ABSTRACT
Toll-like receptor 9 (TLR9)-deficient (TLR9−/−) mice are resistant to periodontitis, a disease characterized by a dysbiotic microbiota and deregulated immune response and resulting in tooth loss and various systemic conditions. However, the mechanisms and biological pathways by which TLR9 instigates periodontal inflammation are yet to be identified. In a ligature-induced model of periodontitis, we demonstrate that TLR9−/− mice exhibited significantly less alveolar bone loss than their wild-type (WT) counterparts. Consistent with the disease phenotype, gingival tissues showed significantly more inflammatory cell infiltration in the WT ligated but not in the TLR9−/− ligated mice compared to the unligated controls. The peritoneal infection model using Porphyromonas gingivalis, a keystone pathogen for periodontitis, revealed reduced neutrophils in TLR9−/− mice on day 1 postinfection compared to the levels in WT mice. Transcriptomics analyses showed increased expression of A20 (tumor necrosis factor alpha [TNF-α]-induced protein 3 [TNFAIP3]), an inhibitor of the NF-κB pathway and a negative regulator of TLR signaling, in ligated TLR9−/− mouse gingival tissues compared to its expression in the WT. Ex vivo, TLR9−/− bone marrow-derived macrophages produced more A20 than WT cells following P. gingivalis challenge. Clinically, A20 was modestly upregulated in human gingival tissue specimens from chronic periodontitis patients, further confirming the biological relevance of A20 in periodontal inflammation. We conclude that TLR9 modulates periodontal disease progression at both the cellular and molecular level and identify A20 as a novel downstream signaling molecule in the course of periodontal inflammation. Understanding the regulation of the TLR9 signaling pathway and the involvement of A20 as a limiting factor of inflammation will uncover alternative therapeutic targets to treat periodontitis and other chronic inflammatory diseases.
KEYWORDS: A20, TNFAIP3, Toll-like receptor, cytokine, inflammation, periodontal disease
INTRODUCTION
A key feature of the innate immune response to infectious microorganisms is pattern recognition receptors (PRRs), which respond to microbial components by activating proinflammatory molecules to limit microbial insult (1). However, deregulated immune responses and the failure of the immune system to resolve inflammation prolong the inflammatory response and lead to various chronic conditions, including autoimmune disorders, persistent infectious and inflammatory disease, aging-related metabolic conditions, and cancer (2–5). Therefore, it is crucial to understand the molecular and biological pathways related to innate sensing and the interaction of these pathways with each other and the resident microbiome to identify effective therapeutic targets to resolve inflammation and restore tissue homeostasis.
Innate sensors, such as Toll-like receptors (TLRs), play a critical role in immunity as a first line of defense. Although each TLR shows ligand specificity, downstream signaling pathways are redundant and likely communicate with each other. For example, except for TLR3, all TLRs signal through the adaptor protein MyD88, which activates downstream cascades and transcription factors that include mitogen-activated protein kinases (MAPKs), the nuclear factor of kappa-light-chain enhancer of activated B cells (NF-κB), activator protein 1 (AP-1), and interferon regulatory factors (IRFs). Together, these signals culminate in a program of gene expression that allows de novo cytokine/chemokine release while also coordinating the development of both the innate and adaptive immune response to eliminate the pathogen. A thorough understanding of the molecular mechanisms leading to the activation, regulation, and postresolution phases of TLR signaling is crucial in the development of therapeutics for inflammatory conditions.
To avoid excessive inflammatory responses and to return tissues to homeostasis, host cells have developed negative-regulation strategies of TLR signaling (6). A number of regulatory programs to inhibit inflammatory signaling cascades have been described, including deubiquitination of enzymes, sequestration of adaptor proteins, activation of phosphatases, and expression of inhibitors (7). Emerging evidence has identified deubiquitinase (DUB) A20, which is encoded by the gene TNFAIP3 (tumor necrosis factor alpha [TNF-α]-induced protein 3), as a powerful negative regulator of TLR signaling and an inhibitor of the NF-κB pathway. A20 restricts inflammation triggered by TLRs by deubiquitinating TNF receptor-associated factor 6 (TRAF6) and preventing its interaction with the E2 ubiquitin-conjugating enzymes Ubc13 and UbcH5c. It also modifies Ubc13 and UbcH5c by the addition of K48-linked ubiquitin chains, thus targeting these proteins for proteasomal degradation. Thus, A20 disrupts TLR signaling by preventing protein-protein interactions necessary for the propagation of downstream signaling complexes and downregulates the production of proinflammatory cytokines. Furthermore, A20 has been implicated as a disease susceptibility gene for many inflammatory and autoimmune disorders (8). Therefore, more studies are now exploring the potential of A20 as a therapeutic target.
Periodontitis is one of the most prevalent oral diseases, affecting almost half of the American population, and is associated with increased risk of systemic conditions, including diabetes, cardiovascular disease, cancer, adverse pregnancy outcomes, gastrointestinal and pulmonary diseases, and rheumatoid arthritis (9–15). While periodontal diseases are initiated by periodontopathic bacteria and dysbiosis, the deregulated host inflammatory response to this dysbiotic microflora leads to destruction of the tooth supporting structures and eventual tooth loss (16). In this regard, disease outcomes are often guided by the polymicrobial nature of the disease, leading to the engagement of multiple innate sensors, the activation of downstream inflammatory signaling cascades, and the inability of the host to resolve inflammation, which in turn provides nourishment to feed the dysbiosis. A number of PRRs have been implicated in periodontal disease pathogenesis, including plasma membrane-associated TLR2, TLR4, complement receptors, and more recently, intracellular innate sensors, such as TLR9 and nucleotide-binding oligomerization domain-containing (NOD) receptors (17–22). The mechanisms by which TLR2 and complement receptors cause tissue damage in periodontitis are well documented, with P. gingivalis, a keystone periodontal pathogen, instigating cross talk between these receptors in neutrophils to prevent the antimicrobial response while allowing the proinflammatory response to ensue (17). Specifically for TLR9, in vivo evidence revealed that TLR9-deficient (TLR9−/−) mice are resistant to periodontitis using an oral gavage model of periodontitis. This provided the first proof-of-concept evidence that nucleic acid sensors are involved in periodontitis (18). Furthermore, TLR9 has been shown to modulate TLR2- and TLR4-triggered inflammation, suggesting cross talk between these sensors, possibly through downstream signaling pathways, in the course of periodontal inflammation (18). Hence, understanding the mechanisms by which TLR9 contributes to periodontal inflammation can provide important insights on how to control aberrant periodontal inflammation and identify therapeutic targets and disease biomarkers critical both for local and systemic outcomes.
The primary goal of the current study is to characterize the mechanisms by which TLR9 regulates periodontal inflammatory responses. For this purpose, we induced periodontal disease in wild-type (WT) and TLR9−/− mice using a ligature-induced periodontitis model and measured global mRNA changes in the gingival tissues using PCR array analysis. Through this methodical study, which includes in vivo, ex vivo, and clinical studies, we report novel findings related to the interplay of neutrophils, macrophages, and A20, a host deubiquinating enzyme and negative regulator of TLR signaling, as it pertains to periodontal disease. To the best of our knowledge, this is the first study to determine the contribution of both A20 and TLR9 signaling in periodontitis.
RESULTS
TLR9 promotes alveolar bone loss in a murine ligature-induced model of periodontitis.
