Abstract
Genomic variation in Indian populations is of great interest due to the diversity of ancestral components, social stratification, endogamy and complex admixture patterns. With an expanding population of 1.2 billion, India is also a treasure trove to catalogue innocuous as well as clinically relevant rare mutations. Recent studies have revealed four dominant ancestries in populations from mainland India: Ancestral North-Indian (ANI), Ancestral South-Indian (ASI), Ancestral Tibeto–Burman (ATB) and Ancestral Austro-Asiatic (AAA). The 1000 Genomes Project (KGP) Phase-3 data include about 500 genomes from five linguistically defined Indian-Subcontinent (IS) populations (Punjabi, Gujrati, Bengali, Telugu and Tamil) some of whom are recent migrants to USA or UK. Comparative analyses show that despite the distinct geographic origins of the KGP-IS populations, the ANI component is predominantly represented in this dataset. Previous studies demonstrated population substructure in the HapMap Gujrati population, and we found evidence for additional substructure in the Punjabi and Telugu populations. These substructured populations have characteristic/significant differences in heterozygosity and inbreeding coefficients. Moreover, we demonstrate that the substructure is better explained by factors like differences in proportion of ancestral components, and endogamy driven social structure rather than invoking a novel ancestral component to explain it. Therefore, using language and/or geography as a proxy for an ethnic unit is inadequate for many of the IS populations. This highlights the necessity for more nuanced sampling strategies or corrective statistical approaches, particularly for biomedical and population genetics research in India.
Keywords: Indian genomic diversity, 1000 Genomes Project, population structure, ancestry, social stratification
Introduction
India, occupying the centre-stage of Palaeolithic and Neolithic migrations, has been under-represented in genome-wide studies of variation (Cann 2001). Being at the cross-roads of migration, Indian populations have undergone complex and ancient admixture events over a long period (Bamshad et al. 2001; Zerjal et al. 2007; Reich et al. 2009; Moorjani et al. 2013; Basu et al. 2016), and have been the melting-pot of disparate ancestries originating from different parts of Eurasia and South-East Asia (Basu et al. 2003, 2016; Sengupta et al. 2006; Abdulla et al. 2009). Genetic evidence suggests that some Indian populations might be amongst the earliest people to leave Africa via the southern exit route (Quintana-Murci et al. 1999; Mellars 2006). Social customs and hierarchies have also led to a complex diversity of largely endogamous populations which tolerate some degree of porosity (Reich et al. 2009; Moorjani et al. 2013; Basu et al. 2016). Demographically, post-agriculture, India has experienced huge recent population expansion. As a consequence, India harbors a huge amount of genomic diversity exceeding the genetic diversity of the whole of Europe (Majumder 1998; Reich et al. 2009). Reich et al. (2009) concluded that most of India is an admixture of two ancestries (the Ancestral North Indian (ANI) and the Ancestral South Indian (ASI) component) (Moorjani et al. 2013). A more recent study exploring Indian genomic diversity showed four major ancestral genetic components in mainland India that included the ANI, ASI as well as the Ancestral Tibeto-Burman (ATB) and Ancestral Austro-Asiatic (AAA) components (Basu et al. 2016).
Although there have been many population genetic studies based on genotyping array data (Xing et al. 2009; Reich et al. 2009; Narang et al. 2011; Moorjani et al. 2013; Basu et al. 2016), there is a scarcity of publicly available whole genome sequence data from the Indian subcontinent. The Phase 3 1000 Genomes Project (KGP) data provides a great resource for studying Indian genomic variation based on whole genome sequence (WGS) data, as it includes five populations from the Indian subcontinent (Auton et al. 2015). These include two populations originating from the North-Western Indian subcontinent (GIH and PJL), two populations from the Southern Indian subcontinent (ITU and STU) and one population from Eastern Indian subcontinent (BEB). It is important to note that three of the five IS populations (GIH, STU and ITU) were sampled from the Houston (USA) and UK. Previous studies have indicated that estimating genetic variation by studying Indian diaspora populations might lead to gross underestimation of the existing genetic diversity. For example, a study based on the genomic analysis of Gujrati Indians from Houston (Rosenberg et al. 2006), a population similar to that representing India in HapMap and KGP, has argued in favor of low divergence in Indian populations. This observation was contradictory to studies based on Indian populations sampled from the Indian-subcontinent (Abdulla et al. 2009; Reich et al. 2009), and therefore diaspora based studies need more critical evaluation. Though a recently published WGS based study on Indian populations includes more populations compared with KGP-IS, and has provided interesting insights into the history and peopling of the Indian subcontinent, the small sample sizes (<10 for most mainland Indian populations) per population restricts the possible use of such data for many population genetic analyses (Mondal et al. 2016). The KGP being the first publicly available large-scale whole genome sequence dataset to represent the Indian subcontinent, and a database aimed to be a reference cataloguing human variation for clinical studies, this dataset is expected to be used widely as a reference Indian population in various human genetic studies as well as advanced understanding of disease biology. Although the KGP populations have a wide ethno-linguistic spread, there is a need to investigate the effect of sampling multiple diaspora populations to assess the genetic diversity of India.
In contrast to largely homogenous non-IS populations in KGP or HapMap, the long-standing social hierarchy and endogamy in IS populations have resulted in scenarios when linguistic groups from a geographic region might include more than one sub-population instead of a single homogenous population. To address this complexity, most population genomic studies on Indian populations have used the caste/tribe information in addition to language and geography to define an ethnolinguistic group (Bamshad et al. 2001; Basu et al. 2003; Brahmachari et al. 2005; Reich et al. 2009; Moorjani et al. 2013). Therefore, it was also necessary to investigate whether the classification of populations based only on language and geography, as used in the KGP, is sufficient to define ethnolinguistic units for the IS populations.
