Abstract
Differences in expression levels are an important source of phenotypic variation within and between populations. MicroRNAs (miRNAs) are key players in post-transcriptional gene regulation that are important for plant development and stress responses. We surveyed expression variation of miRNAs and mRNAs of six accessions from two rice subspecies Oryza sativa L. ssp. indica and Oryza sativa L. ssp. japonica using deep sequencing. While more than half (53.7%) of the mature miRNAs exhibit differential expression between grains and seedlings of rice, only 11.0% show expression differences between subspecies, with an additional 2.2% differentiated for the development-by-subspecies interaction. Expression variation is greater for lowly conserved miRNAs than highly conserved miRNAs, whereas the latter show stronger negative correlation with their targets in expression changes between subspecies. Using a permutation test, we identified 51 miRNA–mRNA pairs that correlate negatively or positively in expression level among cultivated rice. Genes involved in various metabolic processes and stress responses are enriched in the differentially expressed genes between rice indica and japonica subspecies. Our results indicate that stabilizing selection is the major force governing miRNA expression in cultivated rice, albeit positive selection may be responsible for much of the between-subspecies expression divergence.
Keywords: expression variation, microRNA, mRNA, rice
Introduction
MicroRNAs (miRNAs) are short (∼21 nt) non-coding RNAs that regulate target transcripts post-transcriptionally via translational repression or mRNA degradation (Jones-Rhoades et al. 2006; Filipowicz et al. 2008; Bartel 2009). Mature miRNAs are incorporated into RNA-induced silencing complexes (RISCs) and guide RISCs to specific targets through Watson–Crick base-pairing (Jones-Rhoades et al. 2006; Filipowicz et al. 2008; Bartel 2009). A large proportion of miRNAs that were initially discovered in plants regulate transcription factors (Zhang et al. 2006); some of them are highly conserved miRNAs and play important roles in plant development (Carrington and Ambros 2003; Chuck et al. 2009; Carlsbecker et al. 2010), stress adaptation (Lv et al. 2010, Ding et al. 2011), and hormone signaling (Liu et al. 2009). Sequence evolution of miRNAs and their target sites have been extensively explored between closely related plant species (Fahlgren et al. 2010; Ma et al. 2010). For example, rapid sequence evolution of the miR482/2118 gene family has promoted the evolution of resistance genes in the Solanaceae (de Vries et al. 2015). However, little is known about how expression variation in miRNAs may affect variation in gene expression among individuals, which is an important source of phenotypic variation within species (Britten and Davidson 1971; King and Wilson 1975).
Rice is one of the most important crops in the world. The two rice subspecies, Oryza sativa L. ssp. indica (indica) and Oryza sativa L. ssp. japonica (japonica), show significant phenotypic and genetic differentiations (He et al. 2011). To date, 592 precursor miRNAs, representing 334 miRNA families, have been identified in rice (miRBase, v20.0). MiRNAs play a significant role in regulating rice development and stress responses (Campo et al. 2013). For example, regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice (Jiao et al. 2010; Luo et al. 2012) and OsmiR397 substantially enhance grain yield in rice through the repression of OsLAC (Zhang et al. 2013). Moreover, differential expression of miRNAs was reported among japonica cv. Nipponbare and indica cv. 93-11, probably playing roles in heterosis (Chen et al. 2010). However, variation in miRNA expression among rice accessions and its effects on target and global gene expression remain largely unknown.
Here, we surveyed the expression variation in miRNAs and mRNAs among six rice accessions (three from each of indica and japonica subspecies) using RNA-seq. In contrast to extensive differential miRNA expression between developmental stages, only a few miRNAs showed expression differences between indica and japonica subspecies of rice. Highly conserved miRNAs exhibited less expression variation but repressed target gene expression more strongly than lowly conserved miRNAs. Our analysis also identified significantly correlated miRNA–mRNA pairs that differed in expression levels among indica and japonica cultivars, which may be candidates for adaptive regulatory evolution underlying indica–japonica differentiation in cultivated rice.
Results
Small RNA Data Processing and Expression Analysis of Rice miRNAs
Small RNA (sRNA) libraries of rice grains and seedlings were constructed and sequenced for three indica cultivars Khal Dawk Mali 105, Guangluai 4 and Rathuwee, and three japonica cultivars Taipei 309, Heukgyeong and Nipponbare individually. About 9–14 million short reads were obtained for each library, representing an average of 14,849,295 unique reads for indica grain library, 12,978,406 for japonica grain library, 9,924,065 for indica seedling library and 9,395,033 for japonica seedling library (supplementary table S1, Supplementary Material online). About 94% of the sRNAs were 20–24 nucleotides (nts) in length, with 21 and 24 nt as the two major size classes (supplementary fig. S1, Supplementary Material online). In plants, most miRNAs are 21-nt in length while 24-nt sRNAs consist mainly of sRNAs that are associated with repeats and transposable elements (TEs) (Axtell 2013). These results indicate that rice has a highly complex sRNA population to which repetitive sequences are the major contributors (Zhu et al. 2008). Small reads matching rice plastid DNA, structural noncoding RNAs (ncRNAs), and repetitive sequences were removed before miRNA annotation. On average, only a small portion of the total reads were mapped to the chloroplast (6.64%) or mitochondria (1.71%) genome, whereas short reads matching ncRNAs and repetitive sequences accounted for 49.3% and 14% of the total reads, respectively. While the sRNAs belonging to the above categories showed an uniform distribution among all 12 libraries, the proportion of signatures that matched miRNA precursors were consistently higher in seedlings (31.3%) than in grain (3.8%), consistent with a previous report (Xue et al. 2009).
To date, 592 rice miRNAs representing 334 families have been registered in the miRBase database (v20.0). Among them, 22 miRNA families were found conserved in both monocots and eudicots previously (Cuperus et al. 2011). We denoted these miRNA families as highly conserved miRNAs and the others as lowly conserved miRNAs in this study. MiRNAs were considered to be expressed by requiring at least ten raw reads in all three accessions from the same developmental stage and the same subspecies. As a result, a total of 272 mature miRNAs were expressed among the 12 libraries, comprising 111 highly conserved miRNAs and 161 lowly conserved miRNAs (table 1). The expression level of each mature miRNA was measured as Reads Per Million (RPM). A pairwise comparison of log2(RPM) of all miRNAs across the 12 libraries demonstrated the measurement of miRNA expression is highly reproducible between biological replicates in our data (Pearson’s correlation, r = 0.86–0.97; supplementary fig. S2, Supplementary Material online). In addition, we also predicted 20 novel miRNAs using the criteria for plant miRNA annotation (Meyers et al. 2008). Most of these novel miRNAs exhibited developmental stage- or subspecies-specific expression and were predicted to regulate 12 target genes (supplementary table S3, Supplementary Material online).
Table 1.
Summary of Differentially Expressed miRNA Families
| Effect | Highly Conserved (n = 111) | Lowly Conserved (n = 161) | Total (n = 272) |
|---|---|---|---|
| Developmental stage | 64 (57.7%) | 82 (50.9%) | 146 (53.7%) |
| Subspecies | 2 (1.8%) | 28 (17.4%) | 30 (11.0%) |
| Development × Subspecies | 0 (0%) | 6 (3.7%) | 6 (2.2%) |
Note.—MiRNAs with fold change ≥2 and FDR ≤ 0.05 are defined as differentially expressed.
Highly Conserved miRNAs Exhibit More Stable Expression than the Lowly Conserved miRNAs
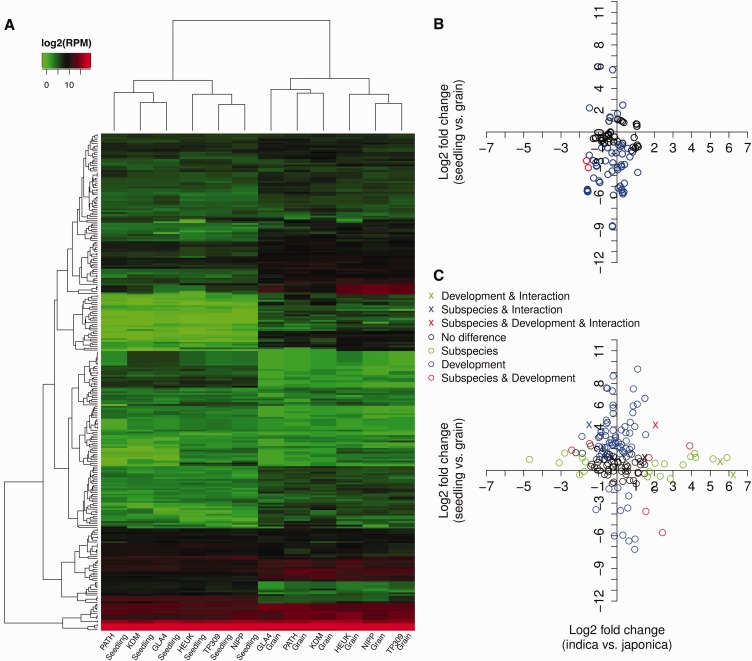
We first explored the miRNA expression variation across the 12 samples by hierarchical clustering (fig. 1A). Dendrogram classification grouped the samples first by subspecies (indica–japonica) and then by developmental stage (grain-seedling), indicating that rice miRNA expression, which is largely remodeled between the different developmental stages, exhibits a relatively small difference between the two subspecies. The mature miRNAs were clustered into several clades (fig. 1A). MiRNAs from the basal clades showed high expression levels with similar expression profiles across rice cultivars from different developmental stages and subspecies; most of them are highly conserved miRNAs, including miR156, miR164, miR166, miR167 and miR168, and two are lowly conserved miR535 and miR5794. In contrast, the majority of miRNAs showed grain- or seedling-biased expression between developmental stages. A number of miRNA families, including miR1861, miR1868, miR319, as well as miR444, exhibited higher expression in grains, whereas other miRNAs, such as members of the miR160, miR169, miR172, and miR529 families, mainly showed increased expression in seedlings. These stage-biased miRNAs are promising candidates for developmental regulation in rice (fig. 1A and supplementary table S2, Supplementary Material online).
