Abstract
Fluorescent proteins (FPs) are well known and broadly used as bio-imaging markers in molecular biology research. Many FP genes were cloned from anthozoan species and it was suggested that multi-copies of these genes are present in their genomes. However, the full complement of FP genes in any single coral species remained unidentified. In this study, we analyzed the FP genes in two stony coral species. FP cDNA sequences from Acropora digitifera and Acropora tenuis revealed the presence of a multi-gene family with an unexpectedly large number of genes, separated into short-/middle-wavelength emission (S/MWE), middle-/long-wavelength emission (M/LWE), and chromoprotein (CP) clades. FP gene copy numbers in the genomes of four A. digitifera colonies were estimated as 16–22 in the S/MWE, 3–6 in the M/LWE, and 8–12 in the CP clades, and, in total, 35, 31, 33, and 33 FP gene copies per individual shown by quantitative PCR. To the best of our knowledge, these are the largest sets of FP genes per genome. The fluorescent light produced by recombinant protein products encoded by the newly isolated genes explained the fluorescent range of live A. digitifera, suggesting that the high copy multi-FP gene family generates coral fluorescence. The functionally diverse multi-FP gene family must have existed in the ancestor of Acropora species, as suggested by molecular phylogenetic and evolutionary analyses. The persistence of a diverse function and high copy number multi-FP gene family may indicate the biological importance of diverse fluorescence emission and light absorption in Acropora species.
Keywords: reef-building corals, copy number variation, fluorescence diversity
Introduction
Fluorescent proteins (FPs), especially green fluorescent protein (GFP), are well known and broadly used as bio-imaging markers in molecular biology research. FP was initially isolated from jellyfish Aequorea victoria (Shimomura 1979; Field et al. 2006). FPs are excited by environmental light and emit longer wavelength fluorescence than the excitation light wavelength (Johnsen 2012). FP emission light is determined by its amino acid sequence (Field et al. 2006). Subsequently, divergent jellyfish FP homologs were cloned from anthozoan species (Matz et al. 1999), marine crustacean copepods (Shagin et al. 2004), and deuterostome chordate amphioxus species (Deheyn et al. 2007; Baumann et al. 2008; Bomati et al. 2009; Yue et al. 2016). These FPs are classified into four groups based on the color of the emitted light: cyan (CFP), green (GFP), yellow (YFP), and red (RFP) (Labas et al. 2002; Alieva et al. 2008). Non-fluorescent chromoprotein (CP) is also classified as a FP gene family member, on account of amino acid sequence similarity (Labas et al. 2002). The FPs of the same fluorescence class have emerged repeatedly and independently during coral evolution. It was reported that CFPs, YFPs, and RFPs have evolved several times in different lineages (Alieva et al. 2008). All known FPs from Acropora species were included in one of the three clades in the FPs from all corals (Alieva et al. 2008). The center of the light emission determinant of FPs comprises tripeptide “–X–Y–G–”, termed chromophore, where X varies in different FPs (Henderson and Remington 2005). FPs comprise a high proportion, 4.5–14%, of the total soluble protein content of FP-expressing anthozoan tissues (Leutenegger et al. 2007; Oswald et al. 2007). Because of this high cellular content, many biological roles have been proposed for FPs. They are considered essential for viability and may play photo-protective and antioxidant roles (Palmer et al. 2009; Roth and Deheyn 2013). However, the detailed biological function of FPs in corals remains elusive.
Multiple FP genes have been reported for a single species. In the amphioxus genome (Branchiostoma floridae), 16 unique GFP-like genes were present and this is the largest known FP gene family in a single organism to date (Bomati et al. 2009). Among anthozoans, four to seven separate genetic loci that encode CFP, GFP, and RFP genes were predicted in Montastraea cavernosa. These genes may have undergone gene conversion events (Kelmanson and Matz 2003). In the genus Acropora, one of the most abundant coral genera in coral reefs of the Indo-Pacific region (Veron 2000), several FP sequences were reported for a single species, as follows: in Anniella pulchra, two FP genes (one CFP and one CP) (D’Angelo et al. 2008); in Artocarpus nobilis, three FP genes (one GFP and two CFP); in Acropora aculeus, three FP genes (two GFP and one CP); and in Acropora hyacinthus, one FP gene (CP) (Alieva et al. 2008). In the studies of Acropora millepora, four (two GFP, one CFP, one RFP) (D’Angelo et al. 2008) and four (each of one GFP, CFP, RFP, and CP) (Alieva et al. 2008) FP sequences were isolated, but a homologous–paralogous relationship between sequences from the same emission light groups has not been identified. Eight copies of RFP genes were deduced based on exon 3 sequences in A. millepora (Gittins et al. 2015), but full-length coding regions were not determined. Analysis of the entire Acropora digitifera genomic nucleotide sequence revealed ten FP-like genes, but nine of these were truncated compared with a typical FP-coding sequence (Shinzato et al. 2012). Studies of anthozoan FP sequences indicated the possibility of high FP gene copy number in their genomes; however, the full FP gene complement in a single coral species is still unidentified.
In this study, we analyzed the FP genes from two stony coral species, A. digitifera that the genome sequences were decoded, and Acropora tenuis. A. tenuis is distantly related to A. digitifera and is located at the basal lineage in the genus Acropora (Fukami et al. 2000; Richards et al. 2013) with different habitats from A. digitifera (Suzuki et al. 2008). We found that A. digitifera encodes the largest set of FP genes and the multi-FP gene family has persisted during the evolution of Acropora species.
Materials and Methods
Specimen Collection and Species Identification
This study was approved by the Aquaculture Agency of Okinawa Prefecture (permits numbers 26–9 and 28–31). Five colonies of A. digitifera and one colony of A. tenuis were collected from field and subsequently maintained in Sesoko Station aquarium (Tropical Biosphere Research Center, University of the Ryukyus). Among five A. digitifera colonies, we did not find clear color differences at photograph level. Additionally, a small piece of one A. digitifera colony was collected from field and preserved in RNAlater (Waltham, MA, USA). These two species were identified based on morphology. We collected planula larvae from each of five colonies of A. digitifera and A. tenuis that were kept in separate aquariums. All specimen information is given in supplementary table S1, Supplementary Material online.
