Abstract
Introduction:
Naive Bayesian networks (NBNs) are one of the most effective and simplest Bayesian networks for prediction.
Objective:
This paper aims to review published evidence about the application of NBNs in predicting disease and it tries to show NBNs as the fundamental algorithm for the best performance in comparison with other algorithms.
Methods:
PubMed was electronically checked for articles published between 2005 and 2015. For characterizing eligible articles, a comprehensive electronic searching method was conducted. Inclusion criteria were determined based on NBN and its effects on disease prediction. A total of 99 articles were found. After excluding the duplicates (n= 5), the titles and abstracts of 94 articles were skimmed according to the inclusion criteria. Finally, 38 articles remained. They were reviewed in full text and 15 articles were excluded. Eventually, 23 articles were selected which met our eligibility criteria and were included in this study.
Result:
In this article, the use of NBN in predicting diseases was described. Finally, the results were reported in terms of Accuracy, Sensitivity, Specificity and Area under ROC curve (AUC). The last column in Table 2 shows the differences between NBNs and other algorithms.
Discussion:
This systematic review (23 studies, 53,725 patients) indicates that predicting diseases based on a NBN had the best performance in most diseases in comparison with the other algorithms. Finally in most cases NBN works better than other algorithms based on the reported accuracy.
Conclusion:
The method, termed NBNs is proposed and can efficiently construct a prediction model for disease.
Keywords: Naive Bayes Algorithms, Naive Bayes Models, Naive Bayesian Network, Naive Bayesian Network and disease prediction
1. INTRODUCTION
Bayesian theory and probability are named after a British 18th century mathematician, Thomas Bayes. Bayesian logic can show the result of a patient’s test with a pre-test probability (of the population), to predict or determine the chance of finding a disease. Bayesian theory implies that Bayes’ theorem can be used as a rule for inferring or updating the amount of ‘belief’ in the light of new information. Bayesian networks can be seen as a substitute for logistic regression models where we can formulate dependency or independency of variables (1, 2).
The NBNs consist only of one parent and several child nodes as in Figure 1 and it is based on Bayes theorem in machine learning. Let D be a training set of database. Each database is represented by an n _ D attribute vector. X includes ‘n’ independent attributes (x1, x2, …, xn). Suppose there are ‘m’ classes such as c1, c2, …, cm, then classification is to derive the maximal p(ci|x). This can be derived from Bayes’ theorem as equation 1:
Figure 1.
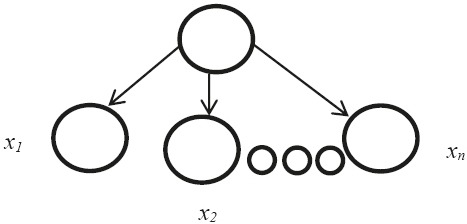
The structure of the naïve Bayes network
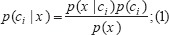
p(x) needs to be maximized because it has equal value for all classes as equation 2:
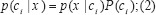
An easy assumption in NBN is that the attributes are conditionally independent (i.e. there is no dependency between all attributes). So, the class assignments of this test samples are according to equations (3) and (4);
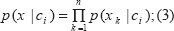
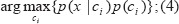
For example, if a new sample posterior probability comes i.e. p (c2|x) has the highest value among the all p (ck|x) for all the k classes, it belongs to c2 class base on the NBN rule (3, 4).
NBNs can estimate the post-test probability given the values of various predictive variables. Astonishingly, the performance of a NBN is somewhat competitive given that this is obviously an unrealistic assumption (5, 6).
The structure of NBNs can be showed with a directed acyclic graph which is showed in Figure 1, where the nodes represent variables and edges between the nodes show dependency among the variables. Within a node, a variable can take many distinctive values, each with a special probability. One major issue in this model is that the root node of the network has a connection to all predictive variables and does not depend on any other variables (7).
In a NBN, as used in this study, among the different feature variables, there is no inter-dependency. They are thus regarded as conditionally independent, hence the definition of ‘naive’. An example of the classification problems of these naïve Bayesian networks is the article published by Price et al. on the classification of cervical cancer patients (1).
This study was designed to perform a comprehensive review of a naïve Bayesian network and its use in predicting diseases. The purpose of the present study was to review published evidence of using NBN in disease prediction to show the power of this method in comparison with the other methods. Moreover, to the best of our knowledge, it is the first study that directly compares NBN with other models for disease prediction.
