ABSTRACT
The human pathogen Helicobacter pylori uses the host receptor α5β1 integrin to trigger inflammation in host cells via its cag pathogenicity island (cag PAI) type IV secretion system (T4SS). Here, we report that the H. pylori ImaA protein (HP0289) decreases the action of the cag PAI T4SS via tempering the bacterium's interaction with α5β1 integrin. Previously, imaA-null mutants were found to induce an elevated inflammatory response that was dependent on the cag PAI T4SS; here we extend those findings to show that the elevated response is independent of the CagA effector protein. To understand how ImaA could be affecting cag PAI T4SS activity at the host cell interface, we utilized the Phyre structural threading program and found that ImaA has a region with remote homology to bacterial integrin-binding proteins. This region was required for ImaA function. Unexpectedly, we observed that imaA mutants bound higher levels of α5β1 integrin than wild-type H. pylori, an outcome that required the predicted integrin-binding homology region of ImaA. Lastly, we report that ImaA directly affected the amount of host cell β1 integrin but not other cellular integrins. Our results thus suggest a model in which H. pylori employs ImaA to regulate interactions between integrin and the T4SS and thus alter the host inflammatory strength.
KEYWORDS: autotransporter proteins, inflammation, pathogenesis, secretion systems, virulence factors
INTRODUCTION
The human pathogen Helicobacter pylori is one of the world's most common pathogens, chronically colonizing the stomachs of at least one-third of the world's population, with the populations of many countries experiencing rates of colonization of over 50% (1, 2). The outcomes of this infection vary on the basis of a combination of bacterial genetics, host genetics, and environmental factors (3). Ten to 15% of those infected go on to develop severe diseases, including ulcers and gastric adenocarcinoma (4–6). H. pylori-triggered diseases cause significant mortality and morbidity worldwide. For example, gastric adenocarcinoma killed over 700,000 people worldwide in 2012, and in the United States, 26,370 people were expected to have been diagnosed with this disease in 2016 (7, 8).
One of the main factors that influences the H. pylori disease outcome is whether a person is infected with a strain that possesses the cytotoxin-associated gene (cag) pathogenicity island (cag PAI). cag PAI-positive strains are associated with severe inflammation, peptic ulcers, and gastric cancer (9). The cag PAI encodes a type IV secretion system (T4SS), a large multiprotein system that triggers a host inflammatory response directly via interactions with the host cells (10) and also via delivery of proinflammatory cargo: the protein CagA and the bacterial molecule peptidoglycan (11, 12).
Given the cost and consequence of producing an active cag PAI T4SS, its function is controlled at several levels (2, 13, 14). While there is some transcriptional modulation (14, 15), the greatest control appears to operate at the assembly and translocation steps. H. pylori constitutively produces the cag PAI T4SS proteins (14) but does not assemble them to form a detectable pilus until H. pylori contacts epithelial cells (16). Furthermore, H. pylori injects only 10 to 30% of its CagA, suggesting that there may be regulation at the translocation step (17). The mechanisms that lead to pilus assembly and translocation are not yet understood, however.
Additional control of cag PAI T4SS-host cell interactions depends on the level of H. pylori-host cell interactions. H. pylori requires α5β1 integrin for the efficient injection of CagA (18). Additional interactions between H. pylori adhesins and other host cell proteins can also promote cag PAI delivery (19–21). A number of H. pylori proteins have been shown to bind to α5β1 integrin, on the basis of the findings of studies that focused on proteins encoded within the cag PAI. CagL was the first H. pylori protein identified to bind to α5β1 integrin (18, 22), with subsequent studies showing that the CagA, CagI, and CagY cag PAI proteins also had this ability (22). Of note, these proteins do not depend on the classical integrin-binding motif, RGD (arginine-glycine-aspartate), for their integrin interactions and thus interact in undefined ways. H. pylori can interact with purified integrin independently of host cells, and estimates suggest that ∼5% of in vitro-grown H. pylori cells are able to interact with α5β1 integrin (17). Host cells respond to the H. pylori-α5β1 integrin interaction, e.g., by activating secretion of the mammalian pH regulation hormone gastrin (23). H. pylori clearly invests significant efforts into interaction with α5β1 integrin, strongly suggesting its importance to H. pylori pathogenesis.
One recently identified protein that modulates the cag PAI-mediated inflammatory response is the outer membrane protein ImaA (HP0289). Mutants lacking imaA induce higher levels of interleukin-8 (IL-8) from infected gastric epithelial cells than wild-type H. pylori does, a response that depends on the cag PAI (24). ImaA is a member of the classical autotransporter family, also called type Va (25). These proteins consist of a long beta-helix domain that places the functional portion at a distance from the bacterial surface. ImaA is quite large and is predicted to adopt a structure that would place the functional domain over 100 nm from the bacterial outer membrane. Autotransporters play numerous roles in bacterial pathogenesis and bacterial physiology (25).
ImaA plays important roles in host colonization. imaA transcription is upregulated in the host in a pH-dependent manner (24, 26). Furthermore, imaA mutants exhibit murine colonization defects (24, 27). ImaA is thus important for murine colonization and inflammation control, but its mode of action remained elusive.
We thus embarked on this work to gain a mechanistic understanding of how ImaA is able to lead to greater cag PAI-dependent inflammation. In this study, we demonstrate that ImaA acts at the level of the cag PAI itself and does not require CagA for its effect. We provide evidence that ImaA antagonizes the action of the cag PAI T4SS and the interaction of H. pylori with α5β1 integrin. We mapped the functional portion of ImaA to a region that shares remote homology with integrin-binding proteins and proteases. Lastly, we show that ImaA may modulate the level of host integrin. Our results support a model in which H. pylori uses ImaA to control initial interactions with α5β1 integrin that precede the full cag PAI-α5β1 integrin interactions and proinflammatory activities.
RESULTS
ImaA operates independently of CagA.