Live Coral Fluorescence Measurements in the Field
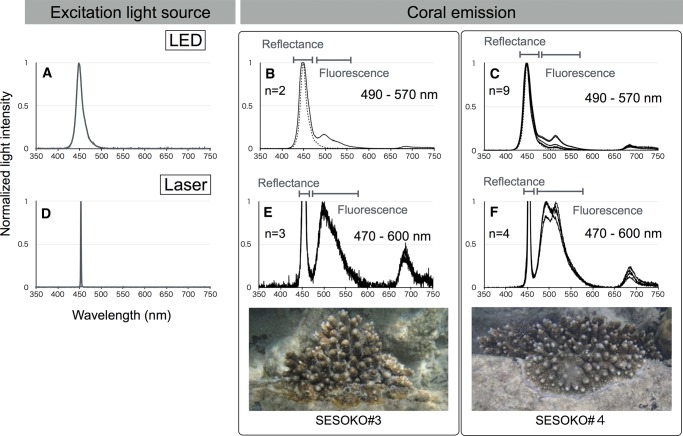
We measured light emission, including reflectance and fluorescence, from five A. digitifera colonies in the vicinity of Sesoko Station (Tropical Biosphere Research Center, University of the Ryukyus). We used two excitation illumination light sources: LED source, with spectrum peak at 448 nm, and laser source, with spectrum peak at 452 nm (fig. 1A and D). The distances of illumination of excitation lights and measurement probe from objects were 6 cm in LED and 1 cm in laser measurements. Measurements were performed in the dark, at night, to avoid the effect of sunlight. Each spectrum was recorded by Jaz Spectrometer (Ocean Optics, Dunedin, FL, USA). In all measurements, emission light longer than 660 nm was not considered as coral fluorescence, because chlorophyll a from the symbiotic dinoflagellate algae living within the coral tissues emits light spectra with a primary peak around 685 nm and a secondary peak at 730 nm (Moisan and Mitchell 2001; Mazel 2003).
Fig. 1.—
Live coral fluorescence measurements in the field. Excitation spectra of different light sources, LED (A) and laser (D). Horizontal and vertical axes indicate wavelengths (nanometers) and normalized light intensities, respectively. A. digitifera emission spectra (solid line) excited by LED (B, C) and laser (E, F) lights from two colonies with different fluorescence emissions (SESOKO#3 and SESOKO#4). LED excitation spectrum is shown by a dotted line (B, C).
RNA Extraction and Sequencing
Total RNAs were extracted from adult specimens of A. digitifera and A. tenuis, and a single A. digitifera larva, each of ∼50 individuals of A. digitifera and A. tenuis larvae, using TRIzol reagent (Thermo Fisher Scientific, MA, USA). RNA libraries were constructed from adult specimens of A. digitifera and A. tenuis, and each of ∼50 individuals of A. digitifera and A. tenuis larvae with NEBNext Poly(A) mRNA Magnetic Isolation Module and NEBNext Ultra RNA Library Prep Kit for Illumina (New England Bio Labs, MA, USA). Short DNA sequences (paired-end 100 bp) were determined from libraries by Illumina HiSeq2000 platform (RNA-seq). After removal of the adaptor sequences and low-quality reads, RNA-seq read assembly was performed using CLC genomic workbench (https://www.qiagenbioinformatics.com/) with auto-word size. Assembled sequences were used for searching a distantly related FP sequence to known Acropora FP genes.
Identification and Cloning of FP-like cDNA Sequences
To identify FP-like sequences, we selected the longest FP sequences (AdiFP2, AdiFP8, and AdiFP10, accession numbers BR000963, AB698751, and BR000970, respectively) from three different clades of A. digitifera FP-like sequences (Shinzato et al. 2012); however, AdiFP8 and AdiFP10 were shorter than a typical FP-coding sequence. To identify the possible previously undetermined 5′ terminal gene sequence, AdiFP8 and AdiFP10 were used to search RNA-seq reads for short sequences with high similarity (>90%) to 5′ termini of these two FP-like sequences. These short sequences were aligned with AdiFP8 and AdiFP10, and 5′ termini of the two FP-like genes were subsequently extended (see supplementary fig. S3, Supplementary Material online). We named these extended sequences AdiFP8L and AdiFP10L. To design primers, we mapped RNA-seq reads from A. digitifera and A. tenuis to reference sequences, AdiFP2, AdiFP8L, and AdiFP10L. Several primer sets were designed in the conserved regions that were identified based on the mapping results to PCR-amplify all FP-like sequences identified in RNA-seq reads.
cDNAs were synthesized from total RNA extracted from adult (n = 1) and larva (n = 1) specimens of A. digitifera, and adult (n = 1) and larvae (n = ∼50) specimens of A. tenuis, using PrimeScript II 1st Strand cDNA Synthesis Kit (Takara, Shiga, Japan). GFP-like cDNAs were amplified using PrimeSTAR Max DNA Polymerase (Takara, Shiga, Japan) and primers FP2_5UHind_F1 and FP2_3UBam_R1 for A. digitifera, and three sets of primers, AdiFP1KpnI_F and AdiFP1XbaI_R, AdiFP2KpnI_F and AdiFP2XbaI_R, and AdiFP4KpnI_F and AdiFP4XbaI_R, for A. tenuis. RFP-like cDNAs were cloned using primers FP10_5UHind_F1 and FP10_3UBam_R1 for A. digitifera, and primers AdiFP10KpnI_F and AdiFP10XbaI_R for A. tenuis. CP-like cDNAs were cloned using primers FP8_5U_F2 and FP8_5U_R1 for A. digitifera, and primers AdiFP8KpnI_F and AdiFP8XbaI_R for A. tenuis. PCR was performed with GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). All primer positions and sequences are given in supplementary fig. S1A and S1B, Supplementary Material online. PCR conditions for amplification of full-length cDNA were as follows: denaturation step for 3 min at 94 °C, followed by 30 cycles of denaturation for 1 min at 94 °C, annealing for 30 s at 55 °C, and extension for 30 s at 72 °C. PCR products were used as templates in a second-round PCR reaction when the quantity of first-round PCR product was insufficient for cloning. PCR products were cloned into T-Vector pMD20 vector (Takara, Shiga, Japan), and the sequences were verified using Applied Biosystems Automated 3130xl Sequencer (Applied Biosystems, Foster City, CA, USA). These FP-like sequences aligned them with the known FP sequences from A. millepora using ClustalW in MEGA ver. 6 (Tamura et al. 2013). We identified the mismatched nucleotides in the aligned FP sequences. When mismatches from the aligned FP sequences occurred less than twice in RNA-seq reads, we defined those mismatches as PCR errors. Sequences containing PCR errors were excluded from further analysis.