The five KGP-IS populations include the HapMap GIH population sample which was shown to have a curious population structure (Reich et al. 2009; Ali et al. 2014; Juyal et al. 2014). Though some authors have suggested that the substructure may be due to novel ancestral components in one of the subgroups, the probable source of the novel ancestral component has not been fully explored. Using WGS data, we confirmed the substructure in the GIH and report significant stratification in two other KGP-IS populations (the Punjabi and Telugu). Moreover, using recent SNP array data from multiple IS populations (Basu et al. 2016), we provide possible explanations for these observations.
Materials and Methods
Datasets
Our analysis utilized the KGP Phase 3 dataset that consists of low-coverage whole genome sequence data for 2,504 individuals representing 26 populations (Auton et al. 2015). PLINK was used for data format conversion and subsequent downstream analyses (Purcell et al. 2007). Of the 26 populations, we focused on the five IS populations (GIH—Gujarati Indian sampled from Houston, Texas; PJL—Punjabi sampled from Lahore, Pakistan; BEB—Bengali sampled from Bangladesh; ITU—Indian Telugu sampled from UK; STU—Sri Lankan Tamil sampled from UK). In addition, one population each from Europe and East Asia viz. CEU (Utah Residents with Northern and Western European Ancestry) and CHB (Han Chinese in Beijing, China) were included. For comparative analyses, we included genotype data for eight populations from HGDP datasets (Burusho, Kalash, Balochi, Hazara, Makrani, Pathan, Sindhi and Brahui) and 20 different IS populations from the Basu et al. study (Cavalli-Sforza 2005; Basu et al. 2016). The details of the IS populations used in this study are listed in supplementary table S1, Supplementary Material online, and their geographic origin is shown in supplementary figure S1, Supplementary Material online. Five caste-stratified populations from Andhra Pradesh (Moorjani et al. 2013) were also included to investigate the role of social hierarchy in the KGP ITU population.
Assessing Population Structure and Admixture
Principal component analysis (PCA) was performed on the KGP-IS population dataset (∼81 million variants, ∼500 individuals) using the “pca” option in PLINK (Chang et al. 2015) and the results were visualized using GENESIS (Buchmann and Hazelhurst 2014). The KGP-IS dataset was pruned using the “indep pairwise” option in PLINK for the removal of variants in high (r2> 0.1) linkage disequilibrium (LD), resulting in a dataset of about 1 million variants. Similarly, another dataset was generated using a MAF cut-off (MAF > 0.01) on the KGP-IS dataset, resulting in ∼7 million variants. Both of these datasets were used for PC analysis.
Genetic ancestry for the samples in KGP-IS populations was estimated using the unsupervised clustering algorithm as implemented in ADMIXTURE (Alexander et al. 2009). Two LD pruned datasets were used for this analysis, the first one included the five KGP-IS populations only, whereas the second included two additional global populations CEU and CHB (both datasets had about 1 million variants). The patterns of population structure were explored by varying the number of ancestral clusters (k= 3 through 6). For each analysis, 50 iterations for each value of k were performed and summarized using CLUMPP (Jakobsson and Rosenberg 2007). The results were also visualized using GENESIS. Similar PCA and ADMIXTURE based analyses were performed on merged KGP-IS and HGDP datasets (∼0.5 million variants) as well as KGP-IS and Basu dataset (∼0.7 million variants).
Fst
Weir and Cockerham’s FST statistic (Weir and Cockerham 1984) was calculated to estimate the genetic differentiation across all populations (GIH, PJL, BEB, STU, ITU, CEU and CHB) using PLINK (Chang et al. 2015). For comparison, we performed FST calculations both with and without LD pruning.
The PCA and ADMIXTURE analysis showed evidence of substructure within three of the five Indian populations (GIH, PJL and ITU), so FST was also calculated within the subgroups of individuals in these three populations. We then randomly partitioned the individuals from GIH, PJL and ITU into two subgroups with the same number of individuals as observed in the subgroups detected using PCA and ADMIXTURE. The FST values within the randomly sampled set were then calculated. The process was repeated 1,000 times and an empirical P value was assigned to the observed FST scores for the subgroups in each population.
Inbreeding Coefficient
The inbreeding coefficient for all the individuals in GIH, PJL and ITU was calculated using the “ibc” option in PLINK (Chang et al. 2015). This analysis calculates two different estimates of inbreeding coefficient: Fhat1 (usual variance-standardized relationship minus 1) and Fhat2 (approximately equal to the –het estimate) (Chang et al. 2015). The distribution of the Fhat1 and Fhat2 values were compared between the subgroups of GIH, PJL and ITU and the significance of the difference between the groups was evaluated using the bootstrap method.