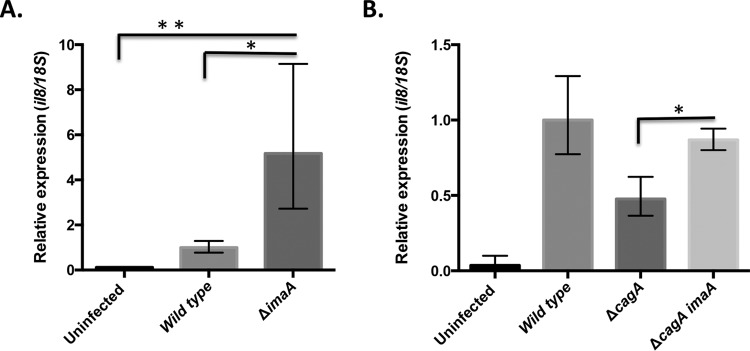
Previous work had shown that imaA mutants cause a strong host inflammatory response that is dependent on the cag PAI (24). The cag PAI has multiple ways to trigger inflammation, including by the delivery of CagA and by direct interactions between its T4SS and the host cell (10, 28, 29). We therefore first addressed which of these attributes were influenced by ImaA by determining whether the imaA phenotype was dependent on CagA. cagA mutants still generate a cag PAI T4SS but cannot deliver CagA (12). We thus generated cagA single mutants and imaA cagA double mutants in H. pylori. Consistent with the findings of a previous analysis of imaA mutants (24), we found that the loss of imaA created strains that triggered elevated levels of Il8 from host cells compared to the levels triggered by the wild type (Fig. 1A). When cagA was eliminated, the strains partially lost the ability to trigger an Il8 increase compared to that of the wild type (Fig. 1B). The loss of imaA in the cagA mutant background resulted in elevated Il8 transcript levels compared to the levels in the cagA single mutant (Fig. 1B). This 2-fold increase was similar to but slightly less than the magnitude of the response for imaA mutants (Fig. 1A) (24). These results suggest that imaA acts in a way that is cag PAI dependent but at least partially CagA independent.
FIG 1.
Loss of ImaA elevates the amount of the Il8 transcript induced by cagA mutants. (A) Il8 transcript response of AGS cells after infection with H. pylori LSH100 and isogenic variants. AGS cells were infected for 2 h with the indicated strains (9 technical replicates were tested in triplicate). RNA was analyzed by quantitative reverse transcription-PCR, differences in the levels of expression were calculated by the ΔΔCT threshold cycle (CT) method, and Il8 levels were normalized to those obtained by the wild-type response (27). *, P < 0.05 by ANOVA; **, P < 0.01 by ANOVA. (B) ΔimaA mutants evoke an elevated inflammatory response independently of cagA. The Il8 transcript responses for the ΔcagA or ΔcagA imaA mutant (10 biological replicates were tested in triplicate) were normalized to the wild-type response, as described in the legend to panel A. *, P < 0.05 by Student's t test.
ImaA antagonizes the action of the cag PAI T4SS.
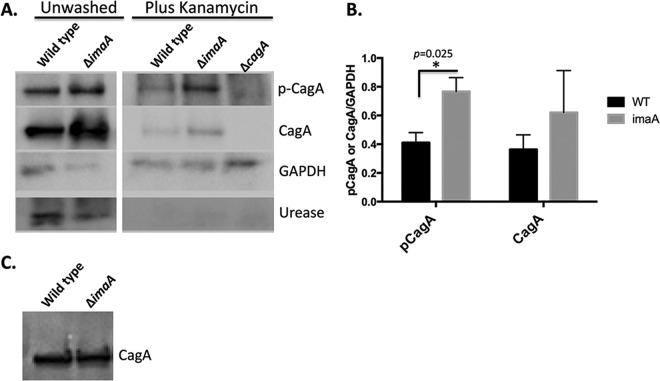
We next asked how ImaA affects the cag PAI function by assessing delivery from the cag PAI T4SS. We used CagA as a readout of cag PAI T4SS delivery, as this protein is the most readily detectable cag PAI T4SS substrate. We infected AGS gastric epithelial cells with either wild-type or imaA mutant H. pylori bacteria and monitored the levels of CagA delivered by examining the amount of tyrosine-phosphorylated CagA, a reaction that happens only inside mammalian cells. AGS cells infected with the imaA mutant contained larger amounts of phosphorylated CagA than those infected with wild-type H. pylori (Fig. 2A and B), suggesting that ImaA mutants either deliver larger amounts of CagA or have higher levels of phosphorylation of CagA. To examine whether a larger amount of CagA was delivered, we employed treatments shown to greatly reduce the amount of extracellular H. pylori: treatment of the bacteria with the antibiotic kanamycin (Km) followed by extensive washing (30). Indeed, this treatment led to a substantial loss of H. pylori, as measured by detection of the H. pylori protein urease (Fig. 2A). Even after removing the H. pylori bacteria, we detected higher levels of total CagA in cells infected with the imaA mutant than in cells infected with wild-type H. pylori (Fig. 2A and B). Thus, our results suggest that imaA mutants have enhanced delivery of cag PAI T4SS substrates.
FIG 2.
imaA mutants deliver more CagA into epithelial cells. (A) AGS cells were infected with either wild-type H. pylori or the imaA mutant for 3 h until treatments were initiated as follows: (i) cells were unwashed, where AGS cells remained unwashed and the infection was allowed to persist for an additional 6 h, or (ii) cells were washed and treated with Km for 6 h, in which AGS cells were washed 3 times and supplemented with medium containing kanamycin for an additional 6 h. After the 6-h period, AGS cells were washed, collected, and analyzed by Western blotting with the antibodies indicated at the right of the blot. p-CagA, phosphorylated CagA. (B) CagA protein levels were quantified from 3 biological replicates using ImageJ pixel densitometry software. *, P < 0.05 by Student's t test. WT, wild type. (C) Lysates from wild-type H. pylori and the imaA mutant grown as described in the legend to panel A were examined for CagA production prior to AGS cell infection using Western blotting. Equal numbers of cells, based on the OD600, were loaded in each lane.
The delivery of elevated amounts of CagA could be due to in part to the overproduction of CagA by the imaA mutant. We therefore analyzed the amount of CagA produced by wild-type and imaA mutant H. pylori bacteria grown in the same manner used for AGS cell infection. Under these conditions, there were no differences in CagA levels between the two strains (Fig. 2C), suggesting that imaA mutants do not express elevated amounts of CagA compared to the wild type. These results thus suggest that imaA mutants deliver large amounts of the cag PAI T4SS substrates to mammalian cells and, concomitantly, that ImaA normally antagonizes the cag PAI T4SS function.
ImaA has a putative functional region that is homologous to integrin-binding proteins and proteases.
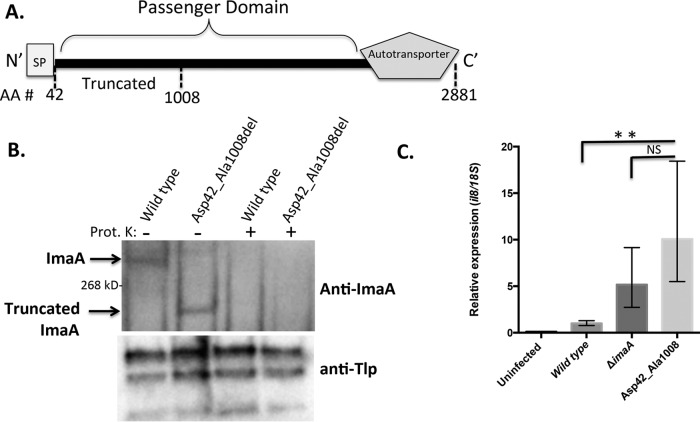
To gain insight into ImaA function, we examined the protein sequence of the predicted passenger/function region in detail. This region of ImaA lacks homology to other proteins when analyzed using standard protein bioinformatic approaches (BLAST, PFAM, and SMART analysis), but with the protein structure prediction program Phyre (31), we found that it exhibited significant structural similarity to the integrin-binding proteins invasin (32, 33) and pertactin (34, 35), as well as several lipases and proteases (Fig. 3A and Fig. S1 in the supplemental material).
