Abstract
Sporothrix globosa is a thermo-dimorphic fungus belonging to a pathogenic clade that also includes Sporothrix schenckii, which causes human and animal sporotrichosis. Here, we present the first genome assemblies of two S. globosa strains providing data for future comparative genomic studies in pathogenic Sporothrix species.
Keywords: Sporothrix globosa, Sporothrix schenckii, Sporotrichosis, Next generation sequencing, de novo assembly
Introduction
Sporotrichosis is a fungal (sub)cutaneous infection of humans and felines that tends to occur in the form of epidemics, sometimes with thousands of individuals. Most cases are reported from East Asia, South America and South Africa (Rodrigues et al. 2014a; Zhang et al. 2015). The disease is caused by a group of dimorphic species of the genus Sporothrix, formerly united in a single taxon, Sporothrix schenckii, and composing a so-called “pathogenic clade” that includes three additional pathogens: Sporothrix brasiliensis, Sporothrix luriei, and Sporothrix globosa (Romeo and Criseo 2013; Zhang et al. 2015). On the other hand, the genus also contains occasional opportunists, such as Sporothrix pallida and Sporothrix mexicana, which have a potential to cause occasional infection in susceptible hosts (Dias et al. 2011; Morrison et al. 2013). Nevertheless, these two species showed a low degree of virulence and pathogenicity if compared with the members of the pathogenic clade and are, usually, better considered as environmental saprophytes (Arrillaga-Moncrieff et al. 2009; Romeo et al. 2011; Rangel-Gamboa et al. 2016).
Among the members of the S. schenckii group, S. brasiliensis was reported as the most pathogenic species followed by S. schenckii, S. luriei and S. globosa (Arrillaga-Moncrieff et al. 2009; Fernández-Silva et al. 2012). In addition, considerable differences have also been reported for other aspects of their biology including epidemiology, antifungal resistance and genetics (Marimon et al. 2008; Teixeira et al. 2014; Zhang et al. 2015; Rangel-Gamboa et al. 2016). Sporothrix schenckii has a worldwide distribution and its population shows high degrees of genetic variability. In contrast, S. brasiliensis and S. globosa appear to consist of genetically homogeneous populations (Zhang et al. 2015; Rangel-Gamboa et al. 2016). S. brasiliensis is restricted to Brazil, whereas most of the S. globosa clinical isolates originate from China where the incidence of this fungus is exceptionally high. The latter species also shows decreased susceptibility to azoles that are commonly used in the treatment of cutaneous sporotrichosis (Marimon et al. 2008). Conversely, clinical isolates of S. brasiliensis appear to be highly susceptible to most of the antifungals tested in vitro (Marimon et al. 2008; Rodrigues et al. 2014b). Moreover, sporotrichosis by S. globosa mostly presents as a fixed cutaneous type, whereas S. schenckii and S. brasiliensis prevalently cause the classical lymphocutaneous form of disease.
Recently the genomes of the most virulent species S. schenckii and S. brasiliensis and the avirulent environmental species S. pallida (D’Alessandro et al. 2016) have been sequenced (Cuomo et al. 2014; Teixeira et al. 2014). In this article, we make available two S. globosa genomes, with the aim to facilitate future studies for understanding the evolution of antifungal drug resistance and virulence among Sporothrix species.
Materials and Methods
Fungal Strains and DNA Extraction
The genome of the type strain S. globosa CBS 120340 from a facial lesion in Spain and that of a clinical strain (named SS01), isolated from a Chinese patient with sporotrichosis who failed itraconazole treatment, were entirely sequenced in this study. The latter strain showed a minimum inhibitory concentration for itraconazole of 8 µg/ml for the mycelial phase and 2 µg/ml for the yeast form.
Genomic DNA was extracted using the Gentra Puregene Yeast/Bact Kit (Qiagen, China) following the manufacturer’s instructions.
Genome Sequencing and Assembly
The whole genomes of the two S. globosa strains were sequenced using the Illumina Hiseq 2000 technology.
