Abstract
The nucleocytoplasmic large DNA viruses (NCLDV) are a group of extremely complex double-stranded DNA viruses, which are major parasites of a variety of eukaryotes. Recent studies showed that certain unicellular eukaryotes contain fragments of NCLDV DNA integrated in their genome, when surprisingly many of these organisms were not previously shown to be infected by NCLDVs. These findings prompted us to search the genome of Acanthamoeba castellanii strain Neff (Neff), one of the most prolific hosts in the discovery of giant NCLDVs, for possible DNA inserts of viral origin. We report the identification of 267 markers of lateral gene transfer with viruses, approximately half of which are clustered in Neff genome regions of viral origins, transcriptionally inactive or exhibit nucleotide-composition signatures suggestive of a foreign origin. The integrated viral genes had diverse origin among relatives of viruses that infect Neff, including Mollivirus, Pandoravirus, Marseillevirus, Pithovirus, and Mimivirus. However, phylogenetic analysis suggests the existence of a yet-undiscovered family of amoeba-infecting NCLDV in addition to the five already characterized. The active transcription of some apparently anciently integrated virus-like genes suggests that some viral genes might have been domesticated during the amoeba evolution. These insights confirm that genomic insertion of NCLDV DNA is a common theme in eukaryotes. This gene flow contributed fertilizing the eukaryotic gene repertoire and participated in the occurrence of orphan genes, a long standing issue in genomics. Search for viral inserts in eukaryotic genomes followed by environmental screening of the original viruses should be used to isolate radically new NCLDVs.
Keywords: nucleo-cytoplasmic large DNA virus, acanthamoeba, lateral gene transfer
Introduction
Viruses have long been only viewed under the angle of human, animal, and plant diseases which considerably restrained our vision of the viral world and its role in global ecology. In this age of virus discovery, we are beginning to appreciate the enormous diversity of viruses, far beyond what we originally thought. Viruses are environmentally ubiquitous, obligate intracellular parasites that infect organisms from all three domains of life, as well as other viruses. Sequencing viral metagenomes from various environments has revealed that viruses are the most numerous and diverse inhabitants on Earth (Breitbart and Rohwer 2005; Angly et al. 2006; Kristensen et al. 2010). In addition to their role in major evolutionary transitions (Forterre 2006; Koonin et al. 2006) and control over host populations, the discovery of many fragments of viral genomes integrated in the genome of their eukaryotic host has recently opened new avenues to further our understanding of viral evolution and of host–virus interactions (Feschotte and Gilbert 2012).
The nucleocytoplasmic large DNA viruses (NCLDV, proposed order Megavirales) form an apparently monophyletic order of highly complex double-stranded DNA viruses, which are major parasites of a variety of eukaryotes (Iyer et al. 2006; Colson et al. 2013). They comprise by far the largest known viruses with genomes scaling from a hundred kilobases (kb) to 2.5 megabases (Philippe et al. 2013), including specimens with capsids of up to 1.5 μm in length (Legendre et al. 2014). So far, about 50 NCLDVs infectious to eukaryotic algae and aquatic protists have been characterized. It is anticipated that all the ∼350,000 marine algal species might have one or more specific viruses (Short 2012), and viral metagenomic studies have highlighted the existence of a huge diversity of marine viruses (Angly et al. 2006), with the NCLDV as the second most abundant virus group after bacteriophages (Kristensen et al. 2010). The largest known specimens of NCLDVs infect Acanthamoeba spp., a genus of cosmopolitan free-living amoebas inhabiting aquatic environments (Aherfi et al. 2016). To date, five phylogenetically related clades of NCLDVs, including giant viruses with genome >500 kb have been isolated from co-culture with Acanthamoeba spp., namely, Mimiviridae, Marseilleviridae, pandoraviruses, Pithovirus sibericum, and Mollivirus sibericum (Raoult et al. 2004; Boyer et al. 2009; Philippe et al. 2013; Legendre et al. 2014, 2015; Reteno et al. 2015). A smaller double stranded DNA virus, the virophage, was shown to co-infect Acanthamoeba spp. together with a mimivirus (La Scola et al. 2008). Thus, Acanthamoeba spp., and especially the Neff strain, is the most prolific model hosts in the discovery of new giant virus families, though other large or giant DNA viruses have been isolated from various aquatic protists and algae.
The Ectocarpus siliculosus virus ESV-1 is a member of NCLDVs that has a lysogenic cycle and integrates the genome of its filamentous brown alga host possibly through an active mechanism involving an integrase activity (Delaroque and Boland 2008). The integrated ESV-1 genome remains latent in vegetative cells but its transcription is reactivated in cells of the reproductive organs under certain stress conditions. A lysogenic life cycle has never been evidenced in other NCLDVs. However, recent genomic studies showed that NCLDV insertions in eukaryotic genomes are more common than previously thought. A number of sequenced eukaryotic genomes were found to contain DNA fragments of various sizes originating from NCLDVs (i.e., up to 400 kb in Hydra magnipapillata; see Filée 2014), but the mechanism of integration and the role of the inserts in virus or host biology remained elusive (Maumus et al. 2014; Sharma et al. 2014; Wang et al. 2014; Blanc et al. 2015). Strikingly, most of the eukaryotes containing vestiges of virus insertions were not previously known as to be infected by NCLDV, including land plants and cercozoan algae (Maumus et al. 2014; Blanc et al. 2015). Paradoxically, only very limited data exists regarding viral integration in organisms that are used as model to study host/NCLDV interactions including Neff, the host of most giant viruses isolated so far. Initial analysis of the 42-Mb Neff genome revealed extensive lateral gene transfers (LGT) with bacteria and to a lower extent with archaea and viruses, including NCLDVs (Clarke et al. 2013). By searching giant virus core genes in sequenced eukaryotic genomes, Filée (2014) identified five gene copies encoding major capsid proteins in the Neff genome. These initial findings prompted us to perform an in-depth study of gene exchanges between amoebas and their viruses.
Material and Methods
Sequence Accessions
Protein and genome sequences of selected amoebozoas and viruses were downloaded from the Genbank assembly database using the following accession numbers: A. astronyxis (GCA_000826245), A. castellanii strain Ma (GCA_000826485), A. castellanii strain Neff (GCA_000313135), A. culbertsoni (GCA_000826265), A. divionensis (GCA_000826405), A. healyi (GCA_000826305), A. lenticulata (GCA_000826285), A. lugdunensis (GCA_000826425), A. mauritaniensis (GCA_000826465), A. palestinensis (GCA_000826325), A. pearcei (GCA_000826505), A. polyphaga (GCA_000826345), A. quina (GCA_000826445), A. rhysodes (GCA_000826385), A. royreba (GCA_000826365), D. discoideum (GCA_000004695), E. histolytica (GCA_000208925), Mollivirus (GCF_001292995), and Pandoravirus (GCF_000911955).
Neff transcriptome datasets were downloaded from the Genbank sequence read archive (SRA) using the following accessions: SRR611709, SRR611787, SRR611788, SRR611789, SRR611790, SRR611791, SRR611792, SRR611793, SRR611795, SRR611796, SRR611797, SRR957287, SRR957291, and SRR957297.