To verify the absence of FP sequences sharing low similarity with the known Acropora FP genes from RNA-seq reads, we mapped RNA-seq reads from A. digitifera and A. tenuis to several cnidarian FP sequences (eqFP611: AY130757, hcriCP: AF363776, hcriGFP: AF420592, pporRFP: DQ206380, KO: AB128820, efasGFP: DQ206385, pporGFP: DQ206391, meleCFP: DQ206382, meleRFP: DQ206386), and AdiFP2, AdiFP8L, and AdiFP10L. Reads showing similarity (>80%, >80 bp) were mapped to query sequences. The same cnidarian FP sequences were used as queries to blastx-search (Gish and States 1993) to verify the absence of FP sequences with low similarity to known Acropora FPs in each of the assembled contigs from A. digitifera and A. tenuis adults and larvae.
Cloning, Purification, and Spectroscopic Analysis of Recombinant FP Proteins
To construct recombinant FP proteins, vectors bearing cloned FP sequences verified by sequencing were used as templates for subcloning into expression vectors. GFP-like full-length cDNAs were amplified using PrimeSTAR Max DNA Polymerase (Takara, Shiga, Japan), with AdiFP2KpnI_F_L or AdiFP2KpnI_F2_L forward primers, and AdiFP2XbaI_R_L reverse primers. RFP-like full-length cDNAs were amplified using AdiFP10L_KpnIF_L forward primer and AdiFP10L_type1_R, AdiFP10L_type2_R, or AdiFP10L_type3_R reverse primers. CP-like full-length cDNAs were amplified using AdiFP8L_KpnIF_L forward primer, and AdiFP8XbaI_R1_L or AdiFP8XbaI_R2_L reverse primers. Full-length FP cDNAs were subcloned into pCold I expression vector (Takara, Shiga, Japan) and then used to transform BL21 Escherichia coli cells (Takara, Shiga, Japan). Each clone was grown in 20 mL of LB medium, supplemented with ampicillin and IPTG, overnight, and the recombinant proteins were extracted by sonication and purified using TALON beads with poly-histidine tags (Takara, Shiga, Japan).
Emission spectra of purified recombinant FP proteins in 50 mmol/l phosphate buffer solution with 500 mmol/l Imidazole, pH 7.0 were measured with USB-4000 Spectrometer (Ocean Optics, Dunedin, FL, USA). Absorption spectra of purified recombinant FP proteins were measured using UV-1800 spectrophotometer (SHIMAZU, Kyoto, Japan). Both measurements were performed three times for each FP protein.
Phylogenetic and Sequence Comparison Analyses
Coding sequences of FP genes were translated into amino acids and aligned with the known FP sequences (AY646067, AY646070, AY646073, AY646075, EU709808, EU709809, EU709810, EU709811, JX258845, JX258846, KC349891, KC411499, and KC411500) from A. millepora using ClustalW in MEGA ver. 6 (Tamura et al. 2013). Phylogenetic analysis was performed using the p-distance method with 1,000 bootstrap replications. Three major clades in phylogenetic tree were termed based on the fluorescent emissions of FPs encoded by FP sequences as follows: S/MWE clade comprised short-wavelength emission (SWE) and middle-wavelength emission (MWE1) clades; M/LWE clade comprised middle-wavelength emission (MWE2) and long-wavelength emission (LWE) clade; and CP clade. Wavelength of these categories was defined as follows: an emission peak less than 500 nm (short), an emission peak from 500 to 530 nm (middle), an emission peak over 570 nm (long), and absorbance with no emission (CP). Ancestral sequences were estimated using the maximum likelihood method with a pre-set tree topology by MEGA ver. 6 (Tamura et al. 2013). DNA fragments of estimated ancestral sequences were then constructed by in vitro mutagenesis (Ho et al. 1989), and recombinant proteins were purified as described above. The aligned FP sequences were used in four-gamete tests to detect recombination events, using an in-house computer program. The nucleotide sequences were deposited in GenBank under accession numbers LC125047–LC125121, LC177540–LC177542 and in the DDBJ Sequenced Read Archive under accession numbers DRX049620–DRX049623.
Estimation of A. digitifera FP Gene Copy Numbers and Identification of High Coverage Regions
FP gene copy numbers in A. digitifera genome were estimated by quantitative PCR (qPCR) from four A. digitifera specimens. qPCR was performed with Thermal Cycler Dice TP800 (Takara, Shiga, Japan). We designed three sets of primers specific to S/MWE, M/LWE, and CP clade sequences to amplify all FP sequences detected in RNA-seq reads. We amplified partial sequences of AdiFP2, AdiFP8L, AdiFP10L, and elongation factor 1 (EF1) gene. All PCR products were cloned into one pMD20 vector (Takara, Shiga, Japan), in tandem, using In-Fusion® HD Cloning Kit (Takara, Shiga, Japan). This plasmid DNA was used as a control for all genes. Primers for the amplification of each gene were as follows: MWE_qPCR_F3 and MWE_qPCR_R1 for AdiFP2 (S/MWE); MLWE_qPCR_F3 and MLWE_qPCR_R1 for AdiFP10L (M/LWE); CP_qPCR_F1 and MiA_CP_e3_R1 for AdiFP8L (CP); and EF1a-qPCR_F1 and EF1a-qPCR_R1 for EF1. qPCR reactions were performed with the same primers that were used for amplification of partial sequences using SYBR® Premix Ex Taq™ II (Takara, Shiga, Japan). A. digitifera genomic DNA was quantified by Qubit® 2.0 Fluorometer (Thermo Fisher Scientific, MA) and used as qPCR template. The number of genome copies in a reaction was calculated from the weight of genomic DNA and genome size (420 Mb) of A. digitifera (Shinzato et al. 2011). A series of diluted plasmid DNA (10 pg, 1 pg, 0.1 pg, 10 fg, and 1 fg, per μL) was used to construct standard curves to estimate FP gene copy numbers/reaction. From these two numbers, the number of FP gene copies per genome was calculated. Differences between the amplification efficiencies of genomic and plasmid DNA were calibrated using a single copy gene, EF1, as standard. qPCR reactions were performed three times for each genomic DNA sample and control plasmid DNA, with all primer sets.
In addition to ten FP-like genes (Shinzato et al. 2012), we identified FP-like gene loci in the A. digitifera genome (Shinzato et al. 2011) by blastn search (Altschul et al. 1990), using AdiFP2, AdiFP8L, and AdiFP10L as query sequences. After locating all FP gene exon sequences within circa 3 kb genomic regions (exons 1–5), we defined those regions as FP genes. In addition to genes containing complete exon sets, some exon sequences were missing in several genes due to un-assembled genomic regions because of the difficulty associated with high copy number gene assembly (Mariano et al. 2015). We defined those partial genes also as FP genes. To estimate the copies of un-assembled FP genes, we mapped short reads of A. digitifera (DRX000980 and DRX000981) to its genome and extracted high coverage regions (p < 0.0001) using CLC Genomics Workbench (https://www.qiagenbioinformatics.com/).