Fig. 1.—
Expression variation of known miRNAs in rice. (A) Heat map and unsupervised hierarchical clustering of known miRNA expression. The color key represented the scale of the relative expression levels of the miRNAs (log2 RPM). KDM: indica cv. Khal Dawk Mali 105; GLA4: indica cv. Guangluai 4; PATH: indica cv. Rathuwee; TP309: japonica cv. Taipei 309; HEUK: japonica cv.Heukgyeong; NIPP: japonica cv.Nipponbare. (B, C) Scatter plot of differentially expressed highly conserved (B) and lowly conserved (C) miRNAs. A generalized Poisson-regression linear model was used to identify the differentially expressed miRNAs for the factors of development, subspecies, and development-by-subspecies interaction. MiRNAs with fold change ≥ 2 and FDR ≤ 0.05 are denoted as significantly differentially expressed. Mature miRNAs that show no differential expression (black) or show significant differential expression between subspecies (green), developmental stage (blue) and both (red) are indicated by circles in different colors, while those with differential expression for the additional factor of development-by-subspecies interaction are indicated by crosses with the same color setting.
Using a Log-linear Poisson Regression model (see Methods), 146 (53.7%), 30 (11.0%), and 6 (2.2%) of the 272 expressed mature miRNAs exhibited significantly differential expression for the factors developmental stage, subspecies and development-by-subspecies interaction (fold change ≥ 2, FDR ≤ 0.05), respectively (table 1). These miRNAs were considered to play important roles in regulating rice development or subspecies differentiation. For example, miR159a-f showed a grain-biased expression (supplementary table S2, Supplementary Material online) and is involved in ABA-mediated responses in the hormonal and abiotic stress signaling networks (Reyes and Chua 2007); members of miR156, miR166, miR169, miR171, and miR444 families showed a leaf-biased expression (supplementary table S2, Supplementary Material online) and are known to play defensive roles against stress by targeting transcription factors (Macovei et al. 2012).
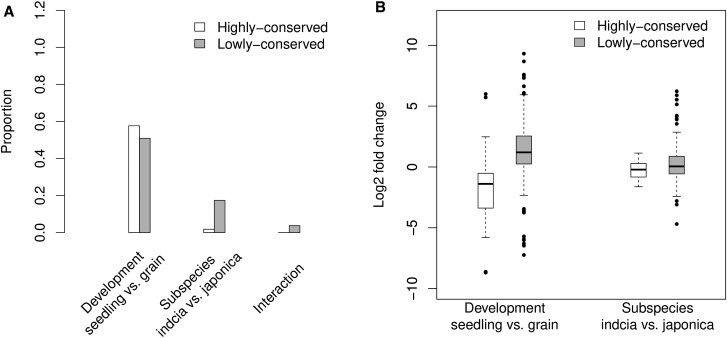
When grouping the mature miRNAs according to their conservation, expression variation was less prominent in highly conserved miRNAs (fig. 1B) than lowly conserved miRNAs (fig. 1C). Only 1.8% (2/111) of highly conserved miRNAs were differentially expressed between subspecies while the proportion was 17.4% (28/161) for lowly conserved miRNAs. An additional 50.9% (82/161) and 3.7% (6/161) of lowly conserved miRNAs were also differentially expressed between developmental stages or development-by-subspecies interaction (table 1 and fig. 2A). Furthermore, magnitudes of the expression fold changes are much higher for lowly conserved miRNAs than highly conserved miRNAs (fig. 1B vs. C and fig. 2B). These results strongly suggest that variation in expression is higher for the less conserved miRNAs than for the more conserved miRNAs found in both monocots and dicots. A possible explanation for this result is that conserved miRNAs are processed better from their foldback structures (Shen et al. 2011). Another explanation could also be that conserved miRNAs are under strong functional constraints (Cuperus et al. 2011). Using qRT-PCR, we validated the differential expression between subspecies for eight out of nine lowly conserved miRNAs in rice seedlings but not for the highly conserved miR166j-5p due to its low expression (supplementary fig. S3A and C).
Fig. 2.—
Expression variation of the highly conserved and lowly conserved miRNAs between subspecies or developmental stages. (A) The proportions of differentially expressed miRNAs in both sets of the highly conserved and lowly conserved miRNAs. A generalized Poisson-regression linear model was used to identify the differentially expressed miRNAs for the factors of development, subspecies, and development-by-subspecies interaction. MiRNAs with fold change ≥ 2 and FDR ≤ 0.05 are denoted as significantly differentially expressed. MiRNAs that show significantly differential expression between subspecies or interactions are enriched in the lowly conserved miRNAs (Fisher's Exact Test, P-value ≪ 0.01). (B) Fold changes in expression of the highly conserved and lowly conserved miRNAs between subspecies or developmental stages. The lowly conserved miRNAs exhibit significantly more variation in expression than the highly conserved miRNAs for both comparisons (Kolmogorov–Smirnov test, P-value ≪ 0.01).
Differential Expression of Transcriptome between Indica and Japonica Cultivars
To understand how miRNA expression may affect mRNA expression, we conducted RNA-seq for each of the seedling samples with 30× coverage (supplementary table S1, Supplementary Material online). We report the results of the whole transcriptome analysis here and analyzed the expression correlation between miRNAs and targets with the transcriptome as a control in the next two sections. All reads were mapped to the japonica cv. Nipponbare genome (Kawahara et al. 2013). Approximately 81% and 86% of the unique reads could be mapped to the reference genome for indica and japonica cultivars, respectively. To allow differential expression analysis between rice subspecies, 32,044 homologous genes were identified between the genomes of japonica cv. Nipponbare and indica cv. 93-11 (see Methods), of which 18,288 were considered to be expressed, i.e., having more than ten raw reads across all three accessions from the same developmental stage and the same subspecies (supplementary table S4, Supplementary Material online). A regression comparison was then performed based on the expression level of these 18,288 expressed homologous genes. Among them, 2,530 genes (13.8%) were differentially expressed (fold-change ≥ 2 and FDR ≤ 5%) between the two rice subspecies, with 1,452 up-regulated in indica and 1,078 in japonica, respectively (supplementary fig. S4, Supplementary Material online). To confirm the results of RNA-seq, we conducted qRT-PCR of ten experimentally validated or predicted miRNA targets and confirmed the differential expression patterns between rice subspecies in all the comparisons (supplementary fig. S3B and C, Supplementary Material online).