Sequence Analyses
To identify genetic exchanges between viruses and Neff, the 15,655 Acanthamoeba annotated proteins (Clarke et al. 2013) were aligned against the Genbank NR (non-redundant) protein database using the BLASTP program and an E-value threshold of 0.001. Because genes of viral origin may be overlooked by gene prediction programs in a eukaryotic genetic background, an additional set of 6,674 ORFs (>100 codons) located between Neff annotated genes was also compiled. The translation products of these ORFs were treated in the same way as the annotated proteins in subsequent analyses. A subset of candidate Neff genes that might have been exchanged with viruses was delineated using an alignment score plot approach (Maumus et al. 2014; Blanc et al. 2015). For each Acanthamoeba protein, the best BLAST score obtained for viral matches were plotted against the best BLAST score obtained for cellular matches. Altogether, 789 proteins had a match in viruses (irrespective of its ranking in the BLASTP result report). For 267 proteins listed in supplementary dataset S1, Supplementary Material online, the alignment score of the match against viruses was higher than the alignment score of the match with cellular organisms. The corresponding genes will be hereinafter referred as to LGT markers. The same procedure was used to identify potential LGT markers in the Entamoeba histolitica and Dictyostelium discoideum genomes. The 267 Neff LGT markers were organized into gene clusters by means of the MCL program (inflation = 2.0) (Coppe et al. 2006), after aligning their protein products using BLASTP. We also compiled a set of 2,355 Neff genes whose protein products had a best match in amoeba species distinct from Acanthamoeba species. This gene set, assumed to contain sequences vertically inherited from an amoebal ancestor, was used as reference in subsequent analysis. Transcriptional activity of Neff genes was determined by mapping transcriptome sequence reads on the Neff genome using the Bowtie 2 program with default parameters (Langmead and Salzberg 2012). The physical location of LGT marker genes across the Neff genome assembly was analyzed using the REEF program (Coppe et al. 2006). A window of 30Kb was slided along the Neff genome assembly with a step of 5 kb. The statistical significance of an excess of LGT marker genes in each window was tested using the Hypergeometric Distribution. We only considered windows that contained at least three LGT markers. The False Discovery Rate was set to 5%.
Phylogenetic Reconstruction
Phylogenetic trees were constructed using the following general pipeline: homologous sequences were first searched in the NR database using the BLAST EXPLORER tool (Dereeper et al. 2010). Multiple-sequence alignment of homologous proteins was then performed using the MAFFT program (Katoh et al. 2005). We removed alignment positions containing >30% gaps. Phylogenetic reconstruction was performed using the PHYML program with the LG + G + F model of amino acid substitution (Guindon et al. 2010). Statistical support for branches was estimated with the approximate likelihood-ratio test. Sequences, alignments and phylogenetic trees of the reference proteins of LGT marker clusters are available in supplementary dataset S2, Supplementary Material online.
Nucleotide Composition Analysis
We used a compositional deviation index (CDI) to analyze the nucleotide composition of coding sequences. The CDI score reflects how much the nucleotide composition of a Coding DNA Sequence (CDS) deviates from that of a reference set of Neff coding sequences. The calculation of the CDI was described in details by Jeanniard et al. (2013). The expectation is CDI = 0 for ORFs with nucleotide compositions that fit with the model for Neff coding nucleotide sequences, whereas ORFs whose nucleotide composition significantly deviates from the model shall have CDI ≠ 0. A five-order non-homogeneous Markov chain model of nucleotide frequency for Neff coding sequences was constructed from a training set containing the 2,355 reference Neff CDSs (supplementary dataset S3, Supplementary Material online).
Molecular Evolution
To investigate the selective constraints acting of Neff CDSs, we first identified and annotated homologous sequences in sequenced Acanthamoeba species genomes using the following procedure: Neff proteins were aligned against the Acanthamoeba species genomes using the TBLASTN program. The top-5 best matching genomic regions and the Neff proteins were passed to the GENEWISE program (Birney et al. 2004) to extract Acanthamoeba species coding sequences. Putative orthologs between Neff genes and Acanthamoeba species coding sequences were identified using the reciprocal best blast hit criterion. A putative Acanthamoeba ortholog could be identified for only 151 (57%) of the 267 LGT marker clusters. In contrast 2,167 (>92%) of the 2,355 Neff reference genes had an identified Acanthamoeba ortholog. The low rate of ortholog identification for LGT markers may be due to a fraction of the viral gene acquisitions that occurred specifically in the Neff lineage and were therefore not shared with the other Acanthamoeba species. Pairs of orthologous coding sequences between Neff and an Acanthamoeba species were aligned on a codon basis using the MAFFT program (Katoh et al. 2005) and an in-house perl script. The resulting alignments were passed to the CODEML program (Yang 2007) to compute the ω = Ka/Ks ratio using the pairwise runmode.
Polymerase Chain Reaction
PCR amplifications of selected virus-like genes were performed using the primers listed in Table S1. PCR reactions were performed using 1 U GoTaq (Promega), 1× GoTaq Flexi Buffer (Promega), 2.5 mM MgCl2, 0.2 mM dNTPs, and 0.4 μM of each forward and reverse primer. A single PCR amplicon of expected size was obtained for each PCR reaction and sent to Eurofins (Germany) for bidirectional sequencing.
Results
Evidence for LGTs between Amoebas and Viruses
Two hundred and sixty seven (267) Neff proteins had a better alignment score with a viral match than with a cellular match, including 103 proteins that only have a detectable match in viruses, but none in cellular organisms. These proteins depicted by dots below the diagonal in the similarity score plot shown in figure 1A, are thus likely to share preferential phylogenetic affinity with virus homologs, suggestive of potential LGT events between viruses and amoeba. The corresponding genes were therefore considered as LGT markers. The number of 267 LGT markers is substantially higher than the 71 Neff genes with best BLAST hits in viruses identified by Clarke et al (2013). Two potential causes can explain this discrepancy. First, our list of LGT markers include 31 ORFs lying between annotated genes that were not considered by Clarke et al. Second, a number of new genomes of amoeba viruses have been published these last years (e.g., pandoraviruses, Mollivirus, and Pithovirus), all of which encode many gene families that were not represented in databases when the Clarke et al.’s study was performed. Thus, LGTs involving those new gene families could not be evidenced at that time. Third, the E-value threshold used for BLAST searches in the present study (E-value < E−3) is less conservative than that used in the Clarke et al.’s study (E-value < E−10). As a result, our estimate of the number of LGT markers may be viewed as an upper bound. Applying a more conservative E-value (<E−10) would have identified 238 LGT markers, which is also significantly higher than the 71 of Clarke and colleagues (supplementary fig. S1, Supplementary Material online). Thus, the major reason for the increased number of LGT markers between the Clarke’s study and our study appears to be a greater availability of reference virus genomes in database.