Nucleotide Divergence between Two Subclades in S/MWE Clade
We estimated nucleotide divergence between two subclades in the S/MWE clade. Each subclade comprised FP sequences from both A. digitifera and A. tenuis. We defined the average number of nucleotide differences per site from all pairwise comparisons between the different groups as the divergence between two groups. The analysis involved 34 FP sequences, namely, for subclade 1, FP4KX_Tl_8, FP2KX_Tl_16, FP2KX_Tl_4, FP4KX_Tl_1, FP2KX_Tl_3, FP2KX_Tl_7, FP4KX_Tl_7, FP2KX_Tl_5, FP2KX_Tl_15, FP2KX_Tl_13, FP2KX_Tl_14, FP2KX_Tl_9, FP4KX_Tl_3, FP4_Tl_11, FP2KX_Tl_6, FP4_Tl_16, FP2KX_Tl_2, FP2KX_Ta_10, FP2_BH_38, FP2_BH_41, FP2R1_Dl1_5, FP2_R1_13, and FP2R1_Dl1_9; and, for subclade 2, FP2_BH_3, FP2_BH_39, FP2_BH_1, FP2_BH_4, FP1KX_Tl_2, FP1_Tl_9, FP1_Tl_12, FP1KX_Tl_1, FP1KX_Tl_3, FP2KX_Ta_7, and FP2_S1603_BH4. Genetic difference between each sequence pair was calculated by MEGA 6 (Tamura et al. 2013). Mean nucleotide difference (p-distance) between A. digitifera and A. tenuis was estimated based on sequence pairs of A. digitifera genome and A. tenuis assembled RNA-seq sequences. We employed reciprocal blastn hit pairing between A. digitifera scaffold and A. tenuis contigs with e-value <e−50 (Altschul et al. 1990). We discarded A. tenuis contigs with a second hit to A. digitifera scaffolds with e-value <e−50 to avoid putative orthologous pairs with single-multicopy relationships.
Results
A. digitifera Emits a Wide Range of Fluorescence in the Sea
Because its entire genome DNA sequence is available, we first focused on one Acropora species, A. digitifera, to reveal the full complement of its FP genes. Before analyzing FP and FP sequences from this species, we measured the fluorescence emitted from colonies of A. digitifera in the sea. We measured the emission light (including reflectance and fluorescence) from five A. digitifera colonies (see supplementary table S1, Supplementary Material online) excited by two excitation light sources, LED (448 nm spectrum peak; fig. 1A) and laser (452 nm spectrum peak; fig. 1D). As shown in fig. 1B and C, and supplementary fig. S2A and C, Supplementary Material online, A. digitifera fluorescence spanned 490–570 nm, as estimated by the subtraction spectrum from excitation (LED) and emission lights. When laser was used as an excitation light, the boundaries between excitation and fluorescence were clear because of the narrower band of laser light in comparison with LED (fig. 1A and D). The fluorescence spanned 470–600 nm (fig. 1E and F and supplementary fig. S2D–F, Supplementary Material online).
FP-like Sequences have Diversified in A. digitifera
To isolate all FP sequences expressed in different developmental stages (larva and adult), RNA sequences (9.4 and 8.0 Gbp, respectively) were determined by Illumina HiSeq2000 platform. To design primers in the conserved regions among mapped-reads, we mapped paired-end sequences from A. digitifera adult and larvae to AdiFP2, AdiFP8L, and AdiFP10L to PCR-amplify all FP sequence types. The average coverage was 37–194 for the adult and 18,032–397,662 for larval reads for the three mapped sequences (see supplementary table S2, Supplementary Material online). Using these primers, in total 22 and 9 FP-like sequences were identified in adult and larva specimen cDNAs, respectively. No FP sequences were identical in the adult and larva. Compared with the known Acropora FP genes, the new sequences did not contain any insertion/deletion frame shifts or premature stop codons.
To verify the phylogenetic relationships between sequences identified in this study and the known FP and FP-like sequences, we included A. millepora FP sequences and A. digitifera FP-like sequences identified in its genome (Shinzato et al. 2012) in the present analysis. As shown in supplementary fig. S4A, Supplementary Material online, the newly isolated FP-like sequences from clades 1–3 clustered with FP sequences that encode proteins with emission peak values 484–512 nm, or 516–599 nm, or with FP sequences that encode proteins with only absorption, respectively. All FP-like sequences identified in A. digitifera genome data were also included in the three clades (see supplementary fig. S4B, Supplementary Material online), even though the number of positions used for phylogenetic tree construction was reduced from 657 to 342 because of sequence truncation. No FP-like sequences clustering with the known CFP sequences encoding proteins with emission peak values 485–495 nm (Alieva et al. 2008) were identified in A. digitifera adult or larva cDNA. We additionally extracted RNA from one A. digitifera colony (ID: S1603) that emitted fluorescence with an emission peak value less than 500 nm in the field. Two sequences that were identified from cDNA of this colony were clustered with the known CFP sequences (see supplementary fig. S4C, Supplementary Material online). We constructed a phylogenetic tree using all the sequences identified in this study. A. digitifera FP-like sequences formed three different monophyletic clades (clades 1–3, fig. 2). To verify the absence of FP sequences that shared low similarity with the known Acropora FP genes in RNA-seq reads, we mapped each RNA-seq read from A. digitifera (adult and larva) and A. tenuis (adult and larvae) that shared similarity (>80%, >80 bp) with several cnidarian FP sequences. Large number of reads (minimum 9,820 reads, maximum 2,776,230 reads) mapped to Acropora FP sequences (AdiFP2, AdiFP8L, and AdiFP10L), whereas six reads mapped to other cnidarian FP sequences. Also, in the assembled contigs from A. digitifera and A. tenuis RNA-seq reads, we only found FP genes highly similar to AdiFP2, AdiFP8L, and AdiFP10L. These results suggest that A. digitifera only possesses FP genes with high similarity to the known FP genes of Acropora species.