2. METHODS
Inclusion and exclusion criteria
Inclusion criteria were determined based on the topic of study and the effects of naïve Bayesian networks in predicting different diseases. The full text of articles needed to be available. The articles written in English language were selected.
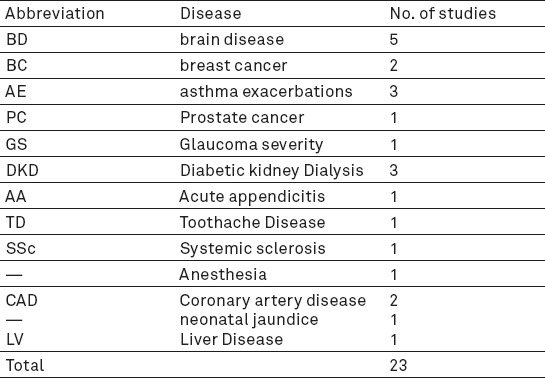
The type of diseases predicted by Naïve Bayesian network and included in this review was shown in Table 1.
Table 1.
frequency of disease
NBNs aim to improve disease treatment and also its diagnosis in early stages for a faster and better treatment. Therefore, any NBNs attempt to:
* Make a faster and more accurate disease prediction.
* Help the physicians for making a reliable decision.
We classified the studies according to disease prediction and Method comparison. Original articles were included in this study. On the other hand, studies that used NBN as a feature selection (n =16) as well as those that reported survival of disease based on NBN (n =2) were excluded.
The titles and abstracts of the articles found were screened based on inclusion criteria as described above. Full texts of published articles were reviewed. Two reviewers independently did the review process and summarized the information in every article. Any disagreements between the reviewers were discussed by the references to the original articles. Exclusion criteria were as the following: a) studies that used the naïve Bayesian networks as a feature selection, b) studies lacking in disease prediction (3) those which used naïve Bayesian networks for predicting anything but the disease. We did not include studies in languages other than English.
As shown in Figure 2, a total of 99 articles were identified based on the search conducted in PubMed. Just one database was selected because it is the reliable medical source. After the duplicates were excluded, we checked titles and abstracts of 94 articles based on our inclusion criteria. Finally, 23 articles met our eligibility criteria and were included in this study.
Figure 2.
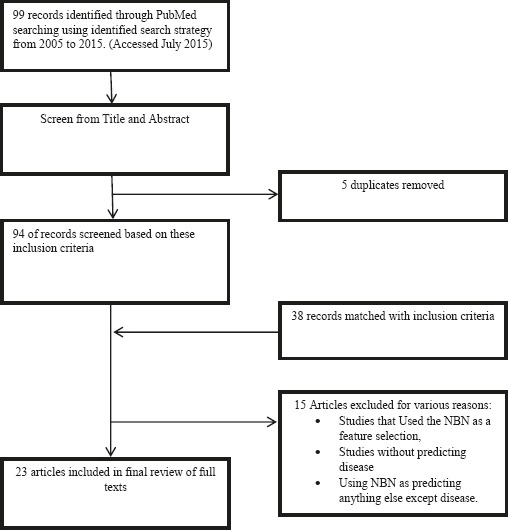
Selection process for study inclusion
2.2 Search strategy
A literature review was done on 26 July 2015 using PubMed database in order to peruse the relevant studies published in the past ten years from 2005 to 2015. We only used one valid database like PubMed in order to receive original and relevant articles and PubMed is the main database that includes several medical articles. The more databases we searched, the more articles but irrelevant we got, so we preferred using only one database.
A combination of the following MESH terms and keywords was used:
((“Naïve Bayes models”[Mesh] or (disease prediction) or (predicting disease), (“Naïve Bayes algorithms” [Mesh]), (“Naïve Bayesian network” [Mesh]).
We also set limits to our search according to the study result and language.
2.3 Data extraction
Titles and abstracts of all selected articles were reviewed independently by two reviewers. Papers categorized as relevant or of irrelevance based on the abstracts were again independently assessed by both reviewers. Any disagreements between the two reviewers were resolved through discussion. Reasons for exclusion were recorded according to the exclusion criteria.
The data extraction and quality assessment of the articles were done by the first author and checked by the coauthor for accuracy to identify any missing information individually.
A data extraction was done independently. This extraction contains the following items:
* Article properties e.g. title and publication year,
* Subject,
* Illnesses,
* Number of Variables (features).