Fig. 1.—
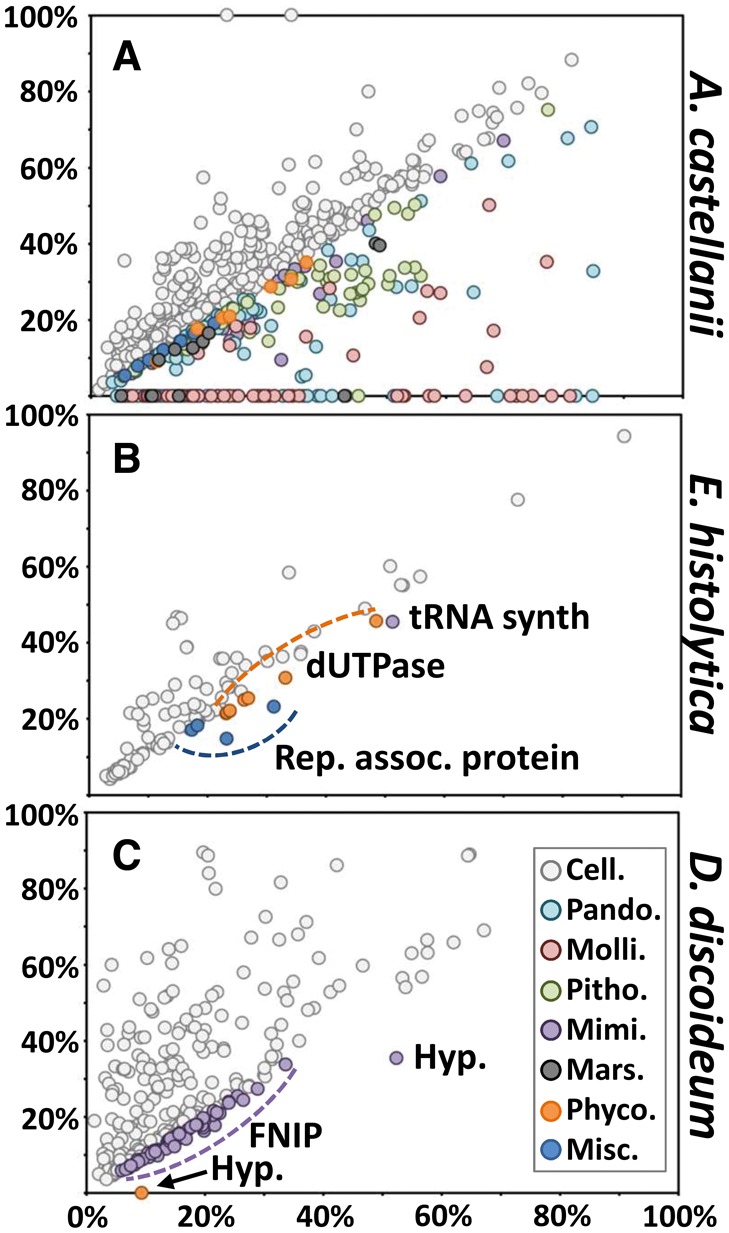
Similarity plots of amoeba proteins against viruses and cellular best hits. Circles represent relative BLASTP scores of amoeba proteins aligned against their best cellular hits in the NR database (y-axis) and their best viral hits (x-axis). When no cellular hit was recorded whereas a viral hit was obtained, the cellular score was set to zero. BLAST scores were normalized by dividing them by the score of the alignment of the query sequence against itself. Circles are colored according to the origin of the best overall scoring hit. Cell.: cellular organisms, Pando.: Pandoraviridae, Molli.: Mollivirus, Pitho,: Pithovirus, Mimi.: Mimiviridae, Mars.: Marseilleviridae, Phyco.: Phycodnaviridae, Misc.: miscellaneous DNA viruses. (A) Similarity plot of A. castellanii. (B) Similarity plot of E. histolytica. (C) Similarity plot of D. discodeum.
The significance of these observations can be appreciated by comparing the Neff score plot to those obtained for two other sequenced Amoebozoa, namely, Entamoeba histolytica and Dictyostelium discoideum. Fourteen E. histolytica and 61 D. discoideum proteins were represented by dots below the diagonal, which is substantially smaller than for Neff. Phylogenetic reconstruction confirmed that the corresponding genes likely underwent lateral transfer between viruses and amoebas. For example, members of the Entamoeba genus contain different copy numbers of a dUTPase gene (eight copies in E. histolytica represented by orange dots in fig. 1B) which is most closely related to homologs in phycodnavirus. The phylogenetic tree shown in supplementary figure S2, Supplementary Material online suggests that the dUTPase gene of a large DNA virus was captured by an Entamoeba ancestor. The same amoeba genus contains duplicated copies of a replication associated protein gene (four copies in E. histolytica represented by light blue dots in fig. 1B) most likely captured from a circular ssDNA virus (supplementary fig. S3, Supplementary Material online). The phylogenetic tree of tyrosyl-tRNA synthetases (supplementary fig. S4, Supplementary Material online) suggests an alternative scenario where the amoeba gene was captured by a member of Mimiviridae. This event is reflected by a purple dot below the diagonal in figure 1B. D. discoideum encodes 60 paralogous FNIP repeat-containing proteins which have a highest similarity with Mimiviridae homologs, resulting in a cloud of purple dots in figure 1C. Phylogenetic reconstruction suggests that at least two amoebal gene copies have been independently captured by early members of Mimiviridae (supplementary fig. S5, Supplementary Material online). In addition, members of the Dictyostelium genus contain a hypothetical protein gene that has been captured by a common ancestor of Mimivirus and Megavirus (supplementary fig. S6, Supplementary Material online; purple dot in fig. 1C). D. discoideum encodes another hypothetical protein (orange dot in fig. 1C) that has detectable homologs only in chloroviruses (Phycodnaviridae) also suggesting LGT between this amoeba and a large DNA virus. Altogether, these results indicate that both the E. histolytica and D. discoideum genomes provide evidence of LGT with viruses but in a much more limited amount in comparison to Neff. This suggests that these amoebal lineages have had direct contacts with large DNA viruses at some point in their evolution, even if E. histolytica and D. discoideum have not been previously shown to be infected by large DNA viruses. However, it is possible that the identified LGTs are vestiges of ancient association between viruses and their amoebal ancestors; these associations may no longer exist in extant species. E. histolytica and D. discoideum will not be considered further in subsequent analysis.
The 267 Neff LGT markers were organized into 146 gene clusters based on shared sequence similarity. The majority of the clusters encode proteins with unknown or generic functions (supplementary dataset S1, Supplementary Material online). However, some LGT markers encode proteins involved in virion structure and assembly—i.e., seven copies of the major capsid protein and 1 copy of the DNA packaging ATPase—strongly suggesting that these genes have a viral origin. Other genes commonly found in large DNA virus genomes are also present in the Neff genome assembly, including genes encoding a viral-type RNA ligase, an endonuclease, a lipase type-3, an mRNA capping enzyme and a transposase. The transcriptional activity of LGT marker genes was investigated by analyzing publicly available RNA-seq transcriptomic datasets generated from Neff cells grown under a variety of culture conditions. Approximately half of the LGT markers (i.e., 52% or 140 genes) showed virtually no transcription (i.e., RPKM < 0.5) in these transcriptomes. For comparison, only 6.7% (i.e., 805 out of 12,008) of those genes that have a best match in eukaryotes (i.e., likely vertically inherited) had RPKM < 0.5, suggesting that genes involved in LGT with large DNA viruses are more likely to be transcriptionally silent either because they are expressed in very specific conditions not covered by the investigated transcriptomic datasets or because they are no longer functional or silenced by the amoeba.
Origins of LGTs
The taxonomic distribution of protein best matches for the 267 Neff LGT markers is shown in figure 2. Remarkably, the great majority of them (94%) belong to viral families that contain amoeba-infecting viruses, including Pandoraviridae, Mimiviridae, Marseilleviridae, Pithovirus, and Mollivirus. More than a third of the protein best matches are encoded by pandoraviruses, the largest ever discovered amoeba-infecting viruses. Overall, the origin of the viral protein best matches is consistent with preferential genetic exchanges between partners that have intimate contacts in the environment. However, 16 best matches are encoded by members of diverse DNA virus families that are not known to infect amoeba. They include bacteriophages, Iridoviridae, Poxviridae, and Phycodnaviridae (supplementary dataset S1, Supplementary Material online). Two alternative hypotheses can explain their occurrence in the Neff genome assembly. First, Clarke et al. (2013) suggested that a relatively high number of Neff genes were acquired from bacteria. Thus, bacteriophage genes can arise from fragments of bacteria genomes containing prophage elements that were incorporated into the Neff genome. Alternatively, viral genes can result from the integration of DNA of as yet unknown amoeba-infecting viruses that share conserved genes with amoeba-noninfecting viruses.