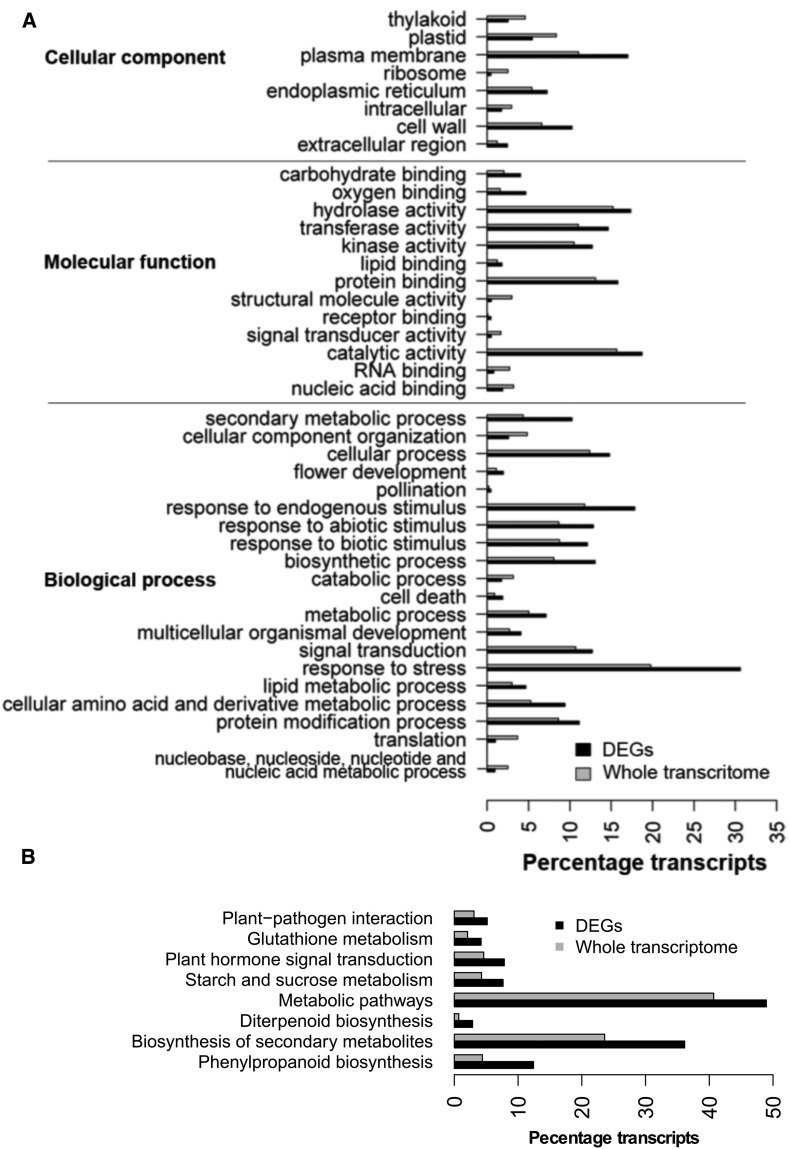
The differentially expressed genes (DEGs) are significantly enriched in Gene Ontology categories, including “plasma membrane,” “cell wall,” “endoplasmic reticulum,” and “extracellular region” for “Cellular Component”, categories of molecular binding (“Hydrocarbon binding”, “Oxyen binding”, “protein binding” etc.) and enzyme activities (“hydrolase activity,” “catalytic activity,” “kinase activity” etc.) for “Molecular function,” and “cellular process,” “flower development,” and many categories of metabolic process (“Secondary metabolic process,” “lipid metabolic process,” etc.) or response to stimuli and stress (“endogenous stimulus,” “abiotic stimulus,” and “biotic stimulus”) for “Biological Process” (fig. 3A). Consistent with the GO analysis, DEGs are significantly enriched in the KEGG pathways related to secondary metabolites, such as “Glutathione metabolism” (supplementary fig. S5) and “starch and sucrose metabolism” (supplementary fig. S6, Supplementary Material online and fig. 3B). The former is known to play important roles in plant stress tolerance (Noctor et al. 1998), while the latter is presumably affecting the varietal difference of soluble sugar in different rice varieties (Yang et al. 2014). The over-representative KEGG pathways in DEGs also included “Plant–pathogen interaction” (supplementary fig. S7, Supplementary Material online) and “Plant hormone signal transduction” (supplementary fig. S8, Supplementary Material online and fig. 3B).
Fig. 3.—
Gene ontology (GO) and KEGG pathway enrichment analyses of DEGs. The DEGs (FDR ≤ 0.05) with a fold change larger than 2 or 1.5 were used for the enrichment analyses of GO terms and KEGG pathways, respectively. The significantly over-represented and under-represented GO terms (A) and KEGG pathways (B) with a FDR ≤ 0.05 were presented. Grey and black bars indicate the percentages of DEGs and the whole transcriptome that were classified into different functional annotations, respectively.
MicroRNAs, Particularly Highly Conserved miRNAs, Show a Negatively-Correlated Expression Pattern with Their Direct Targets
As miRNAs negatively regulated target gene expression, one may expect to see negative correlation between expression of miRNAs and target mRNAs. To test this speculation, we performed correlation analyses either for all the miRNAs and their targets globally using the log2 fold changes of expression between indica and japonica subspecies, or for individual miRNA-target pairs using their expression levels across six rice cultivars. We shall focus on the global analysis in this section and present the results of the individual analysis in the next. Such analyses were based on four target sets, including one predicted target set for all the expressed miRNAs (target set I; see “Methods” for rice miRNA target prediction) and three experimentally verified target sets by high-throughput degradome sequencing for japonica cv. Nipponbare (target sets II, III) or indica cv. 93-11 (target set IV) (Wu et al. 2009; Li et al. 2010; Zhou et al. 2010).
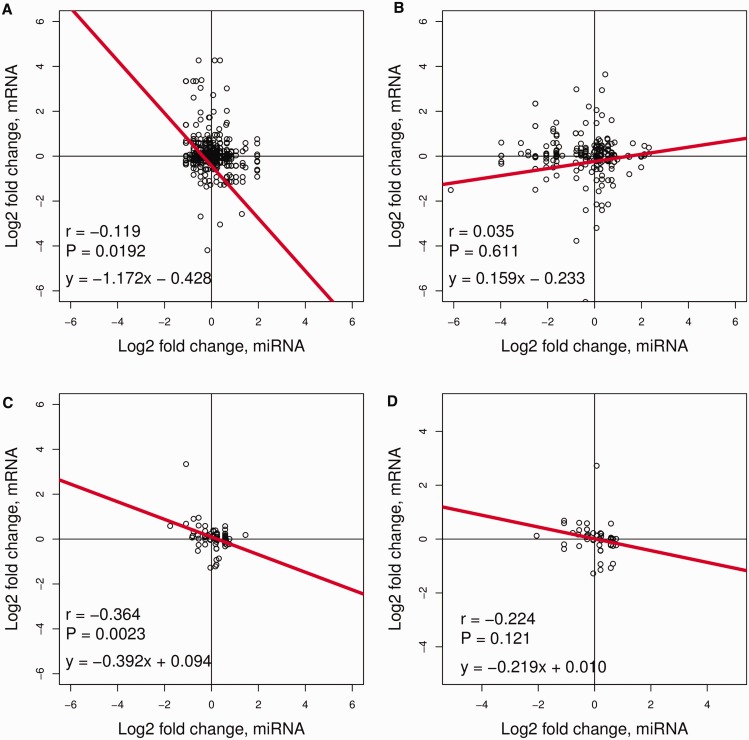
As for the global analysis, no correlation (Pearson’s correlation hereafter, P-value = 0.420, r = −0.033, n = 609) was observed for all the expressed miRNAs and their predicted targets (target set I) (supplementary fig. S9A, Supplementary Material online). However, when separating miRNAs according to their conservation, a weak but significant negative correlation was observed for the highly conserved miRNAs and their predicted targets (P-value = 0.019, r = −0.119, n = 390; fig. 4A), but not for the lowly conserved miRNAs and targets (P-value = 0.611, r = 0.035, n = 219; fig. 4B). The negative correlation in expression fold changes was highly significant for the co-expressed miRNAs and degradome-verified target set IV (Zhou et al. 2010) (P-value = 0.002, r = −0.364, n = 68; fig. 4C), and nearly significant for target set II (Wu et al. 2009) (P-value = 0.121, r = −0.224, n = 49; fig. 4D) and target set III (P-value = 0.119, r = −0.161, n = 95; supplementary fig. S9B, Supplementary Material online) (Li et al. 2010). The extent of negative correlation seemed to be associated with the proportion of targets of highly conserved miRNAs in the analyzed target sets, which is 86%, 81%, and 91% for target set II, III, and IV, respectively. Taken together, the overall expression of miRNAs correlates negatively with the expression of their targets in rice, which is consistent with miRNA functions in guiding mRNA cleavage in plants (Bartel 2004). Highly conserved miRNAs exhibit less variation in expression but repress targets more strongly than lowly conserved miRNAs.
Fig. 4.—
Correlation between the coexpressed miRNAs and their targets in seedlings. (A) highly conserved miRNAs and their predicted targets (390 pairs); (B) lowly conserved miRNAs and their predicted targets (219 pairs); (C) miRNAs and the degradome-verified targets in target set IV (Zhou et al. 2010) (68 pairs) and (D) miRNAs and the degradome-verified targets in target set II (Wu et al. 2009) (49 pairs). The log2 fold changes of miRNA or mRNA expression between rice indica and japonica subspecies in seedlings were used for Pearson’s correlation analysis.
We then compared the indica–japonica expression differentiation of miRNAs relative to that of target genes using transcriptome as a control. Interestingly, the extent of differential expression between rice subspecies was much greater for miRNAs (median absolute fold change = 0.630) than for the whole transcriptome (median absolute fold change = 0.263). Kolmogorov–Smirnov test (KS test) on the cumulative plot shows that the differences are highly significant (P-value ≪ 0.001; supplementary fig. S10A, Supplementary Material online). In contrast, the extent of differential expression was comparable between miRNA targets (median absolute fold change = 0.279) and the transcriptome (median absolute fold change = 0.263, supplementary fig. S10B, Supplementary Material online). Although lowly conserved miRNAs (median absolute fold change = 0.677) were more variable in expression than highly conserved miRNAs (median absolute fold change = 0.457, KS test P-value ≪ 0.001, supplementary fig. S10A, Supplementary Material online), no such expression difference was observed for their targets (KS test, P-value = 0.926, supplementary fig. S10B, Supplementary Material online). It is thus evident that miRNA targets exhibit less variation in expression than miRNAs.
