Abstract
Great genetic variability among teleost immunomes, with gene losses and expansions of central adaptive and innate components, has been discovered through genome sequencing over the last few years. Here, we demonstrate that the innate Myxovirus resistance gene (Mx) is lost from the ancestor of Gadiformes and the closely related Stylephorus chordatus, thus predating the loss of Major Histocompatibility Complex class II (MHCII) in Gadiformes. Although the functional implication of Mx loss is still unknown, we demonstrate that this loss is one of several ancient events appearing in successive order throughout the evolution of teleost immunity. In particular, we find that the loss of Toll-like receptor 5 predates the loss of Mx involving the entire Paracanthopterygii lineage. Using a time-calibrated phylogeny, we show that loss of MHCII and Mx overlap with major paleoclimatic and geological events indicating that these genetic changes were adaptive responses to the changing environment at the time.
Keywords: teleosts, innate immunity, adaptive immunity, Myxovirus resistance (Mx), gene loss, adaptation
Background
Comprehensive characterization of immune gene repertoires has, over the last decade, provided the scientific community with new discoveries that have challenged our perception of the evolution of vertebrate immunity. The detection of variable lymphocyte receptors in jawless vertebrates, functional analogs to immunoglobulins in jawed vertebrates, reveals the presence of several adaptive immune strategies in vertebrates. Lack of Major Histocompatibility Complex (MHC) class II in Atlantic cod (Gadus morhua) and possibly in pipefish (Syngnathus typhle) further indicate that classic adaptive immunity is more flexible than initially believed. Moreover, the discovery of different repertoires of central innate immunity genes reflects great plasticity in the vertebrate innate immune system (Pancer et al. 2005; Han et al. 2008; Star et al. 2011; Boehm et al. 2012; Haase et al. 2013; Buonocore and Gerdol 2016). Recently, Malmstrøm et al. (2016) demonstrated that the loss of central adaptive immunity components found in Atlantic cod (Star et al. 2011) is a common immunological trait in the Gadiformes lineage. Through genome sequencing and draft assembly of 66 novel teleost genomes, they showed that the MHCII pathway was lost approximately 105 Ma (million years ago) in the common ancestor of Gadiformes. This was followed by an independent event resulting in the expansion of MHCI. Moreover, in Atlantic cod, additional gene losses and expansions within the central innate gene family