A total of three different insert size libraries (200 bp, 500 bp, and 6 kb) were constructed for each strain and paired-end sequenced. After sequencing, 1,943 and 2,012 Mb data were produced for strains CBS 120340 and SS01, respectively. Raw reads were processed using FASTX-toolkit (version 0.0.14; http://hannonlab.cshl.edu/fastx_toolkit) to remove adapters, duplicated reads and sequences with low Phred-scores (cutoff: ≥ 20).
Before de novo assembly, k-mer frequency analysis (k = 15) was performed using jellyfish (http://www.cbcb.umd.edu/software/jellyfish) to estimate the size of the genome including the level of heterozygosity and duplication.
The S. globosa genomes were assembled using SOAPdenovo version 2.04 (http://soap.genomics.org.cn). The assembled scaffolds generated by the two strains, including those from other Sporothrix species (GenBank accession numbers: AXCR00000000.1, AWEQ00000000.1, AWTV00000000.1 and JNEX00000000.2), were aligned and oriented with the software MAUVE (http://darlinglab.org/mauve/mauve.html).
Ab-initio Gene Prediction, Analysis of Repetitive DNA Sequences and Transposons
Open reading frames were predicted and annotated using a combination of the two programs AUGUSTUS (Stanke and Morgenstern 2005) and PLAN (He et al. 2007) whereas the tRNA genes were predicted by tRNAscan-SE (Schattner et al. 2005).
DNA repetitive sequences were detected using Tandem Repeat Finder algorithm (version 4.04; http://tandem.bu.edu/trf/trf.html).
Transposable elements (TEs) were predicted using RepeatMasker, RepeatModeler (www.repeatmasker.org) and TransposonPSI (http://transposonpsi.sourceforge.net) and later confirmed manually with BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Mitochondrial Genomes and Phylogenetic Analysis
S. globosa mitochondrial genomes were extracted and identified from the sequencing data of each strain using the GRABb program (Brankovics et al. 2016). A phylogenomic tree, based on a multiple alignment of additional Sporothrix mitogenomes obtained from sequence data deposited in Genbank (AZHD01000000.1; AXCR00000000.1; AWEQ00000000.1; AWTV00000000.1; JNEX00000000.2), was constructed using MEGA 6.0 software (Tamura et al. 2013).
Results
Characteristics of the S. globosa Genome and Gene Content
Genome statistics of the two S. globosa strains examined in this study are shown in table 1
Table 1.
Genome Statistics of the S. globosa CBS 120340 and SS01
| CBS 120340 | SS01 | |
|---|---|---|
| Genome size (bp) | 33,473,551 | 33,493,113 |
| N50 | 5,350,014 | 3,279,759 |
| N90 | 2,936,705 | 1,965,226 |
| GC content (%) | 54.37 | 54.37 |
| Gene number | 7,760 | 7,719 |
| Total length (bp) | 11,553,735 | 11,470,230 |
| Gene average length (bp) | 1,489 | 1,486 |
| Gene length/genome (%) | 34.51 | 34.24 |
| GC content in gene region (%) | 62.40 | 62.44 |
| Intergenic region length (bp) | 21,925,813 | 22,028,824 |
| GC content in intergenic region (%) | 50.14 | 50.17 |
| Intergenic region length/genome (%) | 65.49 | 65.76 |
| Tandem repeat number | 6,836 | 6,868 |
| Total length (bp) | 272,187 | 268,724 |
| Repeat size (bp) | 1-837 | 1-456 |
| Tandem repeat length/genome (%) | 0.8130 | 0.8022 |
| Microsatellite DNA number | 2,267 | 2,218 |
| Total length (bp) | 91,381 | 86,522 |
| Repeat size (bp) | 2-10 | 2-10 |
| Microsatellite DNA length/genome (%) | 0.2729 | 0.2523 |
| tRNA number | 126 | 126 |
The genome of strain CBS 120340 was sequenced at 146× coverage and assembled in 222 contigs which were linked by paired-end reads into 24 scaffolds (larger scaffold: 6,771,991 bp). The estimated genome size was 33,473,551 bp with a G + C content of 54.37%. The number of predicted genes was 7,760 with an estimated total length of 11,553,735 bp which makes up 34.51% of the entire genome. A total of 126 putative tRNA genes decoding standard 20 aminoacids were found by tRNAscan-SE (table 1).