FIG 3.
ImaA has a putative functional region that is important for controlling inflammation. (A) Diagram of ImaA, with the location of the truncation being depicted. AA = amino acid. (B) (Top) Wild-type H. pylori LSH100 and the isogenic imaA Asp42_Ala1008del strain were either left untreated or treated with the protease proteinase K (Prot. K) followed by Western blotting with anti-ImaA; (bottom) the same blot shown in the top panel stripped and reprobed with an antibody (anti-Tlp) that recognizes the inner membrane chemoreceptor proteins TlpA, TlpB, and TlpC (26). (C) AGS cells were infected for 2 h with either wild-type H. pylori LSH100, the ΔimaA isogenic mutant, or the isogenic imaA Asp42_Ala1008del strain. The data for the wild-type and ΔimaA strains represent those for three biological replicates with nine technical replicates, while the data for the imaA Asp42_Ala1008del strain represent those for eight biological replicates with three technical replicates each. RNA was collected and used for quantitative reverse transcription-PCR to analyze the expression of Il8. All reactions were done in triplicate, and differences in the levels of expression were calculated by the ΔΔCT threshold cycle (CT) method (27). **, P < 0.01 compared to the wild type by ANOVA; NS, no significant difference compared to the wild type by ANOVA.
We then asked whether this region of ImaA, referred to as the invasin homology region, was required for its function. We generated a truncated ImaA protein that is missing this region, from Asp42 to Ala1008, covering amino acids from the end of ImaA's signal peptide to the C-terminal end of the predicted homology (Fig. 3A). The gene expressing this truncated ImaA, called ImaA Asp42_Ala1008del, was used to replace the wild-type copy of imaA on the chromosome and thus is expressed from the native promoter in a single copy. Upon generating this strain, we validated that ImaA Asp42_Ala1008del is expressed to wild-type levels using Western blotting (Fig. 3B). We then assessed whether it was assembled at the outer membrane by treating cells with proteinase K to degrade surface-exposed proteins. This is a widespread method for assessing outer membrane protein localization and was previously used to show that ImaA is on the H. pylori surface (24, 36). ImaA Asp42_Ala1008del was degraded by proteinase K, as was the wild-type protein, while control inner membrane proteins were not (Fig. 3B). These results suggest that ImaA lacking amino acids 42 to 1008 is expressed and localizes to the outer membrane.
We next determined whether the imaA Asp42_Ala1008del strain would retain the ImaA function. AGS cells were infected with H. pylori strains bearing imaA Asp42_Ala1008del, and production of the Il8 transcript was enumerated. The increases in Il8 levels in these mutants were similar to those in the full imaA-null mutant (Fig. 3C). This result supports the idea that the invasin homology region of ImaA is critical for the protein's function to control Il8 production.
ImaA antagonizes the interactions of H. pylori with integrin.
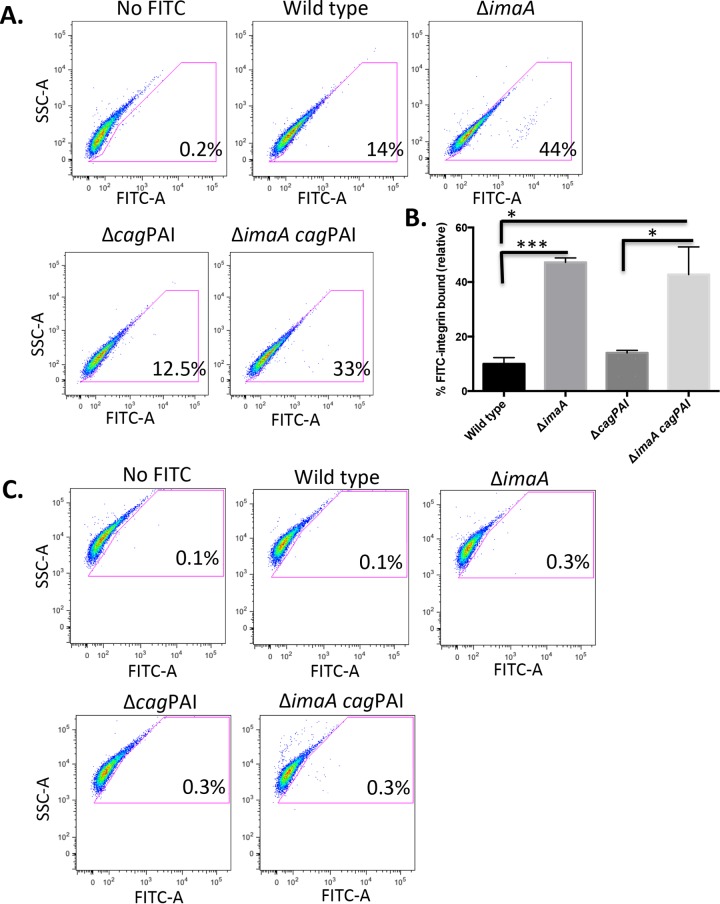
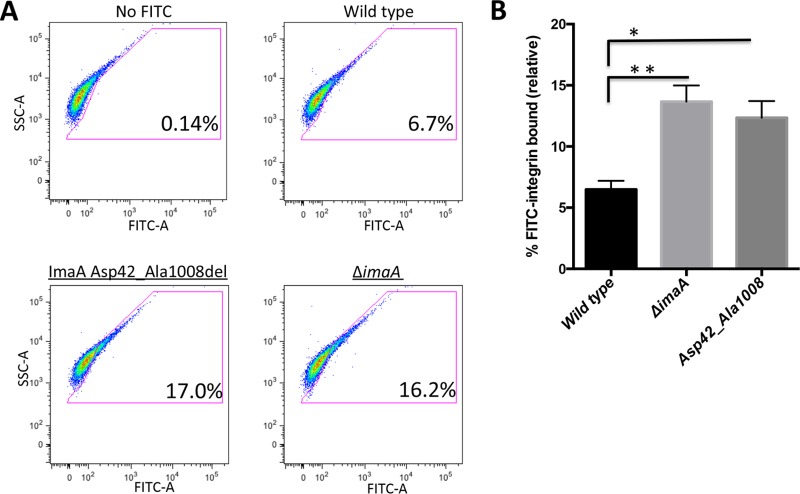
The homology of ImaA with the integrin-binding proteins invasin and pertactin (Fig. S1) prompted us to examine whether ImaA influenced the interactions of H. pylori with α5β1 integrin. We employed an in vitro flow cytometry-based integrin-binding assay which was derived from a method previously used to examine H. pylori interactions with extracellular matrix proteins (37). For this assay, H. pylori bacteria were incubated with fluorescein isothiocyanate (FITC)-labeled α5β1 integrin, fixed, and then analyzed for the amount of integrin bound to the bacteria. Using this method, we detected integrin bound to wild-type H. pylori (Fig. 4A and B). The ΔimaA mutant, surprisingly, displayed a significant increase in integrin binding (Fig. 4A and B). This finding suggests that ImaA normally antagonizes H. pylori-integrin interactions.
