C. albicans cell wall (23, 25). The cantilevers were functionalized based on a protocol from Hermann Gruber, Johannes Kepler University. Briefly, cantilevers were cleaned by rinsing in chloroform, dried with nitrogen gas, and placed into a desiccator purged with argon gas containing 30 μl of APTES and 10 μl of triethylamine, in separate trays, for 2 h to amino functionalize the cantilever tips. After removal of the trays, the desiccator was purged with argon gas, and the tips were left for 2 days. Next, 1 mg of acid-PEG 2500-NHS was added to 0.5 ml of chloroform in a tray and placed into the desiccator, and 30 μl trimethylamine was added to the tray and mixed, followed by purging of the desiccator with argon gas and incubation for 2 h. For coupling Dectin-1 to the acid-PEG 2500-functionalized cantilever tip, 0.4 mg of EDC and 1.1 mg of sulfo-NHS were dissolved in 1 ml of 0.1 M MES buffer at pH 6.1. Droplets of EDC–sulfo-NHS were then placed onto Parafilm, and cantilevers were placed in contact with the droplets for 15 min, followed by rinsing 4 times in PBS. Dectin-1 was diluted to 5.6 × 109 particles per ml in PBS and, droplets were placed onto Parafilm. Cantilevers were then placed in contact with the droplets for 2 h to functionalize the tips with Dectin-1, rinsed 4 times in PBS, and stored in PBS. Force volume maps over a 2-by-2 area of cell surfaces were collected as described above for obtaining elasticity and indentation measurements.
ACKNOWLEDGMENTS
We are grateful to Merck, Kenilworth, NJ, USA, for the contribution of caspofungin.
D.P.A., S.T.R., and M.J.D. acknowledge support from the U.S. DOE Office of Biological and Environmental Research Genomic Science Program under the Plant-Microbe Interfaces Scientific Focus Area at Oak Ridge National Laboratory. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. Department of Energy under contract no. DEAC0500OR22725. We gratefully acknowledge the support of the University of Tennessee-Oak Ridge National Laboratory (UT-ORNL) Joint Institute for Biological Sciences for this project. T.B.R. gratefully acknowledges support from NIAID (NIH-1 R01AL105690).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Calderone RA, Fonzi WA. 2001. Virulence factors of Candida albicans. Trends Microbiol 9:327–335. doi: 10.1016/S0966-842X(01)02094-7. [DOI] [PubMed] [Google Scholar]
- 2.Karkowska-Kuleta J, Kedracka-Krok S, Rapala-Kozik M, Kamysz W, Bielinska S, Karafova A, Kozik A. 2011. Molecular determinants of the interaction between human high molecular weight kininogen and Candida albicans cell wall: identification of kininogen-binding proteins on fungal cell wall and mapping the cell wall-binding regions on kininogen molecule. Peptides 32:2488–2496. doi: 10.1016/j.peptides.2011.10.021. [DOI] [PubMed] [Google Scholar]
- 3.Ammerlaan HS, Harbarth S, Buiting AG, Crook DW, Fitzpatrick F, Hanberger H, Herwaldt LA, van Keulen PH, Kluytmans JA, Kola A, Kuchenbecker RS, Lingaas E, Meessen N, Morris-Downes MM, Pottinger JM, Rohner P, dos Santos RP, Seifert H, Wisplinghoff H, Ziesing S, Walker AS, Bonten MJ. 2013. Secular trends in nosocomial bloodstream infections: antibiotic-resistant bacteria increase the total burden of infection. Clin Infect Dis 56:798–805. doi: 10.1093/cid/cis1006. [DOI] [PubMed] [Google Scholar]
- 4.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39:309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 5.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenneth VI, Rolston M, Bodey GP. 2003. Fungal infections, p 2641–2646. In Kufe DW, Pollock RE, Weichselbaum RR, Bast RC Jr, Gansler TS, Holland JF, Frei E III (ed). Holland Frei cancer medicine, 6th ed, vol 2 BC Decker, Hamilton, Ontario, Canada. [Google Scholar]