of Toll-like receptors (TLRs) have been reported (Star et al. 2011). This TLR repertoire has been found to be extreme compared to other teleosts (Solbakken et al. 2016). In this study, we take advantage of the genome resources and phylogeny generated by Malmstrøm et al. (2016) to further elucidate the evolutionary origin of the immunological strategy common to Gadiformes and to infer our findings in a broader paleontological perspective.
Results and Discussion
An Ancient Loss of Mx
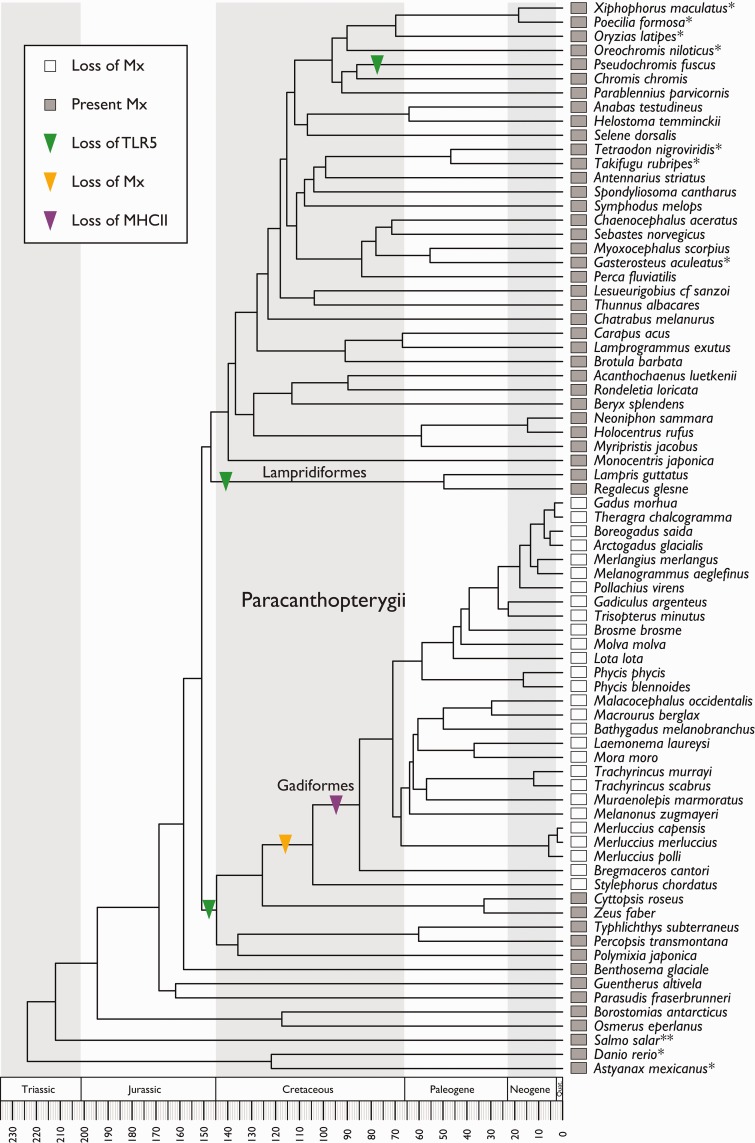
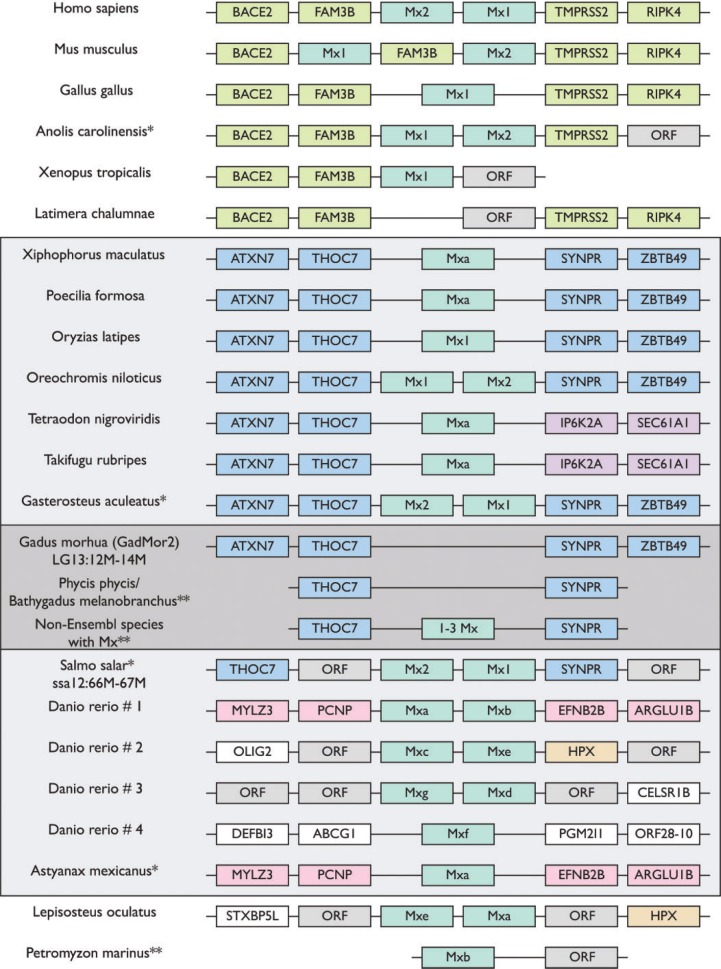
Here we show that the innate Myxovirus resistance (Mx) gene is lost from the Gadiformes and S. chordatus, and this predates the loss of MHCII (fig. 1). Further, we find that the gene copy number of Mx in teleost, which harbor it, lies between 1 and 3 with the exception of 7 in Danio rerio (supplementary table S1, Supplementary Material online). Mx was identified in 38 of the 66 species sequenced by Malmstrøm et al. (2016). Of these 38, it was possible to obtain partial local gene synteny for 15 species, all of which share the same Mx-containing genomic region (supplementary table S1, Supplementary Material online). This partial synteny was then compared to the Mx genomic regions in the fish reference genomes available from Ensembl as well as a selected number of vertebrates (fig. 2) (Cunningham et al. 2015). All teleosts investigated, with the exception of D. rerio and Astyanax mexicanus, share local gene synteny. In D. rerio we find seven copies of Mx that are distributed among four clusters in the genome (fig. 2) where one of them shares synteny with the Mx region in A. mexicanus. Moreover, we find that Lepisosteus oculatus shares synteny with another of the identified Mx regions in D. rerio. As the teleost outgroup L. oculatus share an Mx containing region with D. rerio (HPX/STXBP5L) one could speculate that it is the most likely ancestral organization. However, we found no other partial syntenies within the teleost lineage for other genes than THOC7, SYNPR and IP6K2A (supplementary table S6, Supplementary Material online). Petromyzon marinus’ single Mx is located on a short scaffold without any similarity to the other species investigated. The Mx regions of Homo sapiens, Mus musculus, Gallus gallus, Anolis carolinensis, and Xenopus tropicalis share synteny. However, these Mx regions are dissimilar to the Mx regions found in the investigated teleosts (fig. 2). Finally, we found no Mx in Latimeria chalumnae (fig. 2). The synteny patterns demonstrated are likely related to the vertebrate genome duplications where different Mx genomic regions have been preserved while superfluous genetic material has been discarded throughout evolution (Glasauer and Neuhauss 2014).