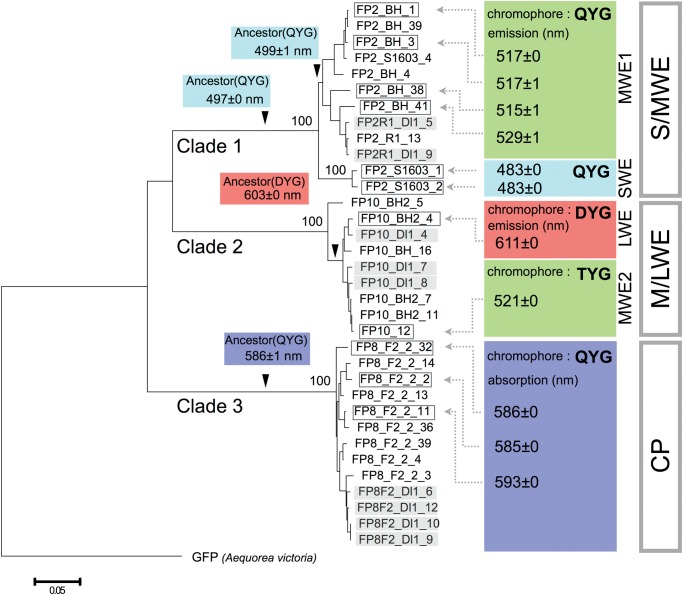
Fig. 2.—
Phylogenetic relationships between A. digitifera FP sequences and FP-encoded functions. The figure shows FP sequence-based phylogenetic tree. The tree was constructed using the neighbor-joining method with the p-distance method. Bootstrap probability for each clade was obtained by 1,000 replicates and is shown next to each node. A. victoria GFP was used as an outgroup. The scale bar represents 0.05 substitutions per site. FP sequences used for recombinant analyses are enclosed in rectangles, and FP sequences from larva are shown in gray. The arrowheads indicate positions of the estimated ancestral sequences. Maximum values for emission or absorption spectra of each recombinant protein are shown at the tails of dotted arrows, in colored branch boxes (ancestral sequences). Chromophores, in colored boxes, are defined by three amino acids.
Functional Analysis of FP Sequences
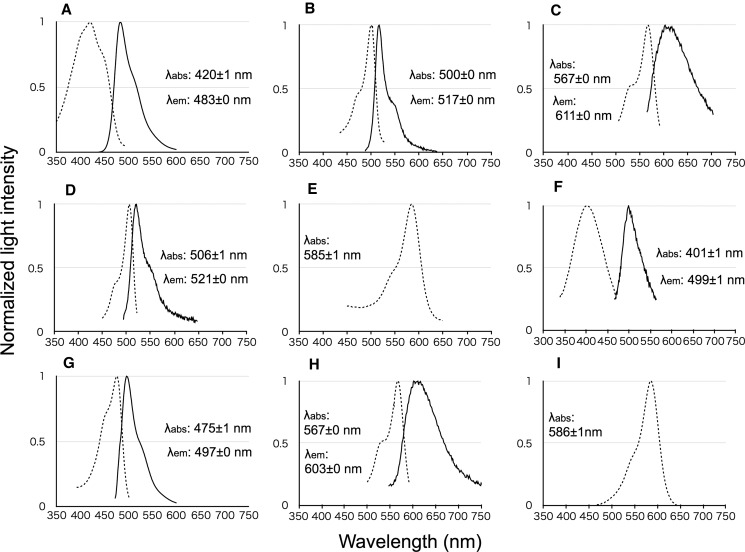
We purified proteins encoded by FP-like sequences and measured their absorption and emission spectra. Clade 1 proteins were characterized by SWE (483 nm) (figs. 2 and 3A, and supplementary fig. S5A, Supplementary Material online) or MWE spectra (515–529 nm) (fig. 3B, and supplementary fig. S5B–D, Supplementary Material online) and were split into two subclades (SWE and MWE1). Clade 3 proteins showed only absorbance (586–593 nm), similarly to the known CPs (figs. 2 and 3E and supplementary figs. S5H and I, Supplementary Material online). Clade 2 was split into two subclades (LWE and MWE2) with DYG and TYG chromophores (see supplementary fig. S6A, Supplementary Material online), respectively, and proteins encoded by sequences from each clade (FP10_BH2_4 and FP10_12) were characterized by LWE (611 nm, FP10_BH2_4) or MWE (521 nm, FP10_12) spectra (figs. 2, 3C and D). Because FP-like sequences from the three clades were highly similar to one other (>95%), we anticipated that all the newly isolated FP-like sequences would encode fluorescence or CP functions. Based on the emission and absorption spectra, we termed clade 1 the S/MWE clade, clade 2 the M/LWE clade, and clade 3 the CP clade. Twelve of the newly isolated A. digitifera sequences belonged to the S/MWE clade, other nine belonged to the M/LWE clade, and 13 to the CP clade.
Fig. 3.—
Emission and absorption spectra of FP recombinant proteins. Absorption (dotted line) and emission (solid line) spectra of recombinants belonging to S/MWE clade (A, FP2_S1603_BH1; and B, FP2_BH_1), M/LWE clade (C, FP10_BH2_4; and D, FP10_12), CP clade (E, FP8_F2_2), MWE1 clade ancestor (F), S/MWE clade ancestor (G), two subclade in M/LWE clade ancestors (H), and CP clade ancestor (I) are shown. Horizontal and vertical axes indicate wavelengths (nanometers) and normalized light intensities, respectively.
We mutated the first amino acid, T, of TYG-chromophore sequence to D (T66D, FP10_12) in a protein from the MWE2 clade (see supplementary fig. S6B, Supplementary Material online) and did not detect light emission. An amino acid difference between FP10_BH2_4 and FP10_12 sequences that resulted in amino acid polarity change was located at position 191 (see supplementary fig. S6B, Supplementary Material online), and, subsequently, an additional mutation (S191P) was introduced in the mutant protein. The fluorescence of the double mutant shifted toward the long wavelengths (see supplementary fig. S6C, Supplementary Material online, λem = 593 nm), but a single change at position 191 (S191P) did not affect the fluorescence emission (see supplementary fig. S6D, Supplementary Material online).
Assessment of Ancestral FP Sequences
We estimated the ancestral sequences for the MWE1 clade, the S/MWE clade, the two subclades of M/LWE clade, and the CP clade (shown by arrowheads in fig. 2). Light emission and absorption of purified proteins encoded by these ancestral sequences were measured (fig. 3F–I). Emission or absorption of the ancestral proteins from MWE1, S/MWE, M/LWE, and CP clades were categorized as SWE (λem = 499 nm), SWE (λem = 497 nm), LWE (λem = 603 nm), and CP (λabs = 586 nm) spectra, respectively.
