ABSTRACT
Aggregatibacter actinomycetemcomitans is associated with aggressive periodontal disease, which is characterized by inflammation-driven alveolar bone loss. A. actinomycetemcomitans activates the p38 mitogen-activated protein kinase (MAPK) and MAPK-activated protein kinase 2 (MK2) stress pathways in macrophages that are involved in host responses. During the inflammatory process in periodontal disease, chemokines are upregulated to promote recruitment of inflammatory cells. The objective of this study was to determine the role of MK2 signaling in chemokine regulation during A. actinomycetemcomitans pathogenesis. Utilizing a murine calvarial model, Mk2+/+ and Mk2−/− mice were treated with live A. actinomycetemcomitans bacteria at the midsagittal suture. MK2 positively regulated the following macrophage RNA: Emr1 (F4/80), Itgam (CD11b), Csf1r (M-CSF Receptor), Itgal (CD11a), Tnf, and Nos2. Additionally, RNA analysis revealed that MK2 signaling regulated chemokines CCL3 and CCL4 in murine calvarial tissue. Utilizing the chimeric murine air pouch model, MK2 signaling differentially regulated CCL3 and CCL4 in the hematopoietic and nonhematopoietic compartments. Bone resorption pits in calvaria, observed by micro-computed tomography, and osteoclast formation were decreased in Mk2−/− mice compared to Mk2+/+ mice after A. actinomycetemcomitans treatment. In conclusion, these data suggest that MK2 in macrophages contributes to regulation of chemokine signaling during A. actinomycetemcomitans-induced inflammation and bone loss.
KEYWORDS: Aggregatibacter actinomycetemcomitans, chemokine receptors, chemokines, mitogen-activated protein kinases, neutrophils, osteoclast
INTRODUCTION
Aggregatibacter actinomycetemcomitans is a Gram-negative capnophilic coccobacillus often isolated from the oral cavity of patients diagnosed with aggressive periodontal disease (1–3). Aggressive periodontal disease has a unique characteristic of preferentially affecting succedaneous incisors and 1st molars (4), whereas chronic periodontal disease can affect all teeth in the oral cavity. Aggregatibacter actinomycetemcomitans has been implicated in extraoral infections, such as infective endocarditis (5), cerebral abscesses (6, 7), bacterial arthritis (8), osteomyelitis (9), and pregnancy-associated septicemia (10). Despite the association of A. actinomycetemcomitans with several diseases, the source of infection is ultimately the oral cavity (11–13). During periodontal disease pathogenesis, the host response disrupts the physiological state of the periodontium, which includes the gingiva and periodontal ligament. This osteoimmunological response leads to net alveolar bone resorption and, eventually, tooth loss.
While there are 6 serotypes of A. actinomycetemcomitans, serotypes a, b, and c have been isolated the most often from periodontal disease sites (12, 14, 15). The host response to A. actinomycetemcomitans is the strongest against serotype b strains when the responses against serotype a, b, and c strains in the United States are compared (16). It has long been known that of the 6 serotypes of A. actinomycetemcomitans, serotype b also increases the risk of development of periodontal disease (17, 18). Aggregatibacter actinomycetemcomitans possesses several virulence factors, including leukotoxin, cytolethal distending toxin (CDT), and lipopolysaccharide (LPS), which have potent immunomodulatory effects. It is thought that serotype b strains of A. actinomycetemcomitans are highly virulent and have potent LPS endotoxin and leukotoxin, which are responsible for significantly increasing the host response compared to the host response to other serotypes of A. actinomycetemcomitans (15, 19).
When mice are used as a model of periodontal disease, they mount an inflammatory response against A. actinomycetemcomitans infection that results in a high abundance of macrophages (20, 21). Peripheral blood and splenic monocytes act as progenitors for osteoclasts, macrophages, and dendritic cells in vitro (22). These monocytes are lymphoid negative and myeloid positive, and they are defined as B220− CD3− NK1.1− CD11b+ Ly6Chi CD115+ CCR2hi CX3CR1+ common lineage cells (22). Hematopoietic system-derived host immune cells include macrophages, neutrophils, dendritic cells, T cells, and osteoclasts, all of which have been shown to respond to A. actinomycetemcomitans in vitro (23). Nonhematopoietic cell types, including epithelial cells, fibroblasts, and osteoblasts, are also involved in A. actinomycetemcomitans pathogenesis. Both hematopoietic and nonhematopoietic systems dually participate in the host immune function.
The chemokine/chemokine receptor signaling axis is also a key regulator of aggressive periodontal disease and is associated with A. actinomycetemcomitans infection. In a clinical study, chemokine ligand 3 (CCL3) levels were enhanced in salivary samples from A. actinomycetemcomitans-infected children who progressed to bone loss (24). This study suggested that CCL3 is a potential prognostic marker of periodontal disease progression in patients infected with A. actinomycetemcomitans (24). Chemokines are necessary to promote the recruitment of host macrophages through their chemokine receptors during infection. Macrophage inflammatory protein 1α (MIP-1α) and MIP-1β, which are also called CCL3 and CCL4, are macrophage-secreted ligands for the chemokine receptors CCR1 and CCR5, respectively. Mature macrophages (F4/80 positive) were shown to express CCR1 and CCR5 on the cell surface (20). Moreover, macrophages had sustained Ccl3 and Ccl4 gene expression in response to oral A. actinomycetemcomitans treatment in a mouse periodontal disease model (21). Additionally, A. actinomycetemcomitans recruited CCR5-, CCR1-, and receptor activator of nuclear factor kappa B ligand (RANKL)-positive cells in an experimental mouse model (20). RANKL is an essential cytokine for osteoclast-driven bone loss that is secreted and expressed on the surface of fibroblasts, osteoblasts, and T cells. These clinical and preclinical studies separately revealed the importance of chemokines in immune function; however, the mechanism of action of chemokines in immune plasticity during inflammatory bone loss has yet to be elucidated.
Once peripheral inflammatory cells reach the local site of infection through the chemokine gradient, A. actinomycetemcomitans induces intracellular signaling cascades that enhance the host immune response. A critical cascade involved in stress signaling is the mitogen-activated protein kinase (MAPK) pathway, composed of 3 MAPKs: p38, Jun N-terminal kinase (JNK), and extracellular regulated kinase (ERK). We have shown that A. actinomycetemcomitans induced the phosphorylation of the JNK, ERK, and p38 MAPKs in macrophages (25). Aggregatibacter actinomycetemcomitans also led to the phosphorylation of MAPK-activated protein kinase 2 (MK2), a direct substrate of the p38α/β MAPK (25). Additionally, histological analysis in a clinical study showed that p38 MAPK levels were the most strongly correlated with periodontal disease severity when the correlation of the three MAPKs with disease severity were compared, but MK2 levels were not assessed (26). Based on these findings, we hypothesized that MK2 signaling regulates the recruitment of inflammatory monocytes through the chemokine/chemokine receptor axis and local monocyte plasticity.
Thus, the role of MK2 in the regulation of the chemokine and chemokine receptor signaling axis has not previously been addressed. This study demonstrates the critical role of MK2 signaling in the hematopoietic and nonhematopoietic cell lineages during regulation of the chemokine/chemokine receptor axis in monocyte plasticity during A. actinomycetemcomitans-induced inflammation and bone loss.
RESULTS
MK2 signaling regulates host inflammatory infiltrate in murine calvarial infection.