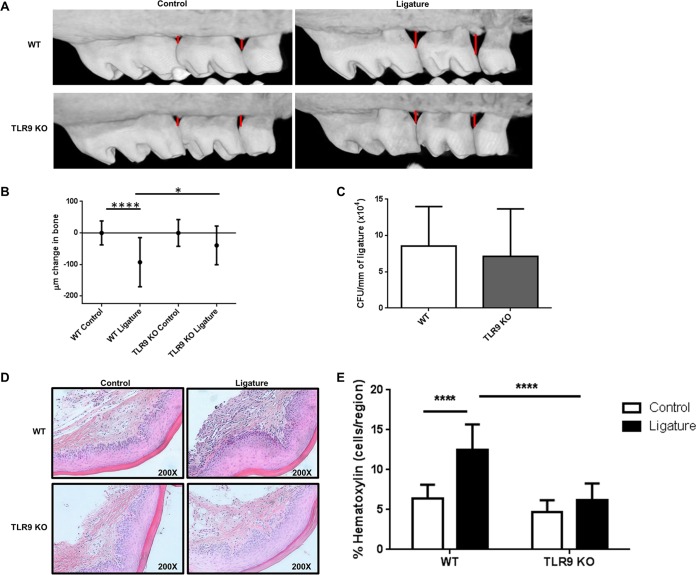
Our previous study identified that in vivo TLR9 signaling facilitates inflammatory and osteoclastogenic cytokine activation and subsequent alveolar bone loss in a chronic periodontitis model using Porphyromonas gingivalis oral gavage (18). To better understand the mechanism by which TLR9 contributes to periodontitis pathogenesis, we utilized the mechanically induced ligature model for periodontal disease in which silk sutures were placed in between all molar teeth. This model represents a more aggressive form of the disease, which is characterized by rapidly progressing bone loss (17, 23). Micro-computed tomography (micro-CT) imaging of alveolar bone levels revealed substantial visible bone destruction in WT ligated mice, while bone loss was not evident in TLR9−/− mice (Fig. 1A). The change in the maxillary bone loss for ligated mice was determined by subtracting the mean cementoenamel junction-alveolar bone crest (CEJ-ABC) distance among unligated controls from the CEJ-ABC distance measured for each ligated mouse. Our data revealed significant bone loss in the WT ligated mice compared to that in WT unligated control sites (Fig. 1B). Although there was slight, insignificant bone loss in the TLR9−/− ligated mice compared to the bone loss in the TLR9−/− controls, WT ligated mice exhibited significantly more bone loss than TLR9−/− ligated mice (Fig. 1B). TLR9 deficiency did not significantly affect the burden of ligature-associated periodontal bacteria compared to that in WT mice (Fig. 1C). As shown by the results in Fig. 1C, the bacterial burden reached 8.5 × 104 CFU per millimeter of ligature in WT mice at the time of euthanasia (day 7). When bacteria in TLR9−/− mice were examined, there were 7.1 × 104 CFU per millimeter of ligature. Spearman test results revealed no correlation between the amount of bone loss and the bacterial load in each group (data not shown). Taken together, these results demonstrate that TLR9 signaling but not a difference in bacterial loads is associated with the induction of bone loss in the murine ligature-induced periodontitis model. It is yet to be determined, however, whether there were any differences in the genotypic composition of the bacteria.
FIG 1.
TLR9 deficiency prevents ligature-induced periodontal bone loss in vivo. On day 7 following ligature placement, WT and TLR9−/− mice were sacrificed, and entire heads were processed for micro-CT imaging. (A) Representative micro-CT images of maxillae from each group of animals. (B) Amounts of bone change in WT and TLR9−/− mice. Negative values represent bone loss in ligated mice relative to the results for the control (unligated) mice. Ten to 18 mice were analyzed. Whiskers show standard deviations. (C) CFU enumeration of periodontal microflora recovered from the ligatures of WT and TLR9−/− mice. The numbers were determined by extracting bacteria from the ligatures and plating serial dilutions of bacterial suspensions onto blood agar plates for anaerobic growth. Ligatures from six to seven mice were included. The average results and standard deviations are shown. (D and E) Gingival tissues were harvested, sectioned, and processed for H&E staining. Representative images of gingival tissue sections from 4 WT and 4 TLR9−/− mice (D) and quantification of inflammatory cells in the tissue sections (E) are shown for ligated and control tissue sections. Tissue specimens from four mice were analyzed in triplicates to sextuplicates in a minimum of 2 independent experiments, and average results and standard deviations are shown. *, P < 0.05; ****, P < 0.0001.
TLR9 deficiency reduces inflammatory cell infiltration in both the ligature-induced periodontitis and P. gingivalis intraperitoneal infection models.
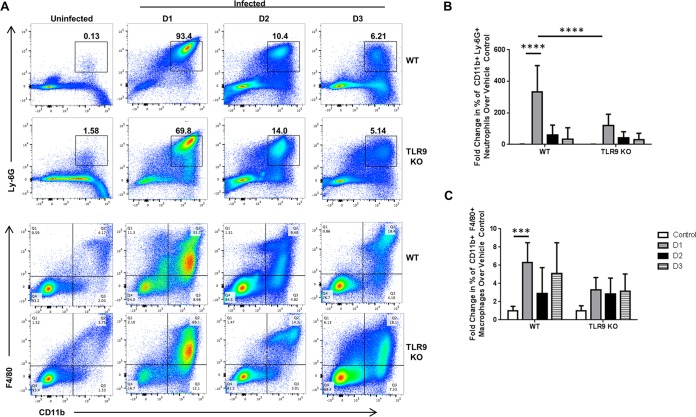
During periodontal disease, deregulated immune and inflammatory responses in reaction to the dysbiotic microflora are responsible for the events leading to bone resorption. Our previous work demonstrated that TLR9−/− mice, compared to their WT controls, exhibited decreased expression of the proinflammatory cytokines interleukin-6 (IL-6) and TNF and the osteoclastogenic molecule RANKL using the oral gavage infection model (18). We therefore hypothesized that TLR9 promotes inflammation by increased immune cell infiltration at the local site of infection. To investigate this, dissected tissues from each group of WT and TLR9−/− mice were processed for hematoxylin-and-eosin (H&E) staining to examine immune cell infiltration using the ligature model. In line with the bone loss data, WT mice exhibited extensive inflammatory cell infiltration, as assessed by hematoxylin-positive nucleated cells in the connective tissue of the gingiva following ligation (Fig. 1D and E). When the infiltration in the ligated TLR9−/− mice was examined, the response was drastically reduced, matching that of the control animals (Fig. 1D and E). Because the inflammatory cell infiltration was less in the gingival tissues of TLR9−/− ligated mice, we next determined whether the lack of TLR9 affects the cellular innate immune response in vivo using the P. gingivalis peritoneal infection model. On day 1 postinfection, the percentage of neutrophils (CD11b+ Ly-6G+) increased 331-fold in WT mice compared to the percentage in noninfected controls, and the increase was significantly less, only half as great (118-fold), in TLR9−/− mice (Fig. 2A and B). By days 2 and 3 postinfection, similar levels of neutrophils were detected for both genotypes. Macrophages are critical to the periodontal lesions, taking part in both innate and adaptive immunity. Therefore, we also determined whether TLR9 deficiency affected the distribution of macrophages in the peritoneal infection model. When we assessed the percentages of cells on days 1, 2, and 3 postinfection, we found that there was no significant difference in the distribution of macrophages (CD11b+ F4/80+) between TLR9−/− mice and their WT counterparts (Fig. 2A and C). Taken together, these results demonstrate that abolishing TLR9 signaling reduces inflammatory cell infiltration in both the ligature and peritoneal infection model. Although neutrophils are the first line of defense, aberrant neutrophil activity is associated with the exacerbated inflammation and catabolic events that lead to periodontal tissue destruction. It is therefore likely that more controlled neutrophil recruitment to the sites in TLR9−/− mice during the early phases of the disease may be critical in regulating the inflammatory response and promoting resolution.
FIG 2.
Decreased inflammatory cell infiltrate in the peritoneal cavities of TLR9−/− mice following Porphyromonas gingivalis challenge. WT and TLR9−/− mice were intraperitoneally injected with 1 × 108 CFU of Porphyromonas gingivalis, and the peritoneal exudate cells were harvested and stained with anti-TCR-β, anti-B220, anti-CD11b, anti-Ly-6G, and anti-F4/80 antibodies. (A) Representative flow cytometry dot plots: the numbers in the panels are representative of the percentage of cells in the respective quadrant or gate. (B and C) Quantification of the percentages of neutrophils (B) and macrophages (C) from the peritoneal lavage exudate samples from mice sacrificed at the indicated time points. The average results and standard deviations are shown. Five to 13 mice were analyzed in a minimum of two independent experiments. ***, P < 0.001; ****, P < 0.0001.
Increased A20 mRNA expression in the gingival tissues of TLR9−/− mice following ligature placement.