FIG 4.
imaA mutants exhibit enhanced binding to α5β1 integrin. (A) Wild-type H. pylori LSH100 and its isogenic mutants, the ΔimaA, Δcag PAI, and Δcag PAI imaA mutants, were incubated with FITC-labeled α5β1 integrin (octyl-β-d-glucopyranoside preparation) for 5 h. Bacteria were washed and fixed in 4% formaldehyde prior to analysis by flow cytometry. Ten thousand bacteria were counted per replicate, with a total of three biological replicates being used per strain. No FITC treatment of the wild type served as a negative control. To establish this control, a gate was created around bacterial cells in the no-FITC treatment group. All cells in the FITC-integrin treatment groups that shifted outside the no-FITC gate were considered positive for integrin binding. (B) Quantification of the amount of bound FITC-α5β1 integrin relative to that for the no-FITC control. Statistical analyses were done using Student's t test. *, P < 0.05; ***, P < 0.0005. (C) BSA conjugated to FITC was incubated with H. pylori to serve as a negative control, demonstrating that the observed population shifts seen with α5β1 integrin are α5β1 integrin specific. SSC, side scatter.
Integrin binding has been shown to be conferred by the T4SS proteins CagL, CagY, CagI, and CagA, although it has not been shown that these proteins are sufficient for integrin binding (18). We thus examined the role of the cag PAI T4SS in the integrin-binding phenotype. We used a mutant that has a complete loss of the cag PAI, to eliminate all the known integrin-binding proteins. Somewhat surprisingly, this mutation did not lower the levels of integrin interactions in this assay (Fig. 4A and B). This result may be explained in part by the fact that there are only low numbers of integrin interactions in the absence of host cells because the cag PAI T4SS machinery is not assembled (17, 38). Control experiments using FITC-bovine serum albumin (BSA) showed that H. pylori does not bind FITC or protein in general, nor is there any effect of ImaA (Fig. 4C). Taken together, these results suggest that ImaA tempers the interactions of H. pylori with integrin.
Integrin-binding modulation requires ImaA's functional region.
We next examined whether the invasin homology region of ImaA is important for the ΔimaA α5β1 integrin-binding phenotype (Fig. 4). We incubated the ImaA Asp42_Ala1008del strain with FITC-labeled α5β1 integrin (Fig. 4). The ImaA Asp42_Ala1008del strain exhibited elevated levels of binding to integrin compared to wild-type H. pylori, similar to the findings for the ΔimaA strain (Fig. 5). This result suggests that ImaA amino acids 42 to 1008 are required for the protein's ability to affect integrin binding.
FIG 5.
α5β1 integrin binding requires ImaA's predicted functional domain. (A) Wild-type H. pylori LSH100 and the isogenic imaA Asp42_Ala1008del strain were incubated with FITC-labeled α5β1 integrin essentially as described in the legend to Fig. 4, with the exception of the use of a Triton X-100 preparation of α5β1 integrin. (B) Gates were generated as described in the legend to Fig. 2, and the amount of bound FITC-α5β1 integrin is relative to that of the no-FITC treatment control. Statistical analyses were done using Student's t test. *, P < 0.05; **, P ≤ 0.009.
ImaA modulates the levels of host cell α5β1 integrin.
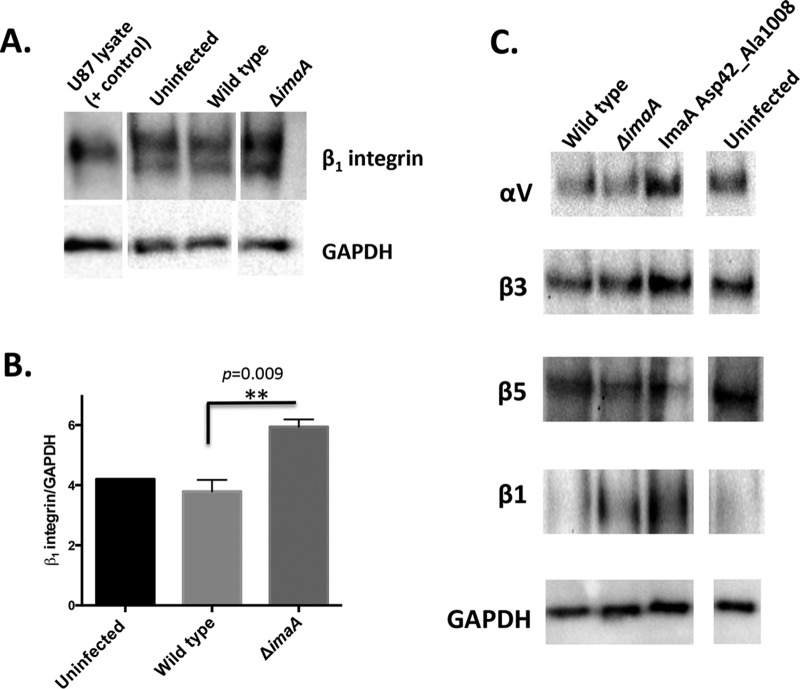
Our results presented above suggest that ImaA inhibits both the H. pylori-integrin interaction and cag PAI translocation. Given that ImaA also displayed remote homology to proteases, we hypothesized that ImaA might degrade integrin; without ImaA, therefore, there would be larger amounts of integrin to bind and an elevated ability of the cag PAI to interact with host cells. To test this idea, we infected AGS gastric epithelial cells with H. pylori and examined the state of β1 integrin using Western blotting. Lysates between AGS cells infected with either wild-type H. pylori or the ΔimaA mutant were compared. We detected larger amounts of β1 integrin in lysates taken from cells infected with ΔimaA mutant bacteria than those infected with the wild type (Fig. 6A and B). β1 integrin has multiple disulfide bonds and also is glycosylated, so it typically appears as a smeared band (39, 40). Control experiments supported the idea that this phenotype was specific to β1 integrin rather than to other integrins and required the putative ImaA functional region (Fig. 6C). These results suggest that the presence of functional ImaA correlates with a reduced amount of host cell integrin.
FIG 6.