Fig. 2.—
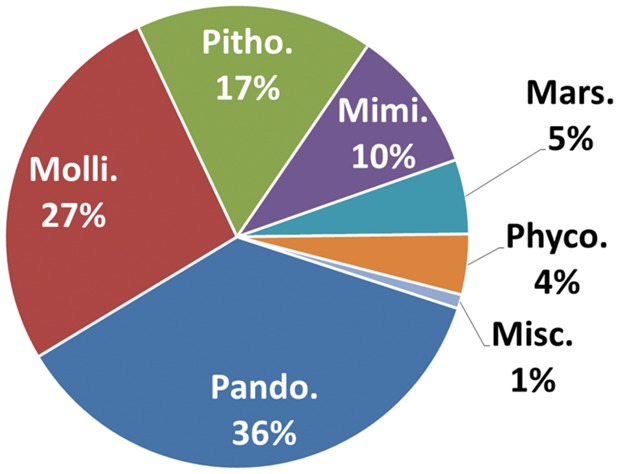
Taxonomic distribution of the viral protein best matches of the 267 A. castellanii LGT markers. Pando.: Pandoraviridae, Molli.: Mollivirus, Pitho,: Pithovirus, Mimi.: Mimiviridae, Mars.: Marseilleviridae, Phyco.: Phycodnaviridae, Misc.: miscellaneous DNA viruses.
The genetic transfers highlighted by LGT markers may have occurred in two alternative directions: from amoeba to virus or from virus to amoeba. To distinguish between the two alternatives, we performed a phylogenomic study of the 146 LGT marker clusters. However, phylogenetic reconstruction for NCLDV proteins can be challenging because viral sequences may evolve rapidly and/or emerge early in the phylogeny, generating long branches and making phylogenetic trees often poorly resolved. This issue may be even reinforced for viral inserts containing decaying pseudogenes that diverge at an accelerated rate. Furthermore, the prediction of LGT directionality requires conservative criterions for the interpretation of phylogenetic trees: schematically, eukaryote-to-virus LGT is inferred when a viral sequence emerges from within a clade of closely related eukaryotes, whereas virus-to-eukaryote LGT is predicted when a eukaryotic sequence emerges from within a viral clade. These criterions were used for interpreting the phylogenetic trees below. However, given the small numbers of sequenced NCLDVs and amoeba species, we are often confronted with a lack of homologs. For all these reasons, only 29% (42) of the 146 LGT marker clusters could be assigned to a LGT scenario based on phylogenetic reconstruction (supplementary dataset S2, Supplementary Material online). Important nodes in some phylogenetic trees were not highly statistically supported and potential hidden paralogy may also blur phylogenetic relationships. Thus, erroneous interpretations of the evolutionary scenario may have punctually happened. Nevertheless, table 1 shows a clear contrast in the counts of interpreted phylogenetic scenarios between transcribed and nontranscribed LGT markers.
Table 1.
Summary of Phylogenetic Tree Interpretations for Families of LGT Markers
| LGT marker family category1 | LGT: Neff->Virus2 | LGT: Virus->Neff3 | Only viral homologs4 | Inconclusive phylogenetic trees |
|---|---|---|---|---|
| Transcribed (52) | 15 (28%) | 5 (9%) | 11 (21%) | 22 (42%) |
| Non-transcribed (93) | 2 (2%) | 20 (22%) | 52 (56%) | 19 (20%) |
1LGT marker families were divided between the transcribed and nontranscribed categories according to the RPKM value of the longest gene in the family.
2,3Trees supporting LGT from amoeba to virus or virus to amoeba, respectively.
4LGT marker families having only homologs in a single viral species.
Compared with transcribed LGT markers, nontranscribed LGT markers had a smaller percentage of trees supporting LGT from amoeba to virus (2% vs. 28% for transcribed markers) and a higher percentage of trees supporting LGT from virus to amoeba (22% vs. 9%). Moreover, a higher number of nontranscribed LGT marker clusters only had homologs in a single viral species (56% vs. 21%); these genes that are more likely to originate from viruses. Thus, although our phylogenetic analysis only encompasses a subset of the LGT markers, these results suggest that nontranscribed LGT markers are more likely to have been acquired by the amoeba.
One of the LGT marker clusters provides an eloquent instance of the intense evolutionary history of interactions between viruses and amoebas. This cluster comprises 60 genes encoding nucleotidyl cyclases consisting of an extracellular receptor domain separated by a transmembrane helix from an intracellular cyclase domain flanked by two serine/threonine kinase domains. This domain configuration, possibly involved in environmental sensing, is present in a number of the amoeba-infecting giant viruses but thus far appears unique for a cellular organism (Clarke et al. 2013). Phylogenetic analysis suggests a eukaryotic origin of the kinase domain, followed by multiple ins and outs between eukaryotes and viruses, involving at least one LGT from eukaryote to virus (initial acquisition by viruses) and two LGTs from viruses to amoeba (supplementary fig. S7, Supplementary Material online). The high level of intertwining between viral and Acanthamoeba sequences suggests that additional LGTs between the two lineages have likely taken place.
Compositional Analysis
The directionality of LGT was also examined using an alternative approach. A compositional analysis relies on the species-specific oligonucleotide frequency of sequences, which can be used as a genomic signature (Dufraigne et al. 2005). Since DNA transfers originate from species that most likely have a compositional signature different from that of the recipient species, significant deviation of a signature between coding sequences (CDS) and the rest of the genome may highlight recently transferred DNA. However, because the nucleotide composition of foreign genes eventually converges toward that of the recipient genome, the compositional analysis approach is generally limited to spotting recently acquired genes. This latter approach was applied here by first reconstructing a five-order nonhomogeneous Markov chain model of nucleotide frequency from a reference set of 2,355 Neff CDSs. This model was used to compute a compositional deviation index (CDI) for sets of Neff CDSs.
The distributions of CDI values shown in figure 3 were not significantly different between the reference CDSs used to construct the oligonucleotide frequency model and the subset of transcribed LGT markers (i.e., Wilcoxon rank sum test P = 0.13). This result suggests that transcribed LGT markers globally have the same genomic signature as bona fide vertically inherited genes. This supports the hypothesis that most transcribed LGT markers have a eukaryotic origin and were lately captured by viruses through LGT (amoeba-to-virus scenario). Alternatively, some transcribed LGT markers may have a viral origin but were captured by an amoeba ancestor at a sufficiently remote period so that they have had enough time to accumulate mutations and adopt the specific nucleotide composition of the amoeba genome. This virus-to-amoeba scenario implies relatively ancient transfer events. Phylogenetic trees of transcribed LGT markers supported either one of the two evolutionary scenarios. For instance, the topologies of phylogenetic trees for the chorismate synthase, major facilitator transporter subfamily protein and translation initiation factor eIF-2B—which are encoded by transcribed LGT marker genes—are compatible with gene acquisitions by viruses (supplementary fig. S8–S10, Supplementary Material online). In contrast, transcribed genes encoding Zinc finger domain proteins, Dyptype peroxidase superfamily proteins, and three clusters of unknown proteins produced phylogenetic trees supporting virus-to-amoeba gene transfers (supplementary fig. S11–S15, Supplementary Material online). The fact that genes captured from viruses are transcribed suggests that they may have acquired a new function in the amoeba. Remarkably, one of unknown protein clusters is broadly distributed among large DNA viruses but has no recognizable counterpart in cellular organisms outside of Neff (supplementary fig. S13, Supplementary Material online), suggesting that its origin is deeply anchored in the viral world and its function was initially involved in virus-specific processes. Thus, these data provide examples of genes that probably emerged in viruses and have subsequently contributed to the gene repertoire and molecular functions of the amoeba.