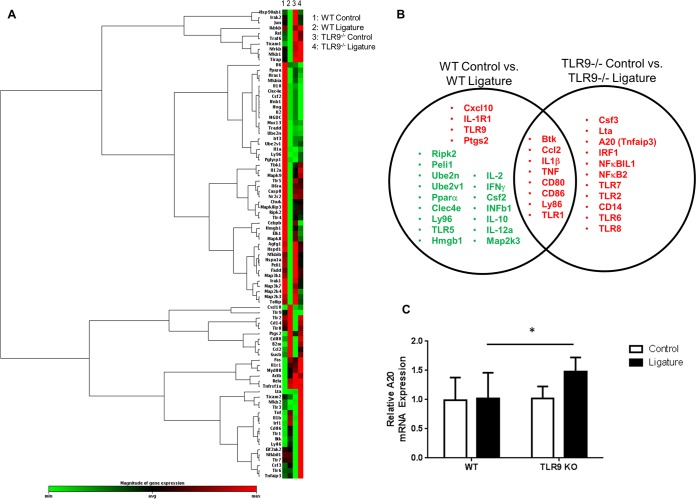
The TLR signaling pathway was examined in order to identify any differences in the expression patterns of signaling molecules and inflammatory mediators which could help explain the mechanism by which TLR9 contributes to periodontal disease pathogenesis. Analysis by a PCR array composed of 84 genes was conducted in control and ligated gingival tissues from WT and TLR9−/− mice (Fig. 3A). When a 1.5-fold difference was considered, we identified a number of genes which were differentially expressed in the gingival tissues of WT versus TLR9−/− mice (Fig. 3B). These differences imply that there might be key downstream molecules regulating TLR9-mediated inflammation in the course of periodontitis that limit the disease in TLR9−/− mice. In fact, A20, a negative regulator of TLR signaling and an inhibitor of the NF-κB pathway, was one of the genes that was significantly upregulated in TLR9−/− ligated tissues but not in WT ligated tissues. Consistent with the results of the arrays, increased A20 expression in the gingival tissues from TLR9−/− ligated mice was further verified using independent primer sets, with the expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control (Fig. 3C). A20 is a newly emerging endogenous regulator of inflammation and is associated with several receptors, downstream molecules, and cellular processes that are involved in the course of periodontal inflammation (8, 24). Previous studies reported that inflammatory cytokine production was decreased in TLR9−/− bone marrow-derived macrophages (BMDM) compared to the level of cytokine production in WT BMDM following P. gingivalis challenge (18). The current findings combined with those of the previous reports prompted us to hypothesize that A20 might be a key downstream molecule limiting periodontal inflammation. Therefore, our subsequent studies focused on A20 as a potential downstream mediator that could explain why TLR9 deficiency ameliorates periodontal disease.
FIG 3.
TLR9−/− gingival tissues exhibit dampened inflammatory responses and increased expression of negative regulators of inflammation following ligature placement. Ligatures were placed in WT and TLR9−/− mice, and the mice were euthanized 7 days later. RNA was extracted from gingival tissues and subjected to qPCR. (A) Heat map analysis of mRNA expression profiles of TLRs, TLR signaling pathways, cytokines, cytokine receptors, and regulators of the TLR pathways using the Qiagen RT2 Profiler PCR array kit. As shown in the color bar, red represents upregulated genes and green represents downregulated genes. (B) Venn diagram showing the unique and overlapping upregulated (red) and downregulated (green) genes in the gingival tissues for each genotype following ligature-induced inflammation. Five to six mice were analyzed. (C) qPCR analysis of A20 (TNFAIP3) in the gingival tissues of control and ligated WT and TLR9−/− mice. Results are reported as fold induction following normalization to the expression of GAPDH. The average results and standard deviations are shown. Tissue specimens from eight to nine mice were analyzed. *, P < 0.05. Abbreviations: Btk, Bruton agammaglobulinemia tyrosine kinase; Ccl2, chemokine (C-C motif) ligand 2; IL-1β, interleukin 1 beta; TNF, tumor necrosis factor; CD80, CD80 antigen; CD86, CD86 antigen; Ly86, lymphocyte antigen 86; TLR1, Toll-like receptor 1; Cxcl10, chemokine (C-X-C motif) ligand 10; IL-1R1, interleukin 1 receptor type I; TLR9, Toll-like receptor 9; Ptgs2, prostaglandin-endoperoxide synthase 2; RipK2, receptor (TNF receptor superfamily [TNFRSF])-interacting serine-threonine kinase 2; Peli1, pellino 1; Ube2n, ubiquitin-conjugating enzyme E2N; Ube2v1, ubiquitin-conjugating enzyme E2 variant 1; Para, peroxisome proliferator-activated receptor alpha; Clec4e, C-type lectin domain family 4, member e; Ly96, lymphocyte antigen 96; TLR5, Toll-like receptor 5; Hmgb1, high mobility group box 1; IL-2, interleukin 2; IFNγ, interferon gamma; Csf2, colony-stimulating factor 2 (granulocyte-macrophage); IFNb1, interferon beta 1, fibroblast; IL-10, interleukin 10; IL-12a, interleukin 12A; Map2k3, mitogen-activated protein kinase kinase 3; Csf3, colony-stimulating factor 3 (granulocyte); Lta, lymphotoxin A; (A20)Tnfaip3, tumor necrosis factor alpha-induced protein 3; IRF1, interferon regulatory factor 1; NF-κBIL1, nuclear factor of kappa-light-chain polypeptide gene enhancer in B-cells inhibitor-like 1; NF-κB2, nuclear factor of kappa-light-chain polypeptide gene enhancer in B-cells 2, p49/p100; TLR2, Toll-like receptor 2; TLR7, Toll-like receptor 7; TLR6, Toll-like receptor 6; TLR8, Toll-like receptor 8; CD14, CD14 antigen.
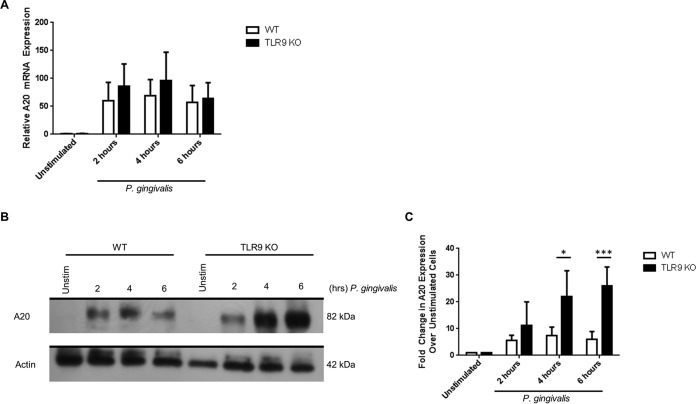
Lack of TLR9 signaling increases A20 protein expression in BMDM challenged with P. gingivalis.
We sought to examine whether the alterations in A20 expression found in the PCR array could explain the functional differences in cytokine responses between TLR9−/− and WT BMDM (18). To address this question, WT and TLR9−/− BMDM were stimulated for 2, 4, or 6 h with P. gingivalis, and A20 expression was examined. The results in Fig. 4A show that, following P. gingivalis challenge, A20 mRNA expression was slightly elevated in TLR9−/− BMDM compared to the level in WT BMDM, but this difference was not statistically significant. In the experiment whose results are shown in Fig. 4B and C, we compared the A20 protein levels between WT and TLR9−/− BMDM. In WT BMDM, A20 increased 5.9-fold over the level in unstimulated cells by 2 h and reached a peak of 7.5-fold by 4 h. When A20 protein in TLR9−/− BMDM was examined, the level increased 11.3-fold compared to the level in unstimulated cells by 2 h. Analyses at the 4- and 6-h time points in TLR9−/− BMDM revealed 22-fold and 26-fold increases in A20 levels compared to the levels in unstimulated controls, and the change was significantly different between WT and TLR9−/− cells. Taken together, our findings imply that abolishing TLR9 signaling results in increased cellular A20 levels and, possibly, increased A20 activity and subsequent regulation of cytokine production in response to bacterial challenge.
FIG 4.