Results
Representation of Indian Genomic Diversity in KGP
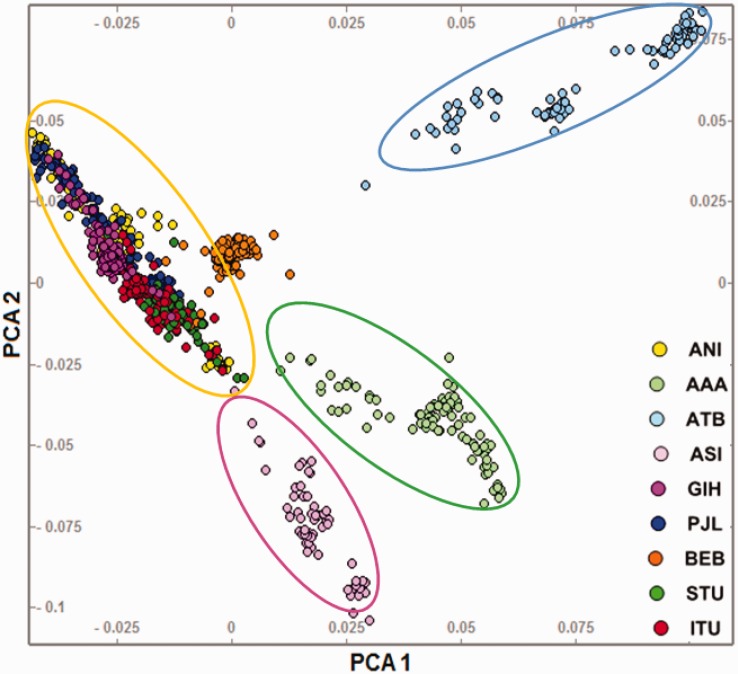
To investigate the extent to which the KGP-IS populations are able to capture the Indian genomic diversity, we merged the 5 KGP-IS populations with 18 mainland IS populations from the Basu et al. study (Basu et al. 2016), and performed a PCA on the merged dataset (fig. 1). The two Andaman and Nicobar island populations (ONG and JRW) were excluded as there is evidence of a long separation from mainland populations with negligible gene flow. The PCA of the merged dataset revealed four ancestral components (similar to the Basu et al. 2016 study), each component forming a distinct cluster and cline (fig. 1). As expected from geographic origin, the north-west (PJL and GIH) and east (BEB) KGP-IS populations clustered with the ANI populations which include the BRG, KSH, and WBR from Basu study (Basu et al. 2016). Interestingly, both the KGP southern IS populations (ITU and STU) were also observed to segregate with the ANI cluster (localizing near the IYR and PLN populations of South India) and distant from the South Indian tribes (KDR, IRL and PNY) which correspond to the ASI cluster (fig. 1). Therefore, despite the geographic spread of the KGP-IS populations, three of the main Indian ancestral components (ASI, ATB and AAA) are not adequately represented in this dataset.
Fig. 1.—
PCA of the 5 KGP-IS populations and 18 mainland Indian populations from the Basu study (Basu et al. 2016). Four distinct ancestral component clusters were observed. For clarity, the populations from the Basu study have been grouped and displayed according to ancestry component (see supplementary table S1, Supplementary Material online). The 5 KGP-IS populations cluster with the ANI populations from the Basu dataset.
Population Structure within KGP–IS Populations
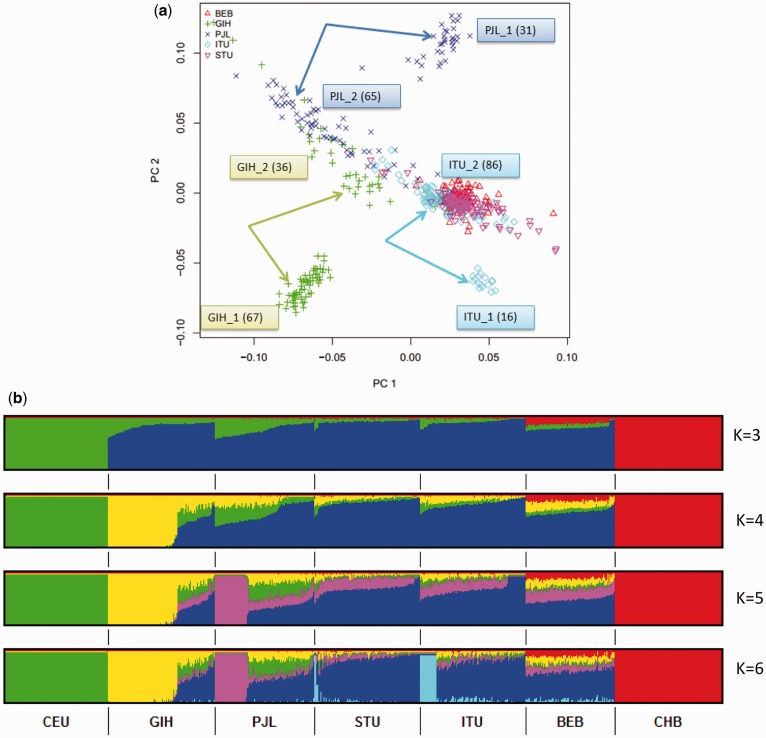
We performed a PCA on the five KGP-IS populations to identify possible population structure. Two IS populations (PJL and ITU) in addition to the GIH (who are known to show substructure; Reich et al. 2009; Ali et al. 2014; Juyal et al. 2014) showed unambiguous bipartite clustering (fig. 2a). PC analysis might be influenced by a number of factors including the total number of SNPs, allele frequency cut-offs and the presence of linkage disequilibrium (LD). To verify the robustness of the observed population stratification, we performed additional PCA studies, using LD pruned, MAF pruned and randomly downsized SNP sets (supplementary fig. S2a and b, Supplementary Material online). The results from all analyses showed the presence of two subclusters with no change in the cluster membership for each of the three populations, suggesting the observed structures to be inherent to the data and not an artifact of the choice of the SNP set or parameters used for PCA. Since the PCA strongly suggested the presence of stratification in the GIH, PJL and ITU populations, we named these subgroups as GIH_1 (67 individuals), GIH_2 (36 individuals), PJL_1 (31 individuals), PJL_2 (65 individuals), ITU_1 (16 individuals) and ITU_2 (86 individuals). The suffix “_1” represents the subgroup that formed an independent cluster away from the main north-south cline in the PCA whereas the suffix “_2” represents the subgroup that clustered along the main cline.