Identification of Individual miRNA-Target Pairs with Significant Expression Correlations
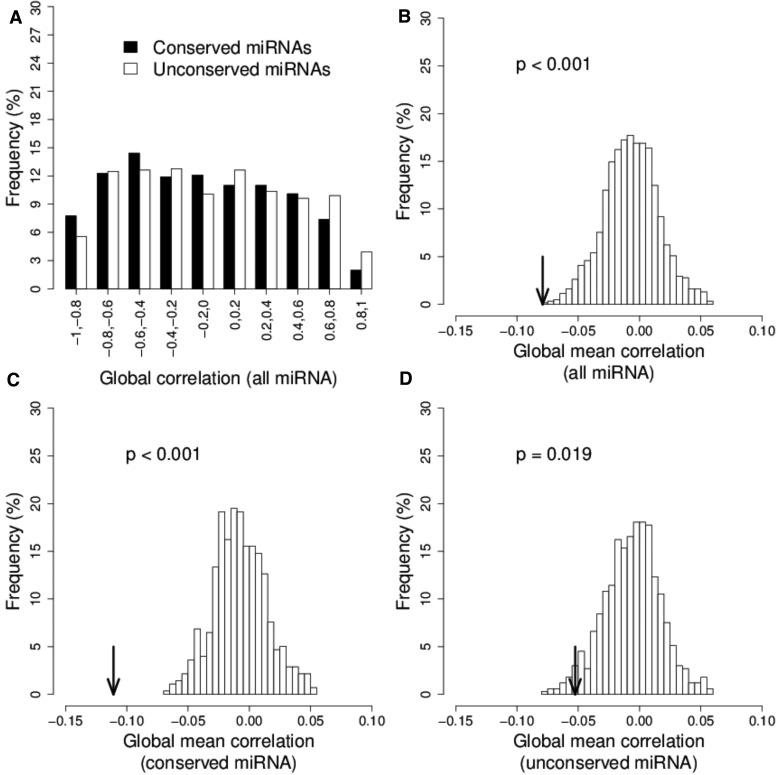
We further examined the individual correlations of the expression of miRNA–mRNA pairs across the six accessions. The average correlation coefficient for the 476 miRNA-target pairs (in target set I) was −0.11. The highly conserved miRNAs tend to be more negatively correlated with their corresponding target mRNAs than the lowly conserved miRNAs in expression variation among accession (KS test, P-value = 0.0008, fig. 5A) but no pair passed the 0.05 FDR level after the multiple-testing correction.
Fig. 5.—
Permutation of miRNA–mRNA target relationships at the lineage level. (A) The empirical distribution of the Pearson’s correlation coefficient values for 436 miRNA–mRNA pairs between expression levels of 96 miRNAs and those of their target mRNAs across 6 lineages. (B) The histogram plot represents the distribution of the global mean correlation values for the expression levels of all miRNA–mRNA pairs for 1,000 permutations, (C) for the highly conserved miRNAs and (D) for the lowly conserved miRNAs. The black arrowhead indicates the true value.
We then asked whether the mean expression correlation among all miRNA-target pairs shifted to the negative end in comparison with the random miRNA-gene pairs. Using a permutation test as previously described (Nunez-Iglesias et al. 2010), we identified 51 significantly correlated miRNA-target pairs, including 29 negatively correlated and 22 positively correlated pairs (table 2). Indeed, the mean correlation of the real miRNA–mRNA pairs is more negative than the permuted pairs (empirical P-value ≪ 0.001, fig. 5B), both for the highly conserved (empirical P-value ≪ 0.001, fig. 5C) and lowly conserved miRNAs (empirical P-value = 0.019, fig. 5D). Therefore, the overall tendency in the individual miRNA-target correlations is towards the negative correlation, which is in contrast to a similar test performed in human brain samples (Nunez-Iglesias et al. 2010). Also, the degradome verified targets are significantly enriched in the correlated pairs (27 verified targets, Chi-squared test P-value ≪ 0.001, table 2), suggesting that those genes are indeed under the control of miRNA regulation.
Table 2.
Significantly Correlated miRNA–mRNA Pairs in the Permutation Test
| miRNA | miRNA FCa | mRNA FCb | Correlationc | W | Target | TIGR Annotationd | Target Verificatione | |
|---|---|---|---|---|---|---|---|---|
| miR156k | 0.080 | 0.142 | 0.953 | 2.082 | LOC_Os01g69830 | OsSPL2-SBP-box gene family member | Y3,4 | |
| miR160a–d | −0.048 | -0.040 | −0.815 | −2.096 | LOC_Os06g47150 | Auxin response factor 18 | Y2,3,4 | |
| miR160e | 0.537 | −0.040 | −0.817 | −1.923 | LOC_Os06g47150 | Auxin response factor 18 | Y2,3,4 | |
| 0.222 | 0.873 | 1.573 | LOC_Os04g43910 | Auxin response factor | Y2,3,4 | |||
| miR160f | 0.379 | 0.222 | 0.702 | 1.593 | LOC_Os04g43910 | Auxin response factor | Y2,3,4 | |
| miR166a–d,f,n | 0.073 | 2.722 | 0.888 | 1.845 | LOC_Os04g48290 | MATE efflux family protein | Y2 | |
| miR166g,h | 0.203 | 0.383 | 0.791 | 1.427 | LOC_Os03g43930 | START domain containing protein | Y3,4 | |
| miR166m | −0.252 | −0.223 | 0.864 | 2.339 | LOC_Os08g34740 | SGT1 protein | N | |
| miR168a | 0.624 | −1.165 | −0.902 | −1.639 | LOC_Os11g44860 | Cysteine-rich receptor-like protein kinase 28 precursor | N | |
| miR169f–g | 0.661 | −0.930 | −0.940 | −1.922 | LOC_Os03g29760 | Nuclear transcription factor Y subunit | Y2,3,4 | |
| miR169h–m | 0.846 | −1.145 | −0.948 | −1.747 | LOC_Os07g41720 | Nuclear transcription factor Y subunit | Y2,3,4 | |
| miR171b–f | 0.242 | −0.126 | −0.774 | −1.880 | LOC_Os05g34460 | OsDegp7 - Putative Deg protease homologue | N | |
| 0.098 | 0.668 | 1.649 | LOC_Os02g44370 | Myosin | Y2,3,4 | |||
| miR171i | −0.426 | −0.375 | 0.882 | 1.933 | LOC_Os03g04300 | Targeting protein-related | N | |
| miR172b | −0.175 | 0.105 | −0.867 | −1.747 | LOC_Os05g03040 | AP2 domain containing protein | Y2,3,4 | |
| 0.321 | −0.772 | −1.593 | LOC_Os03g44420 | Tubulin/FtsZ domain containing protein | N | |||
| miR172c | 0.072 | −0.044 | 0.855 | 1.921 | LOC_Os07g13170 | AP2 domain containing protein | Y2,3,4 | |
| miR319a–b | 0.171 | −0.065 | 0.930 | 2.056 | LOC_Os08g16660 | Aspartic proteinase nepenthesin precursor | N | |
| miR393 | 0.766 | −1.128 | −0.695 | −1.645 | LOC_Os03g36080 | Expressed protein | Y3 | |
| miR395b,d–e,g–n,p–s,y | 1.449 | −0.505 | −0.972 | −1.944 | LOC_Os03g53230 | Bifunctional 3-phosphoadenosine 5-phosphosulfate synthetase | N | |
| miR396d–e | −0.035 | 0.053 | −0.991 | −2.419 | LOC_Os06g02560 | Growth-regulating factor | Y2,3,4 | |
| 0.129 | −0.855 | −1.999 | LOC_Os03g47140 | Growth regulating factor protein | Y3,4 | |||
| −0.325 | −0.812 | −1.887 | LOC_Os02g53690 | Ankyrin repeat domain containing protein | Y3,4 | |||
| 0.241 | −0.749 | −1.690 | LOC_Os04g51190 | Growth-regulating factor | Y3,4 | |||
| −0.008 | −0.693 | −1.654 | LOC_Os11g35030 | Growth regulating factor protein | Y2,3,4 | |||
| miR396f | −0.054 | 0.053 | −0.987 | −2.399 | LOC_Os06g02560 | Growth-regulating factor | Y2,3,4 | |
| 0.129 | −0.870 | −2.030 | LOC_Os03g47140 | Growth regulating factor protein | Y3,4 | |||
| −0.325 | −0.795 | −1.843 | LOC_Os02g53690 | Growth regulating factor protein | Y3,4 | |||
| 0.241 | −0.744 | −1.671 | LOC_Os04g51190 | Growth-regulating factor | Y3,4 | |||
| miR397a | −0.076 | 0.413 | −0.812 | −1.833 | LOC_Os11g48060 | Laccase-22 precursor | Y3 | |
| miR397b | −0.108 | 0.413 | −0.828 | −1.875 | LOC_Os11g48060 | Laccase-22 precursor | Y3 | |
| 1.263 | 0.731 | 1.745 | LOC_Os09g27950 | Galactosyltransferase | N | |||
| miR444b–c | −0.466 | 0.006 | 0.678 | 1.797 | LOC_Os02g49840 | Scarecrow | Y2,3 | |
| miR444f | 1.947 | −0.646 | −0.941 | −1.789 | LOC_Os02g34080 | Targeting protein for Xklp2 | N | |
| miR528 | −0.581 | 0.136 | −0.897 | −1.583 | LOC_Os08g44770 | Copper/zinc superoxide dismutase | N | |
| miR529b | −1.633 | 0.219 | −0.876 | −1.923 | LOC_Os04g28420 | Peptidyl-prolyl isomerase | N | |
| −0.038 | −0.687 | −1.529 | LOC_Os12g08760 | Carboxyvinyl-carboxyphosphonate phosphorylmutase | N | |||
| miR530 | −0.900 | −0.678 | 0.794 | 2.288 | LOC_Os05g09650 | Ubiquinone biosynthesis protein COQ4 | N | |
| miR1847 | −0.376 | −0.150 | 0.912 | 1.924 | LOC_Os01g63190 | Laccase precursor protein | N | |
| miR1856 | −6.129 | −1.509 | 0.827 | 1.785 | LOC_Os09g10274 | Expressed protein | N | |
| miR1860 | 0.174 | −0.069 | 0.920 | 2.125 | LOC_Os01g01030 | Monocopper oxidase | N | |
| miR1861a | 1.984 | 0.021 | 0.796 | 1.590 | LOC_Os03g40020 | PPR repeat containing protein | N | |
| miR1864 | −1.514 | 0.414 | −0.863 | −1.374 | LOC_Os01g14020 | Expressed protein | N | |
| miR1873 | 0.003 | 0.223 | −0.717 | −2.359 | LOC_Os05g01790 | Expressed protein | N | |
| miR1884a | 0.575 | −1.074 | −0.685 | −1.786 | LOC_Os11g34910 | Expressed protein | Y2 | |
| 0.435 | 0.952 | 1.782 | LOC_Os06g14780 | Expressed protein | N | |||
| miR1884b | 0.083 | −0.039 | −0.677 | −2.294 | LOC_Os12g01680 | Macrophage migration inhibitory factor | N | |