- 7.Georgopapadakou NH, Walsh TJ. 1994. Human mycoses: drugs and targets for emerging pathogens. Science 264:371–373. doi: 10.1126/science.8153622. [DOI] [PubMed] [Google Scholar]
- 8.Denning DW. 2003. Echinocandin antifungal drugs. Lancet 362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- 9.White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev 11:382–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat 10:121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kontoyiannis DP, Lewis RE. 2002. Antifungal drug resistance of pathogenic fungi. Lancet 359:1135–1144. doi: 10.1016/S0140-6736(02)08162-X. [DOI] [PubMed] [Google Scholar]
- 12.Bondaryk M, Kurzatkowski W, Staniszewska M. 2013. Antifungal agents commonly used in the superficial and mucosal candidiasis treatment: mode of action and resistance development. Postepy Dermatol Alergol 30:293–301. doi: 10.5114/pdia.2013.38358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loeffler J, Stevens DA. 2003. Antifungal drug resistance. Clin Infect Dis 36:S31–S41. doi: 10.1086/344658. [DOI] [PubMed] [Google Scholar]
- 14.Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med 125:S3–S13. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Sardi JC, Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. 2013. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62:10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 16.Netea MG, Joosten LA, van der Meer JW, Kullberg BJ, van de Veerdonk FL. 2015. Immune defence against Candida fungal infections. Nat Rev Immunol 15:630–642. doi: 10.1038/nri3897. [DOI] [PubMed] [Google Scholar]
- 17.Nemunaitis J, Shannon-Dorcy K, Appelbaum FR, Meyers J, Owens A, Day R, Ando D, O'Neill C, Buckner D, Singer J. 1993. Long-term follow-up of patients with invasive fungal disease who received adjunctive therapy with recombinant human macrophage colony-stimulating factor. Blood 82:1422–1427. [PubMed] [Google Scholar]
- 18.Casadevall A, Pirofski LA. 2001. Adjunctive immune therapy for fungal infections. Clin Infect Dis 33:1048–1056. doi: 10.1086/322710. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Herrera J, Elorza MV, Valentin E, Sentandreu R. 2006. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res 6:14–29. doi: 10.1111/j.1567-1364.2005.00017.x. [DOI] [PubMed] [Google Scholar]
- 20.Klis FM, de Groot P, Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med Mycol 39(Suppl 1):1–8. doi: 10.1080/mmy.39.1.1.8-0. [DOI] [PubMed] [Google Scholar]
- 21.Chaffin WL. 2008. Candida albicans cell wall proteins. Microbiol Mol Biol Rev 72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, Wong SY, Gordon S. 2002. Dectin-1 is a major beta-glucan receptor on macrophages. J Exp Med 196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler RT, Kombe D, Agarwala SD, Fink GR. 2008. Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog 4:e1000227. doi: 10.1371/journal.ppat.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis SE, Hopke A, Minkin SC Jr, Montedonico AE, Wheeler RT, Reynolds TB. 2014. Masking of beta(1-3)-glucan in the cell wall of Candida albicans from detection by innate immune cells depends on phosphatidylserine. Infect Immun 82:4405–4413. doi: 10.1128/IAI.01612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin J, Wester MJ, Graus MS, Lidke KA, Neumann AK. 2016. Nanoscopic cell-wall architecture of an immunogenic ligand in Candida albicans during antifungal drug treatment. Mol Biol Cell 27:1002–1014. doi: 10.1091/mbc.E15-06-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Kirat-Chatel S, Dufrene YF. 2012. Nanoscale imaging of the Candida-macrophage interaction using correlated fluorescence-atomic force microscopy. ACS Nano 6:10792–10799. doi: 10.1021/nn304116f. [DOI] [PubMed] [Google Scholar]