FP Genes are Present in High Copies in the A. digitifera Genome
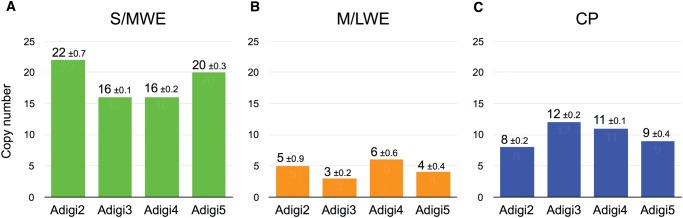
FP gene copy numbers in the genome were quantified with qPCR from four adult specimens of A. digitifera. The copy numbers of FP genes from each clade were 16–22 in the S/MWE clade, 3–6 in the M/LWE clade, and 8–12 in the CP clade (fig. 4A–C).
Fig. 4.—
Individual clade FP gene copy numbers. FP gene copy numbers in S/MWE (A), M/LWE (B), and CP (C) clades from four colonies of A. digitifera are given. Adigi2–Adigi4 indicate colony IDs.
Using three sequences from each of the three FP gene clades (AdiFP2, AdiFP10L, and AdiFP8L) as query sequences, we searched the A. digitifera genome. We treated partial FP sequences with incomplete exon sets as de facto FP genes because of many un-assembled nucleotide regions in these partial FP genes. When we used AdiFP2 as the query, 12 FP genes were identified that shared high similarity with the S/MWE clade sequences on three scaffolds (see supplementary fig. S7A, Supplementary Material online). Similarly, five and eight FP genes were identified as highly similar with M/LWE on three scaffolds, and CP on three scaffolds (see supplementary fig. S7B and C, Supplementary Material online). We identified 25 FP genes in the A. digitifera genome. In addition, we mapped short sequence reads determined from the genome of A. digitifera to its genomic sequence to identify high coverage regions. We found that eight S/MWE, five M/LWE, and seven CP genes, out of 25 FP genes, reside with high coverage regions (see supplementary fig. S7D–F, Supplementary Material online). This suggests the possibility that additional copies of FP genes that had not been identified on account of unassembled genomic DNA sequences may be present in the genome.
FP Genes Have Also Diversified in A. tenuis
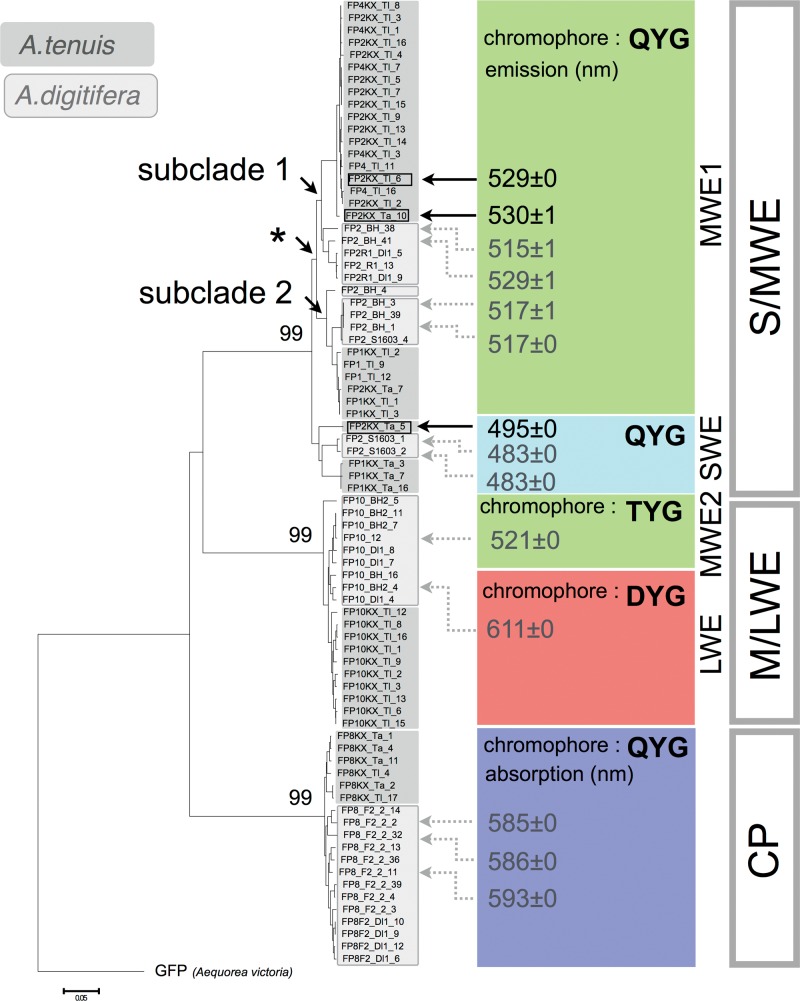
To examine whether FP genes have also diversified within genomes of other Acropora species, we determined FP sequences of A. tenuis. We designed primers (see mapping coverage in supplementary table S2, Supplementary Material online), as described for A. digitifera, and amplified FP sequences from cDNAs of adult and larval specimens of A. tenuis. After accounting for PCR errors, 10 and 34 FP-like sequences were identified in the adult and larvae, respectively. We then constructed a phylogenetic tree comprising FP genes from A. digitifera and A. tenuis. All sequences were clustered in one of the three major clades (fig. 5). We purified proteins encoded by the S/MWE clade sequences and measured their emission and absorbance spectra. SWE (see supplementary fig. S5E, Supplementary Material online) and MWE (see supplementary fig. S5F and G, Supplementary Material online) spectra were recorded (fig. 5). We identified 28 A. tenuis sequences in the S/MWE clade, ten sequences in the M/LWE clade, and six sequences in the CP clade. Sequences encoding TYG chromophore were confirmed in the adult and larva A. tenuis RNA-seq data; however, we were unable to clone and analyze these sequences because their expression was lower than that of other FP sequences in the M/LWE clade. Together with the cloning from A. digitifera, the FP sequence clustering with known CFP sequences was identified from neither A. digitifera nor A. tenuis larval specimens.