Fig. 1.—
Phylogenetic distribution of Mx genes in 76 teleost species. Mx is mapped onto a teleost phylogeny generated by Malmstrøm et al. (2016). The presence of Mx is marked by gray boxes. The loss of Mx is marked by an orange arrow. The losses of MHCII and TLR5 are marked by purple and green arrows, respectively. The absence of Mx is a characteristic of the Gadiformes and S. chordatus and thus predates the loss of MHCII from the Gadiformes. The absence of TLR5 affects the entire Paracanthopterygii superorder together with the Lampridiformes and P. fuscus. The loss of Mx occurs between 126–104 Ma, the loss of MHCII 105–85 Ma, and the loss of TLR5 151–147 Ma.
Fig. 2.—
Local gene synteny analysis of Mx regions in all investigated teleost species in addition to representatives from mammals, birds, reptiles, amphibians, coelacanths above, and non-teleost bony fish (Lepisosteus oculatus) and jawless vertebrates (Petromyzon marinus) below. The dark gray box represents the species derived from Malmstrøm et al. and Atlantic cod, and the light gray box encompasses all teleost species investigated. The synteny is presented with up to two flanking genes both up-stream and down-stream of the Mx region. Due to the fragmented nature of the novel teleost genomes only one flanking gene up- and down-stream of the Mx region is presented (see supplementary table S1, Supplementary Material online, for details). Colors are only for visualization. ORF: open reading frame representing reported gene models in the Ensembl genomes without gene name annotation. *This region has been reversed for presentation purposes. **Only novel teleost species, where local gene synteny was possible, are represented in this syntenic presentation. Also see supplementary tables S4–S6, Supplementary Material online.
Additionally, we examined the presence/absence of another immune gene, TLR5 recently reported to be lost from the Atlantic cod genome (Star et al. 2011; Solbakken et al. 2016). Local gene synteny analyses demonstrated that the TLR5 region appears to be more conserved across vertebrate lineages, i.e., containing a larger set of homologous flanking genes compared to Mx. Furthermore, we find that TLR5 is lost from the entire Paracanthopterygii and Lampridiformes lineages as well as in Pseudochromis fuscus, and thus predates the loss of Mx (fig. 3). Using the time-calibrated phylogeny made by Malmstrøm et al., we were able to date the loss of TLR5 to 151–147 Ma (fig. 1).