For the Chinese SS01 strain, its genome was sequenced at 153× coverage and assembled into 356 contigs organized in 19 scaffolds (larger scaffold: 9,646,222 bp) covering 33,493,113 bp (G + C content: 54.37%). From the genome analysis, we found 7,719 genes (total length 11,470,230 bp; 34.24% of the genome) whereas the number of tRNAs was the same as in CBS 120340 (table 1).
The 15 k-mer frequency distribution followed a typical Poison distribution and showed the absence of an additional peak at ½ of the k-mer depth of the main peak observed (k-mer depth = 31 for CBS 120,340 and 32 for SS01) suggesting that both sequenced fungal genomes do not exhibit high levels of heterozygosity.
DNA Repetitive Sequences and In Silico Analysis of Transposable Elements
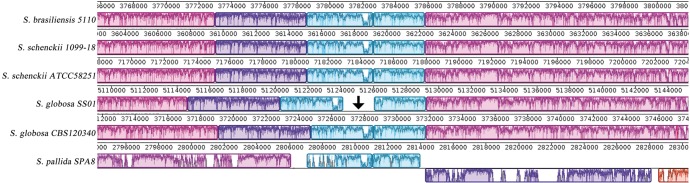
Approximately 0.8% of both S. globosa genomes were represented by repetitive DNA sequences. We found 6,836 and 6,868 tandem repeats in the CBS 120340 and SS01 assemblies, respectively. Of these, over 30% were represented by DNA microsatellites whose global length made over 0.25% of each fungal genome (table 1). In these genomes, differences in transposon content were seen (table 2). In silico analysis showed that CBS 120340 contained 116 TE while the SS01 genome included 86 mobile elements (table 2). One TE element (DDE superfamily; GenBank: ABG26270) is shown in the MAUVE alignment in figure 1 and is located in an intergenic region of the S. globosa SS01 genome (scaffold_4; contig_220). This element is not present in the corresponding DNA region of the CBS 120340 genome (fig. 1). Following a BLAST search in both S. globosa genomes, we found a single copy of this specific TE in SS01 and five copies in CBS 120340, which is in agreement with the observed TE expansion in this strain (table 2).
Table 2.
Mobile Genetic Elements Content in the Two Sequenced S. globosa Genomes
| TEs | Genome copy number |
|
|---|---|---|
| CBS 120340 | SS01 | |
| LTR-Copia-Like | 12 | 12 |
| LTR-Gypsy-Like | 14 | 15 |
| hAT | 37 | 33 |
| Tc1-mariner | 33 | 10 |
| LINE-L1 | 8 | 8 |
| MuLE | 10 | 6 |
| Helitron | 2 | 2 |
| Total | 116 | 86 |
Fig. 1.—
Genomic alignment showing synteny and homology among the members of the pathogenic clade and S. pallida. MAUVE alignment shows the DNA region of the S. globosa SS01 genome (arrow) in which a transposon of the DDE superfamily is inserted.
Characteristics of the Mitochondrial Genomes
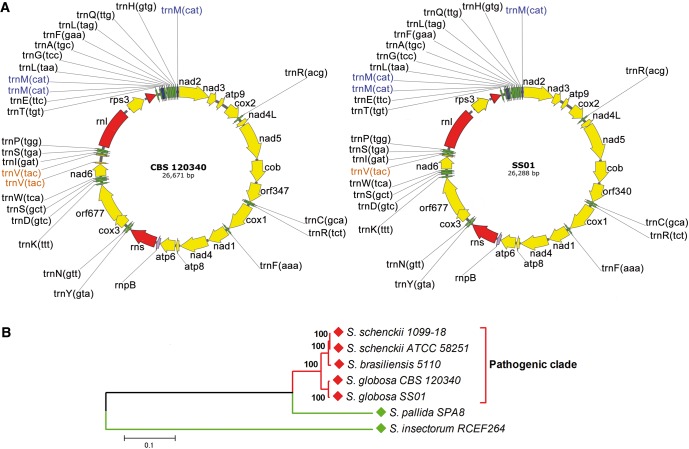
The whole mitochondrial genomes of the two S. globosa strains are shown in figure 2A and consist of a circular DNA molecule ranging from 26,288 (SS01) to 26,671 bp (CBS 120340) in length. They include 44 unique genes subdivided in 24 tRNAs, 17 protein coding genes, 2 rRNAs and the rpnB gene encoding the RNA subunit of the RNaseP, a ribozyme responsible for tRNA maturation. However, three copies of the tRNA gene encoding trnM were found in both genomes while two tRNA trnV loci were observed only in CBS 120340 mtDNA (fig. 2A).