Loss of TLR9 in BMDM increased A20 (TNFAIP3) protein induction following P. gingivalis challenge. WT and TLR9−/− BMDM were stimulated with heat-killed P. gingivalis (MOI of 1:100) for 2, 4, and 6 h. (A) mRNA was isolated, and expression of A20 was determined by qPCR. Results are reported as fold induction relative to expression in unstimulated samples following normalization to expression of GAPDH. The average results and standard deviations are shown. (B and C) Protein lysates were prepared, and Western blot analysis performed. Representative Western blots (B) and quantification of fold increase in A20 expression levels relative to expression in unstimulated samples following normalization to the actin signal (C) are shown. The average results and standard deviations are shown. Results are representative of the results for four mice analyzed in duplicates with a minimum of 2 independent experiments. *, P < 0.05; ***, P < 0.001.
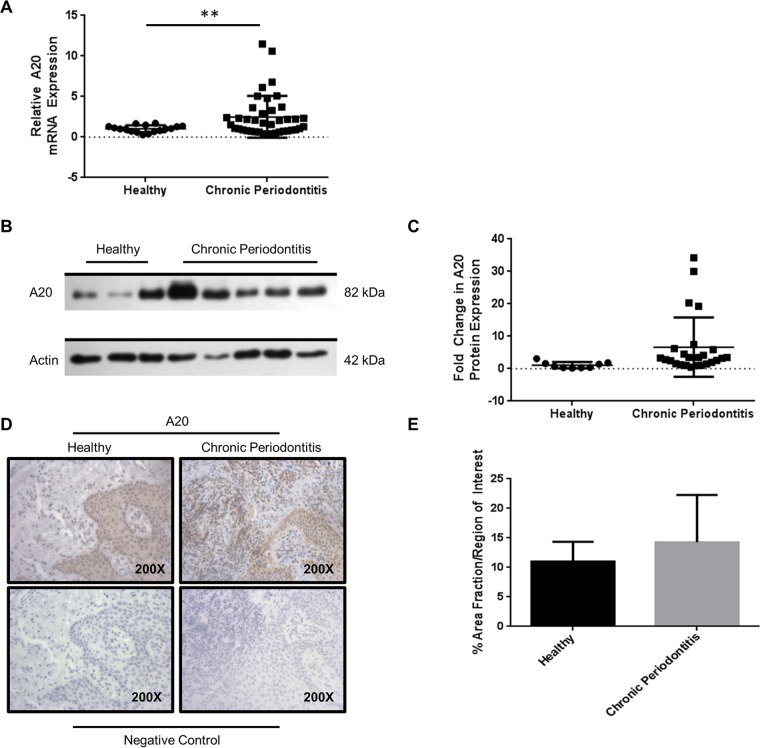
Increased A20 expression in periodontitis patients compared to that in healthy controls.
To our knowledge, A20 expression has not yet been investigated in a well-defined set of periodontitis patients. To provide additional support to the idea that A20 is clinically relevant and is associated with periodontal disease, we analyzed the A20 expression in gingival biopsy specimens from healthy individuals and periodontitis patients. Table 1 summarizes the demographic characteristics of the study population, including age, gender, and race, as well as the clinical parameters at the sampled teeth. Comparison of demographic variables between the healthy and chronic disease groups showed statistically significant differences in gender (P value = 0.03) and age distribution (P < 0.01). After controlling for these differences using multivariate regression analysis, there was a significant increase in A20 mRNA expression in the gingival tissue biopsy specimens of periodontitis subjects compared to its expression in the tissues obtained from healthy controls (P < 0.01) (Fig. 5A). In contrast, there was a modest yet statistically insignificant increase in A20 protein levels in the chronic disease versus the control group (Fig. 5B and C). We also wanted to determine the localization of A20 within the oral mucosa in diseased versus healthy tissues. Further analyses using immunohistochemistry (IHC) revealed that both healthy and diseased patients exhibit low constitutive A20 expression in the gingival epithelium (Fig. 5D). However, strong A20 staining was observed in the underlying gingival connective tissue of chronic periodontitis patients compared to that in the tissue of healthy controls (Fig. 5D). Quantification of 3,3′-diaminobenzidine staining in the epithelium and connective tissues revealed a slight increase in A20 expression in tissue specimens from the diseased patients compared to its expression in tissue specimens from healthy controls (Fig. 5E). The demonstration that A20 expression is clinically present in chronic periodontitis tissues further confirms the clinical relevance of this molecule in periodontal inflammation.
TABLE 1.
Clinical study population demographics
| Variable | Value(s) for group |
P value | |
|---|---|---|---|
| Healthy subjects | Chronic periodontitis patients | ||
| No. male/female | 6/18 | 26/23 | <0.05 |
| Mean age ± SD (yr) | 43.3 ± 17.7 | 57.6 ± 12.4 | <0.01 |
| No. white/black/Asian/Hispanic | 16/6/1/1 | 28/17/1/3 | >0.05 |
| Mean probing depth ± SD (mm) | 2.38 ± 0.72 | 4.88 ± 2.31 | <0.001 |
| Mean attachment loss ± SD (mm) | 1.02 ± 1.10 | 5.56 ± 2.88 | <0.001 |
FIG 5.
Modest increase in A20 (TNFAIP3) mRNA and protein levels in gingival biopsy specimens from chronic periodontitis patients. (A to C) RNA (A) or protein (B and C) was extracted from gingival biopsy specimens from healthy (H) and chronic periodontitis (CP) patients and subjected to qPCR and Western blotting for A20. Results are shown as fold increase relative to the results for healthy patients after normalization to GAPDH (A) or actin (C). Bars and whiskers show mean results and standard deviations. (B) Representative Western blots for individual healthy and diseased patients. (D and E) Immunohistochemical detection (D) and quantification (E) of A20 in CP patients (n = 6) and H controls (n = 5). Sections were analyzed in sextuplicates in a minimum of 2 independent experiments. The average results and standard deviations are shown. **, P < 0.01.
DISCUSSION
In this study, we examined the role and the mechanism of TLR9 in the modulation of periodontal disease using in vivo, ex vivo, and clinical studies. Here, we report five novel observations. First, TLR9 signaling promotes alveolar bone loss in an experimental murine ligature-induced model of periodontitis. Second, A20 expression is significantly increased in TLR9−/− ligated mice compared to its expression in WT ligated mice. Third, the loss of TLR9 diminishes the number of infiltrating neutrophils at the early phases of inflammation following intraperitoneal P. gingivalis infection. Fourth, ex vivo, TLR9−/− BMDMs challenged with P. gingivalis exhibit increased A20 protein expression compared to the level in WT BMDMs. Fifth, human gingival biopsy specimens from chronic periodontitis patients exhibited modest increases in A20 mRNA and protein expression compared to the levels in healthy controls. Together, we believe these results are novel, and they highlight the interplay of multiple cell types and signaling pathways in the progression of periodontal inflammation and identify a novel downstream molecule that may regulate local inflammatory responses.
What are the implications of these findings? Many periodontitis patients develop reoccurring disease because conventional therapies are not effective at controlling host inflammation. Therefore, current research efforts focus on the discovery of new therapeutic avenues. In this regard, prior studies from our laboratory and others have demonstrated that TLR9 signaling contributes to periodontal inflammation and alveolar bone loss (18). We extend these studies, and our results have implications in understanding the immunopathology and the cellular mechanisms by which TLR9 signaling instigates periodontal inflammation.
The first association concerns the role of TLR9 and neutrophils. Neutrophils act as double-edged swords during periodontal inflammation and should be tightly regulated by the host to initiate the immediate immune response to combat infection while limiting prolonged cellular recruitment and activity to prevent tissue injury. It is well documented that neutrophil migration and extravasation in periodontitis are governed by decreased Del-1 and increased IL-17 expression levels (25–27). Our flow cytometry observations of decreased percentages of neutrophils (CD11b+ Ly-6G+) in TLR9−/− mice using the lavage infection model are similar to the observations of aged mice administered Del-1 in the gingival tissues. Hence, it is likely that even a slight shift in neutrophil recruitment/activity in TLR9−/− animals could be responsible for regulating and limiting aberrant tissue responses and, eventually, periodontal inflammation.