Results in terms of accuracy, sensitivity, specificity or Area under ROC (Receiver operating characteristic) curve (AUC) of NBNs.
And finally, the comparison with other methods that is mentioned in the related article.
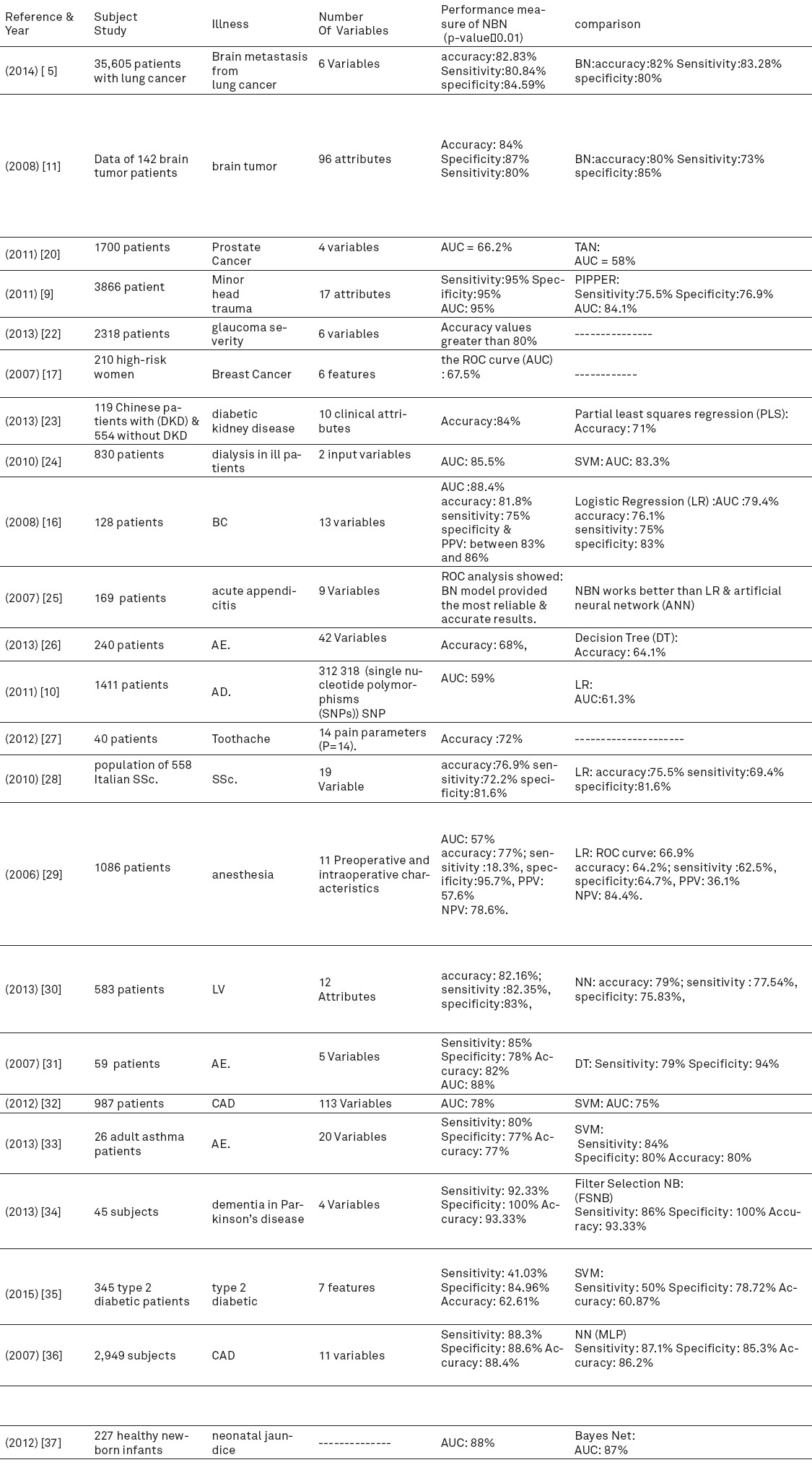
All the extracted data were categorized in Table 2.
Table 2.
Summary of important factors in NBN predictors.PPV: positive predictive value; NPV: negative predictive value
Data-synthesis and analysis
A narrative synthesis was carried out based on the classification of diseases.