Fig. 2.—
S. globosa mitochondrial genomes and phylogenomic analysis of pathogenic and environmental Sporothrix species. (A) Circular maps of S. globosa mitogenomes generated from sequence data. The names of unique (green) and multiple (trnM, blue; trnV, orange) tRNAs, rRNAs (red) and protein-coding genes (yellow) are shown on the outside circle. (B) Phylogenomic tree constructed using whole mitochondrial genomes extracted from sequenced pathogenic (red) and environmental (green) Sporothrix species deposited in Genbank. Evolutionary analysis were conducted in MEGA6 (Tamura et al. 2013) using the Neighbor-Joining method. Bootstrap values are indicated at the nodes.
The two S. globosa mitochondrial genomes share the same gene order and are similar in size to that of S. schenckii ATCC MYA-4821 (26,572 bp) and about 9 Kbp smaller than the genome of S. brasiliensis ATCC MYA-4823 (36,054 bp) and S. pallida CBS 141373 (formerly SPA8) (35,403 bp) (Teixeira et al. 2014; D’Alessandro et al. 2016).
A phylogenomic tree (fig. 2B) placed S. globosa along with S. schenckii and S. brasiliensis in the “pathogenic clade”, with a topology similar to that expected when using nuclear protein-encoding loci (Marimon et al. 2007).
Discussion and Conclusions
There is a growing and strong interest in the study of clinically relevant Sporothrix species due to the high number of infections reported each year worldwide.
A substantial body of scientific literature produced in recent years highlighted important phenotypic and genetic differences not only among the different clades with high or reduced pathogenic potential but also among closely related taxa belonging to a specific group (Fernandes et al. 2013; Romeo and Criseo 2013; Sasaki et al. 2014; Zhao et al. 2015). In fact the degree of virulence observed among members of the pathogenic clade can be defined as S. brasiliensis > S. schenckii > S. luriei > S. globosa (Arrillaga-Moncrieff et al. 2009; Fernández-Silva et al. 2012). This model seems to be inversely proportional to the trend of drug resistance observed in these species which places S. globosa as the most resistant and S. brasiliensis as the most susceptible species (Marimon et al. 2008; Rodrigues et al. 2014b).
A detailed understanding of the molecular mechanisms that underlie evolution of virulence and drug-resistance in these fungi is optimally addressed using truly comprehensive information about their genomes. Therefore, the release of the complete genome sequence of S. globosa represents an important milestone for Sporothrix research, because it offers the possibility to compare the genomes of the three major pathogenic species. The differences in clinical and antifungal data are remarkably large for species that are phylogenetically so closely related, where both S. globosa and S. brasiliensis were suggested to be clonal offshoots of the ancestral species, S. schenckii (Rangel-Gamboa et al. 2016). In addition, future comparative genomic studies will enable to understand what makes the members of the “pathogenic clade” so unique among all other Sporothrix species.
Nucleotide Sequence Accession Numbers
The draft whole-genome sequences of strains S. globosa CBS 120340 and SS01 have been deposited at DDBJ/ENA/GenBank under accession numbers LVYW00000000 and LVYX00000000, respectively. The versions described in this article are the first versions LVYW01000000 and LVYX01000000.