Second, the role of TLR9 in periodontal inflammation was not restricted to regulating the generation of innate immune cells. In contrast to the neutrophil data, no difference in the percentages of macrophages (CD11b+ F4/80+) was detected between WT and TLR9−/− mice with intraperitoneal infections. Still, even though the numbers were not affected, functional changes in the cellular activity would account for the overall inflammatory response. In fact, we reported previously that TLR9−/− BMDMs exhibit decreased TNF and IL-6 production when stimulated with P. gingivalis compared to their WT counterparts (18). We extended these previous studies and determined that there was increased expression of A20 protein in the TLR9−/− macrophages compared to its expression in WT cells, which could possibly explain a mechanism by which TLR9−/− limits inflammation in the various experimental periodontitis models. And yet, future investigations are warranted to fully characterize the observed associations.
A20 is promiscuous in that not only does it target the redundant downstream TLR signaling pathways, it also targets many other inflammatory pathways that are relevant to periodontal inflammation, including TLR2, TLR4, nucleotide-binding oligomerization domain-containing 1 and 2 (NOD1/2) receptors, T cell receptor (TCR), TNF, IL-1, IL-17, and the nucleosome complex. A20 inhibits inflammation through interfering with NF-κB, MAPK, and interferon regulatory factor pathways and regulates cellular functions, including apoptosis, necroptosis, and autophagy, in several models of inflammatory diseases and cell types (28). Furthermore, increased A20 expression or activity has also been associated with dampening numerous disease outcomes (29, 30). Thus, there is great interest in manipulating A20 for therapeutic benefit (31). In regard to the TLR pathways, prior studies have identified cross talk between TLR9, TLR2, and TLR4 in both periodontitis and other disease models (18, 32). For periodontal inflammation, we demonstrated that TLR9−/− BMDM challenged with TLR2 and TLR4 agonists exhibited decreased cytokine production compared to that in WT cells (18). Our observation that A20 is upregulated in TLR9−/− BMDM following P. gingivalis stimulation, which activates multiple innate sensors, strongly argues that A20 might be a key downstream molecule that limits inflammation triggered by the activation of other receptors as well. Furthermore, the additional inflammatory pathways described above have also been associated with periodontal inflammation. Therefore, future studies are warranted to delineate regulatory mechanisms for A20 and how it interacts with multiple signaling pathways, possibly affecting immune cell plasticity in the course of periodontitis.
In regard to the human studies, we detected a significant increase in A20 mRNA expression and a modest but insignificant increase in protein levels in chronic periodontitis patients compared to their expression in healthy controls. Physiologically, A20 is constitutively expressed at low levels and can be further induced by microbial components (e.g., lipopolysaccharide [LPS]) and cytokines (e.g., IL-1, TNF, and IL-17) in many cell types which are relevant to periodontitis. Because the study participants exhibited severe forms of the disease compared to healthy tissues, upregulation of A20 was expected in the diseased tissues as part of the inflammatory cascade. However, why does this increase not resolve inflammation so as to result in changes in the clinical signs of the disease? TLR9 is one of the most upregulated PRRs in chronic periodontitis (19). It is possible that, although it is still a functional protein in the course of periodontitis, A20 expression does not reach sufficient levels to facilitate the resolution of inflammation. Thus, the questions that remain to be answered are how A20 levels and activity are regulated and whether the manipulation of A20 can prevent further periodontal tissue destruction, similar to its effects in other inflammatory diseases (29–31).
Several studies have determined that A20 expression is regulated by microRNAs (miRNAs) (33–35). In diffuse large B cell lymphoma, overexpression of miRNA 125a (miR-125a) and miR-125b has been shown to suppress A20 expression, contributing to NF-κB-mediated inflammation (33). In contrast, studies have also determined that A20 is protected from HuR degradation by miR-29 (36). Another study reported that downregulation of let-7f, another miRNA, during M. tuberculosis infection enhances A20 expression, thereby attenuating inflammatory signaling and facilitating bacterial survival (30). Thus, miRNAs can have an impact on A20 protein expression. Our in vitro findings further support this notion. Even though A20 protein levels were increased in P. gingivalis-challenged BMDMs from TLR9−/− mice compared to the levels in their WT counterparts, the A20 mRNA levels were similar between WT and TLR9−/− mice, suggesting that the regulation of A20 differs between these genotypes, probably at either the posttranscriptional or translational level. However, it is also possible that A20 expression was increased in the earlier times (<1 h) during the infection. In addition, A20 protein has a relatively long half-life, and it has been suggested that steady-state levels of A20 may regulate the magnitude of inflammation, which can provide a further explanation for the differences between mRNA and protein expression ex vivo (37). Similarly, although there was a significant increase in A20 gene expression in diseased tissues, the protein levels were not significantly different between groups. Considering the recent studies reporting differential expression of the miR-29 and miR-125 families between periodontitis patients and healthy controls, it will be important to determine the effects of differential miRNA expression on inflammation in these individuals and whether A20 levels and activity can be regulated with specific miRNA in the course of periodontitis (38). It will also be crucial to determine the interaction of A20 with other components of the host immune system and resident microbiome and how these interactions modulate the activity of the protein.
Consistent with the Western blot analyses, the IHC results also revealed a modest, yet insignificant increase in the A20 protein level in periodontitis tissues compared to the level in healthy tissues. While comparable levels of A20 expression were observed in the gingival epithelium of both healthy and diseased subjects, A20 staining was stronger in specimens of the underlying gingival connective tissue from chronic periodontitis patients than in specimens from healthy controls. These observations imply that in the course of periodontal inflammation, A20 expression varies among different tissues. A20 is indispensable for survival, since complete knockout of A20 in a mouse model leads to multiorgan inflammation and early death within 3 weeks after birth (9). This also corroborates that A20 is important for the resolution of inflammation. Therefore, it will be important to assess whether the variations in A20 expression in different cells affect periodontal disease pathogenesis.
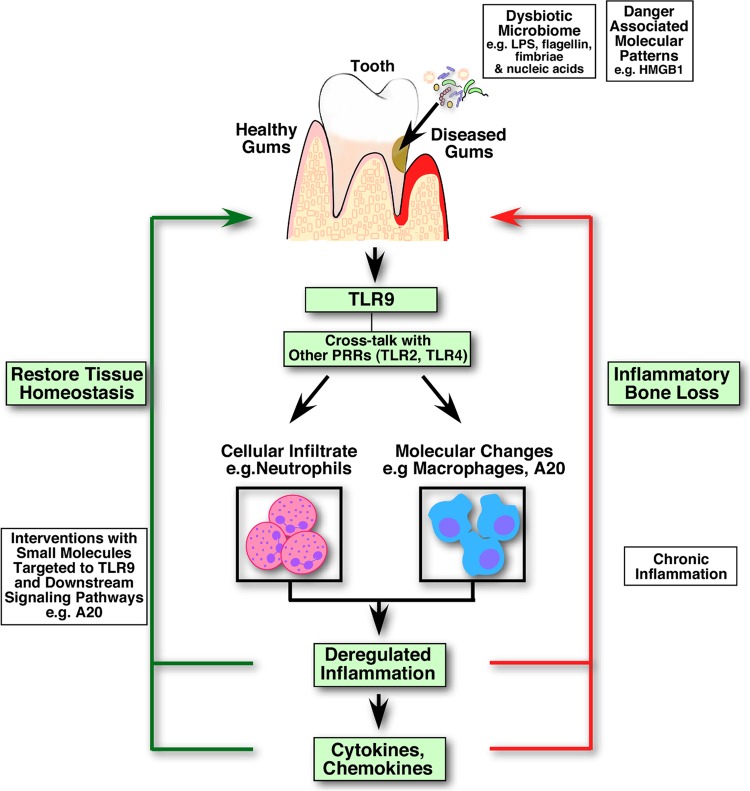
The challenge in studying chronic inflammatory conditions is characterizing the complex network of biological and molecular pathways, how they are regulated, and their ability to communicate with one another and facilitate the proinflammatory, anti-inflammatory, and resolution phases of the disease. TLR9 is unique, as it can engage both microbial nucleic acids and danger-associated molecular patterns (DAMPs) (20) and communicate with other TLRs. The current study, using ligature, intraperitoneal, and ex vivo models, provides experimental evidence that TLR9 modulates periodontal inflammation both at the cellular and molecular level (Fig. 6). We also reveal A20, an endogenous regulator of inflammation, as a new molecule that may play a role in periodontal inflammation. In addition, we report A20 expression in human clinical gingival biopsy specimens, corroborating its expression in the local tissues within the oral cavity. New therapeutic designs are critical to fine-tune the course of inflammation away from damaging, chronic inflammation and toward acute inflammation, which is resolved to restore tissue homeostasis. A20 is a common downstream molecule for all TLR signaling pathways and for additional pathways, including other PRRs, cytokine receptors, lymphocyte receptors, and apoptosis/necroptosis pathways. Therefore, to develop alternative strategies to treat polymicrobial and inflammatory diseases like chronic periodontitis and autoimmune disorders, it will be important to determine the exact role of A20 in periodontal disease pathogenesis and the mechanisms by which innate sensors communicate with each other and A20.