ImaA regulates the availability of α5β1 integrin. (A) AGS cells were infected with either wild-type H. pylori or the imaA mutant or were left uninfected for 3 h. Following infection, cell lysates were collected and analyzed with an anti-β1 integrin antibody via Western blotting. Anti-GAPDH served as a loading control. (B) The levels of β1-integrin were quantified from three biological replicates using the ImageJ pixel densitometry program. **, P ≤ 0.009 by Student's t test. (C) AGS cells were infected with either wild-type H. pylori, the imaA mutant, or the imaA Asp42_Ala1008del mutant. Lysates were probed with a panel of antibodies that detect host cell integrins. Blots are representative of those from two biological replicates.
DISCUSSION
H. pylori has the ability to induce substantial inflammation via its T4SS and delivery of proinflammatory molecules (41). Here, we describe that the ImaA outer membrane protein tempers the bacterium's interactions with the receptor for the T4SS, α5β1 integrin. The loss of ImaA—either via a full null mutation or by elimination of its predicted functional domain—creates H. pylori isolates that exhibited heightened binding to α5β1 integrin. These mutants additionally evoked a strong proinflammatory response, which appears to have been due to increased T4SS interactions and delivery. Lastly, we show that imaA mutants leave a larger amount of integrin intact. Altogether, our data support a model in which ImaA promotes the degradation of integrin, with the result being that H. pylori lessens the host inflammatory response.
Our data presented here suggest that ImaA acts at the level of T4SS delivery. We found that ImaA mutants delivered larger amounts of CagA, as determined by measurement of both host cell-associated total CagA and phosphorylated CagA (Fig. 2). T4SS delivery occurs via several steps, including assembly of the T4SS pilus, binding of the pilus to its host cell target, and active protein secretion (42). The imaA phenotype did not depend on CagA per se, suggesting that ImaA acts at one of the steps that are CagA independent, such as binding or assembly. There is not much known, however, about the molecular steps of cag PAI T4SS assembly, aside from information about which proteins are required for cag PAI T4SS formation and for integrin binding (18, 22, 43). We were able to gain insight into the ImaA function, however, when we found that it normally antagonizes integrin binding. This finding suggests that ImaA acts at the host cell receptor binding stage of T4SS delivery.
We were able to determine that ImaA has a predicted functional domain with a remote similarity to other integrin-binding proteins, including invasin and pertactin (32–35), and to extracellular proteases from Serratia and Pseudomonas species. We show here that this region is the functional region of ImaA. Most autotransporters similarly place their functional domains in the same N-terminal region (25). If it is assumed that ImaA adopts a typical autotransporter architecture (44), the functional domain would be placed ∼200 nm from the bacterial surface and might be poised to be the first point of contact between H. pylori and the host cells. One possible model that is consistent with both our data and bioinformatic predictions of proteolytic function is that ImaA interacts with and either digests or enzymatically alters integrin. We did not detect any small integrin fragments, however, which could mean that ImaA is not a protease or simply that the fragments are not detected by the antibody. Our data support a model in which H. pylori alters the amount of ImaA so as to regulate the number of binding sites for H. pylori integrin-binding proteins. With a low level of ImaA, a larger amount of intact integrin is present, there are more potential points of contact for H. pylori integrin-binding proteins, and the host inflammatory response is increased. Alternatively, it is possible that through an ImaA-α5β1 integrin interaction, a downregulation in the production of β1 integrin by the host cell occurs, also resulting in reduced binding sites for cag PAI T4SS.
Our findings presented here show that ImaA alters the interactions of H. pylori with α5β1 integrin. Integrins are a class of heterodimeric mammalian proteins that play a prominent role in many types of bacterial and viral infections, including acting as the receptor for the H. pylori cag PAI pilus (45, 46). There are many documented cases where bacterial proteins exploit integrins to gain a fitness advantage. Examples of these methods include the use of integrins to promote bacterial uptake inside host cells or the activation of integrins to dysregulate epithelial cell turnover rates (45). Additionally, a single integrin subunit can have multiple binding partners, often spanning species of bacteria. For example, H. pylori binds to β1 integrin to scaffold its T4SS to host cells, while uropathogenic Escherichia coli exploits β1 integrin to become internalized within host cells (22, 47). Thus, it is clear that integrins play a pivotal role at the bacterium-host cell interface.
In summary, we have identified a new mechanism utilized by H. pylori to regulate the bacterium's interactions with α5β1 integrin, the pathogen's conduit for delivering proinflammatory effectors. These results suggest a model in which H. pylori utilizes ImaA to reduce the number of available α5β1 integrin interaction sites that can be bound by the T4SS, which modulates the severity of the immune response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
For these studies, we used H. pylori strain LSH100, a mouse-adapted descendant of clinical isolate G27 (48, 49) (Table 1). H. pylori strains were maintained on Columbia blood agar base (Becton Dickinson) supplemented with 5% defibrinated horse blood (Hemostat Laboratories, Dixon, CA) and 0.2% (wt/vol) β-cyclodextrin plus 5 μg/ml trimethoprim, 8 μg/ml amphotericin B, 50 μg/ml cycloheximide, 10 μg/ml vancomycin, 5 μg/ml cefsulodin, and 2.5 U/ml polymyxin B (CHBA) to inhibit the growth of unwanted microbes under 10% CO2, 7 to 10% O2, and a balance of N2 at 37°C. Liquid H. pylori cultures were grown in brucella broth supplemented with 10% fetal bovine serum (FBS; Life Technologies). The antibiotic chloramphenicol (Cm) was used for selection at a concentration of 13 μg/ml. H. pylori strains were stored at −80°C in brain heart infusion medium supplemented with 10% fetal bovine serum, 1% (wt/vol) β-cyclodextrin, 25% glycerol, and 5% dimethyl sulfoxide. All antibiotics and chemicals were obtained from Sigma, Gold Biosciences, or Fisher.
TABLE 1.
Strains used in this study
| H. pylori strain | Description | Reference or source |
|---|---|---|
| LSH100 | NSH57 with repaired fliM allele | 49 |
| KO1370 | LSH100 ΔimaA::cat | 24 |
| KO1372 | LSH100 ΔcagE::kan | 24 |
| KO1373 | LSH100 ΔcagE::kan imaA::cat | 24 |
| G27 | ΔcagA::cat | 51 |
| G27 | ΔcagPAI::kan | 50 |
| KO1413 | LSH100 Δcag PAI::kan imaA::cat | This study |
| KO1414 | LSH100 Δcag PAI::kan | This study |
| KO1415 | LSH100 ΔcagA::cat | This study |
Generation of H. pylori mutants.