Fig. 3.—
Local gene synteny analysis of TLR5 regions in all investigated teleost species in addition to representatives from mammals, birds, reptiles, amphibians, coelacanths above, and non-teleost bony fish (Lepisosteus oculatus) below. The dark gray box represents the species derived from Malmstrøm et al. and Atlantic cod, and the light gray box encompasses all teleost species investigated. The synteny is presented with up to two flanking genes both up-stream and down-stream of the TLR5 region. Due to the fragmented nature of the novel teleost genomes only one flanking gene up- and down-stream of the TLR5 region is presented (see supplementary table 2, Supplementary Material online, for details). Colors are only for visualization. ORF: open reading frame representing reported gene models in the Ensembl genomes without gene name annotation. *This region has been reversed for presentation purposes. **Only novel teleost species, where local gene synteny was possible, are represented in this syntenic presentation. Also see supplementary tables S4–S6, Supplementary Material online.
The Role of Mx in Teleost Immunity
Although the specific function of Mx is still unknown, the diverse nature of its targets and responses between species indicate that Mx is under strong selection and thus is important in vertebrate innate immunity. From studies using mammals, we know that Mx gene products are interferon-inducible dynamin-like large GTPases that block the early steps of virus replication (Haller et al. 2015). Furthermore, Mx shows broad antiviral activity and the gene is usually present in two copies in mammalian species. However, the known diversity of antiviral targets and responses related to Mx does not correspond to the apparent copy number stability (Mitchell et al. 2015 and references therein). Mx has been studied in various fish species such as Atlantic salmon (Salmo salar), Atlantic halibut (Hippoglossus hippoglossus), gilthead seabream (Sparus aurata), and European eel (Anguilla anguilla), and in these species showed similar function to mammalian Mx confirming a diverse range of Mx targets and responses also in fish (Bergan and Robertsen 2004; Das et al. 2009; Fernandez-Trujillo et al. 2013; Huang et al. 2013). In gilthead seabream the three variants of Mx respond to both RNA and DNA viruses from different families in vitro. However, this species’ response towards DNA viruses cannot be replicated in other fish species (Fernandez-Trujillo et al. 2013, and references therein). Strong diversifying selection combined with lineage-specific exchanges between paralogs conserving key enzymatic and structural characteristics, as well as acquiring new antiviral specificities, have been proposed as the underlying mechanisms (Mitchell et al. 2015, and references therein). A single study reports Mx in Atlantic cod using a cross-reactive polyclonal antibody generated against Atlantic salmon Mx (Das et al. 2008). Conversely in this study, we have demonstrated a loss of Mx in Atlantic cod as well as for all the Gadiformes and S. chordatus (fig. 1). Our findings are in accordance with the proposed lineage-specific adaptation of Mx—in this case observed as a loss instead of diversifying selection promoting subfunctionalization (fig. 1) (Fernandez-Trujillo et al. 2013, and references therein). In a recent publication, Braun et al. (2015) reported on the discovery of an evolutionary loss of function of Mx for toothed whales, where it was suggested that pseudogenization of Mx hinders the entry of virus particles into host cells, i.e., protecting the ancestral toothed whale species against harmful virus outbreaks (Braun et al. 2015). Cumulatively, these findings fit the scenario that lineage-specific gene loss events are adaptive responses towards changes in a species’ environment (Olson 1999).