- 28.Formosa C, Pillet F, Schiavone M, Duval RE, Ressier L, Dague E. 2015. Generation of living cell arrays for atomic force microscopy studies. Nat Protoc 10:199–204. doi: 10.1038/nprot.2015.004. [DOI] [PubMed] [Google Scholar]
- 29.Formosa C, Schiavone M, Martin-Yken H, Francois JM, Duval RE, Dague E. 2013. Nanoscale effects of caspofungin against two yeast species, Saccharomyces cerevisiae and Candida albicans. Antimicrob Agents Chemother 57:3498–3506. doi: 10.1128/AAC.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weisenhorn ALK, Kasas MS, Gotzos V, Butt H-B. 1993. Deformation and height anomoly of soft surfaces studied with an AFM. Nanotechnology 4:106–113. doi: 10.1088/0957-4484/4/2/006. [DOI] [Google Scholar]
- 31.Maivald P, Butt HJ, Gould SAC, Prater CB, Drake B, Gurley JA, Elings VB, Hansma PK. 1991. Using force modulation to image surface elasticities with the atomic force microscope. Nanotechnology 2:103–106. [Google Scholar]
- 32.Allison DP, Hinterdorfer P, Han W. 2002. Biomolecular force measurements and the atomic force microscope. Curr Opin Biotechnol 13:47–51. doi: 10.1016/S0958-1669(02)00283-5. [DOI] [PubMed] [Google Scholar]
- 33.Zlatanova J, Lindsay SM, Leuba SH. 2000. Single molecule force spectroscopy in biology using the atomic force microscope. Prog Biophys Mol Biol 74:37–61. doi: 10.1016/S0079-6107(00)00014-6. [DOI] [PubMed] [Google Scholar]
- 34.Radmacher M, Fritz M, Kacher CM, Cleveland JP, Hansma PK. 1996. Measuring the viscoelastic properties of human platelets with the atomic force microscope. Biophys J 70:556–567. doi: 10.1016/S0006-3495(96)79602-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li QS, Lee GY, Ong CN, Lim CT. 2008. AFM indentation study of breast cancer cells. Biochem Biophys Res Commun 374:609–613. doi: 10.1016/j.bbrc.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 36.El-Kirat-Chatel S, Beaussart A, Alsteens D, Jackson DN, Lipke PN, Dufrene YF. 2013. Nanoscale analysis of caspofungin-induced cell surface remodelling in Candida albicans. Nanoscale 5:1105–1115. doi: 10.1039/C2NR33215A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dammer U, Hegner M, Anselmetti D, Wagner P, Dreier M, Huber W, Guntherodt HJ. 1996. Specific antigen/antibody interactions measured by force microscopy. Biophys J 70:2437–2441. doi: 10.1016/S0006-3495(96)79814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florin E-L, Rief M, Lehmann H, Ludwig M, Dornmair C, Moy VT, Gaub HE. 1995. Sensing specific molecular interactions with the atomic force microscope. Biosens Bioelectron 10:895–901. doi: 10.1016/0956-5663(95)99227-C. [DOI] [Google Scholar]
- 39.Gad M, Itoh A, Ikai A. 1997. Mapping cell wall polysaccharides of living microbial cells using atomic force microscopy. Cell Biol Int 21:697–706. doi: 10.1006/cbir.1997.0214. [DOI] [PubMed] [Google Scholar]
- 40.Hinterdorfer P, Baumgartner W, Gruber HJ, Schilcher K, Schindler H. 1996. Detection and localization of individual antibody-antigen recognition events by atomic force microscopy. Proc Natl Acad Sci U S A 93:3477–3481. doi: 10.1073/pnas.93.8.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinterdorfer P, Dufrene YF. 2006. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods 3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 42.Alsteens D, Van Dijck P, Lipke PN, Dufrene YF. 2013. Quantifying the forces driving cell-cell adhesion in a fungal pathogen. Langmuir 29:13473–13480. doi: 10.1021/la403237f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alsteens D, Garcia MC, Lipke PN, Dufrene YF. 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci U S A 107:20744–20749. doi: 10.1073/pnas.1013893107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beaussart A, Alsteens D, El-Kirat-Chatel S, Lipke PN, Kucharikova S, Van Dijck P, Dufrene YF. 2012. Single-molecule imaging and functional analysis of Als adhesins and mannans during Candida albicans morphogenesis. ACS Nano 6:10950–10964. doi: 10.1021/nn304505s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Kirat-Chatel S, Beaussart A, Alsteens D, Sarazin A, Jouault T, Dufrene YF. 2013. Single-molecule analysis of the major glycopolymers of pathogenic and non-pathogenic yeast cells. Nanoscale 5:4855–4863. doi: 10.1039/c3nr00813d. [DOI] [PubMed] [Google Scholar]