The outcomes were the effects on the following items:
-
a)
Accuracy = TP + TN / (TP + FP + FN + TN),
-
b)
Sensitivity = TP / (TP + FN),
-
c)
Specificity = TN / (TN + FP),
-
d)
Area under ROC curve (AUC): Accuracy is measured by the area under the ROC curve (As shown in Figure 3). A rough guide for classifying the accuracy of a diagnostic test is the traditional academic point system:
Figure 3.
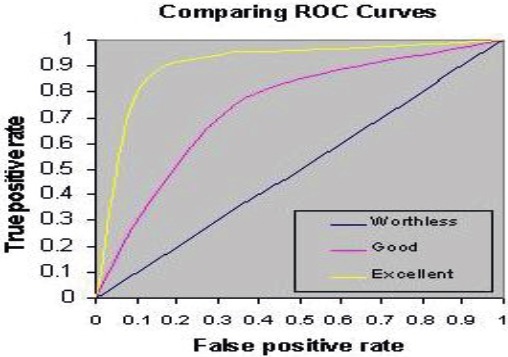
ROC Curves
* 0.9-1 = excellent (A),
* 0.8-0.9 = good (B),
* 0.7-0.8 = fair (C),
* 0.6-0.7 = poor (D),
* 0.5-0.6 = fail (F) (8).
3. RESULTS
3.1 Description of the articles included
Articles were published between 2005 and 2015. A total of 15 articles reported accuracy and 12 of them reported sensitivity and specificity while only 11 studies in 23 studies reported AUC. 5 studies used naïve Bayesian networks for predicting brain disease. Other diseases are presented in Table 1.
Table 1 demonstrates the frequency of diseases applied in the articles. The mean duration was approximately 9 years and the mean of variables or features that NBN used was about 17 cases. Results are explained based on the outcomes of naïve Bayesian networks performance. Table 2 demonstrates the results and important factors that influenced our outcomes briefly. In the proceeding, we will explain the effects of NBNs on predicting different diseases and compare them with other methods like logistic Regression (LR), support vector machine (SVM), neural network (NN), decision tree (DT), tree augmented NBN (TAN), Bayesian network (BN).
In most cases, NBNs work better than the others and it shows that the fundamental algorithm can work well and it is really useful for disease prediction. Maybe, in some cases are not necessary to enhance our method, because of good effects of NBNs in accurate predictions.
3.1.1 Using NBN as a brain disease prediction
21.7 percent of the articles in this study included brain diseases such as brain tumor, metastasis, trauma and Alzheimer (5, 9-11). Computed tomography (CT) is widely accepted as an effective method to diagnose and detect rare but clinically significant in patients suffering minor head injury or brain disease. As such, it has been increasingly utilized as a simple and available test for these patients (12). However, a seminal study conducted by Brenner and Hall (13) warns against its bad effects (particularly for children) due to the radiation exposure associated with CT. Independent CT imaging studies (12, 14, 15) advocate the adoption of a comprehensive approach that targets physicians’ education to decrease the over-reliance on CT imaging for those suffering from brain disease. Therefore, to reduce the harmfulness of CT, it can be beneficial to use data mining methods specially, NBN as a predictor. As shown in Table 2, the final results are presented.
3.1.2 Naïve Bayesian networks predictor in cancer disease
3 articles belonged to this group. Two of them used naïve Bayesian networks for predicting breast cancer (16, 17). It is one of the most important cancer in women (18, 19), and one discussed the most common disease in men that is the prostate cancer (20). Cancer is a widely spread disease that accounts for many mortalities all over the world. In 2008, the World Health Organization (WHO) estimated the number of new cancer diseases in the world to be increasing up to 7.5 million (21). Among all types of cancer, prostate cancer is the most frequent one among men. In 2008, around 900,000 new cases of prostate cancer were diagnosed, and approximately 260,000 men died as a result over the same period (16).
This study considers the use of naïve Bayesian networks techniques in order to improve the prediction of pathological stage in prostate cancer and breast cancer.
The 2 articles that discussed breast cancer chose a different attribute in predicting with NBNs and finally achieved different results as shown in Table 2.
4. DISCUSSION
This systematic review (23 studies, 53,725 patients) indicates that predicting diseases based on NBNs had the best performance in most diseases and the prediction model depends on the attributes defined for NBNs. Finally, the results show that in some cases NBNs work better than other methods that are reported in the last columns in Table 2. However, its use can improve physicians’ decision in disease diagnosis.