Loss of Mx —A Putative Precursor to the Loss of MHCII
Here, combined with findings reported in the literature (Star et al. 2011; Malmstrom et al. 2016), we find a succession of immune-relevant gene losses throughout the evolution of the teleost immune system: TLR5 151–147 Ma, Mx 126–104 Ma, and MHCII 105–85 Ma. The loss of TLR5 in the late Jurassic is encompassing the Paracanthopterygii superorder together with the Lampridiformes and P. fuscus. The loss of Mx in Gadiformes and S. chordatus appears in the early Cretaceous followed by the loss of MHCII in Gadiformes during the transition from the early to the late Cretaceous. Viewing the successive gene losses in light of changes in paleontological climate, oceanography, and major extinctions we see that the loss of TLR5 is close to the Jurassic–Cretaceous (J–K) boundary. There is accumulating evidence of both species extinctions and radiations coinciding with this transition together with an ongoing debate about average global temperatures in the same period (Bambach 2006; Alroy 2010; Benson et al. 2010; Cavin 2010; Price et al. 2013; Benson and Druckenmiller 2014; Korte et al. 2015). This is further supported by the fact that periods of extinctions are often followed by population diversification and subsequent species radiation enabling the invasion of new habitats (Wellborn and Langerhans 2015; Simoes et al. 2016). Habitat wise, the formation of the central Atlantic Ocean in the early Jurassic continued with a subsequent northward expansion in the Early Cretaceous (Melankholina and Sushchevskaya 2015). Thus, if there were large changes in climate, or possibly an unknown larger extinction event, the loss of TLR5 may be associated with adaptation of new species—possibly towards new habitats within the opening Atlantic Ocean.
Dating of the loss of Mx show that it took place close to the early/late Cretaceous boundary and also overlapping one of the global anoxia events within this period approximately 120 Ma. Coincidently, the loss of MHCII also occurred close to the early/late Cretaceous boundary but spanning a second global anoxia event approximately 95 Ma (Wilson and Norris 2001, Sinninghe Damsté et al. 2010). Additionally, these two anoxia events co-occurred with the continued opening northward of the Central Atlantic Ocean expanding the North Atlantic Ocean and the formation of a gateway between the South Atlantic Ocean and the Central Atlantic Ocean (Granot and Dyment 2015; Melankholina and Sushchevskaya 2015). The metabolically taxing anoxic environments, even though some adaptation likely was possible, resulted in the deep seas being depleted of fish (Rogers 2000; Priede and Froese 2013). This is supported by higher extinction rates in the same period (Takashima et al. 2006; Harnik et al. 2012). The anoxic scenario fits with one of several mechanisms proposed to promote loss of MHCII—metabolic cost (Star and Jentoft 2012). Nevertheless, it could also be coupled to post extinction speciation in which new species invade habitats where maintaining MHCII and Mx, in this case, was less favorable.
Our findings can further be linked to the family richness of bony fish species, diversification and extinction rates through evolutionary history. Bony fish species family richness gradually increased from Jurassic to modern time. However, there is a shift from increasing to decreasing richness with the J–K transition following the TLR5 loss event combined with a small increase in extinction rate (Guinot and Cavin 2015). The loss of Mx and the global anoxia event ∼120 Ma are associated with a small increase in extinction rate but otherwise overall higher and stable species richness levels compared to the J–K transition. The loss of MHCII spanning the second global anoxia event ∼95 Ma coincides with a large drop in species richness combined with an increase in extinction rate and a large increase in species diversification rate. As the losses of TLR5, Mx and MHCII are clearly lineage specific and likely responses towards changes in species’ habitats (Olson 1999) the loss of TLR5 can be seen as an adaptation to events in the J–K transition. These events could then have led to extinctions promoting survival and speciation in the subsequent early Cretaceous which is characterized by an increase in species richness and diversification rates (Guinot and Cavin 2015). The loss of Mx spanning a global anoxia event ∼120 Ma does not overlap with any large changes in species richness, extinction or speciation rates. However, after this event, there is an increase in species richness and speciation rate and thus Mx loss can be viewed as a beneficial adaptation in the anoxic environment (Guinot and Cavin 2015). The loss of MHCII spanning the second global anoxia event ∼95 Ma presents a different pattern than TLR5 and Mx. Here, there is an overlap between the gene loss and large drops in species richness and origination rates (Guinot and Cavin 2015). This indicates that the loss MHCII had more adverse effects than the loss of TLR5 and Mx, however, still over time promoting speciation within the Gadiformes lineage (Malmstrom et al. 2016).
Even though the functional implication of TLR5, Mx, and MHCII loss on the teleost immune system remains unclear our data indicates that the J–K transition harbors events central to shaping the teleost immune system initiated by the loss of TLR5. Further, the loss of Mx directly outside of the Gadiformes lineage indicates that this loss might have been a catalyst for the subsequent loss of MHCII. This combined with the increased metabolic cost to maintain the MHCII system in an anoxic environment likely led to the alternate immune system seen in Gadiformes today.