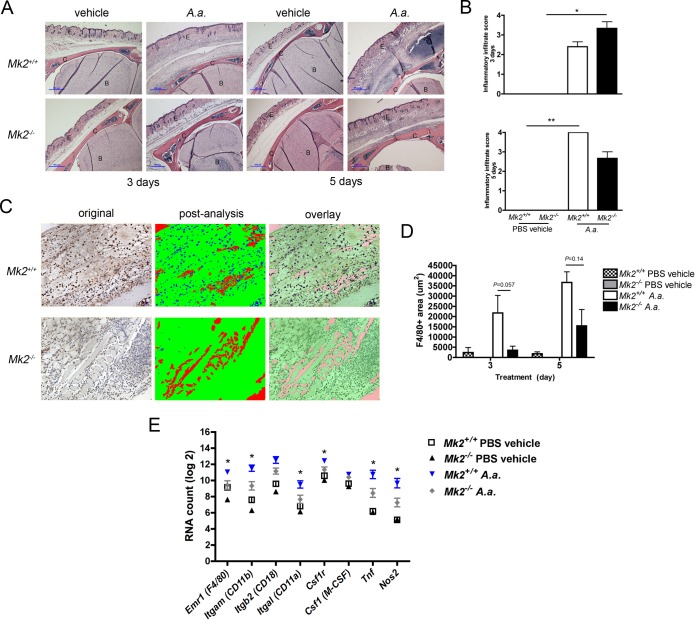
To demonstrate that A. actinomycetemcomitans induced inflammation through MK2 signaling, we assessed the murine calvarium model by histology. After 3 days, A. actinomycetemcomitans induced a significant amount of inflammation in Mk2−/− mice compared to that found in phosphate-buffered saline (PBS) vehicle-treated Mk2−/− mice (P ≤ 0.05), as detected by hematoxylin and eosin (H&E) staining and by use of the inflammatory infiltrate score (Fig. 1A and B). By day 5 of A. actinomycetemcomitans treatment, A. actinomycetemcomitans increased overall inflammation in A. actinomycetemcomitans-treated Mk2+/+ mice compared to PBS-vehicle treated Mk2+/+ mice (P ≤ 0.01; Fig. 1A and B). F4/80 immunohistochemistry was used to identify mature macrophages using digital analysis (Fig. 1C). There were trends toward decreased F4/80-positive staining in A. actinomycetemcomitans-treated Mk2+/+ mice compared to Mk2−/− mice on days 3 and 5 of treatment (Fig. 1D). RNA analysis revealed that the macrophage markers Emr1 (F4/80), Itgam (CD11b), Itgal (CD11a), Csf1r (macrophage colony-stimulating factor [M-CSF] receptor), Tnf, and Nos2 were significantly upregulated in Mk2+/+ mice compared to Mk2−/− mice (P ≤ 0.05) during A. actinomycetemcomitans stimulation (Fig. 1E).
FIG 1.
MK2 signaling regulates A. actinomycetemcomitans-induced inflammatory infiltrate. (A) Representative ×40-magnification images of Mk2+/+ and Mk2−/− mouse calvaria, including the epithelium (E), calvarium (C), and brain (B), treated with PBS vehicle or A. actinomycetemcomitans (A.a.) for 3 or 5 days. Blue scale bars, 100 μm. (B) Inflammatory infiltrate scores for Mk2+/+ and Mk2−/− mice treated with PBS vehicle or A. actinomycetemcomitans for 3 days (top) and 5 days (bottom). (C) Representative ×200-magnification images obtained after F4/80 staining of A. actinomycetemcomitans-treated Mk2+/+ and Mk2−/− mice for 3 days and Visiopharm analysis. (D) F4/80-positive (F4/80+) area of Mk2+/+ and Mk2−/− mice treated with PBS vehicle or A. actinomycetemcomitans for 3 and 5 days. (E) Analysis of macrophage marker RNA in mouse calvarial tissue treated for 3 days by use of the NanoString Technologies immunology panel (n = 3 to 5). Data are expressed as means ± SEMs. *, P ≤ 0.05; **, P ≤ 0.01.
Aggregatibacter actinomycetemcomitans activates the p38/MK2 MAPK signaling pathway.
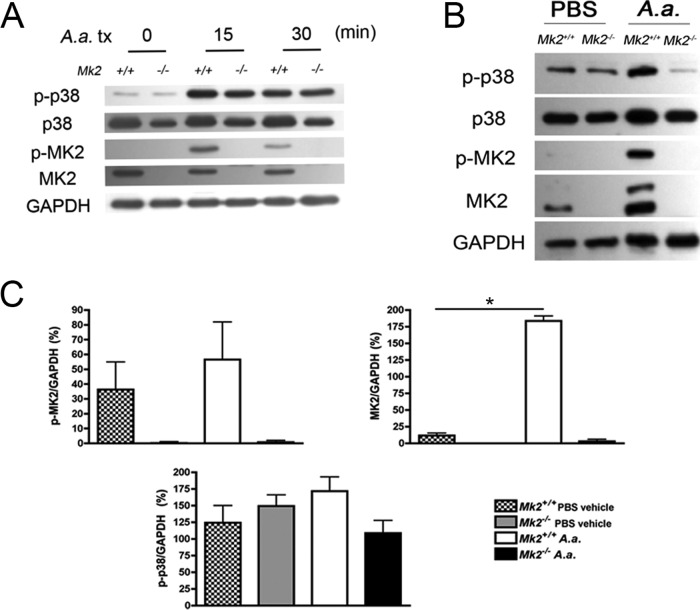
Based on the finding that MK2 signaling regulated local macrophage gene expression during A. actinomycetemcomitans infection, we next investigated whether MK2 signaling in macrophages is engaged upon A. actinomycetemcomitans challenge. Since the p38α/β MAPK acts directly to phosphorylate MK2, we utilized immunoblotting to detect p38 and MK2 phosphorylation levels in bone marrow-derived macrophages (BMDMs) and calvarial tissue. Aggregatibacter actinomycetemcomitans stimulated p38 phosphorylation in both Mk2+/+ and Mk2−/− mouse BMDMs (Fig. 2A), suggesting that MK2 levels do not change p38 phosphorylation upstream. Aggregatibacter actinomycetemcomitans induction of MK2 phosphorylation was recapitulated in calvarial tissue protein (Fig. 2B). Densitometric analysis of the immunoblot showed that during calvarial challenge, A. actinomycetemcomitans significantly increased MK2 levels (P ≤ 0.05), and there was a trend of increasing phospho-MK2 (p-MK2) levels in Mk2+/+ mice compared to vehicle-treated Mk2+/+ mice (Fig. 2C). No differences in phospho-p38 (p-p38) abundance were detected during MK2 deficiency or A. actinomycetemcomitans infection (Fig. 2C). These data show that A. actinomycetemcomitans induced MK2 phosphorylation in vivo and ex vivo in BMDMs.
FIG 2.
Aggregatibacter actinomycetemcomitans stimulates MK2 signaling in macrophages and in vivo. (A) A representative immunoblot of A. actinomycetemcomitans (A.a.) treatment (tx) stimulating p38 and MK2 phosphorylation in murine BMDMs (n = 2). (B) A representative immunoblot of calvarial tissue protein treated with A. actinomycetemcomitans for 3 days. (C) Densitometric analysis of calvarial tissue proteins in which the protein levels were normalized to the levels of GAPDH (n = 3 to 5). Data are expressed as means ± SEMs. *, P ≤ 0.05.
MK2 signaling regulates the chemokine/chemokine receptor axis during A. actinomycetemcomitans challenge.
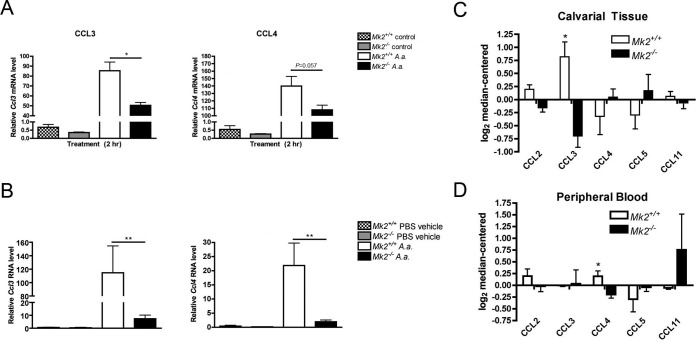
To determine which chemokines or chemokine receptors were regulated by MK2 during A. actinomycetemcomitans calvarial infection, comprehensive quantitative RNA analysis was performed using the NanoString Technologies immunology panel. These results of the mRNA counts are detailed in Table 1. The Ccl3, Ccl4, Ccl5, and Ccl12 chemokine RNAs were significantly upregulated in A. actinomycetemcomitans-treated Mk2+/+ mice compared to A. actinomycetemcomitans-treated Mk2−/− mice (P ≤ 0.05). The chemokine RNA transcripts that were the most highly expressed in Mk2+/+ mice during A. actinomycetemcomitans challenge compared to their levels of expression in vehicle-treated Mk2+/+ mice were Ccl3 and Ccl4. Ccl3 was the only chemokine RNA transcript with a level of expression of greater than 10,000 RNA counts in Mk2+/+ mice challenged with A. actinomycetemcomitans. The mean level of Ccl3 gene expression was 9 times higher in Mk2+/+ mice than Mk2−/− mice challenged with A. actinomycetemcomitans (P = 0.0159). Ccl3, Ccl4, and Ccl5 encode the ligands MIP-1α and MIP-1β, respectively. All three of these chemokines signal through receptors CCR1 and CCR5. There was a significant 2-fold decrease in the level of expression of Ccl12 in A. actinomycetemcomitans-treated Mk2−/− mice compared to A. actinomycetemcomitans-treated Mk2+/+ mice (P = 0.0159). The levels of the Ccr4 and Ccr5 chemokine receptor transcripts were significantly different in Mk2+/+ mice and Mk2−/− mice challenged with A. actinomycetemcomitans. Interestingly, the level of Ccr5 transcription was increased approximately 8 times by A. actinomycetemcomitans treatment in Mk2+/+ mice, but during bacterial challenge the level was significantly reduced 2-fold in Mk2−/− mice compared to that in Mk2+/+ mice (P = 0.0317). Conversely, A. actinomycetemcomitans treatment significantly decreased the level of Ccr4 gene expression in Mk2+/+ mice compared to Mk2−/− mice (P = 0.0317), although the RNA counts for Ccr4 were relatively low compared to those for the other transcripts (Table 1). These data highlight the positive function of MK2 signaling in the CCL3/CCL4/CCR5 signaling axis.