Fig. 2.—
Population structure analysis of the five KGP-IS populations shows evidence of substructure for three IS populations. (a) PCA Plot showing the first two principal components for the five KGP-IS populations. Three of the five KGP-IS populations showed bipartite clustering. (b) Admixture cluster analysis combining the five KGP Phase 3 Indian populations and an European (CEU) and an East Asian (CHB) population. As “k” increases from 4 to 6, the distinct population stratification as shown by GIH, PJL and ITU becomes evident.
Using unsupervised clustering, as implemented in ADMIXTURE (Alexander et al. 2009) we investigated these subgroups further and estimated the ancestry of each individual from the five IS as well as CEU and CHB populations from KGP. The results of ADMIXTURE, visualized using GENESIS (Buchmann and Hazelhurst 2014), are shown in figure 2b. At k= 3, the clusters corresponded to the three main global genetic components (East Asian (red), European (green) and the Indian subcontinent (blue) component). All five IS populations showed varying proportions of “European like” contributions, whereas the BEB, as expected, showed the presence of a significant “East-Asian like” genetic component. These observations are in concordance with previous studies which have shown north Indian populations to harbor higher “European like” ancestry and it decreases in a cline in populations living more southward (Moorjani et al. 2013). The population subgroups for GIH, PJL and ITU separated out as the value of k was increased from 4 to 6, respectively (fig. 2b). We repeated the ADMIXTURE analysis using only the five IS populations, and detected the same pattern of clustering but at lower k values (supplementary fig. S3, Supplementary Material online).
To study the genetic distance between the sub-groups, we estimated the pairwise Weir and Cockerham’s FST statistic (Weir and Cockerham 1984) between the five KGP-IS populations and the subgroups of the three populations (table 1). The observed FST values correspond to geographic distances between the populations. For example, the GIH (west IS) and BEB (east IS) showed higher divergence (mean FST 0.004) compared with STU and ITU (both from southern region of the IS, mean FST 0.001). These values, as expected, were 10-fold lower compared with values observed from intercontinental population comparisons (mean FST range 0.028–0.069). The subgroup pairs of GIH, PJL and ITU showed a higher genetic differentiation between members of the pair than was observed among the five IS populations (table 1). For example, PJL_1 and PJL_2 are more differentiated (FST= 0.009) than the most differentiated Indian populations (GIH and BEB; GIH and STU). We employed a bootstrap analysis to estimate the chances of observing such high FST values when individuals are divided into subgroups randomly. For all three populations, the observed FST differences were highly significant (P < 0.001). These analyses support to our earlier observation of population substructure.
Table 1.
Weighted FST Estimates for Genetic Distance between Selected KGP Populations As Well As the Subgroups of the GIH, PJL and ITU Populations
| Population 1 | Population 2 | Weighted Mean FST |
|---|---|---|
| BEB | GIH | 0.0045 |
| BEB | PJL | 0.0038 |
| BEB | ITU | 0.0024 |
| BEB | STU | 0.0023 |
| GIH | PJL | 0.0036 |
| GIH | ITU | 0.0039 |
| GIH | STU | 0.0044 |
| PJL | ITU | 0.0033 |
| PJL | STU | 0.0037 |
| ITU | STU | 0.0012 |
| GIH_1 | GIH_2 | 0.0059 |
| PJL_1 | PJL_2 | 0.0086 |
| ITU_1 | ITU_2 | 0.0137 |
| BEB | CEU | 0.039 |
| PJL | CEU | 0.0282 |
| ITU | CEU | 0.0395 |
| BEB | CHB | 0.0549 |
| PJL | CHB | 0.0692 |
| ITU | CHB | 0.0673 |
The analysis of SNP-wise FST values for the subgroups in the three populations shows a significant number of SNPs to have high allele frequency divergence within the subgroups. For example, the number of SNPs with Fst Values >0.15 within GIH and PJL subgroups is 8,360 and 33,000, respectively. Though differences in the sample sizes could have potentially introduced errors and inflated the difference (especially for the ITU) a significant portion of these differences can be expected to be real. Interestingly, some of these highly differentiated SNPs are known to be associated to diseases/traits (as inferred from GWAS catalogue; Welter et al. 2014) such as type 2 diabetes, IgG glycosylation, acute lymphoblastic leukaemia (childhood), inflammatory bowel disease, response to cytidine analogues (gemcitabine) among others. However, further in-depth analysis will be required to decipher the phenotypic relevance of the observed allele frequency differences.
Ancestry Differences as a Probable Source of the Observed Population Substructure
To investigate potential novel genetic components acquired from geographically proximal populations, we analysed the five KGP-IS populations together with eight neighboring Pakistani populations from HGDP (Burusho, Kalash, Balochi, Hazara, Makrani, Pathan, Sindhi, and Brahui). ADMIXTURE analysis (supplementary fig. S4, Supplementary Material online) did not show any “novel” genetic components to be shared between the Pakistani populations and the two KGP North-West IS populations (GIH and PJL).