Fig. 3.—
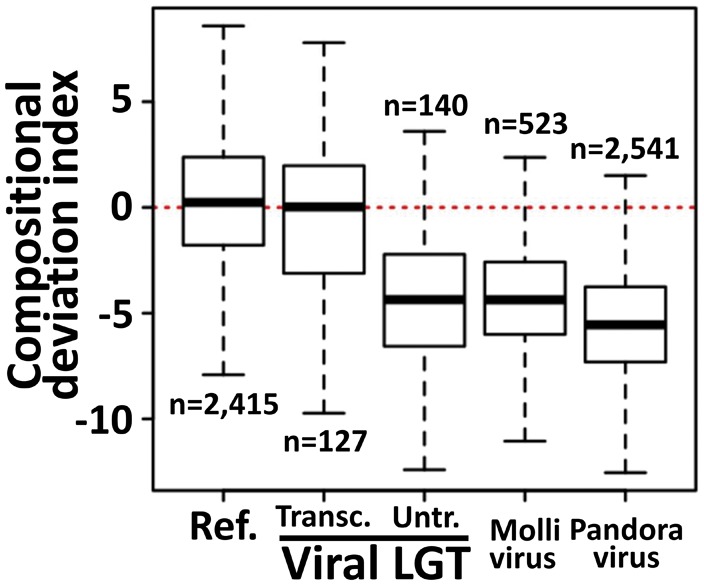
Box-plot representation of the distributions of compositional deviation indexes (CDI) for Neff gene categories. The “Ref” category includes genes used as reference in the calculation of the nucleotide frequency model; the “Transc.” and “Untr.” gene categories include viral LGT marker genes that are transcribed and untranscribed, respectively. The Mollivirus (sibericum) and Pandoravirus (salinus) categories include the entire sets of annotated genes in the respective genomes. The number of genes in a category is given above or below the box-plots. The first and third quartiles of the distributions are represented by the lower and upper edges of the boxes. The thick horizontal lines within boxes indicate the median of the distributions. The lines extending vertically from the boxes indicate variability outside the first and third quartiles as defined in the default version of the R boxplot function.
In contrast, the distributions of CDI values obtained for untranscribed LGT marker genes differed significantly from the reference distribution and the transcribed LGT marker distribution (Kruskal–Wallis rank sum test P < 2.2E−16 and Wilcoxon rank sum test P < 2.2E−16 for each pairwise combination between the untranscribed LGT marker set and the two other sets). Globally, untranscribed LGT markers had lower CDI values meaning that their nucleotide composition tends to exhibit a poorer fit to the Neff oligonucleotide frequency model. For comparison, the CDI distributions obtained for the coding sequences of two Acanthamoeba viruses, namely, Pandoravirus and Mollivirus, were also visually markedly different from the reference distribution, with again a bias toward lower CDI values (Kruskal–Wallis rank sum test P < 2.2E−16 and Wilcoxon rank sum test P < 2.2E−16 for pairwise comparisons). Altogether, our analysis suggests that untranscribed LGT markers include an excess of genes that have been captured from genomes with distinct nucleotide compositional biases and that these events were sufficiently recent so that the differences in nucleotide composition with the Acanthamoeba genome are still visible.
We also compared the distributions of CDI values between LGT markers assigned to the virus-to-cell and cell-to-virus scenarios in table 1. This comparison confirms there is a general agreement between the two methods for predicting LGT directionality: LGT markers inferred as captured from virus tended to have lower CDI values than LGT markers inferred as vertically inherited from eukaryotes (supplementary fig. S16, Supplementary Material online; Wilcoxon rank sum test P < 0.016). However the difference between the two distributions is much less marked than between the CDI distributions of transcribed VS untranscribed LGT markers, possibly because the nucleotide composition of some anciently acquired viral inserts has converged to that of Neff. Interestingly, LGT markers that only have viral homologs in sequence databases have markedly lower CDI values, further supporting the hypothesis that most of them arose from viruses relatively recently.
Selective Constraints
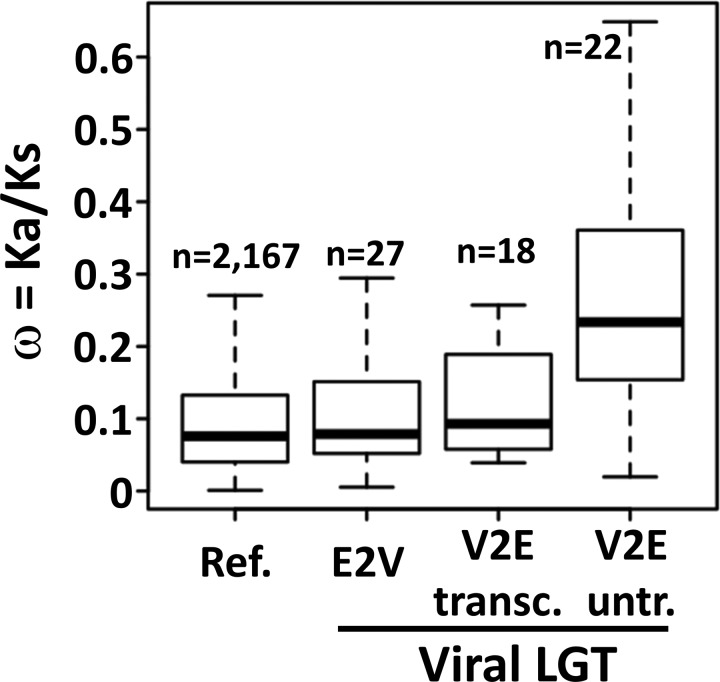
To investigate the selective constraints acting on inserted viral sequences we analyzed the ratio of nonsynonymous to synonymous changes, denoted ω = Ka/Ks, calculated between Neff coding sequences and their putative orthologous sequences identified in other sequenced Acanthamoeba species. The ω ratio is a classical measure of the magnitude and direction of selective constraints acting on coding sequences, with ω = 1, <1, and >1, indicating neutral evolution, purifying selection, and positive diversifying selection, respectively (Yang 2007). The distributions of ω ratios were compared between different sets of Neff genes. Figure 4 shows that Neff reference genes (Ref.), LGT markers transferred to virus (E2V)—all of which have a vertical origin—and the transcribed fraction of LGT markers acquired from viruses (V2E transc.) tended to have smaller ω values (median ω = 0.0758, 0.0788 and 0.09295, respectively) than untranscribed LGT markers acquired from viruses (V2E untr.; median ω = 0.2336). These results suggest two important characteristics of the evolution of inserted viral genes. First, we provide evidence that the subset of inserted viral genes that are transcriptionally active tended to evolve under a purifying selection regime similar in strength to that of vertically inherited genes, which mostly comprise functional genes. This support the hypothesis that at least a fraction of the transcribed viral genes assumed a biological function beneficial for the Neff cell that has been preserved by natural selection. In contrast, the globally higher ω ratios obtained for untranscribed viral genes suggest that these latters tended to evolve under more relaxed selective constraints than indigenous Neff genes and transcriptionally active viral genes. This outcome can arise if following their insertion, viral coding sequences assumed no functional role in the Neff cell and consequently diverged almost freely from selective constraints. However, the fact that a majority of genes had ω ≠ 1 indicates that the viral coding sequences did not probably evolve under absolute no selective constraints or mutational bias.
Fig. 4.—
Box-plot representation of the distributions of ω ratios for Neff gene categories. The “Ref.” category corresponds to the reference Neff gene set; E2V refers to LGT markers assigned to a eukaryote-to-virus scenario by phylogenetic analysis (see table 1, irrespective of the transcriptional status); The V2E category refers to LGT markers assumed to originate from viruses. This later gene set results from the merging of LGT markers assigned to the virus-to-eukaryote scenario by phylogenetic analysis and LGT markers with only viral homologs (table 1). The V2E category was further divided into transcribed (V2E transc.) and untranscribed (V2E untr.) gene sets. See the figure 3 legend for description of the boxplot graphs.