| 0.036 | −0.573 | −1.865 | LOC_Os02g49870 | Expressed protein | N | |||
| −0.231 | 0.575 | 1.640 | LOC_Os01g64520 | Uricase | Y2 | |||
| 0.435 | 0.636 | 1.786 | LOC_Os06g14780 | Expressed protein | N | |||
| miR2097 | 0.543 | 0.787 | 0.779 | 1.585 | LOC_Os08g43920 | Carrier | N | |
aFold change in the log2 ratio of miRNA expression between indica and japonica subspecies in seedlings. The mean expression levels averaged from three accessions were used for the calculation. MiRNAs with significant differentially expressions (unadjusted P-value ≤ 0.05 and Fold change ≥2) between rice subspecies are marked in bold.
bFold change in the log2 ratio of mRNA expression between indica and japonica subspecies in seedlings. The mean expression levels averaged from three accessions were used for the calculation. Genes with significantly differential expression are marked in bold.
cPearson’s correlation coefficient.
dGene annotations from TIGR (Version 6).
eThe Y2,3,4 Target is verified in the corresponding degradome target set II, III, or IV, respectively; N: not yet verified.
Significantly correlated miRNA-target pairs, both negatively and positively, participate in key biological processes such as reproduction, development, pigmentation, and stress responses (supplementary fig. S11, Supplementary Material online). The negatively-correlated pairs included the well-documented miRNA-target pairs, such as miR156:SQUAMOSA promoter binding protein-like (SPL) (Miura et al. 2010); miR169:Nuclear transcription factor Y (NF-Y) (Zhao et al. 2009), miR172-Apetala2 (AP2) (Zhu and Heliwell 2011), and miR396: Growth-regulating factor (GRF) (Debernardi et al. 2012) (table 2). The involvement of the positively correlated miRNA-target pairs in cell cycle, cell death, pollination and the transport process suggest that these pairs also play significant roles in rice development. Positive correlations between the miRNA and target expression levels across rice cultivars may be due to the miRNA-mediated regulatory circuits such as negative feedback loops or incoherent feedforward loops (Wu et al. 2009).
Discussion
We present here the first survey of miRNA expression variation in rice cultivars. Substantial miRNA expression changes were detected between rice grains and seedlings, consistent with the regulatory role of miRNAs in rice seed development (Zhu et al. 2008). In contrast, only a small fraction of miRNAs, mainly lowly conserved miRNAs, differed in expression level between rice indica and japonica subspecies. While miRNA genes are under strong purifying selection (Ehrenreich and Purugganan 2008; Wang et al. 2010), the single nucleotide polymorphisms (SNP) densities of pre-miRNAs and mature miRNAs are significantly higher than their flanking regions in rice (Liu et al. 2013), implying that cis-regulatory mutations affecting miRNA expression level are mostly deleterious and are thus quickly purged from the rice population. The low level of miRNA expression variation coupled with low level of cis-regulatory sequence polymorphism is consistent with the scenario that stabilizing selection commonly uses purifying selection to select against extreme values of characters, i.e., miRNA expression profiles in this study. Indeed, it has been demonstrated both empirically and theoretically that stabilizing selection is the major evolutionary force governing the evolution of gene expression (Denver et al. 2005; Rifkin et al. 2005; Hutter et al. 2008; Bedford and Hartl 2009; Hodgins-Davis et al. 2015). A recent study revealed that a vast majority of miRNAs are under stabilizing selection at the onset of Drosophila metamorphosis (Yeh et al. 2014). Using the same methodology, we estimated that the counts for rice miRNAs with expression variation compatible with particular evolutionary modes are 196, 29, 1, and 46 for stabilizing selection, directional selection, genetic drift and complex scenarios, respectively (see supplementary text, Supplementary Material online). About 72% of miRNAs in this study are not significanlty differentiated within or between rice subspecies, compatible with the evolutionary mode of stabilizing selection. These miRNAs under stabilizing selection are mostly conserved between eudicots and monocots, wherase more than half of the miRNAs under directional selection are species-specific to rice.
It is remarkable that lowly and highly conserved miRNAs showed sharply contrasting patterns in expression variation and regulation strength. The great expression variation of the lowly conserved miRNAs is largely coupled with their high level of sequence polymorphism among cultivated rice (Liu et al. 2013), suggesting they are under weak selection pressures. Such a coupling of expression variation and sequence polymorphism of rice miRNAs is compatible with the correlated divergences between gene sequences and expression patterns during organ evolution in angiosperms (Yang and Wang 2013). The negligible overall correlation between expression changes of the lowly conserved miRNAs, also the lowly expressed miRNAs, and their targets further indicates that these miRNAs exert very modest, if any, repression on target genes. Therefore, the lowly conserved miRNAs are more like young miRNAs, which are expressed lowly or in specialized tissues, evolve rapidly, and tend to be lack of targets (Rajagopalan et al. 2006; Fahlgren et al. 2010; Ma et al. 2010), rather than the old, deeply conserved miRNAs.
While most young miRNAs are evolutionarily transient (Fahlgren et al. 2010), they can occasionally be selectively favored. A good case in point is a new miRNA specific in japonica rice, osa-miR7695, which negatively regulates an alternatively spliced transcript of OsNramp6 (Natural resistance-associated macrophage protein 6), conferring pathogen resistance (Campo et al. 2013). The differentially expressed miRNAs between indica and japonica subspecies, which are resulted from low expression polymorphism within subspecies and high expression divergence between subspecies, may represent a class of miRNAs that are favored by artificial selection in rice domestication and/or improvement. Previous evolutionary analyses have identified rice miRNA genes that are putative candidates of positive selection, including highly conserved MIR164e, MIR395a/b, and MIR399d (Wang et al. 2010), and lowly conserved or rice-specific osa-miR5513, osa-miR818e, osa-miR1847, osa-miR1865, osa-miR160f, osa-miR5143, and osa-miR2118h-l (Liu et al. 2013). Among them, osa-miR395a–v,y and miR399a–d,i–j (unadjusted P-value < 0.05, FDR ≈ 0.2, supplementary table S2, Supplementary Material online) showed significant differential expression between indica and japonica subspecies and the former correlated negatively with a putative target LOC_Os03g53230 in this study. In the Solanaceae, differential expression of the miR482/2188 gene family members that are under different evolutionary constraints also suggested miRNA subfunctionalization between closely related plant species (de Vries et al. 2015).