TABLE 1.
MK2 regulates chemokine/chemokine receptor RNA during A. actinomycetemcomitans pathogenesis
| RNA | RNA counta |
P value | |||
|---|---|---|---|---|---|
| PBS-treated Mk2+/+ mice | PBS-treated Mk2−/− mice | A. actinomycetemcomitans-treated Mk2+/+ mice | A. actinomycetemcomitans-treated Mk2−/− mice | ||
| Ccl2 | 348 ± 112 | 142 ± 12 | 1,655 ± 620 | 1,483 ± 783 | 0.6905 |
| Ccl3 | 114 ± 86 | 104 ± 80 | 17,611 ± 15,783 | 1,905 ± 2,472 | 0.0159b |
| Ccl4 | 46 ± 51 | 31 ± 50 | 6,933 ± 7,276 | 788 ± 1,022 | 0.0317b |
| Ccl5 | 32 ± 13 | 6 ± 6 | 1,226 ± 970 | 463 ± 255 | 0.0317b |
| Ccl6 | 202 ± 87 | 62 ± 27 | 364 ± 344 | 250 ± 149 | 0.8413 |
| Ccl7 | 144 ± 56 | 55 ± 14 | 994 ± 191 | 732 ± 400 | 0.2222 |
| Ccl8 | 1,326 ± 132 | 252 ± 59 | 5,565 ± 2,045 | 6,361 ± 4,105 | 0.5476 |
| Ccl9 | 1,039 ± 345 | 368 ± 24 | 2,510 ± 1,254 | 1,329 ± 559 | 0.1195 |
| Ccl11 | 1,149 ± 267 | 837 ± 177 | 299 ± 101 | 498 ± 219 | 0.2222 |
| Ccl12 | 18 ± 10 | 6 ± 9 | 474 ± 130 | 205 ± 110 | 0.0159b |
| Ccl19 | 398 ± 86 | 213 ± 17 | 474 ± 61 | 592 ± 254 | 0.6905 |
| Ccl20 | 116 ± 32 | 135 ± 39 | 115 ± 53 | 141 ± 30 | 0.6905 |
| Ccl25 | 79 ± 13 | 85 ± 8 | 51 ± 10 | 46 ± 25 | 0.2222 |
| Ccr2 | 898 ± 306 | 359 ± 242 | 2,410 ± 1,330 | 1,233 ± 414 | 0.0952 |
| Ccr4 | 288 ± 38 | 403 ± 83 | 64 ± 50 | 189 ± 97 | 0.0317b |
| Ccr5 | 120 ± 46 | 26 ± 19 | 950 ± 385 | 391 ± 216 | 0.0317b |
| Ccr7 | 12 ± 5 | 5 ± 7 | 40 ± 27 | 17 ± 14 | 0.1508 |
| Ccr9 | 167 ± 49 | 83 ± 8 | 42 ± 23 | 90 ± 43 | 0.0556 |
Values represent the mean RNA count ± SD for each group of PBS vehicle-treated mice (n = 3) and A. actinomycetemcomitans-treated mice (n = 5). RNA counts are from an analysis performed with the NanoString Technologies immunology panel and cavarial tissue from Mk2+/+ and Mk2−/− mice treated with A. actinomycetemcomitans or PBS vehicle for 3 days.
P ≤ 0.05 by comparison of the results for A. actinomycetemcomitans-treated Mk2+/+ mice with those for A. actinomycetemcomitans-treated Mk2−/− mice using a two-tailed Mann-Whitney test.
Notably, MK2 deficiency in BMDMs significantly attenuated Ccl3 (P ≤ 0.05) gene expression levels (Fig. 3A). Reverse transcription-quantitative PCR (RT-qPCR) methods were used to confirm the results for calvarial tissue obtained with the NanoString Technologies immunology panel (Fig. 3B). Mk2+/+ mice challenged with A. actinomycetemcomitans showed a significant increase in Ccl3 (P ≤ 0.01) and Ccl4 (P ≤ 0.01) gene expression levels compared to Mk2−/− mice (Fig. 3B). Protein analysis by a multiplex assay showed that MK2 signaling also significantly upregulated CCL3 (P ≤ 0.05) in calvarial tissue after 3 days of A. actinomycetemcomitans infection (Fig. 3C). Consistent with the gene expression data, CCL3 was the most abundant chemokine (Fig. 3C). Interestingly, MK2 deficiency downregulated only circulating CCL4 levels (P ≤ 0.05; Fig. 3D). From these results, we show that MK2 signaling differentially regulates chemokines locally and peripherally in mice during local bacterial challenge.
FIG 3.
MK2 regulates chemokines in A. actinomycetemcomitans-challenged macrophages and in vivo. (A) RT-qPCR of Ccl3 and Ccl4 gene expression of Mk2+/+ and Mk2−/− BMDMs stimulated with 10 CFU of A. actinomycetemcomitans (A.a.) per cell or untreated control Mk2+/+ and Mk2−/− BMDMs (n = 3 to 4). (B) RT-qPCR of Ccl3 (left) and Ccl4 (right) gene expression in mouse calvarial tissue challenged with A. actinomycetemcomitans or PBS vehicle for 3 days (n = 3 to 7). (C) Multiplex assay of mouse tissue chemokines after 3 days of A. actinomycetemcomitans treatment (n = 4). (D) Multiplex assay of mouse peripheral blood chemokines after 3 days of A. actinomycetemcomitans treatment (n = 4). Data are expressed as means ± SEMs. *, P ≤ 0.05; **, P ≤ 0.01.
MK2 signaling does not regulate circulating chemokine receptors.
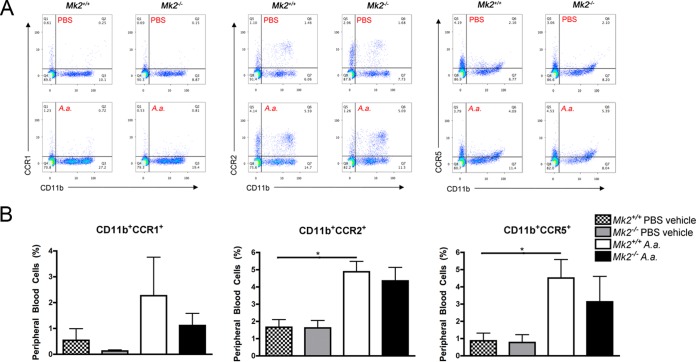
Since chemokines act as chemoattractant proteins for recruiting circulating host immune cells during infection, we assessed chemokine receptors in the peripheral blood of Mk2+/+ and Mk2−/− mice by flow cytometry (Fig. 4A). There was no significant difference in the percentage of CD11b+ CCR1+ peripheral blood cells between vehicle- and A. actinomycetemcomitans-treated mice (Fig. 4B). However, there was a significant increase in CD11b+ CCR2+ (P ≤ 0.05) and CD11b+ CCR5+ (P ≤ 0.05) circulating peripheral blood cells in Mk2+/+ mice treated with A. actinomycetemcomitans compared to vehicle-treated Mk2+/+ mice (Fig. 4B). These results suggest that local A. actinomycetemcomitans challenge systemically regulates circulating CD11b+ CCR2+ cells, which are classically considered the circulating inflammatory monocyte subset. In our study, MK2 signaling did not regulate the CCR1, CCR2, or CCR5 chemokine receptor on CD11b+ monocytic cells.
FIG 4.