The results of studies were categorized in 6 groups as shown in Table 2. This study follows clear search methods as mentioned before.
Our findings show that 12 of the 15 (80%) articles that reported the accuracy had an accuracy higher than 75% and it showed that NBN has a good performance. 6 of the 11 articles (54.5%) has the AUC higher than 80%. Although previous studies explained that using machine learning for predicting diseases specially NBNs can be useful and can help physicians for a better diagnosis (38).
Additionally, our review shows that it helps to use NBNs as a predictor especially for diseases categorized in Table 1.
Prediction models are increasingly important in clinical practice (39, 40), as indicated by the number of new publications which is described their development. One of their purposes is to aid clinical decision-making by merging patient characteristics in order to calculate the probability of a certain disorder or problem (diagnosis and prognosis) (41, 42). Our findings recommend that using NBNs can improve disease prediction better than other methods.
5. CONCLUSION
A lot of machine learning strategies are currently used in predicting diseases. This systematic review has shown that one type called NBNs as the simplest one can be useful for predicting diseases and actually in some way can be better than other methods and this model can help health practitioners to make decisions more confidently.
Acknowledgements
The authors have contributed to the following: a) the conception and design of the study, or interpretation of data; b) drafting the article critically for important content; and c) have made final approval of the version to be submitted. Two persons who qualify as authors have been listed in this article.
Footnotes
• Limitations: The articles used in this study as a systematic review were selected exclusively from PubMed based on the search query described. In some cases, we could not access the full texts which is another limitation considered in this study.
• Declaration of conflicting interests: The authors declared no potential contraction of interest with respect to the research, authorship and publication of this systematic review article.
• Funding: The authors received no financial support for the research, authorship and publication of this article.
REFERENCES
- 1.Price GJ, Mccluggage WG, Morrison ML, Mcclean G, Venkatraman L, Diamond J, et al. Computerized diagnostic decision support system for the classification of preinvasive cervical squamous lesions. Human Pathology. 2003 Nov 1;34(11):1193–203. doi: 10.1016/s0046-8177(03)00421-0. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Assuli O, Leshno M. Assessing electronic health record systems in emergency departments: Using a decision analytic Bayesian model. Health informatics journal. 2015 doi: 10.1177/1460458215584203. [DOI] [PubMed] [Google Scholar]
- 3.Nir Friedman DG. Bayesian Network Classifiers. Machine Learning. 1997;29(2-3):131–63. [Google Scholar]
- 4.Sharma RK, Sugumaran V, Kumar H, Amarnath M. A comparative study of naïve Bayes classifier and Bayes net classifier for fault diagnosis of roller bearing using sound signal. International Journal of Decision Support Systems. 2015 Jan 1;1(1):115–29. [Google Scholar]
- 5.Wang KJ, Makond B, Wang KM. Modeling and predicting the occurrence of brain metastasis from lung cancer by Bayesian network: a case study of Taiwan. Comput Biol Med. 2014 Apr; 47:147–60. doi: 10.1016/j.compbiomed.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Wolfson J, Bandyopadhyay S, Elidrisi M, Vazquez-Benitez G, Vock DM, Musgrove D, et al. A Naive Bayes machine learning approach to risk prediction using censored, time-to-event data. Statist Med. 2015 Sep 20;34(21):2941–57. doi: 10.1002/sim.6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthias Benndorf EK. Development of an online, publicly accessible naive Bayesian decision support tool for mammographic mass lesions based on the American College of Radiology (ACR) BI-RADS lexicon. European Radiology. 2015;25(6) doi: 10.1007/s00330-014-3570-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florkowski CM. Sensitivity, Specifity, Reciever-Operating Characteristic (ROC) Curves and Likelihood Ratios: Communicating the Performance of Diagnostic Tests. Clin Biochem Rev. 2008 Aug;29(Suppl 1):S83–7. [PMC free article] [PubMed] [Google Scholar]