Acknowledgments
This work was supported by the National Nature Foundation of China (grant number 81371746); and the Guangdong Natural Science Foundation Committee (grant number 8151008901000187).
Literature Cited
- Arrillaga-Moncrieff I, et al. 2009. Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect. 15:651–655. [DOI] [PubMed] [Google Scholar]
- Brankovics B, et al. 2016. GRAbB: Selective assembly of genomic regions, a new niche for genomic research. PLoS Comput Biol. 12:e1004753.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, et al. 2014. Genome sequence of the pathogenic fungus Sporothrix schenckii (ATCC 58251). Genome Announc. 2:e00446–e00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandro E, et al. 2016. Draft genome sequence of the dimorphic fungus Sporothrix pallida, a nonpathogenic species belonging to Sporothrix, a genus containing agents of human and feline sporotrichosis. Genome Announc. 4:e00184–e00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias NM, Oliveira MM, Santos C, Zancope-Oliveira RM, Lima N. 2011. Sporotrichosis caused by Sporothrix mexicana, Portugal. Emerg Infect Dis. 17:1975–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes GF, et al. 2013. Characterization of virulence profile, protein secretion and immunogenicity of different Sporothrix schenckii sensu stricto isolates compared with S. globosa and S. brasiliensis species. Virulence 4:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Silva F, Capilla J, Mayayo E, Guarro J. 2012. Virulence of Sporothrix luriei in a murine model of disseminated infection. Mycopathologia 173:245–249. [DOI] [PubMed] [Google Scholar]
- He J, Dai X, Zhao X. 2007. PLAN: a web platform for automating high-throughput BLAST searches and for managing and mining results. BMC Bioinformatics 8:53.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Cano J, Gené J, Cano J, Sutton DA, Kawasaki M, Guarro J. 2007. Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol. 45:3198–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marimon R, Serena C, Gené J, Cano J, Guarro J. 2008. In vitro antifungal susceptibilities of five species of Sporothrix. Antimicrob Agents Chemother. 52:732–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AS, Lockhart SR, Bromley JG, Kim JY, Burd EM. 2013. An environmental Sporothrix as a cause of corneal ulcer. Med Mycol Case Rep. 2:88–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel-Gamboa L, Martínez-Hernandez F, Maravilla P, Arenas-Guzmán R, Flisser A. 2016. Update of phylogenetic and genetic diversity of Sporothrix schenckii sensu lato. Med Mycol. 54:248–255. [DOI] [PubMed] [Google Scholar]
- Rodrigues AM, de Hoog GS, Zhang Y, de Camargo ZP. 2014a. Emerging sporotrichosis is driven by clonal and recombinant Sporothrix species. Emerg Microbes Infec. 3:e32.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues AM, et al. 2014b. Genetic diversity and antifungal susceptibility profiles in causative agents of sporotrichosis. BMC Infect Dis. 14:219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo O, Criseo G. 2013. What lies beyond genetic diversity in Sporothrix schenckii species complex? New insights into virulence profiles, immunogenicity and protein secretion in S. schenckii sensu stricto isolates. Virulence 4:203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo O, Scordino F, Criseo G. 2011. New insight into molecular phylogeny and epidemiology of Sporothrix schenckii species complex based on calmodulin-encoding gene analysis of Italian isolates. Mycopathologia 172:179–186. [DOI] [PubMed] [Google Scholar]
- Sasaki AA, et al. 2014. Chromosomal polymorphism in the Sporothrix schenckii complex. PLoS One 9:e86819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. 2005. Augustus: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33:465–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, et al. 2014. Comparative genomics of the major fungal agents of human and animal Sporotrichosis: Sporothrix schenckii and Sporothrix brasiliensis. BMC Genomics 15:943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, et al. 2015. Phylogeography and evolutionary patterns in Sporothrix spanning more than 14,000 human and animal case reports. Persoonia 35:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao MD, Zhou X, Liu TT, Yang ZB. 2015. Morphological and physiological comparison of taxa comprising the Sporothrix schenckii complex. J Zhejiang Univ Sci B. 16:940–994. [DOI] [PMC free article] [PubMed] [Google Scholar]