MK2 signaling does not regulate circulating monocytes during A. actinomycetemcomitans pathogenesis. (A) Representative flow cytometry plots of peripheral blood CD11b+ CCR1+, CD11b+ CCR2+, and CD11b+ CCR5+ cells from Mk2+/+ and Mk2−/− mice treated with PBS vehicle or A. actinomycetemcomitans (A.a.) for 3 days. Q1 to Q4, quadrants 1 to 4, respectively. (B) Quantification of percent positive CD11b+ CCR1+, CD11b+ CCR2+, and CD11b+ CCR5+ cells from Mk2+/+ and Mk2−/− mice treated with PBS vehicle or A. actinomycetemcomitans for 3 days (n = 4). Data are expressed as means ± SEMs. *, P ≤ 0.05.
MK2 signaling differentially regulates hematopoietic system- and nonhematopoietic system-mediated early inflammatory cell recruitment and chemokines.
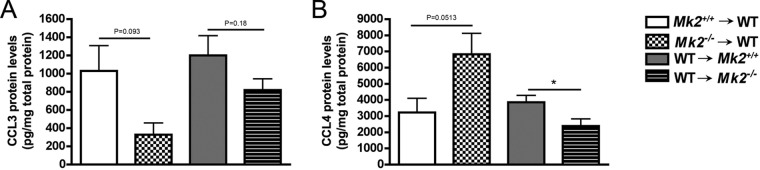
Since cells of nonhematopoietic and hematopoietic origin secrete chemokines, we next aimed to determine the role of MK2 signaling in the hematopoietic system versus the nonhematopoietic system. CCL3 and CCL4 chemokines were quantified in air pouch exudates from four groups of mice treated with A. actinomycetemcomitans. There were no significant differences in CCL3 protein levels (Fig. 5A). Trends of decreasing CCL3 protein levels were observed in wild-type (WT) mice with Mk2−/− mouse hematopoietic cells compared to the levels in the WT control group (WT mice into which Mk2+/+ mouse cells were transplanted) (P = 0.093; Fig. 5A). Opposite from the findings for CCL3, there was a trend toward an increase in CCL4 protein levels in chimeric mice with Mk2−/− mouse hematopoietic cells compared to the WT control group (WT mice into which Mk2+/+ mouse cells were transplanted) (P = 0.0513) when the mice were challenged with A. actinomycetemcomitans (Fig. 5B). The Mk2−/− mouse nonhematopoietic compartment of recipient mice with WT hematopoietic cells had decreased CCL4 levels compared to the WT control group (WT mice into which Mk2+/+ mouse cells were transplanted) during A. actinomycetemcomitans challenge (P ≤ 0.05; Fig. 5B). These results suggest that MK2 differentially regulates chemokines in the hematopoietic and nonhematopoietic compartments during A. actinomycetemcomitans pathogenesis.
FIG 5.
MK2 differentially regulates chemokines CCL3 and CCL4. (A) ELISA quantification of CCL3 in air pouch exudate from A. actinomycetemcomitans-treated mice (n = 6 to 10). (B) ELISA quantification of CCL4 in air pouch exudate from A. actinomycetemcomitans-treated mice (n = 6 to 10). Data are expressed as means ± SEMs compared to the results for the WT control groups. *, P ≤ 0.05.
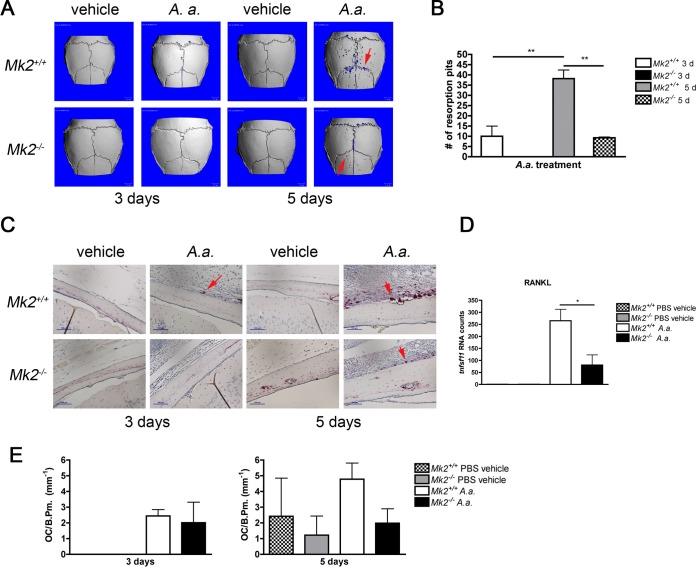
MK2 signaling is critical for A. actinomycetemcomitans-induced bone loss.
To determine the role of MK2 signaling in A. actinomycetemcomitans-induced inflammatory bone loss, we quantified calvarial pit formation and osteoclastogenesis by micro-computed tomography (μCT) and histological analysis. There was a significant increase in the number of resorption pits in A. actinomycetemcomitans-treated Mk2+/+ mice compared to Mk2+/+ mice on days 3 and 5 (P ≤ 0.05; Fig. 6A and B). As anticipated, vehicle treatment did not cause any resorption pit formation (Fig. 6A). Tartrate-resistant acid phosphatase (TRAP)-positive cells adjacent to bone were enumerated as osteoclasts, but the difference in the number of osteoclasts between Mk2+/+ mice and Mk2+/+ mice did not reach statistical significance (Fig. 6C and E). Interestingly, analysis of calvarial tissue RNA revealed that tnfsf11 (RANKL) RNA counts were reduced approximately 3.5-fold in A. actinomycetemcomitans-treated Mk2−/− mice compared to Mk2+/+ mice at day 3 (P ≤ 0.05; Fig. 6D). Furthermore, we elucidated that MK2 signaling did not alter the percentage of circulating common monocyte progenitor cells, marked by CD11b+ Ly6Chi CCR2hi (see Fig. S1A and B in the supplemental material), suggesting that the differences in bone loss detected were not due to a decrease in peripheral osteoclast progenitor cells.
FIG 6.
A. actinomycetemcomitans induces murine calvarial bone loss through MK2. (A) Representative three-dimensional reconstruction of μCT results for mouse calvaria. Red arrows, areas of calvarial resorption that have been magnified 6×. (B) Calvaria resorption pit enumeration for A. actinomycetemcomitans-treated mice (n = 3 to 5). (C) Representative ×200-magnification images of mouse calvaria for TRAP staining for osteoclasts. Red arrows, osteoclasts in contact with bone surface. (D) Results of analysis of tnfsf11 (RANKL) RNA counts from mouse calvarial tissue treated for 3 days (n = 3 to 5) by use of the NanoString Technologies immunology panel. (E) Enumeration of the number of osteoclasts (OC) per calvarial bone perimeter (B.Pm.) for mice treated for 3 days and 5 days (n = 3 to 5). Data are expressed as means ± SEMs. *, P ≤ 0.05; **, P ≤ 0.01.
DISCUSSION
In the present study, our data clearly showed that MK2 signaling regulates the inflammation and bone loss induced by A. actinomycetemcomitans in the calvarial and air pouch murine bacterial challenge models. Interestingly, MK2 signaling did not regulate inflammatory infiltrate, but by taking a closer look at the constituents of the inflammatory infiltrate, we showed that MK2 positively regulates macrophage polarization during A. actinomycetemcomitans challenge. MK2 deficiency caused a trend toward a decrease in the level of the F4/80-positive mature macrophage infiltrate during A. actinomycetemcomitans challenge. Furthermore, MK2 deficiency led to significant reductions in the levels of the following macrophage RNA transcripts: Emr1, Itgam, Itgal, Csf1r, Tnf, and Nos2. This suggests that MK2 positively regulated macrophage differentiation during A. actinomycetemcomitans pathogenesis. In particular, Itgal encodes CD11a, which is an integrin component of LFA-1. LFA-1 is considered a receptor for A. actinomycetemcomitans leukotoxin and is required for A. actinomycetemcomitans internalization, which initiates cell death (27). Since MK2 deficiency downregulated Itgal, the decreases in the amount of macrophage infiltrate seen were not likely due to A. actinomycetemcomitans-induced monocyte apoptosis. By looking at both proapoptosis and antiapoptosis genes, we also confirmed that A. actinomycetemcomitans did not induce apoptosis through MK2 signaling (see Fig. S2A and B in the supplemental material). MK2 signaling also did not alter live bacterial counts at the end of treatment (Fig. S2C). These results highlight that MK2 deficiency is required for attenuation of the host inflammatory response without changing bacterial growth.