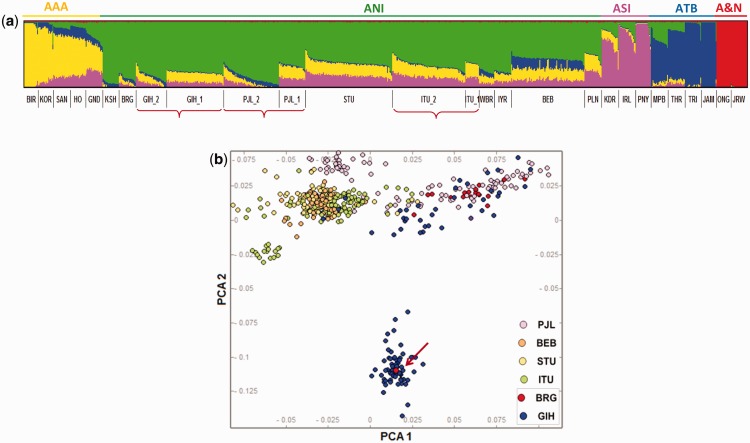
Since neither the HGDP nor KGP dataset included populations with predominant ASI or AAA components, we analysed the five KGP-IS populations with the 20 populations from Basu dataset and performed admixture analysis using the merged dataset (fig. 3a). For k= 5, the five genetic components corresponding to the ANI, ASI, AAA, ATB and Andaman archipelago (A&N) were observed. Interestingly, whereas the admixture plot based only on the KGP-IS populations showed distinct clusters for the different sub-groups (supplementary fig. S3, Supplementary Material online), the admixture plot as shown in figure 3a (includes both KGP as well as 20 populations from Basu study) did not show such clear differentiation between the subgroups at similar values of k. However, the subgroups were distinguishable in terms of the distribution of various ancestry proportions. This can be explained by the inclusion of the ancestral components like ASI and AAA from the Basu study (Basu et al. 2016) that were not represented in KGP-IS dataset. To illustrate this, we have generated another set of admixture plots (supplementary fig. S5, Supplementary Material online), in which we have merged the KGP-IS populations and three of the five ancestral component populations from Basu study (ANI, ATB and A&N). The aim of this analysis was to investigate whether the distinguishable clusters reappear at slightly higher values of k in the absence of AAA and ASI. Since KGP-IS populations are predominantly ANI, at k = 3, we observe three prominent clusters corresponding to the three ancestral components (ATB= Red, ANI-Green and A&N-Blue). As we increase the value of k to 6, the subgroups for each of the three populations reappear. In addition, we could note a difference in the proportion of ANI component (yellow at k = 6, as evident from KSH and BGR) with PJL_2 and GIH_2 showing a much higher proportion of ANI than their counterparts.
Fig. 3.—
Differences in the level of ancesty proportions in the three KGP IS population subgroups. (a) Admixture clustering analysis combining five KGP-IS populations and 20 Indian populations from the Basu study (Basu et al. 2016). At k= 5, the colour codes green, yellow, magenta, blue and red to represent the ANI, AAA, ASI, ATB and the Andamanese archepelago ancestries. (b) PCA showing five KGP Phase 3 Indian populations and an upper caste Gujrati Brahmin (BRG) from Basu study (Basu et al. 2016). The figure shows that one of the BRG individuals (shown by the arrow) clusters with the isolated GIH sub-group (GIH_1), whereas the other BRG individuals overlap with other north Indian populations (including the GIH_2 sub-group).
The proportion of inferred ancestral component for each subgroup was estimated using ADMIXTURE (table 2) which clearly showed differences in the proportions of ancestral components for the two north-western IS populations. PJL_1 and GIH_1 (subgroups that cluster outside the main north-south cline in the PCA), both had a higher proportion of AAA and ASI than their corresponding subgroups (GIH_2 and PJL_2). This shows that PJL_1 has almost 3 times more ASI and AAA proportion than PJL_2. Similarly, GIH_1 has a higher AAA proportion that GIH_2. Differences in the level of AAA ancestries between upper and lower caste populations have been shown using mtDNA (Basu et al. 2003) and Y haplogroup data (Thanseem et al. 2006). The upper caste has been shown to demonstrate significantly higher ANI ancestry in comparison to lower castes from the same geographic region suggesting a relationship between the history of caste-formation and admixture among ancestral components (Reich et al. 2009). This suggests that the stratification observed in GIH and PJL might also, in part, be the result of social hierarchy. Interestingly, no such difference was observed between the ITU_1 and ITU_2.
Table 2.