- 46.El-Kirat-Chatel S, Dufrene YF. 2016. Nanoscale adhesion forces between the fungal pathogen Candida albicans and macrophages. Nanoscale Horiz 1:69–74. doi: 10.1039/C5NH00049A. [DOI] [PubMed] [Google Scholar]
- 47.Chen YL, Montedonico AE, Kauffman S, Dunlap JR, Menn FM, Reynolds TB. 2010. Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol Microbiol 75:1112–1132. doi: 10.1111/j.1365-2958.2009.07018.x. [DOI] [PubMed] [Google Scholar]
- 48.Herrero AB, Magnelli P, Mansour MK, Levitz SM, Bussey H, Abeijon C. 2004. KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot Cell 3:1423–1432. doi: 10.1128/EC.3.6.1423-1432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meaden P, Hill K, Wagner J, Slipetz D, Sommer SS, Bussey H. 1990. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1-6)-beta-d-glucan synthesis and normal cell growth. Mol Cell Biol 10:3013–3019. doi: 10.1128/MCB.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lonergan NE, Britt LD, Sullivan CJ. 2014. Immobilizing live Escherichia coli for AFM studies of surface dynamics. Ultramicroscopy 137:30–39. doi: 10.1016/j.ultramic.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 51.Heinisch JJ, Lipke PN, Beaussart A, El Kirat Chatel S, Dupres V, Alsteens D, Dufrene YF. 2012. Atomic force microscopy—looking at mechanosensors on the cell surface. J Cell Sci 125:4189–4195. doi: 10.1242/jcs.106005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beckmann MA, Venkataraman S, Doktycz MJ, Nataro JP, Sullivan CJ, Morrell-Falvey JL, Allison DP. 2006. Measuring cell surface elasticity on enteroaggregative Escherichia coli wild type and dispersin mutant by AFM. Ultramicroscopy 106:695–702. doi: 10.1016/j.ultramic.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Mortensen NP, Fowlkes JD, Sullivan CJ, Allison DP, Larsen NB, Molin S, Doktycz MJ. 2009. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir 25:3728–3733. doi: 10.1021/la803898g. [DOI] [PubMed] [Google Scholar]
- 54.Doktycz MJ, Sullivan CJ, Hoyt PR, Pelletier DA, Wu S, Allison DP. 2003. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97:209–216. doi: 10.1016/S0304-3991(03)00045-7. [DOI] [PubMed] [Google Scholar]
- 55.Ene IV, Walker LA, Schiavone M, Lee KK, Martin-Yken H, Dague E, Gow NA, Munro CA, Brown AJ. 2015. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. mBio 6:e00986-15. doi: 10.1128/mBio.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahimou F, Touhami A, Dufrene YF. 2003. Real-time imaging of the surface topography of living yeast cells by atomic force microscopy. Yeast 20:25–30. doi: 10.1002/yea.923. [DOI] [PubMed] [Google Scholar]
- 57.Gillum AM, Tsay EY, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler RT, Fink GR. 2006. A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog 2:e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Touhami A, Nysten B, Dufrene YF. 2003. Nanoscale mapping of the elasticity of microbial cells by atomic force microscopy. Langmuir 19:4539–4543. doi: 10.1021/la034136x. [DOI] [Google Scholar]
- 60.Briscoe BJ, Fiori L, Pelillo E. 1998. Nano-indentation of polymeric surfaces. J Appl Phys 31:2395–2405. [Google Scholar]
- 61.A-Hassan E, Heinz WF, Antonik MD, D'Costa NP, Nageswaran S, Schoenenberger CA, Hoh JH. 1998. Relative microelastic mapping of living cells by atomic force microscopy. Biophys J 74:1564–1578. doi: 10.1016/S0006-3495(98)77868-3. [DOI] [PMC free article] [PubMed] [Google Scholar]