Viral Genomic Islands
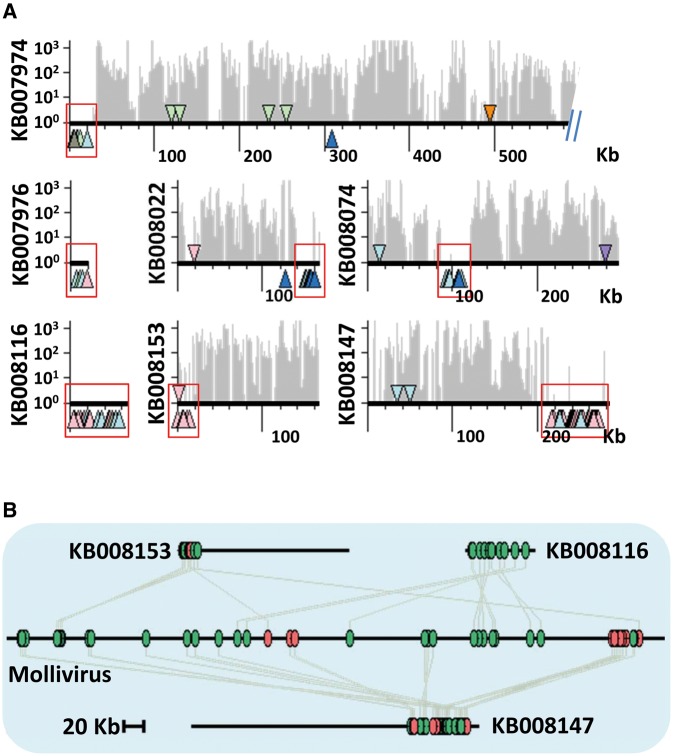
A statistical analysis of gene positions in the Neff genome based on a Hypergeometric Distribution (Coppe et al. 2006) identified seven genomic clusters containing a significant excess of LGT gene markers (regions highlighted by shaded boxes in fig. 5A). Remarkably these regions had virtually no transcriptional activity. This suggests that transcribed markers are randomly scattered across the genome, whereas a subset of the untranscribed markers tend to cluster at specific loci (supplementary fig. S17, Supplementary Material online). Some of these viral regions are contained in larger scaffolds and surrounded by amoebal genes—i.e., transcribed genes that have a best match in eukaryotes—as for example in scaffolds KB007974, KB008022, KB008074, KB008153, and KB008147. The size of the two largest viral regions on scaffolds KB008116 and KB008147 is approximately 60 Kb; however, their boundaries could not be precisely determined because no specific sequence features such as inverted terminal repeats or insertion site duplication marked the transition between the eukaryotic and viral DNA. Other viral regions encompass whole scaffolds. These scaffolds do not contain any eukaryotic genes and are entirely transcriptionally inactive (e.g., KB007976 and KB008116). The remaining untranscribed marker genes are scattered on other scaffolds, either isolated amid amoebal genes or forming smaller gene clusters, the size of which is below statistical significance. Their relative isolation may be explained by gene-scale lateral transfers or relocation of genes derived from larger viral regions.
Fig. 5.—
Viral regions of the Neff genome. (A) Schematic representation of scaffolds containing a significant local excess of untranscribed LGT markers. Scaffolds are represented by thick horizontal line. LGT markers are shown by triangles colored according to the taxonomy of the best protein match. The color code is the same as in figure 1. Transcribed and untranscribed genes are show above and below the scaffold line, respectively. The levels of transcription along scaffolds are shown by grey bars (logarithmic scale on the left hand side of the graph). (B) Residual gene co-linearity between Neff viral regions and the Mollivirus sibericum genome. Genes shared between Neff and Mollivirus genomes are shown by colored ovals and bound by a grey line. The red and green colors indicate genes on the same and opposite strands respectively.
Remarkably, three viral regions on scaffolds KB008116, KB008153 and KB008147 show residual gene co-linearity (i.e., series of homologous genes in the same or opposite order interrupted by co-linearity breakage) with the genome of Mollivirus sibericum (fig. 5B), a recently sequenced amoeba giant virus resurrected from a sample of 30,000 year old Siberian permafrost (Legendre et al. 2015). The three viral regions have a cumulated size of 133 KB (each region were delimited by the first and last genes matching a Mollivirus homolog) and are predicted to encode 153 ORFs, including 73 genes arising from the Neff genome annotation and 80 ORFs with >90 codons identified between annotated genes. Fifty-six of these ORFs (including 11 ORFs lying between annotated genes) were found to match 42 homologous genes in the Mollivirus genome with sequence similarities ranging from 23% to 85% identity at the protein level. Scaffold KB008147 carries a gene encoding a MCP (ACA1_363120) that has 64% sequence identity with the MCP of Mollivirus sibericum (see below). Furthermore, 29 genes shared between Mollivirus and the three viral regions have no identifiable homolog in public databases. Thus, these genes that were previously classified as orphan at the time of the first Neff genome analysis have now an identified viral origin. There is no overlap between the gene complements of the three viral regions suggesting that they correspond to distinct segments of a same genome. In addition, six ORFs matched with other NCLDVs (ie., pandoraviruses or Marseillevirus) and four more ORFs had a best match with cellular organisms (i.e., ACA1_363310, ACA1_324780, ACA1_134100, and ACA1_324960). Eighty-seven predicted ORFs had no match in the NR database, including 19 annotated genes (46–723 codons) and 68 intergenic ORFs (90–433 codons). These coding sequences may encode orphan proteins of the original donor virus and represent 57% of the viral region gene repertoire, a fraction comparable to the 64% orphan genes predicted in the Mollivirus genome (Legendre et al. 2015). Altogether, these results suggest that the A. castellanii genome contains DNA remains of a genome that originally belonged to a virus closely related to Mollivirus sibericum. This virus is known to begin its replication cycle by a nuclear phase where the virus early genes are transcribed by the host transcription machinery (Abergel and Claverie 2016). Thus, during the early replication phase, the genomes of the host and virus are in close physical contact, which may facilitate the integration of the viral DNA.
Altogether these observations indicate that the Neff genome contains large DNA regions harboring transcriptionally inactive genes of probable viral origin. These regions most likely result from the integration of fragments of virus genomes. Alternatively, the viral regions may have arisen by contamination during genome sequencing. However, two types of arguments invalidate this hypothesis. First, PCR amplification and sequencing of three tested virus-like genes—encoding a mRNA capping enzyme on scaffold KB008116 and two MCPs on scaffolds KB008022 and KB008147—confirmed the presence of these sequences in the Neff genome. Second, similarity searches and subsequent phylogenetic analyses for major capsid proteins and DNA packaging ATPases indicate that homologous viral genes are not only present in the Neff genome assembly but also in the genome assemblies of newly sequenced Acanthamoeba species that are closely related to Neff (see below). This indicates that the integration of the corresponding virus DNA occurred in an Acanthamoeba ancestor that existed before the split of the sequenced species.
Evidence for an Unknown Family of NCLDV Infecting Amoebas
Interestingly, the MCP phylogenetic tree shows that Neff contains two taxonomically distinct types of MCP genes (fig. 6A). One of these MCPs (i.e., ACA1_363120)—whose gene was confirmed by PCR amplification—is closely related to the MCP encoded by Mollivirus sibericum, further supporting the probable molliviral origin of the corresponding region (fig. 5B). Similarity search in other Acanthamoeba spp. genome assemblies failed to identify putative orthologs for this Mollivirus-like MCP gene, nor for the surrounding genes, suggesting that the insertion of the corresponding viral DNA fragment occurred in the Neff genome after its split with the other sequenced Acanthamoeba species. Two other Neff MCPs clustered together with closely related homologs in other sequenced Acanthamoeba species whose lineages emerged after the virus insertion event. Quite remarkably the Acanthamoeba proteins branched outside of MCP clades formed by diverse known families of Megavirales. This suggests that the original virus which donated a fragment of its genome was a member of a yet undiscovered Megavirales clade. Interestingly, the phylogenetic tree of packaging ATPases shows a similar topology (fig. 6B), with the amoebal proteins separated from the known Megavirales families. Although the ATPase and MCP protein genes lie on distinct Neff contigs, they might originate from the same virus. This hypothesis is further supported by the observation that orthologs of the ATPase and MCP genes lie on the same contig in the A. lenticulata genome assembly (i.e., contig id: CDFG01077627), suggesting that this genomic segment has conserved the organization of the original viral insertion. Thus, beside the five families of giant-viruses recently isolated from Acanthamoeba cocultures, our analysis provides compelling evidence for the existence of a sixth family of Megavirales probably infecting Acanthamoeba species; however, this suggestion awaits confirmation when a first specimen of this hypothetical new family will be isolated. Unfortunately, no close homologs for the new ATPase and MCP families could be found in public metagenomic datasets.