H. pylori ΔimaA::cat, ΔcagE::kan, and ΔcagE::kan ΔimaA::cat mutants were generated as previously described (24) and are listed in Table 1. The LSH100 mutants Δcag PAI::kan and Δcag PAI::kan imaA::cat were generated by naturally transforming H. pylori with genomic DNA (gDNA) kindly provided by Nina Salama (50). The LSH100 ΔcagA::cat and Δcag::cat imaA::kan mutants were generated by naturally transforming H. pylori with gDNA kindly provided by Manuel Amieva (51).
The LSH100 imaA Asp42_Ala1008del strain, which features a deletion corresponding to amino acids 42 to 1008 at ImaA's N terminus (31), was generated through the insertion of the counterselectable kan-sacB cassette at the imaA locus and through the subsequent removal of the cassette with a homologous suicide plasmid, which excluded the chromosomal region of interest. First, we generated strain LSH100 imaA::kan-sacB by introducing the kan-sacB cassette into the imaA open reading frame between nucleotides 295285 and 296253 (strain G27 genome numbering) (52), using splicing by overlap extension (SOE) PCR, with the primers imaASOE#1 (5′-TGAATGAAACGCTGCAACCT), imaASOE#2 (5′-GCCCGGGCTCGAGGGGGGGGGTAATAGGGCCAAGGCCC), imaASOE#3 (5′-GGGCCCCCCCTCGAGCCCGGG), imaASOE#4 (5′-CGGTGGCGGCCGCTCTAGAC), imaASOE#5 (5′-GGGTCTAGAGCGGCCGCCACAATGCACAGGTACGACTAA), and imaASOE#6 (5-CGCTATTGTTAAAGTTAGC). The presence of the cassette was confirmed through PCR and bacterial sensitivity to sucrose and resistance to kanamycin. Meanwhile, the plasmid pBS-imaA, which contains a 3,369-nucleotide region of imaA spanning nucleotides 293670 to 297039, using strain G27 genome numbering, was generated using the primers imaAStart (5′-CTAGGAATTCATGAAAAAGTTTAAAAAGAAA) and imaAint (5′-CTAGCTCGAGTTTGCCCGGTGAATGTTTGA). This plasmid served as the template for inverse PCR (iPCR), where we deleted nucleotides 293793 to 296688, with the primers imaASigPR (5′-TCCATTCGCATACACCCCAC) and imaAHS9plus3 (5′-CATGGGGCAAGCAGTGAAAA). This plasmid was then used to transform the imaA::kan-sacB strain, removing the kan-sacB cassette and creating a 2,895-nucleotide deletion at the 5′ end of imaA. Confirmation of the deletion was achieved through PCR and Western blotting.
Cell culture.
AGS (ATCC CRL 1739) human gastric epithelial cells were obtained directly from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium (DMEM) (Lonza, Walkersville, MD) containing 10% FBS (Invitrogen) at 37°C under 10% CO2. To assay interleukin-8 (IL-8) production, AGS cells were seeded at 1 × 105 cells/ml in 24-well tissue culture dishes and incubated for 24 h. After this period, H. pylori cells that had been cultured for ∼20 h on CHBA were scraped from the plate, resuspended in sterile DMEM-FBS to a concentration of 1 × 107 CFU/ml, and then used at a multiplicity of infection (MOI) of 100. H. pylori concentrations were determined by measurement of the optical density at 600 nm (OD600), assuming 3 × 108 bacteria/ml/OD600 unit (24, 53). AGS cells were infected for 2 h under H. pylori incubation conditions (10% CO2, 5% O2, balance N2). After a 2-h incubation, the culture supernatant was removed, AGS cell monolayers were washed twice in 1× phosphate-buffered saline (PBS), and then the cells were resuspended in TRIzol for RNA isolation.
For CagA translocation, AGS cells seeded at 1 × 105 cells/ml in 24-well dishes were infected at an MOI of 100 for 3 h with either wild-type H. pylori or the imaA isogenic mutant under the same conditions described above. However, after the third hour of infection, AGS cells underwent various treatments, as follows: (i) the cells were not washed, in which AGS cells remained unwashed and the infection was allowed to persist for an additional 6 h; (ii) the cells were washed and not treated with kanamycin (Km), in which AGS cells were washed 3 times with ice-cold 10 mM sodium orthovanadate and replenished with fresh medium for an additional 6 h; and (iii) washed and treated with Km (400 μg/ml) for 6 h, in which AGS cells were washed 3 times with ice-cold 10 mM sodium orthovanadate and supplemented with medium containing kanamycin for an additional 6 h. After the 6-h period, the AGS cells were washed, collected, and analyzed by Western blotting with antibodies.
RNA preparation.
RNA was isolated from AGS cells using the TRIzol reagent (Invitrogen). Briefly, 1 ml TRIzol reagent per 10 cm2 of the culture dish surface was added directly to cells in the culture dish. The cells were lysed directly in the culture dish by pipetting the cells up and down several times. For RNA isolation, 200 μl of chloroform was added to the TRIzol resuspensions. Samples were then centrifuged (12,000 × g, 15 min, 4°C), and the aqueous layer was removed and placed into new tubes. RNA was precipitated by combining 500 μl of isopropanol with the aqueous layer and incubating the mixture at room temperature for 10 min, followed by a centrifugation as described above. The RNA pellet was washed with 75% ethanol and then dried and resuspended in RNase-free water. Concentrations were then quantified on a NanoDrop spectrophotometer (NanoDrop). RNA was immediately transcribed into cDNA (see below), and the remaining sample was stored at −80°C.
cDNA synthesis and quantitative real-time PCR.
Total RNA served as a template for cDNA synthesis, performed using a Tetro cDNA synthesis kit (Bioline, London, UK). Synthesis was carried out following the manufacturer's protocol, starting with 0.5 to 1 μg total RNA, 50 ng random hexamers, and 10 mM deoxynucleoside triphosphates per 20-μl reaction mixture. The mixture was incubated at 65°C for 10 min before being combined with 10 μl of master mix, which included the reverse transcriptase enzyme (200 U/μl). The reaction proceeded for 1 h at 42°C until the reverse transcriptase enzyme was inactivated by heating at 70°C for 15 min. Quantitative real-time PCR was performed using a CFX Connect real-time cycler (Bio-Rad, Hercules, CA) and SYBR green supermix reagents (Bioline, London, UK). The levels of Il8 expression (54) from AGS cells were normalized to the levels of 18S rRNA expression (54).
FITC labeling of α5β1 integrin.