Aggregatibacter actinomycetemcomitans caused robust MK2 and p38 phosphorylation in BMDMs and in murine calvarial tissues. We observed that total p38 MAPK levels were lower in untreated and A. actinomycetemcomitans-treated Mk2−/− mouse BMDMs. These results are consistent with previous findings showing that during MK2 deficiency total p38 is destabilized but p-p38 levels remain unchanged (28–30). Phosphorylation of MK2 was recapitulated in vivo and was similar to the MK2 phosphorylation by A. actinomycetemcomitans in BMDMs, indicating that macrophages contribute to MK2 protein levels at the site of infection.
Next, we identified the chemokines and their receptors that were regulated by A. actinomycetemcomitans and MK2 signaling. MK2 signaling was essential for expression of the chemokine receptors Ccr4 and Ccr5 and chemokines Ccl3, Ccl4, Ccl5, and Ccl12. During infection, Mk2−/− mice had increased levels of Ccr4 RNA, which encodes the receptor for CCL2, -4, -5, -17, and -22. This increase in Ccr4 RNA levels can be correlated with trends of increased CCL4 and CCL5 protein levels in Mk2−/− mouse calvarial tissue compared to those in Mk2+/+ mouse calvarial tissues. We identified in vivo a novel RNA transcript, Ccl12, that A. actinomycetemcomitans had not yet been shown to activate. Ccl12 encodes the protein monocyte chemotactic protein 5 (MCP-5), which is known to bind only to CCR2 (31). While it was clear that MK2 signaling regulated chemokine gene expression, MK2 was also critical for the presence of the CCL3 protein in calvarial tissue after 3 days of A. actinomycetemcomitans infection, yet no differences in CCL4 protein levels in the presence or absence of MK2 were detected. These data revealed that MK2 signaling significantly regulates the CCL3/CCR5 signaling axis.
We gained further novel insight into the mechanisms of MK2 regulation during A. actinomycetemcomitans pathogenesis by utilizing the bone marrow transplant mouse model. Trends that led to a 2-fold reduction in CCL3 protein levels and a 2-fold increase in CCL4 levels in Mk2−/− mice compared to those in WT mice were observed in the Mk2−/− mouse hematopoietic cells, suggesting a possible compensatory role of chemokines during A. actinomycetemcomitans challenge. These results were consistent with the total protein levels measured in the murine calvarial model, where we observed a significant decrease in local CCL3 levels in Mk2−/− mice compared to Mk2+/+ mice during bacterial challenge. However, in the calvarial model, MK2 did not regulate CCL4 in the tissue but instead regulated circulating CCL4. Thus, we determined that global MK2 deficiency in mice differentially regulated chemokines in the local and peripheral compartments. MK2 signaling also proved to significantly regulate CCL4 in the nonhematopoietic system-derived cells during the air pouch challenge with A. actinomycetemcomitans. Given that these data strongly support the notion that although no differences may be detected in initial studies utilizing globally deficient mice, it is crucial to tease apart the roles of the hematopoietic and nonhematopoietic systems.
Despite the differences in the amounts of chemokines that were detected, there were no changes in the amounts of circulating CD11b+ CCR5+ cells, which bind redundant CCL3 and CCL4 ligands, potentially because MK2 does not regulate all of the ligands for the CCR5 receptor. While it is apparent that MK2 regulated the chemokine/chemokine receptor axis, chemokine ligand redundancy may contribute to the delayed macrophage activation in Mk2−/− mice by day 5 of A. actinomycetemcomitans challenge. For example, while we detected a significant decrease in CCL3, which is a CCR5 agonist in MK2-deficient mice, MK2 deficiency also caused a trend toward increasing CCL5 levels, which bind to CCR5 as well. These results can be explained by studies showing that during A. actinomycetemcomitans infection CCL3-deficient mice had numbers of CCR5- and CCR1-positive cells similar to the numbers in non-CCL3-deficient mice, which was attributed to the biologically homologous roles of CCL4 or CCL5 interactions with CCR5/CCR1 receptors (20). Our results show that the functionally redundant chemokines CCL3 and CCL4 were regulated by MK2 during A. actinomycetemcomitans infection, but the overall decrease was not robust enough to regulate circulating monocytes and their overall recruitment to the local site of infection.
Next, we demonstrated that MK2 signaling regulated pathogenic bone loss. Previous findings showed that CCL3 upregulated RANKL, a critical bone-resorbing cytokine (32), and both CCL3 and RANKL were regulated by MK2 in our murine calvarial bone loss model. Furthermore, MK2 signaling was essential for physiological RANKL-induced osteoclast formation (30, 33) and LPS-induced osteoclast formation (34). The murine calvarial model was an exceptional way to study periopathogenic influences on bone remodeling because the calvariae, maxilla, and mandible are all formed by intramembranous ossification. Osteoclast formation indeed increased from days 3 to 5 during A. actinomycetemcomitans treatment in Mk2+/+ mice but not in Mk2−/− mice. There was a trend toward decreasing numbers of osteoclasts per bone perimeter surface in Mk2−/− mice compared to Mk2+/+ mice at 5 days after infection. Osteoclast levels correlated with increased numbers of resorption pits in Mk2+/+ mice. In the absence of MK2, bone loss was significantly reduced, further confirming the positive regulatory role of MK2 signaling in bone resorption. We determined that osteoclast deficiencies in Mk2−/− mice were not due to a basal deficiency in circulating CD11b+ Ly6Chi CCR2hi osteoclast/macrophage common progenitors. These data suggest that MK2 signaling affects calvarial skeletal homeostasis, resulting in net bone loss at the local site of infection.
Our study determined that MK2 signaling regulates macrophages and osteoclasts. Both cells types are derived from monocyte progenitors in the supraphysiological host response associated with A. actinomycetemcomitans-induced inflammation and bone loss. Most interestingly, we identified the role of MK2 signaling in the regulation of the chemokine gradient during A. actinomycetemcomitans pathogenesis. Chemokines CCL3 and CCL4 were highly regulated by MK2 signaling during A. actinomycetemcomitans pathogenesis. These findings provide novel insight into the systemic role of MK2 signaling in A. actinomycetemcomitans-induced inflammation and bone loss related to mechanisms of periodontal disease pathogenesis. MK2 deficiency did not affect bacterial viability by impeding the host response, suggesting that intervention using MK2 inhibitors may still have therapeutic value, despite the presence of bacterial infection. In conclusion, MK2 proved to be a critical osteoimmunoregulatory protein involved in chemokine regulation and monocyte plasticity during A. actinomycetemcomitans pathogenesis.
MATERIALS AND METHODS
Ethics statement.
Eight- to 12-week-old C57BL/6 Mk2+/+ and Mk2−/− mice were obtained by material transfer from Germany (28), bred at the Medical University of South Carolina Animal Facility, and maintained in accordance with NIH guidelines. Animals were euthanized via CO2 asphyxiation, followed by cervical dislocation. Mice were subject to food and tap water ad libitum, and they were maintained under normal 12-h light and 12-h dark cycles. All experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the Medical University of South Carolina under protocol number 2718.
Bacterial culture.
Aggregatibacter actinomycetemcomitans strain VT1729, serotype b, expressing the green fluorescent protein (obtained from the University of Vermont, Burlington, VT, USA) was originally derived from A. actinomycetemcomitans clinical isolate SUNY 465 (35). This serotype b strain was used in this study because of its high pathogenicity and high prevalence in periodontal disease in the United States (15, 16). Following previously described methods, A. actinomycetemcomitans was initially plated on 3% Trypticase soy broth and 0.6% yeast extract (TSBYE) 1.5% agar plates with 100 μg/ml ampicillin (35). Aggregatibacter actinomycetemcomitans was then grown for 2 days in a 10% CO2 incubator at 37°C and expanded in TSBYE broth containing 100 μg/ml ampicillin overnight in 10% CO2 at 37°C on a shaker. The optical density of Aggregatibacter actinomycetemcomitans at a 495-nm wavelength was measured, and an optical density of 0.3 equated to 6 × 108 CFU/ml. Aggregatibacter actinomycetemcomitans was washed with PBS to remove the growth medium and centrifuged at a 1,500 relative centrifugal force for 10 min.
Murine calvarial model.