Ancestry Proportion Estimates of Five KGP-IS Populations and 20 Indian Populations from the Basu et al. (2016) Study Populations with Five Ancestral Components (K = 5)
| Populations | ANI | ASI | AAA | ATB | A&N |
|---|---|---|---|---|---|
| GIH_1 | 0.762 | 0.094 | 0.138 | 0.003 | 0.002 |
| GIH_2 | 0.807 | 0.071 | 0.085 | 0.031 | 0.005 |
| PJL_1 | 0.657 | 0.147 | 0.173 | 0.017 | 0.006 |
| PJL_2 | 0.837 | 0.054 | 0.052 | 0.05 | 0.007 |
| ITU_1 | 0.621 | 0.17 | 0.207 | 0.001 | 0.001 |
| ITU_2 | 0.649 | 0.17 | 0.165 | 0.01 | 0.005 |
| STU | 0.61 | 0.193 | 0.178 | 0.012 | 0.007 |
| BEB | 0.557 | 0.123 | 0.172 | 0.14 | 0.008 |
To highlight the possible differences in distribution of the different subgroups further, we generated “zoomed in” versions of the PCA using the merged KGP-IS and Basu datasets (supplementary fig. S6a–c, Supplementary Material online). Supplementary figure S6a, Supplementary Material online, shows the GIH_1 and GIH_2 subgroups along with one representative population (least admixed) from each ancestral group from Basu 2016 study (the ASI is represented by PNY, ATB by JAM, AAA by BIR and the ANI by KSH) along with STU and BEB samples from KGP. It has already been shown that the GIH_1 exhibits more overall admixture, with a stable proportion of AAA and ASI components (table 2). The PC analysis clearly demonstrates that the two GIH subgroups show a difference in distribution across the IS north-south cline (supplementary fig. S6a, Supplementary Material online). It is interesting to note that when the KGP-IS populations (predominately ANI) were studied using PCA (fig. 2a), the GIH_1 subgroup clustered away from the north-south cline, however, when the other three ancestral components are added to the analysis, the GIH_1 subgroup shows a relative shift towards the AAA and ASI component in the north-south cline. We performed a similar analysis for the PJL and ITU subgroups separately. Similar to GIH_1, the PJL_1 subgroup shows shifts towards the AAA/ASI components in the PCA (supplementary fig. S6b, Supplementary Material online). However, the ITU subgroups, once again correlating to the admixture analysis results, do not show any separation along the north-south cline (supplementary fig. S6c, Supplementary Material online). These results suggest that the factors responsible for the observed structures might differ for these three populations.
To investigate whether social hierarchical stratification in Indian populations corresponds to the distinctive clustering in any of the 3 KGP-IS populations, we performed a PCA based analysis of the five KGP-IS populations along with the upper caste Gujrati Brahmin population (BRG) (Basu et al. 2016). Interestingly, we observed that 19 of the 20 individuals from BRG completely align with the GIH_2 subgroup, whereas the remaining individual clustered within the GIH_1 subgroup (fig. 3b). This observation, validates the structure previously observed in the diaspora Gujrati population (KGP and HapMap) using a Gujrati population sampled from India. It also suggests an inherent social complexity that was observed in two independent datasets.
Differences in Relatedness and Heterozygosity Levels
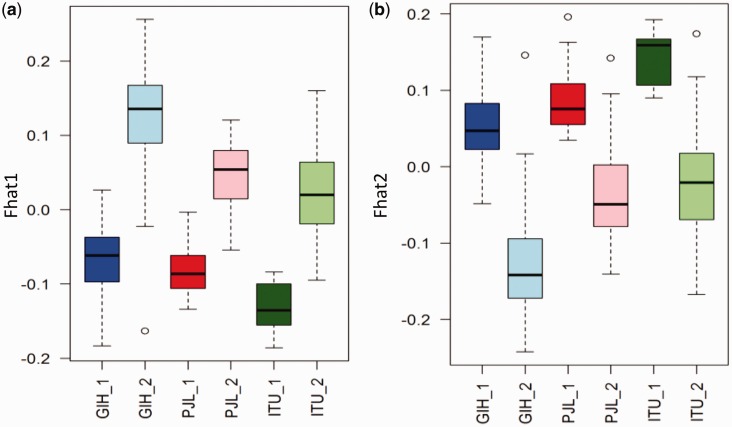
The higher degree of relatedness within a subset of individuals from a population can also result in an observable structure in population genetic analyses. To explore any possible role of relatedness in the observed structures, we computed the inbreeding coefficient (F) parameters that provide an estimate of the level of relatedness between individuals, The Fhat1 values were calculated for all the GIH, PJL and ITU individuals, and their distribution within the subgroups was compared. The distribution of Fhat1 values in GIH individuals is shown in figure 4a and supplementary figure S7a, Supplementary Material online. The individuals from GIH_1 predominately showed negative Fhat1 values while most of the individuals from GIH_2 showed positive Fhat1 values. Since a negative Fhat1 value corresponds to lower relatedness, the results indicate that the individuals in the GIH_1 subgroup are less related to each other in comparison to individuals in the GIH_2 subgroup. The same trend was observed for the PJL and ITU populations (fig. 4a) where the subgroups clustering out of the main north-south cline (PJL_1 and ITU_1) showed lower Fhat1 values than the subgroups closer to the main cline (PJL_2 and ITU2).
Fig. 4.—
Differences in the level of relatedness among the subgroups of three KGP-IS populations (GIH, PJL and ITU). Boxplots represent the distributions of (a) Fhat1 scores and (b) Fhat2 scores.
We also calculated the Fhat2 values, which give an estimate of the heterozygosity of the individual. As expected, we observed a trend opposite to that observed for the Fhat1, since less related individuals are expected to have greater heterozygosity. The Fhat2 values were found to be predominately positive for the subgroups that cluster outside the main cline (GIH_1, PJL_1 and ITU_1) and negative for the subgroups clustering along the main cline (GIH_2, PJL_2 and ITU_2). The comparison of the distribution of the Fhat2 values within the subgroups of the GIH, PJL and ITU populations is summarized in figure 4b (and supplementary fig. S7b, Supplementary Material online) and shows lower heterozygosity for all populations that cluster on the cline.
Bootstrap analyses were performed to assess the significance of the observed differences in the distribution of Fhat1 and Fhat2 values in two sub-groups of the three populations. For each comparison, the differences in the distribution of the statistic were found to be statistically significant (P < 0.001). These results taken together suggest that the subgroups are characterized by inherent differences in population histories (admixture and relatedness) and/or culture (social hierarchy).