Fig. 6.—
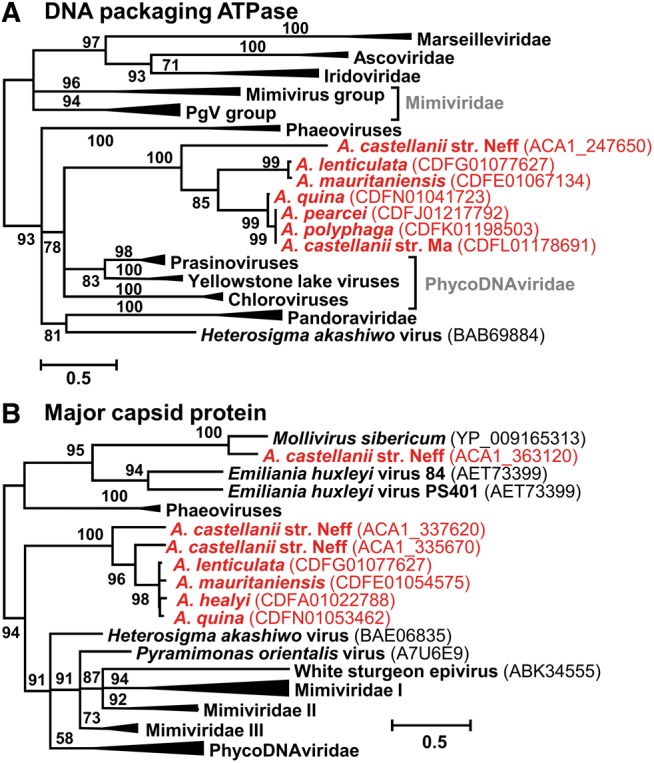
Maximum likelihood phylogenetic trees of conserved viral proteins. (A) DNA packaging ATPase. (B) Major capsid protein. Statistical supports for branch (approximate likelihood-ratio test) are given beside nodes in percentage. Branches with support less than 50% were collapsed. Scale bar indicate the number of substitution per sites. Sequences, alignments and phylogenetic trees are available in supplementary dataset S2, Supplementary Material online.
Discussion
The Fate of Viral Insertions
Our study provides compelling evidence that past interactions between large DNA viruses and amoebas have left recognizable traces in the host genome in the form of DNA regions reaching up to several tens of kb and encoding hallmark proteins of viruses, such MCPs and a DNA packaging ATPase. Insertions of Megavirales DNA were initially identified in the genomes of various eukaryotes, including protists, algae and even some land plants (Cock et al. 2010; Filée 2014; Maumus et al. 2014; Sharma et al. 2014; Wang et al. 2014; Blanc et al. 2015), many of these organisms were actually not known to be infected by NCLDVs. By analyzing the genome of Neff—one of the most popular hosts for giant viruses—we confirm that viral DNA insertion in the host genome is a common feature of NCLDVs, and this phenomenon has substantially impacted genome evolution in the Acanthamoeba genus. However, we still do not know if viral DNA integration results from an active process (i.e., as a result of a virus-encoded integrase activity) or an accidental incorporation of viral DNA floating inside the cell (i.e., as a result of an aborted infection).
Neff LGT markers that clustered in large viral regions did not show evidence of transcription. Furthermore, they have a nucleotide composition significantly departing from that of bona fide Acanthamoeba genes, suggesting that they have been recently acquired by lateral gene transfer and did not have enough time to adopt the compositional signature of the recipient genome. We also provided evidence that transcriptionally inactive viral coding sequences tended to evolve under relaxed constraint suggesting that at least a substantial fraction of them may no longer be functional. Inversely a fewer number of genes with inferred viral origin were found to be transcriptionally active and exhibited evidence of strong purifying selection suggesting that they are functional. In contrast to untranscribed virus-like genes, they are scattered in the Neff genome and have a nucleotide composition resembling that of the rest of the genome, hinting at more ancient gene transfer events. Altogether, these observations suggest a model of evolution for inserted viral sequences: after insertion, viral genes probably get immediately transcriptionally inactive either because they are not recognized by the host transcription machinery or because they are actively silenced by the host. Thus, the most likely fate of recently acquired viral sequences is to evolve under relaxed selection much like junk DNA, with a progressive decay by accumulation of mutations and breaking down of large viral regions into smaller segments and genomic relocation through rearrangements such as inversion or translocation. However, in some probably rare events, an inserted viral gene can escape this fate by recovering transcriptional activity in its new cellular context—e.g., as a result of gene relocation close to a host regulatory sequence—and evolving a new function beneficial to the amoebal host. Following this scenario, a viral gene can eventually get fixed in the host population, leading to its domestication by a cellular lineage. On the long term, domesticated virus-like genes accumulate neutral and adaptive mutations leading their nucleotide composition to converge toward that of the host genome. Given the number of organisms in which NCLDV insertions have already been discovered, it is possible that the eukaryotic gene repertoire has been recurrently fertilized by large DNA viruses in such a way. Another hypothetical role for inserted virus sequences is to serve as template in sequence-based antiviral defense mechanisms such as for example small RNA-directed RNA degradation pathway or RNA-directed DNA methylation, two gene silencing pathways that play a critical role in plant and animal resistance against both RNA and DNA viruses (Wang et al. 2012; Tanguy and Miska 2013).
Viruses Shaped the Amoebal Gene Repertoire
All organisms sequenced so far contain typically 10–30% orphan genes (Khalturin et al. 2009), which are genes that have no recognizable homologs in sequence databases or are restricted to specific taxonomic level. Clarke and colleagues identified 4,156 orphan genes (27%) in the Neff genome (Clarke et al. 2013). Moreover, a substantial proportion of orphan genes are often not shared even between very closely related organisms, suggesting that they arose recently. Understanding the origin and function of orphan genes is one of the great challenges of biology. Although not directly addressing the question of how the gene first arose, HGT has also been suggested as a source for orphan genes, but it has only been barely quantified in orphan gene study (Wissler et al. 2013). Our studies and others demonstrated than insertion of viral DNA fragments are common in eukaryotic genomes, giving rise to new genes in the recipient genome. Although most of the viral genes may not be longer functional in their new cellular habitat, they include a prominent proportion of orphan genes that can be subsequently counted in orphan-gene studies. Thus, viral genome insertions should be considered as a potential mechanism to explain a sudden appearance of orphan genes, especially in certain lineages of protists and eukaryotic microalgae where this phenomenon appears to be relatively frequent. For example, 103 Neff genes only matched viral sequences, and these genes would have been considered as orphan a few years ago before Mimivirus and its fellow giant viruses were sequenced. In fact, the subset of Neff orphan genes with a viral origin is certainly larger than found in this study because the sources of the acquired genes have not yet been sequenced.