Materials and Methods
The generation of teleost sequences, assemblies and time-calibrated phylogeny is described in detail in Malmstrøm et al. (2016) and briefly in Supplementary Material.
In the Ensembl reference species, all Mx genes were characterized by extracting genes annotated with corresponding gene name and using the online BLAST tool at Ensembl.org to detect Mx in the remaining species with default parameters. These collectively were used as query Mx protein sequences (Ensembl v.82) (supplementary tables S3 and S4, Supplementary Material online) (Cunningham et al. 2015). The NCBI BLAST tool was used to search the Salmo salar genome (ICSASG_v2, GCA_000233375.4) with default settings using the Mx protein sequences obtained from Ensembl. For TLR5, query sequences were obtained from Ensembl in the same way as Mx (supplementary table S3, Supplementary Material online). All Mx/TLR5 sequences were used as queries in a BLAST+ v. 2.2.26 TBLASTN search against the non-reference teleost assemblies with an E-value cutoff of 1e−10 on our in-house computing servers (Camacho et al. 2009). The novel teleost genome resources are generated from a low-coverage strategy resulting in highly fragmented genomes, however genes are readily detected (Malmstrom et al. 2016). Here, we first targeted the unitigs which are assembled more conservatively than contigs and overall contain more of the raw sequencing data (Myers et al. 2000). In species with no hits for Mx and/or TLR5, we also blasted against the singletons which contain the sequence information that did not get assembled into unitigs (E-value cutoff 1e−1). The reported top targets for Mx were aligned against queries using MEGA5 to eliminate hits from other GTPase genes (especially Dynamin) sharing a similar domain with Mx which often was reported in the BLAST output. Due to large differences in mismatch numbers and other alignment quality metrics this filtering was done manually. The same alignments were used to establish Mx copy number (alignments are available in the GitHub repository) (Tamura et al. 2011). This was not necessary for TLR5. To establish synteny, genes flanking Mx and TLR5 in all Ensembl vertebrate genomes were noted and homolog sequences were extracted from the Ensembl (supplementary table S5, Supplementary Material online). These sequences were used in TBLASTN searches as described above but with options “outfmt 6” and “sseq” and were readily detected in the unitig datasets. Partial synteny was obtained for 15 of 38 non-reference teleosts harboring Mx and for two of 25 species not harboring Mx (not counting Atlantic cod as the new version of the Atlantic cod genome was investigated; Tørresen et al. 2016). The same approach was also applied for TLR5. Furthermore, for TLR5 the leader domain and TIR domain were used as queries alone in addition to the full length TLR5 sequence as these domains often were located to other unitigs than the main part of the query sequence (supplementary tables S3 and S4, Supplementary Material online). Partial synteny was found for Thunnus albacares and Helostoma temminckii (contains TLR5) as well as in Laemonema laureysi (no TLR5) (not counting Atlantic cod as the new version of the Atlantic cod genome was investigated; Tørresen et al. 2016). Finally, for TLR5 we extracted the TLR5 sequences from species neighboring P. fuscus, Lampris guttatus, and Regalecus glesne to ensure that our original query TLR5 sequences did not miss any potential TLR5 genes in these species.
All novel teleost sequence and genome resources are available at European Nucleotide Archive (ENA) and the Dryad digital repository, submitted by the Malmstrøm et al. (2016). All raw data (sequencing reads) are available at ENA with study accession number PRJEB12469 (sample identifiers ERS1199874–ERS1199939). Genome assemblies, available at Dryad, exist in two versions (UTGs and scaffolds) under DOI: doi:10.5061/dryad.326r8. All additional resources needed to generate the findings presented herein are available in our GitHub repository including scripts and BLAST output files: https://github.com/MonicaSolbakken/Mx (last accessed October 20, 2016).