Another pattern that we observed is the prevalent positive correlation between individual miRNA-target pairs in the cultivated rice, of which many target genes have been experimentally verified by degradome data. Given miRNA negatively regulated target expression, this observation may reflect the composite effect of miRNA-mediated circuits, such as negative feedback loops (FBLs) and incoherent feedforward loops (FFLs) (Alon 2007). miRNA-mediated FBLs and FFLs are recurrent network motifs in animals (Tsang et al. 2007; Wu et al. 2009), and also play significant roles in plants as evidenced by both experiments (Xie et al. 2003; Bari et al. 2006; Vaucheret et al. 2006) and computational analyses (Meng et al. 2009; Wu et al. 2009). From the network perspective, miRNAs can thus play a role in expression buffering and biological robustness, as well recognized in animal systems (Wu et al. 2009; Herranz and Cohen 2010; Ebert and Sharp 2012; Pelaez and Carthew 2012). We found that targets of rice miRNAs overall have less expression variation in comparison with the transcriptome, albeit miRNAs themselves are more variable in expression. These results are consistent with the notion that miRNAs act as dampers in buffering target expression variation (Fei et al. 2013) or target sequence diversity (de Vries et al. 2015). In contrast, miRNA targets compared with non-targets overall have higher level of expression variation in human, despite a small number of highly expressed targets with decreased expression variation, suggesting the dual function of miRNA regulation (Lu and Clark 2012).
Functional enrichment tests of the DEGs highlight that expression changes of genes involved in various metabolism processes and stress responses are important for the indica-japonica differentiation. Genes involved in plant–pathogen pathways are the most prominent examples, since the interaction between rice–host cultivar (genotype) and parasite population is shown to be critical in determining parasite affecting (Huang et al. 2012). Interestingly, we identified a target (LOC_Os02g30900, PBS1) of a lowly conserved miRNA, miR1857, which is involved in this pathway (supplementary fig. S7, Supplementary Material online). The orthologous PBS1 gene in Arabidopsis encodes a putative serine-threonine kinase, which is required for specific recognition of the bacterial protein AvrPphB (Swiderski and Innes 2001). Another related example has also been recently reported in Arabidopsis, where a peptide derived from the plant pathogen Pseudomonas syringae induces the expression of the host miR393 that targets auxin receptor (TIR1, AFB2, and AFB3), and consequently inhibits the growth of the bacteria (Navarro et al. 2006). It would be interesting to explore the role of miRNAs in mediating biotic stress response in the context of differentiation between indica and japonica subspecies.
Our permutation analysis provides a promising approach to further classify the important miRNA regulation pairs in rice. For example, besides the well documented miRNA-target pairs mentioned previously, recent study revealed that osa-miR171c controls the floral transition and maintenance of shoot apical meristem (SAM) indeterminacy in rice by targeting GRAS (GAI-RGA-SCR), a plant-specific transcription factors (LOC_Os02g44370, also for OsHAM2, table 2) (Fan et al. 2015); In addition, miR166-mediated post-transcriptional gene silencing of rice Class III HD-Zip genes (LOC_Os03g43930, also for OsHB5) is reported to be responsible for the auxin signals to regulate leaf and shoot development (Itoh et al. 2008; Toriba et al. 2010). There are also many miRNAs with un-verified targets in our list of correlated miRNA-target pairs (table 2). Further studies are necessary to confirm the functional contribution of these miRNAs to the process of rice development and/or differentiation. The joint miRNA–mRNA data and permutation analyses used in this study provide a novel idea to study miRNA regulatory relationships in rice. Our results may provide a valuable resource for further investigation of miRNA functions in rice developmental regulation, stress responses, and biomass yields under domestication.
Methods
Plant Materials
Seeds of O. sativa L. ssp. indica and ssp. japonica accessions as listed in table 3 were obtained from the International Rice Research Institute (IRRI, Manila, Philippines). For grains, the husks of the seeds were removed before RNA extraction. For seedlings, rice seeds were sterilized and germinated in Petri dishes containing distilled water at 37 °C under dark conditions for 2 days. The uniformly germinated seeds were transferred into Yoshida nutrient solution and grown under a 16-h light (28 °C)/8-h dark (25 °C) photoperiod for one week. The samples were collected and rinsed with double distilled water three times, and then immediately frozen in liquid nitrogen until use.
Table 3.
Summary of the Reads Mapping of Small RNAs and Transcriptomes by RNA Sequencing
| Rice variety |
Small RNAs |
Transcriptome |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Tissue | Subspecies | Accession Name | Accession No.a | No. of Sequences Generated | No. of Sequences Matching the Rice Genome | No. of Unique Sequences | No. of Sequences Generatedb | No. of Sequences Matching the Rice Genomec | Unique Mapped Readsc |
| Seedling | Indica | Khao Dawk Mali 105 | IRGC 27748 | 9,985,373 | 9,758,028 (97.7%) | 2,786,744 (27.9%) | 20,326,850 | 34,063,783 (83.79%) | 33,569,936 (82.6%) |
| Guangluai 4 | IRGC 114900 | 9,788,880 | 9,558,792 (97.7%) | 3,048,520 (31.1%) | 20,216,686 | 32,972,884 (81.54%) | 32,494,623 (80.4%) | ||
| Rathuwee | IRGC 8952 | 9,997,943 | 9,701,803 (97.0%) | 3,403,862 (34.0%) | 19,876,912 | 32,789,681 (82.48%) | 32,328,952 (81.3%) | ||
| Japonica | Taipei 309 | IRGC 42576 | 8,221,032 | 8,051,214 (97.9%) | 2,282,686 (27.8%) | 19,094,551 | 33,286,973 (87.16%) | 32,855, 807 (86.0%) | |
| Heukgyeong | IRGC 55530 | 9,371,607 | 8,988,554 (95.9%) | 2,203,943 (23.5%) | 19,889,198 | 34,863,682 (87.64%) | 34,372,869 (86.4%) | ||
| Nipponbare | IRGC 12731 | 10,592,461 | 10,416,128 (98.3%) | 3,010,461 (28.4%) | 18,818,192 | 32,995,013 (87.67%) | 32,592,2 62 (86.6%) | ||
| Grain | Indica | Khao Dawk Mali 105 | IRGC 27748 | 14,396,308 | 13,994,396 (97.2%) | 5,408,929 (37.6%) | NA | NA | NA |
| Guangluai 4 | IRGC 114900 | 14,433,295 | 14,137,205 (98.0%) | 4,056,622 (28.1%) | NA | NA | NA | ||
| Rathuwee | IRGC 8952 | 15,718,282 | 15,185,127 (96.6%) | 5,740,388 (36.5%) | NA | NA | NA | ||
| Japonica | Taipei 309 | IRGC 42576 | 14,846,530 | 14,293,090 (96.3%) | 2,786,744 (18.8%) | NA | NA | NA | |
| Heukgyeong | IRGC 55530 | 10,225,740 | 9,512,313 (93.0%) | 3,081,796 (30.1%) | NA | NA | NA | ||
| Nipponbare | IRGC 12731 | 13,862,947 | 13,356,435 (96.4%) | 5,014,395 (36.2%) | NA | NA | NA | ||
| Total | 141,440,398 | 136,953,085 (96.8%) | 42,825,090 (30.3%) | NA | NA | NA | |||
aInternational Rice Germplasm Collection at IRRI in the Philippines (http://archive.irri.org/GRC/requests/requests.htm).
bNumber of pairs of 75-nt paired-ends sequencing reads.
cThe proportion was calculated as the number of mapped reads versus the number of total reads.
NA, Not applicable.
RNA Isolation and Preparation of Sequencing Libraries
Total RNA were extracted from rice grains and seedlings using TRIzol Reagent (Invitrogen), and evaluated using an Agilent 21100 Bioanalyzer (Agilent Technologies). Small RNA and transcriptome libraries were prepared using standard protocols of the Illumina Small RNA Sample Prep Kit or the Illumina TruSeq RNA Sample Prep kit, and sequenced using an Illumina Genome Analyzer (Illumina, San Diego, CA, USA) at BGI (Shenzhen, China). As we did not obtain enough quality RNA in grain, only the seedling samples were used for RNA-seq.