Eight- to 12-week-old male mice were treated with A. actinomycetemcomitans or PBS vehicle. Live A. actinomycetemcomitans bacteria were centrifuged, washed with PBS, and resuspended at a volume of 6.67 × 107 CFU A. actinomycetemcomitans/μl PBS. Thirty microliters of the PBS vehicle or 30 μl of 2 × 109 CFU of live A. actinomycetemcomitans bacteria mixed with PBS was injected subcutaneously and supraosteally at the calvarial midsagittal suture into Mk2+/+ and Mk2−/− mice of strain Mapkapk2tm1Mgl (28). This reproducible injection site was at the midpoint between the eyes and ears. Injections were repeated every 24 h until harvest on either day 3 or day 5 of injection. Tissues were harvested 18 h after the final injection.
Immunohistochemistry and analysis.
After fixation and dissection, calvaria were decalcified in 0.5 M EDTA, pH 8.0, with the solution being replaced 6 times in 2 weeks. Decalcified calvaria were embedded in paraffin followed by preparation of 7-μm cryostat sections that were placed on glass slides. A pathologist blind to the treatment used an ordinal scale to score slides with hematoxylin and eosin (H&E) staining. The following scale was used to measure inflammatory cell infiltrate: 0, no significant infiltrate; 1, mild infiltrate (cell number, <500); 2, moderate infiltrate (cell number, 501 to 1,000); 3, severe infiltrate (cell number, 1,001 to 1,500); and 4, extremely severe infiltrate (cell number, >1501). For mature macrophage F4/80 staining, an antigen retrieval protocol was performed by incubating slides at 60°C overnight in 0.2 M boric acid, pH 7.0, retrieval buffer. The slides were blocked for exogenous peroxidase activity and nonspecific secondary antibody interactions using hydrogen peroxide block and 10% normal goat serum. Primary F4/80 rat anti-mouse antibody (Abcam, Cambridge, MA, USA) was applied overnight at 4°C in a humidified chamber. The slides were incubated in biotinylated goat anti-rat immunoglobulin secondary antibody for 1 h at room temperature. 3,3-Diaminobenzidine (DAB) substrate was used to visualize the stain. Subsequently, the slides were counterstained using 15% hematoxylin, dehydrated in alcohol, and mounted with Cytoseal mounting medium. Quantification of F4/80-positive regions in the original ×200-magnification tiff files was performed using Visiopharm analysis software, version 4.4.8.201 (Hoersholm, Denmark). The images were segmented into 3 classes to distinguish between the DAB-stained area, the hematoxylin-stained area, and the white background.
nCounter analysis.
RNA was isolated from calvarial tissue from PBS vehicle-treated or A. actinomycetemcomitans-treated 8- to 12-week-old male Mk2+/+ and Mk2−/− mice. Two hundred nanograms of RNA, quantified using a NanoDrop spectrophotometer, was used as the input into the NanoString Technologies immunology panel. Gene expression was quantified using the nCounter mouse immunology gene expression code set with the nCounter analysis system (NanoString Technologies, Seattle, WA, USA) following the manufacturer's protocol. The levels of RNA in the samples were normalized to the geometric mean level of seven housekeeping genes, Rpl19, Tubb5, Alas1, Tbp, Gusb, Gapdh, and Oaz1. Each housekeeping gene had a low coefficient of variance of less than 37%. Results were analyzed using nSolver analysis software, version 2.0.
Cell sorting.
Bone marrow cells were harvested from 8- to 12-week-old male Mk2+/+ and Mk2−/− mouse tibia, femur, and humerus using sterile technique. Cells were plated at a density of 1 mouse/10-cm tissue culture-treated dish in alpha minimal essential medium (Invitrogen, Carlsbad, CA, USA) containing 1% penicillin-streptomycin and 10% fetal bovine serum (Atlanta Biologicals, Inc., Flowery Branch, GA, USA). Bone marrow cells were incubated overnight in 5% CO2 at 37°C. The cells remaining in suspension (hematopoietic stem cells [HSCs]) were incubated with anti-CD11b−-conjugated magnetic beads prior to sorting with an AutoMACS system (Miltenyi Biotec Inc., San Diego, CA, USA). Cells were sorted into CD11b+ and CD11b− populations. Sorted cells were plated at a density of 2 × 106 cells/cm2 and treated with 25 ng/ml recombinant mouse (rm) macrophage colony-stimulating factor (M-CSF; R&D Systems Inc., Minneapolis, MN, USA) for 6 days. Cytokines were replaced every 2 days.
Protein isolation and immunoblotting.
In vitro, sorted CD11b+ bone marrow cells were differentiated into bone marrow-derived macrophages (BMDMs) by treatment with M-CSF (25 ng/ml) for 6 days. Cytokines were replaced every 2 days. Cells were collected and lysed in radioimmunoprecipitation assay (RIPA) buffer at 0, 15, and 30 min after stimulation with A. actinomycetemcomitans at a concentration of 10 CFU of A. actinomycetemcomitans per cell.
In vivo, the calvarial tissue overlying the injection site was harvested from mice that had been treated with A. actinomycetemcomitans for 3 days, snap-frozen in liquid nitrogen, and stored at −80°C until further processing. The tissue was then homogenized in 500 μl RIPA buffer and sonicated at maximum speed for 40 s with a 1-s on and 1-s off pulse. Twenty to 30 μg of protein was run on a 10% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane using a Trans-Blot Turbo transfer system (Bio-Rad Laboratories Inc., Hercules, CA, USA), and blocked in 5% skim milk for 1 h at room temperature prior to incubation in primary antibody. Membranes were incubated at a 1:1,000 dilution overnight at 4°C in primary antibodies to the following: p-MK2 (Thr334; rabbit monoclonal antibody [MAb] 27B7), MK2 (rabbit polyclonal antibody), p-p38 (Thr180/Tyr182; rabbit polyclonal antibody), p38 (rabbit polyclonal antibody), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; rabbit MAb 14C10) (Cell Signaling Technology Inc., Beverly, MA, USA). The blots were incubated at room temperature for 1 h in anti-rabbit IgG-horseradish peroxidase secondary antibody (Cell Signaling Technology Inc., Beverly, MA, USA). Protein was visualized by use of a chemiluminescence substrate (Thermo Scientific, Pittsburgh, PA, USA). Densitometric analysis was performed by taking images on a Gel-Doc XR system and analyzing the images with Quantity One (version 4.6.1) software (Bio-Rad Laboratories Inc., Hercules, CA, USA).
RNA isolation and quantitative PCR (qPCR).
RNA was isolated from BMDMs and mouse soft tissue near the midsagittal suture of the mouse calvaria overlying the A. actinomycetemcomitans or PBS injection site. Mouse tissue was snap-frozen in liquid nitrogen and stored at −80°C until RNA isolation. RNA was immediately extracted from BMDMs and from thawed mouse tissue using an RNeasy Miniprep kit (Qiagen Inc., Valencia, CA, USA) following the manufacturer's protocol. Mouse tissue was homogenized in 600 μl lysis buffer. BMDMs were washed once with ice-cold PBS and were harvested in 350 μl lysis buffer. Subsequently, lysates were processed using the RNeasy spin columns. RNA was quantified using a NanoDrop spectrophotometer, and reverse transcription was performed using 300 ng of RNA from BMDMs and 1,000 ng of RNA from calvarial tissue (Applied Biosystems Inc., Foster City, CA, USA). BMDM samples were diluted 3-fold, and calvarial tissue RNA was diluted 10-fold with ultrapure water. Samples were run with primers specific for Ccl3, Ccl4, and Gapdh (housekeeping) using a StepOnePlus real-time PCR system (Applied Biosystems Inc., Foster City, CA, USA). The level of RNA was normalized to the level of Gapdh RNA, and relative mRNA levels were determined using the ΔΔCT threshold cycle (CT) values and are presented as the fold change relative to the levels for control Mk2+/+ mice or cells.
Flow cytometry.
Mouse bone marrow cells and peripheral blood were harvested from the femurs of 12-week-old male mice treated with the PBS vehicle or A. actinomycetemcomitans. Peripheral blood was isolated using 3 mg/ml EDTA and 2% dextran in PBS. Subsequently, the remaining red blood cells were lysed in lysis buffer and enumerated with trypan blue dead cell exclusion dye. Cells were stained with commercially available antibodies, including anti-CD11b−-allophycocyanin (APC) (clone M1/70, rat IgG2b; Miltenyi Biotec Inc., San Diego, CA, USA), anti-CD11b−-fluorescein isothiocyanate (clone M1/70, rat IgG2b; Miltenyi Biotec Inc., San Diego, CA, USA), anti-mouse CCR1-APC (clone 643854, rat IgG2b; R&D Systems, Minneapolis, MN, USA), anti-mouse CD195/CCR5-peridinin chlorophyll protein-Alexa Fluor 710 (clone 7A4, Armenian hamster IgG; eBioscience, Inc., San Diego, CA, USA), and anti-mouse CCR2-APC (clone 475301, rat IgG2b; R&D Systems, Minneapolis, MN, USA) following previously described methods (36). Prior to analysis, debris was gated out using forward scatter and side scatter plots, and dead cells were excluded using propidium iodide staining.