- 9.Klement W, Wilk S, Michalowski W, Farion KJ, Osmond MH, Verter V. Predicting the need for CT imaging in children with minor head injury using an ensemble of Naive Bayes classifiers. Artif Intell Med. 2012 Mar;54(3):163–70. doi: 10.1016/j.artmed.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Wei W, Visweswaran S, Cooper GF. The application of naive Bayes model averaging to predict Alzheimer’s disease from genome-wide data. J Am Med Inform Assoc. 2011 Aug;18(4):370–5. doi: 10.1136/amiajnl-2011-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazmierska J, Malicki J. Application of the Naïve Bayesian Classifier to optimize treatment decisions. Radiother Oncol. 2008 Feb;86(2):211–6. doi: 10.1016/j.radonc.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 12.Smits M, Dippel Nederkoorn PJ, Dekker HM, Vos PE, Kool DR, et al. Minor head injury: CT-based strategies for management - a cost-effectiveness analysis. Radiology. 2010;254:532–40. doi: 10.1148/radiol.2541081672. [DOI] [PubMed] [Google Scholar]
- 13.Brener DJ, Hall EJ. Computed tomography - an increasing source of radiation exposure. N Engl J Med. 2007;357:2277–84. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 14.Bairstow PJ, Mendelson R, Nguyen L. Reducing inappropriate diagnostic practice through education and decision support. Int J Qual Health Care. 2010;22:194–200. doi: 10.1093/intqhc/mzq016. [DOI] [PubMed] [Google Scholar]
- 15.Broder JS. CT utilization: the emergency department perspective. Pediatr Radiol. 2008;38(Suppl. 4):664–9. doi: 10.1007/s00247-008-0892-z. [DOI] [PubMed] [Google Scholar]
- 16.Lee SM, Park JH, Park HJ. Implications of systematic review for breast cancer prediction. Cancer Nurs. 2008 Oct;31(5):E40–6. doi: 10.1097/01.NCC.0000305765.34851.e9. [DOI] [PubMed] [Google Scholar]
- 17.Chun J, Schnabel F, Ogunyemi O. Assessing a Bayesian risk prediction model in a high-risk breast cancer population. AMIA Annu Symp Proc. 2007:913. [PubMed] [Google Scholar]
- 18.Langarizadeh M, Mahmud R, Ramli AR, Napis S, Beikzadeh MR, Rahman WEZWA. Improvement of digital mammogram images using histogram equalization, histogram stretching and median filter. J Med Eng Technol. 2011 Feb;35(2):103–8. doi: 10.3109/03091902.2010.542271. [DOI] [PubMed] [Google Scholar]
- 19.Langarizadeh M, Mahmud R, Ramli AR, Napis S, Beikzadeh MR, Rahman WWA. Effects of enhancement methods on diagnostic quality of digital mammogram images. Iranian Journal of Cancer Prevention. 2012;3(1):36–41. [Google Scholar]
- 20.Regnier-Coudert O, McCall J, Lothian R, Lam T, McClinton S, N'dow J. Machine learning for improved pathological staging of prostate cancer: a performance comparison on a range of classifiers. Artif Intell Med. 2012 May;55(1):25–35. doi: 10.1016/j.artmed.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 21.International Agency for Research on Cancer-GLOBOCAN. 2008. [accessed: 21.09.10]. http://globocan.iarc.fr/
- 22.Sacchi L, Tucker A, Counsell S, Garway-Heath D, Swift S. Improving predictive models of glaucoma severity by incorporating quality indicators. Artif Intell Med. 2014 Feb;60(2):103–12. doi: 10.1016/j.artmed.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 23.Leung RKK, Wang Y, Ma RCW, Luk AOY, Lam V, Ng M, et al. Using a multi-staged strategy based on machine learning and mathematical modeling to predict genotype-phenotype risk patterns in diabetic kidney disease: a prospective case-control cohort analysis. BMC Nephrol. 2013;14:162. doi: 10.1186/1471-2369-14-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verplancke T, Van Looy S, Steurbaut K, Benoit D, De Turck F, De Moor G, et al. A novel time series analysis approach for prediction of dialysis in critically ill patients using echo-state networks. BMC Med Inform Decis Mak. 2010;10:4. doi: 10.1186/1472-6947-10-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakai S, Kobayashi K, Nakamura J, Toyabe S, Akazawa K. Accuracy in the diagnostic prediction of acute appendicitis based on the Bayesian network model. Methods Inf Med. 2007;46(6):723–6. [PubMed] [Google Scholar]