Expression Analysis of miRNAs
For all small RNA libraries, after trimming prime adaptors and filtering low quality or adaptor contaminated reads, clean reads within the length range of 19–30 nt were retained for further analysis. These reads were searched against the Rfam database (Griffiths-Jones et al. 2003) and the RepBase database (Jurka et al. 2005) using the SOAP software (Li et al. 2008) with two mismatches, in order to remove reads matching structural RNAs, including rRNA, tRNA, small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA), and repeats/transposons. The remaining reads were mapped to the precursors of all known rice miRNAs registered in miRBase (Release 20, http://www.mirbase.org/index.shtml; last accessed 3 September 2015) using SOAP (Li et al. 2008). We grouped all the variants of rice mature miRNAs into 553 distinct mature miRNAs since some miRNA precursors produced more than one different mature sequences and some identical mature sequences were generated from distinct precursors. Read counts corresponding to individual mature miRNA sequences were normalized and rescaled as Reads Per Million (RPM), which divided the raw read count of each mature miRNA by the total mapped read count in each library and multiplied it by 1 million. Subsequent heatmap clustering and differential expression analysis were performed based on the average RPM of each distinct mature miRNA. An unsupervised, two-dimensional, hierarchical clustering was conducted using R package gplots (Warnes et al. 2009). For differential expression analysis, a generalized Poisson-regression linear model was fitted in the R package edgeR (Nikolayeva and Robinson 2014) to identify the differentially expressed miRNAs for the factors of development, subspecies, and development-by-subspecies. MiRNAs with fold change ≥ 2 and FDR ≤ 0.05 are denoted as significantly mi-expressed.
Expression Analysis of mRNAs
For the transcriptome libraries, after removing adaptors and low-quality reads from raw reads, clean reads were mapped to the reference genome of rice (japonica cv. Nipponbare) using the Bowtie software package (Langmead et al. 2009). Reads that could be mapped equally well to multiple locations without mismatch or with up to two mismatches were randomly assigned to one position and were retained for further analyses as previously described (Wang et al. 2009). According to the TIGR 6.0 gff3 file (Kawahara et al. 2013), reads matching gene or genomic region were recovered and expression level of each transcript was measured as numbers of reads per kilobase of exon region in a gene per million mapped reads (RPKM) as previously described (Mortazavi et al. 2008).
To identify homologous genes between rice subspecies, the longest isoform of each gene model of japonica cv. Nipponbare was aligned to the genome of indica cv. 93-11 by GMAP (Wu and Watanabe 2005). We considered a Nipponbare gene and its counterpart in the 93-11 genome as an homologous gene pair if they share at least 95% sequence identity in at least 95% of their longest isoforms (He et al. 2010). The resulting 32,044 homologous gene pairs were further filtered by requiring at least 10 raw reads in all the three accessions sequenced for at least one developmental stage of a subspecies. These expressed homologous genes were then used for the subsequent differential expression analysis. The generalized Poisson-regression linear model was fitted and the likelihood ratio test was performed with lmtest (Zeileis and Hothorn 2002) to identify the mRNAs with differential expression between rice subspecies. Genes with fold-change ≥2 and FDR ≤ 5% were denoted as significantly differentially expressed.
Gene Functional Annotation
The gene ontology classification developed in the TIGR Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/; last accessed 10 September 2015) was used to assign genes to a hierarchical biological process using the criteria of the Gene Ontology (GO) Consortium databases (Ashburner et al. 2000; Kawahara et al. 2013), and KEGG pathway annotation was identified according to KEGG database (http://www.genome.jp/kegg/; last accessed 10 September 2015) (Kanehisa and Goto 2000; Kanehisa et al. 2016). The GO and KEGG enrichment test (FDR ≤ 0.05) was performed by using the CORNA and clusterProfiler package, respectively (Wu and Watson 2009; Yu et al. 2012). KEGG pathway rendering of the expressed genes was conducted by the R package Pathview (Luo and Brouwer 2013). The DEGs with a fold change larger than 2 were used for the GO enrichment test and 1.5 for the KEGG pathway enrichment test, with a FDR ≤ 0.05 was used as the significance threshold.
Identification of Novel miRNAs
After removing the reads mapped to the structural RNAs, the known miRNAs from miRBase and repeats/transposons, the remaining sequences were mapped on the rice genome and analyzed by an adjusted miRDeep script for novel plant miRNA prediction (Wen et al. 2012). The following criteria for miRNA annotation (Wang et al. 2009) were further applied to filter the novel miRNAs, including: (1) a secondary structure must have a hairpin with at least 18 paired nucleotides in its stem region; (2) the hairpin must have free energy less than or equal to −18 kCal/mol and no more than two central loops; (3) the miRNA and miRNA* form a duplex with a two-nucleotide overhang; (4) fewer than four mismatches exist in the miRNA/miRNA* duplex; and (5) ≥75% of the sRNAs mapped onto a miRNA precursor are derived from the miRNA or miRNA* region. The candidate sequences were extracted and folded with Vienna RNAFold (Lorenz et al. 2011). The miRNAs with more than 10 raw reads matching the mature sequences in at least two replicate sRNA libraries were considered to be expressed.
MiRNA Target Prediction and Integration
A search for the miRNA target genes was performed for all known miRNAs and newly identified miRNA sequences on the japonica cv. Nipponbare cDNA dataset (TIGR 6.0) using psRNAtarget (Dai and Zhao 2011) with a maximum expectation score of 2.5 to reduce the false discovery rate (Klevebring et al. 2009). The predicted target set was referred to as target set I in this study. Three additional sets of miRNA targets that are verified by degradome analysis were also used, which include target set II (Wu et al. 2009) and target set III (Li et al. 2010) for rice japonica cv. Nipponbare, and target set IV for rice indica cv. 93-11 (Zhou et al. 2010).
miRNA Target Permutation Analysis
The permutation test and weighted shift for the miRNA-target correlations were processed as described previously (Nunez-Iglesias et al. 2010). In short, the miRNA-target pairs can be considered a bipartite graph, with nodes representing the miRNAs on one side and the targets on the other, and the edges represent the target prediction relationships. The nodes have associated expression measurements. Correlation statistics were then computed for each miRNA-target pair across the 6 rice lineages. Finally, the network was permuted by shuffling the edges, re-computing the statistics, and then repeating this processes 1000 times. From these, we can obtain an empirical P-value and a “weighted correlation shift”, W, which was defined as the difference between the true value and the mean permuted value divided by the standard deviation of the permuted values: W = (r − r0)/S0.
Quantitative miRNA and mRNA Analysis by qRT-PCR
For miRNA quantification, total RNA from 1 week old seedlings was subjected to stem-loop reverse transcription (RT) (Chen et al. 2005) followed by Taqman PCR (Applied Biosystems) using the miRNA UPL (Roche Diagnostics) probe assay as described previously (Varkonyi-Gasic et al. 2007; He et al. 2016). 5.8S rRNA was used as an internal control. Three biological replicates were examined. For mRNA quantification, the same total RNA was first treated with TURBO DNA-free kit (Ambion) to remove potential genomic DNA contamination and then used for revers transcription with SuperScript III First-Stand Synthesis System (Invitrogen). qRT-PCR was performed with Platinum SYBR Green qPCR SuperMix (Invitrogen) according to the manufacturer’s instructions. Actin was used as an internal control. Three biological replicates with two technique repeats each were examined to ensure reproducibility. The relative levels of miRNA or mRNA were calculated using the 2−▵▵CT method (Livak and Schmittgen 2001). The sequences of primers used are listed in supplementary table S5, Supplementary Material online.
Data Availability
The expression data generated by this study are available in the NCBI Gene Expression Omnibus (GEO) under accession GSE71925.
Supplementary Material
Supplementary text, tables S1–S5, and figures S1–S11 materials are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank International Rice Research Institute for providing rice germplasm. We thank Han Liang for reading the early version of this manuscripts and providing instructive comments. This work was supported by National Science Foundation of China (31170308 and 91231117 to T.T., and 41130208 to S.S.), the Science Foundation for Outstanding Young Teachers in Higher Education of Guangdong (Yq2013005) and the Fundamental Research Funds for the Central Universities (16lgjc75) to T.T., and the General Financial Grant from the China Postdoctoral Science Foundation (2013M531981) to M.W.