Purified full-length human α5β1 integrin, an octyl-β-d-glucopyranoside formulation (0.418 mg/ml), or a Triton X-100 formulation (0.4 mg/ml), the latter two of which were obtained from Millipore, was conjugated to FITC using a Calbiochem FITC labeling kit. We switched to the Triton X-100 formulation because Millipore has discontinued the octyl-β-d-glucopyranoside formulation. Prior to labeling, the entire integrin sample (50 μl) was dialyzed overnight at 4°C with 1 liter of 1× carbonate buffer (Calbiochem) containing 1% Triton X-100 (Fisher) in a Thermo 2,000-molecular-weight-cutoff Slide-A-Lyzer minidialyzer. The dialyzed protein sample was then incubated with 10 μl of the Calbiochem FITC mixture at room temperature for 2 h in the dark. The freshly conjugated FITC-integrin sample was then dialyzed again overnight at 4°C with 300 μl of 1× PBS containing 1% Triton X-100 in the Thermo minidialyzer unit. Finally, the dialyzed FITC-α5β1 integrin sample (∼45 μl) was removed from the unit and stored at −20°C until use.
Flow cytometry.
Flow cytometry experiments were conducted by a method similar to that described previously for H. pylori-laminin binding (37). H. pylori cells that had been growing for ∼18 h were scraped from plates and diluted to an OD600 of 1.1 in 1× PBS with 2% BSA. One-milliliter cell suspensions were rotated at room temperature in 1× PBS with 2% BSA for 30 min to block nonspecific protein binding. Following this step, each H. pylori sample was given 2 μl of the FITC-α5β1 integrin conjugation mixture, with 5 replicates being used for each strain. The cells were then rotated in the dark at room temperature for 5 h. Following the incubation, the cells were centrifuged at 8,000 × g for 3 min, the supernatant was removed, and the cells were washed in 1× PBS with 2% BSA and then spun down again at 8,000 × g for 3 min. The cell pellets were then resuspended in 1 ml 4% formaldehyde and stored overnight at 4°C. Flow cytometry was conducted on a BD LSR II flow cytometer (BD Biosciences), with 10,000 cells per sample being counted. Samples included H. pylori cells that had been incubated with FITC alone. All analyses were done using FlowJo software (BD Biosciences), and statistical analysis was done using Student's t test.
Western blotting.
Proteins for Western blot analysis were resuspended in 4× NuPAGE sample buffer (Invitrogen, Carlsbad, CA) with 0.025% 2-mercaptoethanol, and the mixture was heated at 70°C for 15 min. Samples were separated on 3 to 8% NuPAGE Tris-acetate gradient gels for 60 min at 150 V. Following electrophoresis, the proteins were transferred to methanol-activated polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA) with a Bio-Rad semidry transfer cell for 35 min at 16 V.
Antibodies for detection of phosphorylated CagA (anti-phosphorylated-Tyr; catalog number PY99), total CagA (anti-CagA; catalog number B-300), and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; anti-GAPDH; catalog number G-9) were obtained from Santa Cruz Biotech and used at dilutions of 1:200, 1:400, and 1:500, respectively. Antibodies from an integrin antibody sampler pack (catalog number 4749; Cell Signaling Technology) were used at 1:1,000 dilutions. The remaining H. pylori proteins were detected with a 1:300 dilution of anti-ImaA-1 antibody or a 1:2,000 dilution of anti-glutathione S-transferase (GST)-TlpA22 antibody (24, 55). For visualization, blots were incubated with either goat anti-rabbit or goat anti-mouse immunoglobulin antibodies conjugated to horseradish peroxidase (Santa Cruz Biotech) at a dilution of 1:2,000 for 1 h, followed by incubation with luminol, p-coumaric acid, and hydrogen peroxide. Luminescent blots were visualized by exposure to Ultra Cruz autoradiography film (Santa Cruz Biotech).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Manuel Amieva (Stanford University) and Nina Salama (Fred Hutchison Cancer Research Center) for providing the cagA and cag PAI island mutants, to Stefan Backert for providing advice about phospho-CagA analyses, and to Elliot Carter for providing experimental support. We also thank the University of California, Santa Cruz, Flow Cytometry Facility in the Institute for the Biology of Stem Cells, particularly Bari Nazario, for providing training and support with the flow cytometry work.
The project described here was supported by grant number AI5103624 (to K.M.O.) from the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health, funds from the University of California, Santa Cruz, Graduate Division (to W.E.S.), and funds from the ARCS Foundation (to W.E.S.).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00450-16.
REFERENCES
- 1.Eusebi LH, Zagari RM, Bazzoli F. 2014. Epidemiology of Helicobacter pylori infection. Helicobacter 19(Suppl 1):1–5. doi: 10.1111/hel.12165. [DOI] [PubMed] [Google Scholar]
- 2.Fischer W. 2011. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J 278:1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 3.Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev 19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrera V, Parsonnet J. 2009. Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect 15:971–976. doi: 10.1111/j.1469-0691.2009.03031.x. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco C, Rappuoli R. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol 2:457–466. doi: 10.1038/35073084. [DOI] [PubMed] [Google Scholar]
- 6.Suerbaum S, Michetti P. 2002. Helicobacter pylori infection. N Engl J Med 347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 7.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. 2015. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 8.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA (ed). 1975-2011. SEER cancer statistics review. National Cancer Institute, Bethesda, MD. [Google Scholar]
- 9.Blaser MJ, Berg DE. 2001. Helicobacter pylori genetic diversity and risk of human disease. J Clin Invest 107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorrell RJ, Guan J, Xin Y, Tafreshi MA, Hutton ML, McGuckin MA, Ferrero RL, Kwok T. 2013. A novel NOD1- and CagA-independent pathway of interleukin-8 induction mediated by the Helicobacter pylori type IV secretion system. Cell Microbiol 15:554–570. doi: 10.1111/cmi.12055. [DOI] [PubMed] [Google Scholar]
- 11.Brandt S, Kwok T, Hartig R, Konig W, Backert S. 2005. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc Natl Acad Sci U S A 102:9300–9305. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, Athman R, Memet S, Huerre MR, Coyle AJ, DiStefano PS, Sansonetti PJ, Labigne A, Bertin J, Philpott DJ, Ferrero RL. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol 5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 13.Terradot L, Waksman G. 2011. Architecture of the Helicobacter pylori Cag-type IV secretion system. FEBS J 278:1213–1222. doi: 10.1111/j.1742-4658.2011.08037.x. [DOI] [PubMed] [Google Scholar]
- 14.Vannini A, Roncarati D, Spinsanti M, Scarlato V, Danielli A. 2014. In depth analysis of the Helicobacter pylori cag pathogenicity island transcriptional responses. PLoS One 9:e98416. doi: 10.1371/journal.pone.0098416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson EM, Gaddy JA, Voss BJ, Hennig EE, Cover TL. 2014. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun 82:3457–3470. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson EM, Gaddy JA, Cover TL. 2012. Alterations in Helicobacter pylori triggered by contact with gastric epithelial cells. Front Cell Infect Microbiol 2:17. doi: 10.3389/fcimb.2012.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jimenez-Soto LF, Haas R. 2016. The CagA toxin of Helicobacter pylori: abundant production but relatively low amount translocated. Sci Rep 6:23227. doi: 10.1038/srep23227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862–866. doi: 10.1038/nature06187. [DOI] [PubMed] [Google Scholar]