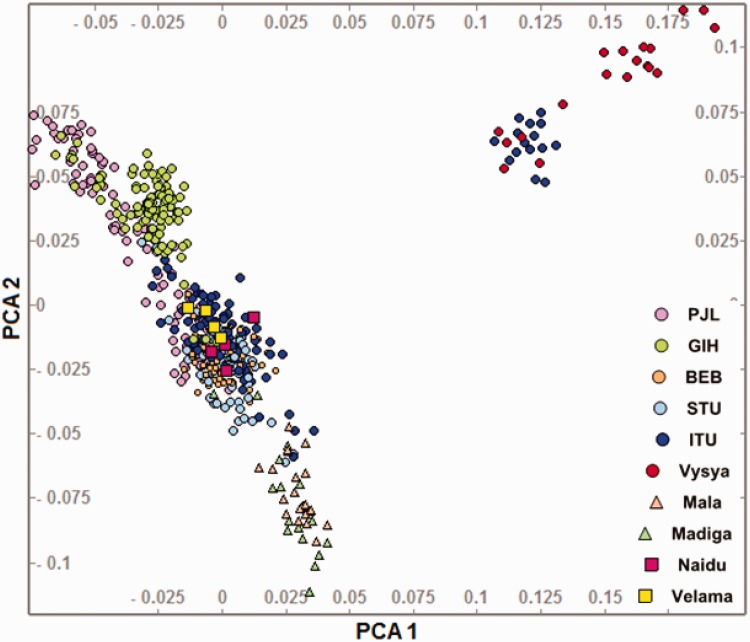
Caste as a Source of the Observed Population Structure
The differences in the proportion of ancestral components as well as differences in the level of relatedness/endogamy indirectly suggested social hierarchy/caste to be a source of the observed substructure in one or more of the three KGP-IS populations. This would also explain the difference in the level of relatedness as a reflection of differences in the level of endogamy known to exist among various caste populations in the IS (Khan et al. 2007; Basu et al. 2016). This possibility was further supported by the observation of complete overlap of one of the two sub-groups of GIH with BRG (the upper caste Gujrati Brahmin community). However, the role of social hierarchy could not be explored further using the Basu study as we required caste-stratified populations from Gujrat, Punjab and Andhra Pradesh (AP), the geographic origins of the three KGP-IS populations (GIH, PJL and ITU, respectively) that showed substructures. A scrutiny of the literature identified five caste stratified AP populations from Moorjani et al. study (Moorjani et al. 2013). Of these five populations, Mala and Madiga are lower caste, Naidu and Velama are upper caste and Vysya is a middle caste population. We merged the five KGP-IS and five AP populations and performed PCA on the merged dataset (fig. 5). As evident from the plot, the two distinct ITU subgroups overlap completely with two different caste groups. While the ITU_2 subgroup (that aligns with the main north south cline in fig. 2a) overlaps completely with the Upper caste population (Naidu and Velama) from AP, the ITU_1 subgroup (the isolated cluster in fig. 2a) clusters with the middle caste population (Vysya) from Reich dataset. It is interesting to note that the Vysya from AP have been shown to be highly endogamous and to have experienced negligible gene flow from neighboring groups in India for an estimated 3,000 years (Reich et al. 2009).
Fig. 5.—
Role of social hierarchy in the population substructure observed in the ITU. PCA showing five KGP Phase 3 Indian populations and five caste stratified populations from Andhra Pradesh from Moorjani study (Moorjani et al. 2013). The two ITU subgroups clearly overlap with two different castes from the same geographic region.
As the ITU_1 and ITU_2 subpopulations were not found to differ significantly in ancestry proportions but only in the level of endogamy, the observations taken together suggest that endogamy alone might be sufficient to cause population substructure in IS populations. While the clusters for the other two populations show similar features and it may be reasonable to speculate that they also result from social hierarchy or caste stratification, it is not possible to assess the role of caste as the basis for the observed subgrouping conclusively because of the unavailability of caste stratified data from these regions.
Discussion
Genetic diversity on the Indian subcontinent is represented by four distinct ancestry components: ANI, ASI, ATB and AAA. In addition, a distinctive ancestry component was observed in isolated populations from the Andaman archipelago. The KGP-IS dataset, consisting of ∼500 WGS is the most comprehensive dataset, both in terms of sample size per population and an unbiased genome-wide view of genetic diversity, available to date for studying Indian population diversity (Auton et al. 2015). Based on comparisons with SNP array data from a wider range of Indian populations from Basu et al. (2016), we have demonstrated that the five populations in the KGP, although aimed at representing the spectrum of Indian genetic diversity by wide geographic sampling, primarily captures genetic diversity from the ANI group, leaving the other three groups largely unrepresented. Therefore, in order to capture Indian genetic diversity, WGS data from the IS populations, especially those representing the other genetic components (ASI, ATB and AAA) is needed.
The population stratification in the HapMap GIH population was first reported by Reich et al. (2009), however, the source for the observed substructure was not fully explored. They proposed that the GIH subgroup that falls outside the main cline of Indian groups, might harbor novel ancestry in addition to ASI and ANI ancestries but had no comparative data to test this hypothesis. They were correct in their speculation, as we have shown that the varied admixture levels of the AAA component (which was not included in their study) are likely to be largely responsible for the substructure of the GIH population.
The overlap between the GIH_2 subgroup and the upper-caste Gujrati population (BRG) suggests that the observed structure in the GIH might be related, in part, to social hierarchy. The upper caste populations of India generally follow a stricter endogamy (Khan et al. 2007; Basu et al. 2016). These factors concur with the observed differences in relatedness and heterozygosity for the two subgroups of the Gujrati population. Social hierarchy could not be explored in the PJL due to lack of comparative data, however, we demonstrate clear variation in AAA ancestry proportions (similar to GIH) which likely contribute to the observed population substructure.