Murine bone marrow chimera and air pouch model.
Mk2+/+ and Mk2−/− mice expressing CD45.2 and C57BL/6J CD45.1 wild-type (WT) mice (stock number 002014) purchased from The Jackson Laboratory were used for the bone marrow chimera experiments. A maximum of one 8- to 12-week-old sex-matched donor mouse was used for every 5 irradiated recipient mice. Six- to 8-week-old male and female recipient mice received total body irradiation consisting of 2 sublethal doses of 550 cGy separated by 4 h in a JL Shepherd model 143/137 cesium irradiator. Each recipient mouse received 2 × 106 red blood cell-depleted donor cells 24 h after the first dose of radiation by tail vein injection. We transplanted bone marrow from Mk2+/+ mice (CD45.2, control) or Mk2−/− mice (CD45.2, Mk2−/− in the hematopoietic compartment) into irradiated 6- to 8-week-old recipient CD45.1 WT mice and transplanted bone marrow from CD45.1 WT mice into irradiated 6- to 8-week-old CD45.2 Mk2+/+ (control) or CD45.2 Mk2−/− (Mk2−/− in the nonhematopoietic compartment) recipient mice. Complete chimerism was detected by flow cytometry using CD45.1 and CD45.2 antibodies (data not shown).
The air pouch model was used to study monocyte chemotaxis, which is a critical first step in the inflammatory process. Mice were injected subcutaneously, using a 3-ml syringe and a 27.5-gauge needle, with 3 ml of air to create a dorsal cavity at 8 weeks after bone marrow transplantation, followed by another injection of 1.5 ml of air 4 days later. A. actinomycetemcomitans strain VT1729 (2 × 109 CFU) was diluted in 1 ml PBS and injected into the dorsal airspace 7 days after the initial air injection. Mice were euthanized 18 h after the bacterial infection and injected 3 times with 1 ml PBS to retrieve air pouch exudate utilizing a 20-gauge needle. Air pouch exudate was stored at −80°C until further processing.
Multiplex assay and ELISA.
For the multiplex assay, peripheral blood plasma was collected, using lithium heparin separation, from 8- to 12-week-old male Mk2+/+ and Mk2−/− mice after calvarial treatment with PBS vehicle or A. actinomycetemcomitans. The mouse plasma was stored at −80°C until further use. Fifty microliters of plasma and 900 μg/ml of tissue protein were utilized for the Bio-Plex mouse cytokine 23-plex assay (Bio-Rad Laboratories Inc., Hercules, CA, USA) following the manufacturer's protocol described in Bio-Rad Technical Bulletins 10014905 and 10024985. In brief, plasma was diluted 4-fold and tissue was diluted 2-fold. Assay plates were read using a Luminex plate reader.
Air pouch exudate was collected by removal of cells and debris by centrifugation, stored at −80°C, and thawed once for use in the mouse CCL3/MIP-1α Quantikine and the mouse CCL4/MIP-1β Quantikine enzyme-linked immunosorbent assays (ELISAs; R&D Systems, Minneapolis, MN, USA). Air pouch exudates were processed following the manufacturer's protocol (R&D Systems, Minneapolis, MN, USA). Plates were read using a VersaMax plate reader (Molecular Devices, Sunnyvale, CA, USA) at an optical density of 450 nm. Chemokine levels were normalized to the total protein level, which was quantified using a Pierce bicinchoninic acid protein assay kit (Thermo Scientific, Pittsburgh, PA, USA).
Micro-computed tomography.
Calvaria were harvested from 12-week-old male mice treated with PBS vehicle or A. actinomycetemcomitans. Whole heads were fixed for 3 days at room temperature in 10% formalin in PBS, followed by dissection of calvaria with soft tissue. Calvarial scanning and analysis were performed using a Scanco Medical μCT 40 system (Scanco USA, Wayne, PA, USA). Calvaria were reoriented to align the midsagittal suture along the y axis. The volume of interest (VOI) was defined as 999 slices in the X and Y directions about the midsagittal suture and 590 slices in the Z direction. Segmentation values were set between 292 and 1,000 to distinguish bone from nonmineralized tissue within the respective ROI.
Osteoclast histology.
Tartrate-resistant acid phosphatase (TRAP) staining of calvarial tissue slides was performed to identify osteoclasts. Formalin-fixed, paraffin-embedded tissue sections were deparaffinized and rehydrated in PBS. The slides were incubated at 37°C in freshly prepared TRAP buffer for 30 min as described in BD Bioscience Technical Bulletin 445. Next, the slides were counterstained using 15% hematoxylin, dehydrated in alcohol, and mounted with Cytoseal mounting medium. For analysis, 3 random ×200-magnification images per sample were captured, and osteoclasts were enumerated as TRAP-positive cells adjacent to calvarial bone.
Statistical analysis.
Data were analyzed with GraphPad Prism software, version 4.0. A two-tailed Mann-Whitney test was used to compare values between 2 groups. A Kruskal-Wallis test followed by a Dunn's multiple-comparison post hoc test was used to compare data from multiple groups. For the RT-qPCR experiments, untransformed ΔCT values were compared by use of a one-tailed Mann-Whitney test for statistical analysis. Results are expressed as the mean ± standard error of the mean (SEM) for at least three (n = 3) biological replicates from each experiment.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00552-16.
REFERENCES
- 1.Slots J, Reynolds HS, Genco RJ. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun 29:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slots J. 1976. The predominant cultivable organisms in juvenile periodontitis. Scand J Dent Res 84:1–10. [DOI] [PubMed] [Google Scholar]
- 3.Newman MG, Socransky SS, Savitt ED, Propas DA, Crawford A. 1976. Studies of the microbiology of periodontosis. J Periodontol 47:373–379. doi: 10.1902/jop.1976.47.7.373. [DOI] [PubMed] [Google Scholar]
- 4.Van der Weijden GA, Timmerman MF, Reijerse E, Wolffe GN, Van Winkelhoff AJ, Van der Velden U. 1994. The prevalence of A. actinomycetemcomitans, P. gingivalis and P intermedia in selected subjects with periodontitis. J Clin Periodontol 21:583–588. [DOI] [PubMed] [Google Scholar]
- 5.Nakano K, Inaba H, Nomura R, Nemoto H, Tamura K, Miyamoto E, Yoshioka H, Taniguchi K, Amano A, Ooshima T. 2007. Detection and serotype distribution of Actinobacillus actinomycetemcomitans in cardiovascular specimens from Japanese patients. Oral Microbiol Immunol 22:136–139. doi: 10.1111/j.1399-302X.2007.00332.x. [DOI] [PubMed] [Google Scholar]
- 6.Stepanovic S, Tosic T, Savic B, Jovanovic M, K'Ouas G, Carlier JP. 2005. Brain abscess due to Actinobacillus actinomycetemcomitans. APMIS 113:225–228. doi: 10.1111/j.1600-0463.2005.apm1130312.x. [DOI] [PubMed] [Google Scholar]
- 7.Zijlstra EE, Swart GR, Godfroy FJ, Degener JE. 1992. Pericarditis, pneumonia and brain abscess due to a combined Actinomyces-Actinobacillus actinomycetemcomitans infection. J Infect 25:83–87. doi: 10.1016/0163-4453(92)93633-2. [DOI] [PubMed] [Google Scholar]
- 8.Cuende E, de Pablos M, Gomez M, Burgaleta S, Michaus L, Vesga JC. 1996. Coexistence of pseudogout and arthritis due to Actinobacillus actinomycetemcomitans. Clin Infect Dis 23:657–658. doi: 10.1093/clinids/23.3.657. [DOI] [PubMed] [Google Scholar]
- 9.Kapopara PR, von Felden J, Soehnlein O, Wang Y, Napp LC, Sonnenschein K, Wollert KC, Schieffer B, Gaestel M, Bauersachs J, Bavendiek U. 2014. Deficiency of MAPK-activated protein kinase 2 (MK2) prevents adverse remodelling and promotes endothelial healing after arterial injury. Thromb Haemost 112:1264–1276. doi: 10.1160/TH14-02-0174. [DOI] [PubMed] [Google Scholar]
- 10.Shalini S, Ganesh P, Anand AR. 1995. Actinobacillus actinomycetemcomitans septicemia during pregnancy. Int J Gynaecol Obstet 51:57–58. doi: 10.1016/0020-7292(95)80010-A. [DOI] [PubMed] [Google Scholar]