Supplementary Material
Supplementary tables S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgment
This work was supported by The Research Council of Norway (Grant number 222378/F20 to KSJ/SJ). The majority of the genomes used were assembled using the Abel Cluster, owned by the University of Oslo and the Norwegian metacenter for High Performance Computing (NOTUR), and operated by the Department for Research Computing at USIT, the University of Oslo IT-department. http://www.hpc.uio.no/ (last accessed October 20, 2016).
Literature Cited
- Alroy J. 2010. The shifting balance of diversity among major marine animal groups. Science 329(5996):1191–1194. [DOI] [PubMed] [Google Scholar]
- Bambach RK. 2006. Phanerozoic biodiversity mass extinctions. Annu Rev Earth Planet Sci. 34:127–155. [Google Scholar]
- Benson RB, Butler RJ, Lindgren J, Smith AS. 2010. Mesozoic marine tetrapod diversity: mass extinctions and temporal heterogeneity in geological megabiases affecting vertebrates. Proc Biol Sci. 277(1683):829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson RB, Druckenmiller PS. 2014. Faunal turnover of marine tetrapods during the Jurassic-Cretaceous transition. Biol Rev Camb Philos Soc. 89(1):1–23. [DOI] [PubMed] [Google Scholar]
- Bergan V, Robertsen B. 2004. Characterization of Atlantic halibut (Hippoglossus hippoglossus) Mx protein expression. Dev Comp Immunol. 28(10):1037–1047. [DOI] [PubMed] [Google Scholar]
- Boehm T, McCurley N, Sutoh Y, Schorpp M, Kasahara M, Cooper MD. 2012. VLR-based adaptive immunity. Annu Rev Immunol. 30:203–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BA, Marcovitz A, Camp JG, Jia R, Bejerano G. 2015. Mx1 and Mx2 key antiviral proteins are surprisingly lost in toothed whales. Proc Natl Acad Sci U S A. 112(26):8036–8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore F, Gerdol M. 2016. Alternative adaptive immunity strategies: coelacanth, cod and shark immunity. Mol Immunol. 69:157–169. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavin L. 2010. Diversity of Mesozoic semionotiform fishes and the origin of gars (Lepisosteidae). Naturwissenschaften 97(12):1035–1040. [DOI] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, et al. 2015. Ensembl 2015. Nucleic Acids Res. 43(Database issue):D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BK, Ellis AE, Collet B. 2009. Induction and persistence of Mx protein in tissues, blood and plasma of Atlantic salmon parr, Salmo salar, injected with poly I:C. Fish Shellfish Immunol. 26(1):40–48. [DOI] [PubMed] [Google Scholar]
- Das BK, Urquhart K, Ellis AE, Collet B. 2008. Induction of Mx protein in Atlantic cod with poly I:C: immuno-cross reactive studies of antibodies to Atlantic salmon Mx with Atlantic cod. Fish Shellfish Immunol. 25(3):321–324. [DOI] [PubMed] [Google Scholar]
- Fernandez-Trujillo MA, Garcia-Rosado E, Alonso MC, Castro D, Alvarez MC, Bejar J. 2013. Mx1, Mx2 and Mx3 proteins from the gilthead seabream (Sparus aurata) show in vitro antiviral activity against RNA and DNA viruses. Mol Immunol. 56(4):630–636. [DOI] [PubMed] [Google Scholar]
- Glasauer SM, Neuhauss SC. 2014. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 289(6):1045–1060. [DOI] [PubMed] [Google Scholar]
- Granot R, Dyment J. 2015. The Cretaceous opening of the South Atlantic Ocean. Earth Planet Sci Lett. 414:156–163. [Google Scholar]
- Guinot G, Cavin L. 2015. ‘Fish’ (Actinopterygii and Elasmobranchii) diversification patterns through deep time. Biol Rev Camb Philos Soc. 25:2314–2318 [DOI] [PubMed] [Google Scholar]
- Haase D, Roth O, Kalbe M, Schmiedeskamp G, Scharsack JP, Rosenstiel P, Reusch TB. 2013. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: evidence from deep transcriptome sequencing. Biol Lett. 9(2):20130044.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, Kochs G. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol. 23(3):154–163. [DOI] [PubMed] [Google Scholar]