- 19.Backert S, Clyne M, Tegtmeyer N. 2011. Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori. Cell Commun Signal 9:28. doi: 10.1186/1478-811X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N, Salama N, Churin Y, Meyer TF. 2013. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol 15:1896–1912. doi: 10.1111/cmi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Boren T, Haas R, Sasakawa C, Mimuro H. 2011. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J Biol Chem 286:25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. 2009. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog 5:e1000684. doi: 10.1371/journal.ppat.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedemann T, Hofbaur S, Tegtmeyer N, Huber S, Sewald N, Wessler S, Backert S, Rieder G. 2012. Helicobacter pylori CagL dependent induction of gastrin expression via a novel alphavbeta5-integrin-integrin linked kinase signalling complex. Gut 61:986–996. doi: 10.1136/gutjnl-2011-300525. [DOI] [PubMed] [Google Scholar]
- 24.Sause WE, Castillo AR, Ottemann KM. 2012. The Helicobacter pylori autotransporter ImaA (HP0289) modulates the immune response and contributes to host colonization. Infect Immun 80:2286–2296. doi: 10.1128/IAI.00312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo AR, Woodruff AJ, Connolly LE, Sause WE, Ottemann KM. 2008. Recombination-based in vivo expression technology identifies Helicobacter pylori genes important for host colonization. Infect Immun 76:5632–5644. doi: 10.1128/IAI.00627-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radin JN, Gaddy JA, Gonzalez-Rivera C, Loh JT, Algood HM, Cover TL. 2013. Flagellar localization of a Helicobacter pylori autotransporter protein. mBio 4:e00613-12. doi: 10.1128/mBio.00613-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Takeuchi H, Nishioka M, Morimoto N, Kamioka M, Kumon Y, Sugiura T. 2009. Relationship of IL-8 production and the CagA status in AGS cells infected with Helicobacter pylori exposed to low pH and activating transcription factor 3 (ATF3). Microbiol Res 164:180–190. doi: 10.1016/j.micres.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree JE, Farmery SM, Lindley IJ, Figura N, Peichl P, Tompkins DS. 1994. CagA/cytotoxic strains of Helicobacter pylori and interleukin-8 in gastric epithelial cell lines. J Clin Pathol 47:945–950. doi: 10.1136/jcp.47.10.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsugawa H, Suzuki H, Saya H, Hatakeyama M, Hirayama T, Hirata K, Nagano O, Matsuzaki J, Hibi T. 2012. Reactive oxygen species-induced autophagic degradation of Helicobacter pylori CagA is specifically suppressed in cancer stem-like cells. Cell Host Microbe 12:764–777. doi: 10.1016/j.chom.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 31.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 32.Isberg RR, Van Nhieu GT. 1995. The mechanism of phagocytic uptake promoted by invasin-integrin interaction. Trends Cell Biol 5:120–124. doi: 10.1016/S0962-8924(00)88962-X. [DOI] [PubMed] [Google Scholar]
- 33.Leong JM, Morrissey PE, Marra A, Isberg RR. 1995. An aspartate residue of the Yersinia pseudotuberculosis invasin protein that is critical for integrin binding. EMBO J 14:422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leininger E, Roberts M, Kenimer JG, Charles IG, Fairweather N, Novotny P, Brennan MJ. 1991. Pertactin, an Arg-Gly-Asp-containing Bordetella pertussis surface protein that promotes adherence of mammalian cells. Proc Natl Acad Sci U S A 88:345–349. doi: 10.1073/pnas.88.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruoslahti E. 1996. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol 12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 36.Lenz JD, Lawrenz MB, Cotter DG, Lane MC, Gonzalez RJ, Palacios M, Miller VL. 2011. Expression during host infection and localization of Yersinia pestis autotransporter proteins. J Bacteriol 193:5936–5949. doi: 10.1128/JB.05877-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senkovich OA, Yin J, Ekshyyan V, Conant C, Traylor J, Adegboyega P, McGee DJ, Rhoads RE, Slepenkov S, Testerman TL. 2011. Helicobacter pylori AlpA and AlpB bind host laminin and influence gastric inflammation in gerbils. Infect Immun 79:3106–3116. doi: 10.1128/IAI.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog 7:e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law ME, Ferreira RB, Davis BJ, Higgins PJ, Kim JS, Castellano RK, Chen S, Luesch H, Law BK. 2016. CUB domain-containing protein 1 and the epidermal growth factor receptor cooperate to induce cell detachment. Breast Cancer Res 18:80. doi: 10.1186/s13058-016-0741-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pan L, Zhao Y, Yuan Z, Qin G. 2016. Research advances on structure and biological functions of integrins. Springerplus 5:1094. doi: 10.1186/s40064-016-2502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaoka Y, Kita M, Kodama T, Sawai N, Kashima K, Imanishi J. 1997. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut 41:442–451. doi: 10.1136/gut.41.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Backert S, Tegtmeyer N, Fischer W. 2015. Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system. Future Microbiol 10:955–965. doi: 10.2217/fmb.15.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD, Cover TL. 2016. Molecular and structural analysis of the Helicobacter pylori cag type IV secretion system core complex. mBio 7:e02001-1. doi: 10.1128/mBio.02001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hartmann MD, Grin I, Dunin-Horkawicz S, Deiss S, Linke D, Lupas AN, Hernandez Alvarez B. 2012. Complete fiber structures of complex trimeric autotransporter adhesins conserved in enterobacteria. Proc Natl Acad Sci U S A 109:20907–20912. doi: 10.1073/pnas.1211872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauck CR, Borisova M, Muenzner P. 2012. Exploitation of integrin function by pathogenic microbes. Curr Opin Cell Biol 24:637–644. doi: 10.1016/j.ceb.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Stewart PL, Nemerow GR. 2007. Cell integrins: commonly used receptors for diverse viral pathogens. Trends Microbiol 15:500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. 2007. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog 3:e100. doi: 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci U S A 90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lowenthal AC, Hill M, Sycuro LK, Mehmood K, Salama NR, Ottemann KM. 2009. Functional analysis of the Helicobacter pylori flagellar switch proteins. J Bacteriol 191:7147–7156. doi: 10.1128/JB.00749-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan S, Tompkins LS, Amieva MR. 2009. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog 5:e1000407. doi: 10.1371/journal.ppat.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amieva MR, Salama NR, Tompkins LS, Falkow S. 2002. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol 4:677–690. doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- 52.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. 2009. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol 191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salama NR, Otto G, Tompkins L, Falkow S. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect Immun 69:730–736. doi: 10.1128/IAI.69.2.730-736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A. 2002. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res 30:e37. doi: 10.1093/nar/30.9.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun 75:3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.