Fig. 5.—
Phylogenetic relationships of A. tenuis FP sequences and FP-encoded functions. This figure contains FP sequence-based phylogenetic tree. The tree was constructed using the neighbor-joining method with the p-distance method. Bootstrap probability for each clade was obtained by 1,000 replicates and is shown next to each node. The scale bar represents 0.05 substitutions per site. A. victoria GFP was used as an outgroup. FP sequences from A. tenuis and A. digitifera are shown in dark and light gray, respectively. A. tenuis FP sequences used for recombinant analyses are indicated by black arrows. Maximum values of emission spectra of each recombinant protein are shown at the tails of black arrows. Asterisk indicates the divergence of subclades 1 and 2 in S/MWE clade. Chromophores, in colored boxes, are defined by three amino acids.
Recombination between FP Sequences from Each Major Clade
We aligned A. digitifera and A. tenuis FP sequences from MWE1, M/LWE, and CP clades using ClustalW in MEGA ver. 6 (Tamura et al. 2013). FP sequences from A. digitifera and A. tenuis shared nucleotide changes at several synonymous sites in each clade (see supplementary fig. S8A–C, Supplementary Material online). Recombination of sequences within extant species and between species, meaning within common ancestral species (hereafter, between species), was detected in each major clade by four-gamete tests (see supplementary fig. S9, Supplementary Material online).
The Nucleotide Divergence between Two Subclades in MWE1 Clade
The nucleotide divergence between two subclades in the MWE1 clade (marked in fig. 5 by an asterisk) was 0.068. This value was slightly higher than the mean nucleotide divergence between A. digitifera and A. tenuis (0.065) calculated from sequence pairs between the A. digitifera genomic sequence and A. tenuis assembled RNA-seq sequences.
Discussion
Multi-Member FP Gene Family Underlies A. digitifera Fluorescence
Before analyzing A. digitifera FP and FP sequences, we measured the fluorescence emitted by the colonies of this species in the sea, because, to the best of our knowledge, these data have not been available. We applied a very narrow band laser light for excitation, and the fluorescence emitted by corals was determined to span 470–600 nm, with clear separation from excitation light.
How many FPs contribute to this fluorescence? To uncover the genetic basis of A. digitifera fluorescence, we determined the sequences of FP genes from adult and larva cDNAs. No sequence was identical between them, suggesting life stage-specific FP gene expression. The sequence difference between the adult and larva may reflect these life stage-specific fluorescence patterns. However, we cannot exclude the possibility that individual, specimen-related differences were responsible for these sequence differences. The types of FPs expressed in adults and larvae of both species might be different; FPs in SWE, MWE1, MWE2, LWE, and CP clades in adults and in MWE1, MWE2, LWE, and CP clades from larvae. The lack of FPs in the SWE clade in larvae was similar to the blue shift of fluorescence from larvae to adults observed in Seriatopora hystrix (Roth et al. 2013). The difference of the number of FP sequences isolated from a single larva of A. digitifara and multiple larvae of A. tenuis originated from five colonies may reflect the variation of the expression difference of FP genes or that of FP genes among individuals of A. tenuis larvae.
Based on the number of FP sequences from an adult A. digitifera cDNA and the fact that it is a diploid organism (Shinzato et al. 2011), we estimated that a minimum of four MWE, three M/LWE, and five CP genes are present in the genome of an adult individual, considering that all adult sequences are allelic variations. Next, we estimated FP gene copy numbers in four adult A. digitifera specimens by qPCR. The total gene numbers per A. digitifera genome were two times higher than the previously reported largest set of FP genes, 16 GFP-like genes, in the amphioxus genome (Bomati et al. 2009). These high copy numbers in A. digitifera genome could account for the number of FP sequences determined from cDNAs. The differences between copy numbers, with low standard deviation between experimental replicates (fig. 4; <1 copy in each clade), indicated the presence of copy number variations between individuals. The S/MWE clade contained the highest number of FP genes, which was in agreement with the major fluorescence emission from FPs in the S/MWE clade, with a peak around 500 nm, from live A. digitifera (fig. 1), although the excitation light (LED: 448 nm spectrum peak and laser: 452 nm spectrum) matched with FPs in the S/MWE clade (λabs = 461–508 nm). Hence, the A. digitifera FP genes form a multi-gene family that is larger than previously reported for other organisms, and this FP multi-gene family could comprise the genetic basis of coral fluorescence.
Our homology search for FP genes in the A. digitifera genomic sequence data supports the existence of a multi-gene FP family as well, since multiple genes were detected in the assembled genome sequences. High copy number genes with high similarity are difficult to assemble (Mariano et al. 2015). Therefore, many genes identified in the assembled genomic sequence data were incomplete due to un-assembled genomic regions, suggesting a possibility that some FP genes were missing from the assembly. This possibility can be evaluated by focusing on high coverage regions in short read mapping. If a proportion of multi-copy genes is not individually assembled, and at least one gene copy is identified in the genomic sequence, the corresponding genomic region should be mapped with high coverage by short reads. Indeed, high coverage regions were detected in all three major clades, suggesting higher copy numbers of each major clade gene than those estimated from the assembled genomic sequence. The copy numbers of FP genes in the M/LWE clade estimated by qPCR were equal to those in the genome sequence data. The high coverage regions in M/LWE clade genes may stem from many un-assembled regions in those genes.
Fluorescence spectra measured for purified FPs revealed that the newly isolated FP sequences could be categorized into three clades, S/MWE, M/LWE, or CP. The FPs in S/MWE and M/LWE clades of fluorescence can cover the A. digitifera fluorescence. The purified proteins encoded by CP sequences absorbed the light, and proteins encoded by CP clade may affect the excitation and emission light of FPs in S/MWE and M/LWE clades by absorbing the light. Accordingly, a multi-FP gene family could generate the summative coral fluorescence.
Multi-Member FP Gene Family Has Evolved with Functional Diversity in the Genus Acropora
To examine whether FP genes of other Acropora species also comprise a multi-gene family, we cloned and determined FP sequences from A. tenuis cDNA. As shown in figure 5, we identified 28 sequences in the S/MWE clade, ten sequences in the M/LWE clade, and six sequences in the CP clade from an adult and larval A. tenuis cDNAs. This result suggests that FP genes from A. tenuis also form a multi-gene family composed of three major clades. Phylogenetically, A. tenuis is located at the basal lineage in the genus Acropora (Fukami et al. 2000; Richards et al. 2013), suggesting that a multi-member FP gene family of three major clades existed in the common ancestor of Acropora species. The estimation of eight copies of A. millepora RFP gene using exon 3 sequence (Gittins et al. 2015) also supports the existence of a multi-FP gene family in the genus Acropora. Subclades 1 and 2 in MWE1 clade comprised A. digitifera and A. tenuis sequences, which indicates that an emergence of these two subclades occurred before the divergence of these species. The nucleotide divergence between the two subclades (0.068) was slightly greater than the mean nucleotide divergence between the two species (0.065), and is in agreement with the older divergence.