FIG 6.
A representative model of TLR9-mediated periodontal inflammation and targeted therapeutics. Periodontal tissues are exposed to various microbe-associated molecular patterns (MAMPs) and danger-associated molecular patterns (DAMPs), which are associated with a dysbiotic oral microbiome and/or host-derived factors. Engagement of nucleic acids with TLR9 in conjunction with the activation of other innate sensors (e.g., TLR2 and TLR4) by their respective agonists triggers cytokine/chemokine production and recruitment of inflammatory cells, including neutrophils and macrophages, to the site of infection. Failure to resolve inflammation efficiently and in a timely manner is detrimental to the host tissues and leads to tissue destruction and tooth loss. Blocking TLR9 signaling results in reduced neutrophil recruitment at early phases of infection and increased expression of A20, an endogenous regulator of inflammation which may limit the extent and duration of inflammation at the affected tissues. Targeted therapies to TLR signaling and downstream molecule A20 can potentially provide promising new therapeutic options to balance the tissue homeostasis and help to resolve deregulated inflammation, preventing adverse clinical outcomes.
MATERIALS AND METHODS
Human subjects and gingival tissue specimens.
The experimental protocol for the study of human gingival tissue specimens was approved by the Institutional Review Board of Virginia Commonwealth University, and a written consent was obtained from each patient. Twenty-four healthy (H) subjects (6 males and 18 females) and 49 chronic periodontitis (CP) patients (26 males and 23 females) between the ages of 20 and 84 years old were included. The periodontitis subjects had probing depths (PD) of >4 mm, clinical attachment loss (CAL) of >2 mm, bleeding on probing (BOP), and radiographic evidence of alveolar bone loss for at least two teeth (excluding third molars) per quadrant. Healthy subjects had PD of <4 mm, CAL of <2 mm without bleeding on probing, no radiographic evidence of alveolar bone loss, and a history of periodontitis (19). Individuals were excluded from the study for the following reasons: (i) pregnancy; (ii) current or former smoker (quit smoking <15 years ago); (iii) history of alcoholism, hepatitis, AIDS, or human immunodeficiency virus infection, diabetes, or radiation therapy; (iv) use of immunosuppressive medications, antibiotics, or nonsteroidal anti-inflammatory drugs within the past 6 months; and (v) history of previous periodontal surgeries or localized drug delivery at the site to be sampled. Gingival biopsy specimens were removed from each predetermined site and bisected longitudinally to generate samples for placement into two separate vials containing either an RNA-stabilizing agent or 10% formalin.
Mice.
All studies involving mice have been approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Commonwealth University. BALB/c wild-type (WT) mice were purchased from Jackson Laboratory (Bar Harbor, ME). TLR9−/− mice on the periodontitis-susceptible BALB/c background were obtained from Denis Klinmann (National Cancer Institute, Bethesda, MD) with the permission of Shizuo Akira (Osaka University). All animals were housed in a sterile, specific-pathogen-free room in individual ventilated cages.
Bacteria.
P. gingivalis (strain ATCC 33277) was grown under anaerobic conditions using brain heart infusion broth supplemented with 0.5% yeast extract, 5-μg/ml hemin, 0.5-μg/ml vitamin K, and 0.1% cysteine as previously described (39). The bacteria were killed by heating at 80°C for 10 min following incubation on agar plates for verification (40).
Ligature-induced periodontitis model.
Periodontal inflammation and bone loss were mechanically induced by ligature placement following previously published protocols (22). Briefly, 8- to 9-week-old mice were administered kanamycin (1 g/liter) in the drinking water, followed by 5 to 7 days of antibiotic-free water. Mice were intraperitoneally anesthetized with a 200-μl mixture of ketamine (10 mg/ml) and xylazine. A black braided silk 5-0 thread was placed at the left side of the maxilla interdentally between the first and second maxillary molars. The right side of the jaw was used as a control. Seven days after ligature placement, the mice were euthanized by isoflurane inhalation and cervical dislocation.
P. gingivalis peritoneal cavity infection model.
WT and TLR9−/− mice were intraperitoneally infected with 1 × 108 CFU of live P. gingivalis (ATCC 33277) cells in 500 μl of sterile phosphate-buffered saline (PBS). On days 1, 2, and 3 postinfection, mice were euthanized as described above. To isolate peritoneal exudate cells (PECs), 5 ml of complete medium containing RPMI 1640 supplemented with 10% fetal calf serum (FCS), 10 mM HEPES, 1 mM sodium pyruvate, 4,500-mg/liter glucose, and 0.05 mM mercaptoethanol was injected into the peritoneal cavities of mice and the abdomens of the mice were massaged. PECs were recovered, and red blood cells were osmotically lysed using ammonium-chloride-potassium (ACK) lysis buffer (Quality Biological) and resuspended in complete medium. For bone marrow cell isolation, both femurs and tibias were flushed with complete medium. Red blood cells were osmotically lysed as described above and then washed and resuspended in complete medium.
Bone marrow-derived macrophage (BMDM) propagation and bacterial challenge.
BMDM were generated according to standard protocols with minor adjustments (18). Briefly, bone marrow cells were isolated as described above and seeded in 100-mm petri dishes (non-tissue culture treated) at a concentration of 2 × 106 cells in 10 ml of bone marrow macrophage growth medium (BMM; Dulbecco's modified Eagle medium [DMEM] containing 4.5-g/liter glucose, 100-mg/liter sodium pyruvate, 10% FCS, glutamine, 0.05 mM mercaptoethanol, and 25% supernatant from L929 cells [ATCC CCL-1]). Cells were fed on day 3 by gently adding 10 ml of BMM. On day 6, the macrophage monolayer was washed by aspirating the medium and gently adding PBS to remove any nonadherent or dead cells. Adherent macrophages were harvested by adding PBS to the monolayer, incubating at 4°C for 10 min, and gently pipetting and scraping to remove cells. Macrophages were seeded in a 12-well plate at a concentration of 2 × 106 cells in 1 ml of BMM without L929 but containing supernatant for 18 h prior to stimulation. Heat-killed P. gingivalis cells were used to challenge BMDM at a multiplicity of infection (MO1) of 1:100. Unstimulated cells were used as negative controls.
Surface staining and flow cytometry.
In this study, the following antibodies were used: anti-mouse Ly-6G (clone 1A8)-fluorescein isothiocyanate (FITC) antibody, anti-mouse TCR-β (clone H57-597)-allophycocyanin (APC) antibody, anti-mouse B220 (clone RA3-6B2)-APC antibody, anti-mouse/human CD11b (clone M1/70)-Brilliant Violet 711 (BV711) antibody, anti-mouse CD11c (clone N418)-APC-Cy7 antibody, and anti-mouse F4/80 (clone BM8)-phycoerythrin (PE)-Cy7 antibody. All antibodies were purchased from BioLegend. Surface staining was performed by incubating cells in a 1:100 dilution of antibody in 2% fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 2% FCS) for 30 min on ice. After washing three times in FACS buffer, cells were fixed in 2% paraformaldehyde. Samples were acquired on a BD LSRFortessa instrument and analyzed using FlowJo software.
Determination of periodontal bone loss.