- Han BW, Herrin BR, Cooper MD, Wilson IA. 2008. Antigen recognition by variable lymphocyte receptors. Science 321(5897):1834–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnik PG, Lotze HK, Anderson SC, Finkel ZV, Finnegan S, et al. 2012. Extinctions in ancient and modern seas. Trends Ecol Evol. 27(11):608–617. [DOI] [PubMed] [Google Scholar]
- Huang B, Huang WS, Nie P. 2013. Characterization of four Mx isoforms in the European eel, Anguilla anguilla. Fish Shellfish Immunol. 35(3):1048–1054. [DOI] [PubMed] [Google Scholar]
- Korte C, Hesselbo SP, Ullmann CV, Dietl G, Ruhl M, et al. 2015. Jurassic climate mode governed by ocean gateway. Nat Commun. 6:10015.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom M, Matschiner M, Torresen OK, Star B, Snipen LG, et al. 2016. Evolution of the immune system influences speciation rates in teleost fishes. Nat Genet. 48(10):1204–1210. [DOI] [PubMed] [Google Scholar]
- Melankholina EN, Sushchevskaya NM. 2015. Development of passive volcanic margins of the Central Atlantic and initial opening of ocean. Geotectonics 49(1):75–92. [Google Scholar]
- Mitchell PS, Young JM, Emerman M, Malik HS. 2015. Evolutionary analyses suggest a function of MxB immunity proteins beyond lentivirus restriction. PLoS Pathog. 11(12):e1005304.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, et al. 2000. A whole-genome assembly of Drosophila. Science 287(5461):2196–2204. [DOI] [PubMed] [Google Scholar]
- Olson MV. 1999. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 64(1):18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, et al. 2005. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci U S A. 102(26):9224–9229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Twitchett RJ, Wheeley JR, Buono G. 2013. Isotopic evidence for long term warmth in the Mesozoic. Sci Rep. 3:1438.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priede IG, Froese R. 2013. Colonization of the deep sea by fishes. J Fish Biol. 83(6):1528–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AD. 2000. The role of the oceanic oxygen minima in generating biodiversity in the deep sea. Deep-Sea Research Part II. Top Stud Oceanogr. 47(1–2):119–148. [Google Scholar]
- Simoes M, Breitkreuz L, Alvarado M, Baca S, Cooper JC, Heins L, Herzog K, Lieberman BS. 2016. The evolving theory of evolutionary radiations. Trends Ecol Evol. 31(1):27–34. [DOI] [PubMed] [Google Scholar]
- Sinninghe Damsté JS, van Bentum EC, Reichart GJ, Pross J, Schouten S. 2010. A CO2 decrease-driven cooling and increased latitudinal temperature gradient during the mid-Cretaceous Oceanic Anoxic Event 2. Earth Planet Sci Lett. 293(1–2):97–103. [Google Scholar]
- Solbakken MH, Torresen OK, Nederbragt AJ, Seppola M, Gregers TF, et al. 2016. Evolutionary redesign of the Atlantic cod (Gadus morhua L.) Toll-like receptor repertoire by gene losses and expansions. Sci Rep. 6:25211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Jentoft S. 2012. Why does the immune system of Atlantic cod lack MHC II? Bioessays 34(8):648–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, et al. 2011. The genome sequence of Atlantic cod reveals a unique immune system. Nature 477(7363):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima R, Nishi H, Huber BT, Leckie M. 2006. Greenhouse World and the Mesozoic Ocean. Oceanography 19:82–92. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tørresen OK, Star B, Jentoft S, Reinar WB, Grove H, et al. 2016. An improved genome assembly uncovers a prolific tandem repeat structure in Atlantic cod. bioRxiv http://dx.doi.org/10.1101/060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellborn GA, Langerhans RB. 2015. Ecological opportunity and the adaptive diversification of lineages. Ecol Evol. 5(1):176–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Norris RD. 2001. Warm tropical ocean surface and global anoxia during the mid-Cretaceous period. Nature 412(6845):425–429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.