The widespread existence of nuclear copies of viral genes raises a new problem in phylogenetic studies, especially when wanting to trace the origin of virus genes (Forterre and Gaïa 2016). Different views circulate among virologists to explain gigantism in large DNA viruses. It has been suggested that Megavirales have an ancient cellular origin and evolved by reductive evolution from a more complex ancestral cellular genome (Raoult et al. 2004; Legendre et al. 2012; Abergel et al. 2015). Others think that viral gigantism is best explained by the gene-pickpocket theory, which postulates that most viral genes have been robbed from cellular hosts. In support to this view, some authors have produced phylogenic trees in which viral genes branch close to eukaryote homologs (Moreira and López-García 2009; Williams et al. 2011). Although not arguing in favor of one or the other evolutionary scenarios, our finding suggests that this type of phylogenetic argument should now be double-checked when inferring the origin of gene transfers. Given the small number of giant-virus genes with an identified host origin (Abergel et al. 2015), it is likely that the horizontal gene transfer flow from virus to Acanthamoeba was more important in numerical terms than from Acanthamoeba to virus.
Exploration of the Megavirales Biodiversity
The biodiversity of NCLDVs is thought to be immense however its characterization has progressed at a low pace until recently. Historically, isolation of large DNA viruses infecting eukaryotic algae or aquatic protists has proceeded by coculturing a host together with a virus sampled from the environment. In this experimental approach the eukaryotic host is chosen a priori for its capacity of being infected by a virus and adapted to lab culture prior virus isolation. Recently, the metagenomic approach has accelerated the rate at which new viruses are brought to light. However this approach suffers from two main shortcomings: first, viral sequences assembled from metagenomic data are generally short, encompassing often only a few genes at best. Second, the hosts of the identified viruses are unknown. The identification of viral insertions in eukaryotic genomes offers a third way in the characterization of NCLDV biodiversity that has some advantages over metagenomics. Although the remains of the original virus genome are often degraded, rearranged, and/or incomplete, it is not uncommon to find viral inserts encompassing several tens or even hundreds of kb. These large regions encoding up to several hundreds of viral genes can help to raise hypothesis on the original virus gene repertoire, metabolic capabilities and/or lifestyle, as well as providing molecular markers for phylogenetic analysis. Furthermore, the virus host is in all likelihood identified (i.e., the sequenced organism) and already adapted to cultivation in laboratory (except for genomes sequenced by the single cell sequencing approach). This offers the possibility to apply a classical co-cultivation method to fish out close relatives of the virus that gave rise to the viral inserts. Environment samples may be prescreened for presence of the desired virus by PCR amplification using primers designed from an inserted viral region or by alignment against metagenomic data. Our identification of DNA remains of what is possibly of a member a yet-undiscovered family of NCLDVs in genomes of Acanthamoeba spp. offers the opportunity to implement this strategy using a well-established system for giant virus isolation (i.e., co-culturing with A. castellanii or A. polyphaga).
Supplementary Material
Supplementary table S1, datasets S1–S2, and figures S1–S17 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Chantal Abergel and her team for kindly providing Neff DNA and lab space for PCR analysis. This work was partially supported by the A*MIDEX project (ANR-11-IDEX-0001-02) funded by the « Investissements d'Avenir » French Government program, managed by the French National Research Agency (ANR). The IGS laboratory is supported by the Centre National de la Recherche Scientifique and Aix-Marseille University. We acknowledge the use of the PACA-Bioinfo Platform, supported by IBISA and France-Génomique (ANR-10-INBS-0009).
Literature Cited
- Abergel C, Claverie J-M. 2016. Giant viruses diversity. Virologie 20:61–63. [DOI] [PubMed] [Google Scholar]
- Abergel C, Legendre M, Claverie J-M. 2015. The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiol Rev. 39:779–796. [DOI] [PubMed] [Google Scholar]
- Aherfi S, Colson P, La Scola B, Raoult D. 2016. Giant viruses of amoebas: an update. Front Microbiol. 7:349.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, et al. 2006. The marine viromes of four oceanic regions. PLOS Biol. 4:e368.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R. 2004. GeneWise and Genomewise. Genome Res. 14:988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Gallot-Lavallée L, Maumus F. 2015. Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc Natl Acad Sci U S A. 112:E5318–E5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, et al. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc Natl Acad Sci U S A. 106:21848–21853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278–284. [DOI] [PubMed] [Google Scholar]
- Clarke M, et al. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, et al. 2010. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature 465:617–621. [DOI] [PubMed] [Google Scholar]
- Colson P, et al. 2013. ‘Megavirales’, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 158:2517–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe A, Danieli GA, Bortoluzzi S. 2006. REEF: searching REgionally Enriched Features in genomes. BMC Bioinformatics 7:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaroque N, Boland W. 2008. The genome of the brown alga Ectocarpus siliculosus contains a series of viral DNA pieces, suggesting an ancient association with large dsDNA viruses. BMC Evol Biol. 8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie J-M, Blanc G. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufraigne C, Fertil B, Lespinats S, Giron A, Deschavanne P. 2005. Detection and characterization of horizontal transfers in prokaryotes using genomic signature. Nucleic Acids Res. 33:e6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 13:283–296. [DOI] [PubMed] [Google Scholar]
- Filée J. 2014. Multiple occurrences of giant virus core genes acquired by eukaryotic genomes: the visible part of the iceberg? Virology 466–467:53–59. [DOI] [PubMed] [Google Scholar]
- Forterre P. 2006. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 117:5–16. [DOI] [PubMed] [Google Scholar]
- Forterre P, Gaïa M. 2016. Giant viruses and the origin of modern eukaryotes. Curr Opin Microbiol. 31:44–49. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156–184. [DOI] [PubMed] [Google Scholar]
- Jeanniard A, et al. 2013. Towards defining the chloroviruses: a genomic journey through a genus of large DNA viruses. BMC Genomics 14:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TCG. 2009. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25:404–413. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Dolja VV. 2006. The ancient Virus World and evolution of cells. Biol Direct. 1:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, et al. 2008. The virophage as a unique parasite of the giant mimivirus. Nature 455:100–104. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, et al. 2015. In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci U S A. 112:E5327–E5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, et al. 2014. Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A. 111:4274–4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Arslan D, Abergel C, Claverie J-M. 2012. Genomics of Megavirus and the elusive fourth domain of Life. Commun Integr Biol. 5:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maumus F, Epert A, Nogué F, Blanc G. 2014. Plant genomes enclose footprints of past infections by giant virus relatives. Nat Commun. 5:4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, López-García P. 2009. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol. 7:306–311. [DOI] [PubMed] [Google Scholar]
- Philippe N, et al. 2013. Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science 341:281–286. [DOI] [PubMed] [Google Scholar]
- Raoult D, et al. 2004. The 1.2-megabase genome sequence of mimivirus. Science 306:1344–1350. [DOI] [PubMed] [Google Scholar]
- Reteno DG, et al. 2015. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol. 89:6585–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Colson P, Giorgi R, Pontarotti P, Raoult D. 2014. DNA-dependent RNA polymerase detects hidden giant viruses in published databanks. Genome Biol Evol. 6:1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short SM. 2012. The ecology of viruses that infect eukaryotic algae. Environ Microbiol. 14:2253–2271. [DOI] [PubMed] [Google Scholar]
- Tanguy M, Miska EA. 2013. Antiviral RNA interference in animals: piecing together the evidence. Nat Struct Mol Biol. 20:1239–1241. [DOI] [PubMed] [Google Scholar]
- Wang L, et al. 2014. Endogenous viral elements in algal genomes. Acta Oceanol Sin. 33:102–107. [Google Scholar]
- Wang M-B, Masuta C, Smith NA, Shimura H. 2012. RNA silencing and plant viral diseases. Mol Plant Microbe Interact. 25:1275–1285. [DOI] [PubMed] [Google Scholar]
- Williams TA, Embley TM, Heinz E. 2011. Informational gene phylogenies do not support a fourth domain of life for nucleocytoplasmic large DNA viruses. PLoS One 6:e21080.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E. 2013. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 5:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.