Alveolar bone loss around the maxillary molars was determined using micro-CT as described previously (41). Briefly, for the ligature-induced periodontitis model, the entire head was harvested and fixed in 4% formaldehyde. The Bruker SkyScan 1173 high-energy micro-computed tomography technology scanner (SkyScan NV; Kontich, Belgium) was used to acquire three-dimensional images in 1,200 projections by 360° rotation. A 250-ms exposure per projection, at 80 kV and 80 μA, with an average image pixel of 16.87 μm and a rotation step of 0.2 degrees, was used to obtain an average scan time of 40 min. Images were reconstructed using NRecon software (Kontich, Belgium) with a Gaussian smoothing kernel of 2. Linear measurements for the distance from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) were obtained using DataViewer (version 1.5.1; SkyScan NV) with a 1,092- by 1,092-pixel size in all three spatial dimensions and setting the sagittal plane parallel to the X-ray beam axis. The CEJ-ABC distance for each treated mouse was subtracted from the mean CEJ-ABC distance of control mice. The results are expressed in micrometers, with negative values indicating bone loss relative to the results for control mice. Three-dimensional analysis was determined using CTvox for volume rendering (version 3.0, SkyScan NV).
Determination of bacterial counts.
The ligature-associated-bacterium counts were determined by previously published protocols (42). Briefly, ligatures were removed from euthanized mice and washed gently in PBS to eliminate excess food. The washed ligatures were then placed in 1 ml of PBS, vortexed gently for 2 min, serially diluted, and plated onto blood agar plates. After anaerobic growth at 37°C for 7 days, bacteria were enumerated. The results were normalized by dividing the total CFU by the corresponding length in millimeters of the ligature.
Immunohistochemistry. (i) Human tissues.
The levels of A20 protein expression in human gingival tissue specimens were determined by immunohistochemistry analysis of tissue specimens from 5 healthy (H) and 6 chronic periodontitis (CP) patients as previously described (19). Briefly, formalin-fixed gingival tissue sections collected from CP and H patients were deparaffinized, rehydrated, and submerged in 3% hydrogen peroxide. After washing with PBS, the slides were blocked with 5% horse serum for 1 h at room temperature. The slides were incubated with anti-A20 (TNFAIP3) antibody (Abcam) in a sealed humidifying tray overnight at 4°C. The following day, tissues were incubated with biotin-conjugated anti-rabbit IgG for 1 h at room temperature. After washing in PBS, slides were incubated with avidin-peroxidase reagent, followed by staining with diaminobenzidine solution and counterstaining with hematoxylin (Vectastain ABC kit). The slides were dehydrated and permanently mounted. Negative controls which omitted the primary antibody were run in parallel. The staining intensity and distribution were determined by light microscopy. To quantify the percentage of diaminobenzidine-positive areas within a region, cellSens software was used, and six fields of view at a magnification of ×200 were analyzed for each patient.
(ii) Mouse tissues.
Explanted gingival tissues from WT (n = 4) and TLR9−/− (n = 4) mice were fixed in 4% formaldehyde, sectioned, and stained with hematoxylin and eosin (H&E) using standard protocols. Histology images were acquired using the QColor 5 imaging system from Olympus Microscopy with a ×10-magnification objective lens. The cellSens software was used to quantify the percentages of nucleated cells (hematoxylin-positive) cells in the gingival tissue specimens. Six regions/fields of view were analyzed for each mouse.
(iii) Western blotting.
Murine BMDM samples were lysed in 100 μl of ice-cold NP-40 lysis buffer (25 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 1 mM EDTA, pH 8) supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 1 mM dithiothreitol, and 1× protease inhibitor cocktail (Sigma-Aldrich) by repeated pipetting. For murine and human gingival tissues, samples were minced and homogenized in the lysis buffer described above. All samples were sonicated twice at 50% pulse for 30 s each time using an ultrasonic sonicator (BioLogics, Inc., Cary, NC). The protein concentration was determined using the bicinchoninic acid (BCA) protein assay kit (Thermo Scientific). Cell lysates were subjected to electrophoresis on 4-to-15% Mini-Protean TGX gels (Bio-Rad) and transferred to nitrocellulose membranes. Membranes were probed with anti-A20 (TNFAIP3) antibody (D13H3; Cell Signaling Technologies) according to the manufacturer's protocol. Bands were visualized using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific), followed by exposure to X-ray film. Densitometric analyses were performed using the G:BOX Chem XX6 system and GeneTools image analysis software (Syngene). To determine the fold increase in A20 expression for BMDM experiments, all samples were first normalized to the levels of actin. The fold increase was calculated by dividing the normalized A20 value for the stimulated sample by the value for the corresponding unstimulated negative control.
(iv) RNA isolation, PCR array, and quantitative real-time PCR (qPCR).
Total RNA was extracted from mouse or human gingival tissues and BMDM using the RNeasy kit (Qiagen) and genomic DNA (gDNA) eliminator spin columns. For array analysis, 500 ng of RNA was used to synthesize cDNA using the RT2 first strand kit (Qiagen) according to the manufacturer's protocol. The expression of genes in the TLR signaling pathway was determined by real-time PCR using the RT2 Profiler PCR array (PAMM-018Z; Qiagen) with the Applied Biosystems 7500 real-time PCR system. Analysis of the PCR array data set was performed on the manufacturer's website.
For all other quantitative real-time PCR analysis, cDNA was synthesized from 1 μg of RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's protocol. The quantitative real-time PCR with cDNA was performed with the Applied Biosystems 7500 real-time PCR system using SYBR green master mix (SABiosciences) and the following specific primer sets: GAPDH (mouse), forward, 5′ AGG TCG GTG TGA ACG GAT TTG 3′, and reverse, 5′ GGG GTC GTT GAT GGC AAC A 3′; TNFAIP3/A20 (mouse), forward, 5′ GAA CAG CGA TCA GGC CAG G 3′, and reverse, 5′ GGA CAG TTG GGT GTC TCA CAT T 3′; GAPDH (human), forward, 5′ CAA TGA CCC CTT CAT TGA CC 3′, and reverse, 5′ TTG ATT TTG GAG GGA TCT CG 3′; and A20/TNFAIP3 (human), forward, 5′ TTG TCC TCA GTT TCG GGA GAT 3′, and reverse, 5′ ACT TCT CGA CAC CAG TTG AGT T 3′. The GAPDH gene was used as an internal control. The relative amount of each mRNA was calculated using the cycle threshold (2−ΔΔCT) method, where ΔCT = (CT mRNA − CT GAPDH).
Statistical analysis.
Data were analyzed by one-way analysis of variance (ANOVA) and the Tukey multiple-comparison test or unpaired t test with Mann-Whitney correction using the InStat program (GraphPad Software, San Diego, CA). The Spearman test was performed to detect the correlation between bacterial load and bone loss. In the analyses of clinical samples, demographic variables including race, gender, and age were compared between the healthy and chronic periodontitis groups using the two-sample t test and Fisher's exact test. Multivariate regression analysis was used to determine the effect of the disease on the A20 level. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
The study was supported by National Institutes of Health grants number DE025037, DE022836, and KL2TR000057-03 to S.E.S. and K12GM093857 for support of K.E.C.
We declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
REFERENCES
- 1.Janeway CA Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Pieterse E, van der Vlag J. 2014. Breaking immunological tolerance in systemic lupus erythematosus. Front Immunol 5:164. doi: 10.3389/fimmu.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagdonas E, Raudoniute J, Bruzauskaite I, Aldonyte R. 2015. Novel aspects of pathogenesis and regeneration mechanisms in COPD. Int J Chron Obstruct Pulmon Dis 10:995–1013. doi: 10.2147/COPD.S82518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao L, Liu Y, Wang N. 2014. New paradigms in inflammatory signaling in vascular endothelial cells. Am J Physiol Heart Circ Physiol 306:H317–H325. doi: 10.1152/ajpheart.00182.2013. [DOI] [PubMed] [Google Scholar]
- 5.Karin M, Clevers H. 2016. Reparative inflammation takes charge of tissue regeneration. Nature 529:307–315. doi: 10.1038/nature17039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondo T, Kawai T, Akira S. 2012. Dissecting negative regulation of Toll-like receptor signaling. Trends Immunol 33:449–458. doi: 10.1016/j.it.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Anwar MA, Basith S, Choi S. 2013. Negative regulatory approaches to the attenuation of Toll-like receptor signaling. Exp Mol Med 45:e11. doi: 10.1038/emm.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catrysse L, Vereecke L, Beyaert R, van Loo G. 2014. A20 in inflammation and autoimmunity. Trends Immunol 35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Eke PI, Thornton-Evans GO, Wei L, Borgnakke WS, Dye BA. 2010. Accuracy of NHANES periodontal examination protocols. J Dent Res 89:1208–1213. doi: 10.1177/0022034510377793. [DOI] [PubMed] [Google Scholar]
- 10.Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madianos PN, Bobetsis YA, Offenbacher S. 2013. Adverse pregnancy outcomes (APOs) and periodontal disease: pathogenic mechanisms. J Clin Periodontol 40(Suppl 14):S170–S180. doi: 10.1111/jcpe.12082. [DOI] [PubMed] [Google Scholar]
- 12.Sahingur SE, Yeudall WA. 2015. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol 6:214. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schenkein HA, Loos BG. 2013. Inflammatory mechanisms linking periodontal diseases to cardiovascular diseases. J Clin Periodontol 40(Suppl 14):S51–S69. doi: 10.1111/jcpe.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song JY, Kim HH, Cho EJ, Kim TY. 2014. The relationship between gastroesophageal reflux disease and chronic periodontitis. Gut Liver 8:35–40. doi: 10.5009/gnl.2014.8.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taylor JJ, Preshaw PM, Lalla E. 2013. A review of the evidence for pathogenic mechanisms that may link periodontitis and diabetes. J Periodontol 84(4 Suppl):S113–S134. doi: 10.1902/jop.2013.134005. [DOI] [PubMed] [Google Scholar]
- 16.Lamont RJ, Hajishengallis G. 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med 21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abe T, Hosur KB, Hajishengallis E, Reis ES, Ricklin D, Lambris JD, Hajishengallis G. 2012. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J Immunol 189:5442–5448. doi: 10.4049/jimmunol.1202339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim PD, Xia-Juan X, Crump KE, Abe T, Hajishengallis G, Sahingur SE. 2015. Toll-Like receptor 9-mediated inflammation triggers alveolar bone loss in experimental murine periodontitis. Infect Immun 83:2992–3002. doi: 10.1128/IAI.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahingur SE, Xia XJ, Voth SC, Yeudall WA, Gunsolley JC. 2013. Increased nucleic acid receptor expression in chronic periodontitis. J Periodontol 84:e48–e57. doi: 10.1902/jop.2013.120739. [DOI] [PubMed] [Google Scholar]
- 20.Crump KE, Sahingur SE. 2016. Microbial nucleic acid sensing in oral and systemic diseases. J Dent Res 95:17–25. doi: 10.1177/0022034515609062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchesan J, Jiao YZ, Schaff RA, Hao J, Morelli T, Kinney JS, Gerow E, Sheridan R, Rodrigues V, Paster BJ, Inohara N, Giannobile WV. 2016. TLR4, NOD1 and NOD2 mediate immune recognition of putative newly identified periodontal pathogens. Mol Oral Microbiol 31:243–258. doi: 10.1111/omi.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao YZ, Darzi Y, Tawaratsumida K, Marchesan JT, Hasegawa M, Moon H, Chen GY, Nunez G, Giannobile WV, Raes J, Inohara N. 2013. Induction of bone loss by pathobiont-mediated Nod1 signaling in the oral cavity. Cell Host Microbe 13:595–601. doi: 10.1016/j.chom.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maekawa T, Abe T, Hajishengallis E, Hosur KB, DeAngelis RA, Ricklin D, Lambris JD, Hajishengallis G. 2014. Genetic and intervention studies implicating complement C3 as a major target for the treatment of periodontitis. J Immunol 192:6020–6027. doi: 10.4049/jimmunol.1400569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beyaert R, Heyninck K, Van Huffel S. 2000. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem Pharmacol 60:1143–1151. doi: 10.1016/S0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 25.Hajishengallis G, Sahingur SE. 2014. Novel inflammatory pathways in periodontitis. Adv Dent Res 26:23–29. doi: 10.1177/0022034514526240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S, Ciero PA, Krauss JL, Li F, Rauner M, Hofbauer LC, Choi EY, Chung KJ, Hashim A, Curtis MA, Chavakis T, Hajishengallis G. 2012. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat Immunol 13:465–473. doi: 10.1038/ni.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajishengallis G, Chavakis T. 2013. Endogenous modulators of inflammatory cell recruitment. Trends Immunol 34:1–6. doi: 10.1016/j.it.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onizawa M, Oshima S, Schulze-Topphoff U, Oses-Prieto JA, Lu T, Tavares R, Prodhomme T, Duong B, Whang MI, Advincula R, Agelidis A, Barrera J, Wu H, Burlingame A, Malynn BA, Zamvil SS, Ma A. 2015. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat Immunol 16:618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Shi XW, Qian T, Li J, Tian ZQ, Ni B, Hao F. 2015. A20 overexpression alleviates pristine-induced lupus nephritis by inhibiting the NF-kappa B and NLRP3 inflammasome activation in macrophages of mice. Int J Clin Exp Med 8:17430–17440. [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar M, Sahu SK, Kumar R, Subuddhi A, Maji RK, Jana K, Gupta P, Raffetseder J, Lerm M, Ghosh Z, van Loo G, Beyaert R, Gupta UD, Kundu M, Basu J. 2015. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kappaB pathway. Cell Host Microbe 17:345–356. doi: 10.1016/j.chom.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Pfoh R, Lacdao IK, Saridakis V. 2015. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocr Relat Cancer 22:T35–T54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews K, Abdelsamed H, Yi AK, Miller MA, Fitzpatrick EA. 2013. TLR2 regulates neutrophil recruitment and cytokine production with minor contributions from TLR9 during hypersensitivity pneumonitis. PLoS One 8:e73143. doi: 10.1371/journal.pone.0073143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. 2012. MicroRNAs miR-125a and miR-125b constitutively activate the NF-kappaB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20). Proc Natl Acad Sci U S A 109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH. 2011. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun 411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 35.Gantier MP, Stunden HJ, McCoy CE, Behlke MA, Wang D, Kaparakis-Liaskos M, Sarvestani ST, Yang YH, Xu D, Corr SC, Morand EF, Williams BR. 2012. A miR-19 regulon that controls NF-kappaB signaling. Nucleic Acids Res 40:8048–8058. doi: 10.1093/nar/gks521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, London CA, Kraybill W, Perrotti D, Croce CM, Keller C, Guttridge DC. 2013. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal 6(286):ra63. doi: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garg AV, Gaffen SL. 2013. IL-17 signaling and A20: a balancing act. Cell Cycle 12:3459–3460. doi: 10.4161/cc.26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kebschull M, Papapanou PN. 2015. Mini but mighty: microRNAs in the pathobiology of periodontal disease. Periodontol 2000 69:201–220. doi: 10.1111/prd.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahingur SE, Xia XJ, Alamgir S, Honma K, Sharma A, Schenkein HA. 2010. DNA from Porphyromonas gingivalis and Tannerella forsythia induce cytokine production in human monocytic cell lines. Mol Oral Microbiol 25:123–135. doi: 10.1111/j.2041-1014.2009.00551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sahingur SE, Xia XJ, Schifferle RE. 2012. Oral bacterial DNA differ in their ability to induce inflammatory responses in human monocytic cell lines. J Periodontol 83:1069–1077. doi: 10.1902/jop.2011.110522. [DOI] [PubMed] [Google Scholar]
- 41.Park CH, Abramson ZR, Taba M, Jin Q, Chang J, Kreider JM, Goldstein SA, Giannobile WV. 2007. Three-dimensional micro-computed tomographic imaging of alveolar bone in experimental bone loss or repair. J Periodontol 78:273–281. doi: 10.1902/jop.2007.060252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe T, Shin J, Hosur K, Udey MC, Chavakis T, Hajishengallis G. 2014. Regulation of osteoclast homeostasis and inflammatory bone loss by MFG-E8. J Immunol 193:1383–1391. doi: 10.4049/jimmunol.1400970. [DOI] [PMC free article] [PubMed] [Google Scholar]