Though a similar difference in ancestry proportion was not observed in the ITU populations, a comparative analysis with caste stratified populations from the same geographic region clearly demonstrates a correlation between the observed structure and caste.
Previous studies have suggested that sampling from diaspora populations might not represent IS populations comprehensively (Abdulla et al. 2009; Reich et al. 2009) and it was possible that diaspora sampling in the KGP dataset could have resulted in population structures in some of these populations. However, the existence of population-structure in one of the in situ populations (PJL sampled in Lahore) and its absence in one of the diaspora populations (STU sampled in UK) suggests the observed structures are probably intrinsic to IS populations (when pooled on the basis of language) and are reflected in the diaspora.
These results emphasize the complexity of population structure in the Indian subcontinent and suggest that it is a combination of factors rather than a single factor that contribute to the observed stratification. Based on our analysis, we have robustly identified at least two factors that contribute to the observed population substructure. The first is the difference in proportions of AAA and ASI ancestry components in the two subgroups, and the second is population history and cultural differences (such as population size and dynamics and extent of assortative mating due to social hierarchical structure).
The KGP is a highly accessed and analysed dataset (the KGP Phase 1 and Phase 3 publications have been cited 1,949 and 116 times (Abecasis et al. 2012; Auton et al. 2015), respectively, as per PubMed accessed in September 2016). Therefore, a careful scrutiny and in-depth understanding of the data is critical in informing various population genetic and bioinformatic analyses based on it. This study shows that only a subset of the Indian genetic diversity is captured in the KGP dataset. Moreover, we demonstrate that population structure, as has previously been reported for the GIH population, is inherent to and shared by several Indian populations. We have proposed two factors: variation in ancestry component and population history/culture that likely contribute to the observed structure. We also wish to highlight that biomedical and computational studies performed using the KGP dataset for these three populations should take the intrinsic population structure into account. From a broader perspective, the results suggest that using language or geography as a proxy for an ethnic unit for some of these populations might be inadequate and a more nuanced sampling or careful corrective statistical measures are advisable.
Supplementary Material
Supplementary figures S1–S7 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
D.S. is a postdoctoral fellow supported by National Research Foundation of South Africa and A.C. is a postdoctoral fellow supported by the National Human Genome Research Institute of the National Institutes of Health under Award Number U54HG006938 (H3Africa Consortium). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or NRF. M.R. is supported by the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa.
Literature Cited
- Abdulla MA, et al. 2009. Mapping human genetic diversity in Asia. Science 326:1541–1545. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, et al. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, et al. 2014. Characterizing the genetic differences between two distinct migrant groups from Indo-European and Dravidian speaking populations in India. BMC Genet. 15:86.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, et al. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, et al. 2001. Genetic evidence on the origins of Indian caste populations. Genome Res. 11:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, et al. 2003. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 13:2277–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Sarkar-Roy N, Majumder PP. 2016. Genomic reconstruction of the history of extant populations of India reveals five distinct ancestral components and a complex structure. Proc Natl Acad Sci. 113:201513197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachari SK, et al. 2005. The Indian Genome Variation database (IGVdb): a project overview. Hum Genet. 118:1–11. [DOI] [PubMed] [Google Scholar]
- Buchmann R, Hazelhurst S. 2014. Genesis manual. Johannesburg: University of the Witwatersrand. Available from: http://www.bioinf.wits.ac.za/software/genesis/Genesis.pdf.
- Cann RL. 2001. Genetic clues to dispersal in human populations: retracing the past from the present. Science 291:1742–1748. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. 2005. The Human Genome Diversity Project: past, present and future. Nat Rev Genet. 6:333–340. [DOI] [PubMed] [Google Scholar]
- Chang CC, et al. 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4:7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. [DOI] [PubMed] [Google Scholar]
- Juyal G, et al. 2014. Population and genomic lessons from genetic analysis of two Indian populations. Hum Genet. 133:1273–1287. [DOI] [PubMed] [Google Scholar]
- Khan F, et al. 2007. Genetic affinities between endogamous and inbreeding populations of Uttar Pradesh. BMC Genet. 8:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PP. 1998. People of India: biological diversity and affinities. Evol Anthropol Issues News Rev. 6:100–110. [Google Scholar]
- Mellars P. 2006. Going east: new genetic and archaeological perspectives on the modern human colonization of Eurasia. Science 313:796–800. [DOI] [PubMed] [Google Scholar]
- Mondal M, et al. 2016. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat. Genet. 48:1066–1070. [DOI] [PubMed] [Google Scholar]
- Moorjani P, et al. 2013. Genetic evidence for recent population mixture in India. Am J Hum Genet. 93:422–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang A, et al. 2011. Recent admixture in an Indian population of African ancestry. Am J Hum Genet. 89:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, et al. 1999. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 23:437–441. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. 2009. Reconstructing Indian population history. Nature 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, et al. 2006. Low levels of genetic divergence across geographically and linguistically diverse populations from India. PLoS Genet. 2:e215.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, et al. 2006. Polarity and temporality of high-resolution Y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 78:202–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I, et al. 2006. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet. 7:42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population structure. Evolution (N. Y.) 38:1358–1370. [DOI] [PubMed] [Google Scholar]
- Welter D, et al. 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, et al. 2009. Fine-scaled human genetic structure revealed by SNP microarrays fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 19:815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerjal T, et al. 2007. Y-chromosomal insights into the genetic impact of the caste system in India. Hum Genet. 121:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.