Literature Cited
- Alon U. 2007. Network motifs: theory and experimental approaches. Nat Rev Genet. 8(6):450–461. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet. 25(1):25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 64:137–159. [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141(3):988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T, Hartl DL. 2009. Optimization of gene expression by natural selection. Proc Natl Acad Sci U S A. 106(4):1133–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. 1971. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 46(2):111–138. [DOI] [PubMed] [Google Scholar]
- Campo S, Peris-Peris C, Sire C, Moreno AB, Donaire L, et al. 2013. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 199(1):212–227. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, et al. 2010. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465(7296):316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. 2003. Role of microRNAs in plant and animal development. Science 301(5631):336–338. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, et al. 2005. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 33(20):e179.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FF, He GM, He H, Chen W, Zhu XP, et al. 2010. Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J Integr Plant Biol. 52(11):971–980. [DOI] [PubMed] [Google Scholar]
- Chuck G, Candela H, Hake S. 2009. Big impacts by small RNAs in plant development. Curr Opin Plant Biol. 12(1):81–86. [DOI] [PubMed] [Google Scholar]
- Cuperus JT, Fahlgren N, Carrington JC. 2011. Evolution and functional diversification of miRNA genes. Plant Cell 23(2):431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XB, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39:W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S, Kloesges T, Rose LE. 2015. Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae. Genome Biol Evol. 7(12):3307–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Rodriguez RE, Mecchia MA, Palatnik JF. 2012. Functional specialization of the plant miR396 regulatory network through distinct microRNA-target interactions. PLoS Genet. 8(1):e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Streelman JT, Kim SK, Lynch M, et al. 2005. The transcriptional consequences of mutation and natural selection in Caenorhabditis elegans. Nat Genet. 37(5):544–548. [DOI] [PubMed] [Google Scholar]
- Ding YF, Chen Z, Zhu C. 2011. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J Exp Bot. 62(10):3563–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. 2012. Roles for microRNAs in conferring robustness to biological processes. Cell 149(3):515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Purugganan MD. 2008. Sequence variation of MicroRNAs and their binding sites in Arabidopsis. Plant Physiol. 146(4):1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlgren N, Jogdeo S, Kasschau KD, Sullivan CM, Chapman EJ, et al. 2010. MicroRNA gene evolution in Arabidopsis lyrata and Arabidopsis thaliana. Plant Cell 22(4):1074–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan T, Li X, Yang W, Xia K, Ouyang J, et al. 2015. Rice osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS One 10(5):e0125833.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. 2013. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25(7):2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 9(2):102–114. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. 2003. Rfam: an RNA family database. Nucleic Acids Res. 31(1):439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Zhu X, Elling AA, Chen L, Wang X, et al. 2010. Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22(1):17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Xie M, Huang J, Zhang T, Shi S, et al. 2016. Efficient and specific inhibition of plant microRNA function by anti-microRNA oligonucleotides (AMOs) in vitro and in vivo. Plant Cell Rep. 35(4):933–945. [DOI] [PubMed] [Google Scholar]
- He ZW, Zhai WW, Wen HJ, Tang T, Wang Y, et al. 2011. Two evolutionary histories in the genome of rice: the roles of domestication genes. PLoS Genet. 7(6):e1002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H, Cohen SM. 2010. MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24(13):1339–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins-Davis A, Rice DP, Townsend JP. 2015. Gene expression evolves under a house-of-cards model of stabilizing selection. Mol Biol Evol. 32(8):2130–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Whitlock R, Press MC, Scholes JD. 2012. Variation for host range within and among populations of the parasitic plant Striga hermonthica. Heredity 108(2):96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Saminadin-Peter SS, Stephan W, Parsch J. 2008. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol. 9(1):R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y. 2008. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147(4):1960–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YQ, Wang YH, Xue DW, Wang J, Yan MX, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 42(6):541–544. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. 2006. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 57:19–53. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, et al. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 110(1–4):462–467. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44(D1):D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (NY) 6(1):4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188(1126):107–116. [DOI] [PubMed] [Google Scholar]
- Klevebring D, Street NR, Fahlgren N, Kasschau KD, Carrington JC, et al. 2009. Genome-wide profiling of populus small RNAs. BMC Genomics 10:620.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. 2008. SOAP: short oligonucleotide alignment program. Bioinformatics 24(5):713–714. [DOI] [PubMed] [Google Scholar]
- Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, et al. 2010. Transcriptome-wide identification of microRNA targets in rice. Plant J. 62(5):742–759. [DOI] [PubMed] [Google Scholar]
- Liu Q, Wang H, Zhu L, Hu H, Sun Y. 2013. Genome-wide identification and analysis of miRNA-related single nucleotide polymorphisms (SNPs) in rice. Rice (N Y) 6(1):10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zhang YC, Wang CY, Luo YC, Huang QJ, et al. 2009. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 583(4):723–728. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408. [DOI] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, et al. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6:26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark AG. 2012. Impact of microRNA regulation on variation in human gene expression. Genome Res. 22(7):1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Li WQ, Miura K, Ashikari M, Kyozuka J. 2012. Control of tiller growth of rice by OsSPL14 and strigolactones, which work in two independent pathways. Plant Cell Physiol. 53(10):1793–1801. [DOI] [PubMed] [Google Scholar]
- Luo W, Brouwer C. 2013. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29(14):1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv DK, Bai X, Li Y, Ding XD, Ge Y, et al. 2010. Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459(1–2):39–47. [DOI] [PubMed] [Google Scholar]
- Ma Z, Coruh C, Axtell MJ. 2010. Arabidopsis lyrata small RNAs: transient MIRNA and small interfering RNA loci within the Arabidopsis genus. Plant Cell 22(4):1090–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei A, Gill SS, Tuteja N. 2012. microRNAs as promising tools for improving stress tolerance in rice. Plant Signal Behav. 7(10):1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Huang F, Shi Q, Cao J, Chen D, et al. 2009. Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta 230(5):883–898. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, et al. 2008. Criteria for annotation of plant microRNAs. Plant Cell 20(12):3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, et al. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 42(6):545. U102. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, et al. 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312(5772):436–439. [DOI] [PubMed] [Google Scholar]
- Nikolayeva O, Robinson MD. 2014. edgeR for differential RNA-seq and ChIP-seq analysis: an application to stem cell biology. Methods Mol Biol. 1150:45–79. [DOI] [PubMed] [Google Scholar]
- Noctor G, Arisi ACM, Jouanin L, Kunert KJ, Rennenberg H, et al. 1998. Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot. 49(321):623–647. [Google Scholar]
- Nunez-Iglesias J, Liu CC, Morgan TE, Finch CE, Zhou XJ. 2010. Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS One 5(2):e8898.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaez N, Carthew RW. 2012. Biological robustness and the role of microRNAs: a network perspective. Curr Top Dev Biol. 99:237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 20(24):3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. 2007. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49(4):592–606. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP. 2005. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature 438(7065):220–223. [DOI] [PubMed] [Google Scholar]
- Shen Y, Lv Y, Huang L, Liu W, Wen M, et al. 2011. Testing hypotheses on the rate of molecular evolution in relation to gene expression using microRNAs. Proc Natl Acad Sci U S A. 108(38):15942–15947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiderski MR, Innes RW. 2001. The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J. 26(1):101–112. [DOI] [PubMed] [Google Scholar]
- Toriba T, Suzaki T, Yamaguchi T, Ohmori Y, Tsukaya H, et al. 2010. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22(5):1452–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. 2007. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell 26(5):753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. 2007. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Mallory AC, Bartel DP. 2006. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22(1):129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Elling AA, Li XY, Li N, Peng ZY, et al. 2009. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21(4):1053–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen D, Bo S, Chen H, Zheng J, et al. 2010. Sequence variation and selection of small RNAs in domesticated rice. BMC Evol Biol. 10:119.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, et al. 2009. gplots: Various R programming tools for plotting data. R Package Version 2(4): 447. [Google Scholar]
- Wen M, Shen Y, Shi S, Tang T. 2012. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinformatics 13:140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T. 2009. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome Res. 19(5):734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Zhang Q, Zhou H, Ni F, Wu X, et al. 2009. Rice MicroRNA effector complexes and targets. Plant Cell 21(11):3421–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21(9):1859–1875. [DOI] [PubMed] [Google Scholar]
- Wu X, Watson M. 2009. CORNA: testing gene lists for regulation by microRNAs. Bioinformatics 25(6):832–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC. 2003. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol. 13(9):784–789. [DOI] [PubMed] [Google Scholar]
- Xue LJ, Zhang JJ, Xue HW. 2009. Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res. 37(3):916–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Wang X. 2013. Organ evolution in angiosperms driven by correlated divergences of gene sequences and expression patterns. Plant Cell 25(1):71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Rao Y, Xu J, Shao G, Leng Y, et al. 2014. Genetic analysis of sugar-related traits in rice grain. South Afr J Bot. 93:137–141. [Google Scholar]
- Yeh SD, von Grotthuss M, Gandasetiawan KA, Jayasekera S, Xia XQ, et al. 2014. Functional divergence of the miRNA transcriptome at the onset of Drosophila metamorphosis. Mol Biol Evol. 31(10):2557–2572. [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeileis A, Hothorn T. 2002. Diagnostic checking in regression relationships. R News 2(3):7–10. [Google Scholar]
- Zhang BH, Pan XP, Cobb GP, Anderson TA. 2006. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 289(1):3–16. [DOI] [PubMed] [Google Scholar]
- Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, et al. 2013. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol. 31(9):848.. [DOI] [PubMed] [Google Scholar]
- Zhao BT, Ge LF, Liang RQ, Li W, Ruan KC, et al. 2009. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol Biol. 10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Gu L, Li P, Song X, Wei L, et al. 2010. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica). Front Biol. 5(1):67–90. [Google Scholar]
- Zhu QH, Helliwell CA. 2011. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 62(2):487–495. [DOI] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, et al. 2008. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Res. 18(9):1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The expression data generated by this study are available in the NCBI Gene Expression Omnibus (GEO) under accession GSE71925.