- 11.van Winkelhoff AJ, Overbeek BP, Pavicic MJ, van den Bergh JP, Ernst JP, de Graaff J. 1993. Long-standing bacteremia caused by oral Actinobacillus actinomycetemcomitans in a patient with a pacemaker. Clin Infect Dis 16:216–218. doi: 10.1093/clind/16.2.216. [DOI] [PubMed] [Google Scholar]
- 12.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Gunsolley J. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J Clin Microbiol 45:3859–3869. doi: 10.1128/JCM.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zambon JJ. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol 12:1–20. doi: 10.1111/j.1600-051X.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 14.Bandhaya P, Saraithong P, Likittanasombat K, Hengprasith B, Torrungruang K. 2012. Aggregatibacter actinomycetemcomitans serotypes, the JP2 clone and cytolethal distending toxin genes in a Thai population. J Clin Periodontol 39:519–525. doi: 10.1111/j.1600-051X.2012.01871.x. [DOI] [PubMed] [Google Scholar]
- 15.Kawamoto D, Ando ES, Longo PL, Nunes AC, Wikstrom M, Mayer MP. 2009. Genetic diversity and toxic activity of Aggregatibacter actinomycetemcomitans isolates. Oral Microbiol Immunol 24:493–501. doi: 10.1111/j.1399-302X.2009.00547.x. [DOI] [PubMed] [Google Scholar]
- 16.Celenligil H, Ebersole JL. 1998. Analysis of serum antibody responses to periodontopathogens in early-onset periodontitis patients from different geographical locations. J Clin Periodontol 25:994–1002. doi: 10.1111/j.1600-051X.1998.tb02404.x. [DOI] [PubMed] [Google Scholar]
- 17.Zambon JJ, DeLuca C, Slots J, Genco RJ. 1983. Studies of leukotoxin from Actinobacillus actinomycetemcomitans using the promyelocytic HL-60 cell line. Infect Immun 40:205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang HW, Asikainen S, Dogan B, Suda R, Lai CH. 2004. Relationship of Actinobacillus actinomycetemcomitans serotype b to aggressive periodontitis: frequency in pure cultured isolates. J Periodontol 75:592–599. doi: 10.1902/jop.2004.75.4.592. [DOI] [PubMed] [Google Scholar]
- 19.Diaz-Zuniga J, Monasterio G, Alvarez C, Melgar-Rodriguez S, Benitez A, Ciuchi P, Garcia M, Arias J, Sanz M, Vernal R. 2015. Variability of the dendritic cell response triggered by different serotypes of Aggregatibacter actinomycetemcomitans or Porphyromonas gingivalis is Toll-like receptor 2 (TLR2) or TLR4 dependent. J Periodontol 86:108–119. doi: 10.1902/jop.2014.140326. [DOI] [PubMed] [Google Scholar]
- 20.Repeke CE, Ferreira SB Jr, Claudino M, Silveira EM, de Assis GF, Avila-Campos MJ, Silva JS, Garlet GP. 2010. Evidences of the cooperative role of the chemokines CCL3, CCL4 and CCL5 and its receptors CCR1+ and CCR5+ in RANKL+ cell migration throughout experimental periodontitis in mice. Bone 46:1122–1130. doi: 10.1016/j.bone.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 21.Garlet GP, Avila-Campos MJ, Milanezi CM, Ferreira BR, Silva JS. 2005. Actinobacillus actinomycetemcomitans-induced periodontal disease in mice: patterns of cytokine, chemokine, and chemokine receptor expression and leukocyte migration. Microbes Infect 7:738–747. doi: 10.1016/j.micinf.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Jacome-Galarza CE, Lee SK, Lorenzo JA, Aguila HL. 2013. Identification, characterization, and isolation of a common progenitor for osteoclasts, macrophages, and dendritic cells from murine bone marrow and periphery. J Bone Miner Res 28:1203–1213. doi: 10.1002/jbmr.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbert BA, Novince CM, Kirkwood KL. 2016. Aggregatibacter actinomycetemcomitans, a potent immunoregulator of the periodontal host defense system and alveolar bone homeostasis. Mol Oral Microbiol 31:207–227. doi: 10.1111/omi.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Donnelly R, Gunsolley J. 2009. Macrophage inflammatory protein-1alpha: a salivary biomarker of bone loss in a longitudinal cohort study of children at risk for aggressive periodontal disease? J Periodontol 80:106–113. doi: 10.1902/jop.2009.080296. [DOI] [PubMed] [Google Scholar]
- 25.Dunmyer J, Herbert B, Li Q, Zinna R, Martin K, Yu H, Kirkwood KL. 2012. Sustained mitogen-activated protein kinase activation with Aggregatibacter actinomycetemcomitans causes inflammatory bone loss. Mol Oral Microbiol 27:397–407. doi: 10.1111/j.2041-1014.2012.00656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Travan S, Li F, D'Silva NJ, Slate EH, Kirkwood KL. 2013. Differential expression of mitogen activating protein kinases in periodontitis. J Clin Periodontol 40:757–764. doi: 10.1111/jcpe.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur M, Kachlany SC. 2014. Aggregatibacter actinomycetemcomitans leukotoxin (LtxA; Leukothera) induces cofilin dephosphorylation and actin depolymerization during killing of malignant monocytes. Microbiology 160:2443–2452. doi: 10.1099/mic.0.082347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk HD, Gaestel M. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol 1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 29.Carayol G, Giron-Michel J, Azzarone B, Castagna L, Cambier N, Mishal Z, Bourhis JH, Chouaib S, Caignard A. 2000. Altered natural killer cell differentiation in CD34+ progenitors from chronic myeloid leukemia patients. Oncogene 19:2758–2766. doi: 10.1038/sj.onc.1203584. [DOI] [PubMed] [Google Scholar]
- 30.Herbert BA, Valerio MS, Gaestel M, Kirkwood KL. 2015. Sexual dimorphism in MAPK-activated protein kinase-2 (MK2) regulation of RANKL-induced osteoclastogenesis in osteoclast progenitor subpopulations. PLoS One 10:e0125387. doi: 10.1371/journal.pone.0125387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarafi MN, Garcia-Zepeda EA, MacLean JA, Charo IF, Luster AD. 1997. Murine monocyte chemoattractant protein (MCP)-5: a novel CC chemokine that is a structural and functional homologue of human MCP-1. J Exp Med 185:99–109. doi: 10.1084/jem.185.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsubaki M, Kato C, Manno M, Ogaki M, Satou T, Itoh T, Kusunoki T, Tanimori Y, Fujiwara K, Matsuoka H, Nishida S. 2007. Macrophage inflammatory protein-1alpha (MIP-1alpha) enhances a receptor activator of nuclear factor kappaB ligand (RANKL) expression in mouse bone marrow stromal cells and osteoblasts through MAPK and PI3K/Akt pathways. Mol Cell Biochem 304:53–60. doi: 10.1007/s11010-007-9485-7. [DOI] [PubMed] [Google Scholar]
- 33.Braun T, Lepper J, Ruiz Heiland G, Hofstetter W, Siegrist M, Lezuo P, Gaestel M, Rumpler M, Thaler R, Klaushofer K, Distler JH, Schett G, Zwerina J. 2013. Mitogen-activated protein kinase 2 regulates physiological and pathological bone turnover. J Bone Miner Res 28:936–947. doi: 10.1002/jbmr.1816. [DOI] [PubMed] [Google Scholar]
- 34.Li Q, Yu H, Zinna R, Martin K, Herbert B, Liu A, Rossa C Jr, Kirkwood KL. 2011. Silencing mitogen-activated protein kinase-activated protein kinase-2 arrests inflammatory bone loss. J Pharmacol Exp Ther 336:633–642. doi: 10.1124/jpet.110.172395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang G, Kitten T, Munro CL, Wellman GC, Mintz KP. 2008. EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect Immun 76:2316–2324. doi: 10.1128/IAI.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valerio MS, Herbert BA, Griffin AC III, Wan Z, Hill EG, Kirkwood KL. 2014. MKP-1 signaling events are required for early osteoclastogenesis in lineage defined progenitor populations by disrupting RANKL-induced NFATc1 nuclear translocation. Bone 60:16–25. doi: 10.1016/j.bone.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.