- 26.Farion KJ, Wilk S, Michalowski W, O'Sullivan D, Sayyad-Shirabad J. Comparing predictions made by a prediction model, clinical score, and physicians: pediatric asthma exacerbations in the emergency department. Appl Clin Inform. 2013;4(3):376–91. doi: 10.4338/ACI-2013-04-RA-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chattopadhyay S, Davis RM, Menezes DD, Singh G, Acharya RU, Tamura T. Application of Bayesian classifier for the diagnosis of dental pain. J Med Syst. 2012 Jun;36(3):1425–39. doi: 10.1007/s10916-010-9604-y. [DOI] [PubMed] [Google Scholar]
- 28.Beretta L, Santaniello A, Cappiello F, Chawla NV, Vonk MC, Carreira PE, et al. Development of a five-year mortality model in systemic sclerosis patients by different analytical approaches. Clin Exp Rheumatol. 2010 Apr;28(2 Suppl 58):S18–27. [PubMed] [Google Scholar]
- 29.Peng SY, Wu KC, Wang JJ, Chuang JH, Peng SK, Lai YH. Predicting postoperative nausea and vomiting with the application of an artificial neural network. Br J Anaesth. 2007 Jan;98(1):60–5. doi: 10.1093/bja/ael282. [DOI] [PubMed] [Google Scholar]
- 30.Kumar Y, Sahoo G. Prediction of different types of liver diseases using rule based classification model. Technol Health Care. 2013;21(5):417–32. doi: 10.3233/THC-130742. [DOI] [PubMed] [Google Scholar]
- 31.Elazmeh W, Matwin W, O'Sullivan D, Michalowski W, Farion W. Insights from predicting pediatric asthma exacerbations from retrospective clinical data. Evaluation Methods for Machine Learning II–Papers from 2007 AAAI Workshop. 2007 [Google Scholar]
- 32.Lu P, Chen J, Zhao H, Gao Y, Luo L, Zuo X, et al. In silico syndrome prediction for coronary artery disease in traditional chinese medicine. Evid Based Complement Alternat Med. 2012;2012:142584. doi: 10.1155/2012/142584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finkelstein J, Wood J. Predicting asthma exacerbations using artificial intelligence. Studies in health technology and informatics. 2013;190:56–8. [PubMed] [Google Scholar]
- 34.Morales DA, Vives-Gilabert Y, Gómez-Ansón B, Bengoetxea E, Larrañaga P, Bielza C, et al. Predicting dementia development in Parkinson’s disease using Bayesian network classifiers. Psychiatry Research: NeuroImaging. 2013;213(2):92–8. doi: 10.1016/j.pscychresns.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 35.Huang GM, Huang KY, Lee TY, Weng JTY. An interpretable rule-based diagnostic classification of diabetic nephropathy among type 2 diabetes patients. BMC bioinformatics. 2015;16(1):1. doi: 10.1186/1471-2105-16-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Q, Li G, Leong T-Y. Proceedings of the 12th World Congress on Health (Medical) Informatics; Building Sustainable Health Systems. IOS Press; 2007. Predicting coronary artery disease with medical profile and gene polymorphisms data MEDINFO 2007. [PubMed] [Google Scholar]
- 37.Ferreira D, Oliveira A, Freitas A. Applying data mining techniques to improve diagnosis in neonatal jaundice. BMC medical informatics and decision making. 2012;12(1):143. doi: 10.1186/1472-6947-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samy GN, Ahmad R, Ismail Z. Security threats categories in healthcare information systems. Health Informatics Journal. 2010 Sep 1;16(3):201–9. doi: 10.1177/1460458210377468. [DOI] [PubMed] [Google Scholar]
- 39.Seixas FL, Zadrozny B, Laks J, Conci A, Saade DCM. A Bayesian network decision model for supporting the diagnosis of dementia, Alzheimer’s disease and mild cognitive impairment. Computers in biology and medicine. 2014;51:140–58. doi: 10.1016/j.compbiomed.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 40.Bellazzi R, Zupan B. Predictive data mining in clinical medicine: current issues and guidelines. International journal of medical informatics. 2008;77(2):81–97. doi: 10.1016/j.ijmedinf.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Barbini P, Barbini E, Furini S, Cevenini G. A straightforward approach to designing a scoring system for predicting length-of-stay of cardiac surgery patients. BMC Med. Inf. Decis. Making. 2014 Oct 13;14(1):89. doi: 10.1186/1472-6947-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JH, Podchiyska T, Altman RB. OrderRex: Clinical order decision support and outcome predictions by data-mining electronic medical records. Journal of the American Medical Informatics Association. 2015:ocv091. doi: 10.1093/jamia/ocv091. [DOI] [PMC free article] [PubMed] [Google Scholar]