Because the FP genes are tandemly arrayed in the A. digitifera genome (Shinzato et al. 2012), we tested the possibility of recombination (unequal crossing over) between the FP sequences. We analyzed synonymous mutations in FP sequences in both species to avoid the effect of functional convergence. For closely related lineages, independent synonymous substitutions are generally thought as a rare event due to very low mutation rate. Synonymous mutations in each clade occurred with different sharing patterns between sequences of the two species, suggesting recombination events. Recombination of sequences between species may have occurred in the genome of the common ancestral Acropora species. Recombination events were also supported by four-gamete tests.
To reveal the function of FPs in the ancestral lineages, we estimated the ancestral sequences from MWE1, S/MWE, and CP clades, and a common ancestor of the two subclades in M/LWE clade in A. digitifera. Fluorescence spectra of the purified ancestral proteins were categorized into SWE, CP, and LWE spectra, suggesting that different FP functions have been already acquired in the ancestral lineages of each clade. Because the ancestral lineages of each clade could be traced back to the common ancestor of the Acropora genus, the ancestral species of this genus may have already possessed a functionally diverse FP gene set. FP fluorescence is mainly determined by three amino acid residues known as chromophores (Henderson and Remington 2005). M/LWE group sequences were split into two clades, an ancestral DYG chromophore-coding type and a derived TYG-chromophore-coding type. Fluorescence spectra of DYG- and TYG-chromophore sequences were categorized into LWE (λem = 611 nm) and MWE (λem = 521 nm), respectively. Mutating two amino acids, in the chromophore (T66D) and position 191 (S191P), shifted the FP emission peak from 521 to 593 nm, suggesting that FP sequences with TYG-type chromophores have evolved into MWE from an ancestral LWE. Although their expression was too low to allow cloning, we verified the existence of TYG-chromophore-type sequences in RNA-seq reads from A. tenuis, indicating that TYG chromophore has emerged before A. digitifera and A. tenuis divergence and persisted in these species. Hence, the multi-member FP gene family has evolved to maintain a functional diversity in Acropora species. The adults and larvae of A. digitifera and A. tenuis expressed FP sequences from each of three major clades. FPs from three major clades might possess an important biological function such as photo-protective and antioxidant roles in adults (Palmer et al. 2009; Roth and Deheyn 2013) and possible roles in larval settlement behavior and long-range dispersal in larvae (Kenkel et al. 2011; Strader et al. 2016).
Similar to multi FP gene family in genus Acropora, at least five amphioxus FP genes in two species of genus Branchiostoma have maintained during their evolution (Yue et al. 2016), indicating the importance of gene copy multiplicity of FP genes. Multi-gene families are present in genomes of many organisms, and the functional importance of gene copy multiplicity is well known (Walsh 2008). Compared with previous studies of multi-gene families, the most enigmatic issues concerning the multi-FP gene family are (1) the purpose for multiple FP genes in corals, and (2) the biological role of these multiple copies. Considering the known roles for multi-gene families, the importance of multi-FP gene family may be of dual roles. The first role is associated with the amount of FPs in the tissues of Acropora species. Increased gene copy number can increase transcript levels, as, for example, for the gene family encoding ribosomal RNA (Weider et al. 2005). Indeed, FPs comprise a high proportion of the total soluble protein content in anthozoan tissues (Leutenegger et al. 2007; Oswald et al. 2007). Production of the large amount of FPs may have been essential for survival during the evolution of Acropora species. The second role is linked with the distinct FP function. As shown in this study, the emissions of short-, middle- and long-wavelength light by FPs in S/MWE and M/LWE clades and absorption of the light by FPs in CP clade have been maintained during the evolution of Acropora species. These different functions encoded by FP genes in the genomes of two analyzed Acropora species may have been important during their evolution.
In this study, we identified the complement of A. digitifera FP genes. Whereas the association between the amount of FP proteins, FP gene copy number, and the precise roles of these proteins remains unresolved, knowing the numbers of genes in each major FP clade and their different functions will facilitate the understanding of the biological roles of FPs in future studies.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S9 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Authors’ Contributions
S.T.K.: research concept, fluorescence measurements, all experiments, data analysis, and manuscript preparation.
J.G.: molecular evolutionary analysis and manuscript editing.
Y.S.: helpful discussions, four-gamete tests, and manuscript editing.
K.S.: species identification and measurements of fluorescence.
Y.T.: research concept, research planning, fluorescence measurements, some recombinant protein and qPCR experiments, data analysis, and manuscript preparation.
Supplementary Material
Acknowledgments
This work was supported by an internal SOKENDAI grant to Y.T., and the Center for the Promotion of Integrated Sciences (CPIS) of SOKENDAI grant to Y.T. We thank Drs. Yinqiang Zheng and Imari Sato (National Institute of Informatics, Japan) for helpful discussions of fluorescence measurements, Masayuki Hatta (Ochanomizu University) for his help with sampling, Mutsumi Nishida (University of the Ryukyus) for arranging the fluorescence measurements from live corals, and Mori Jinza (University of the Ryukyus) for his help with the measurements.
Literature Cited
- Alieva NO, et al. 2008. Diversity and evolution of coral fluorescent proteins. PLoS ONE 3:e2680.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Baumann D, et al. 2008. A family of GFP-like proteins with different spectral properties in lancelet Branchiostoma floridae. Biol Direct. 3:28.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomati EK, Manning G, Deheyn DD. 2009. Amphioxus encodes the largest known family of green fluorescent proteins, which have diversified into distinct functional classes. BMC Evol Biol. 9:77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo C, et al. 2008. Blue light regulation of host pigment in reef-building corals. Mar Ecol Prog Ser. 364:97–106. 10.3354/meps07588. [Google Scholar]
- Deheyn DD, et al. 2007. Endogenous green fluorescent protein (GFP) in amphioxus. Biol Bull. 213:95–100. [DOI] [PubMed] [Google Scholar]
- Field SF, Bulina MY, Kelmanson IV, Bielawski JP, Matz MV. 2006. Adaptive evolution of multicolored fluorescent proteins in reef-building corals. J Mol Evol. 62:332–339. [DOI] [PubMed] [Google Scholar]
- Fukami H, Omori M, Hatta M. 2000. Phylogenetic relationships in the coral family acroporidae, reassessed by inference from mitochondrial genes. Zool Sci. 17:689–696. [DOI] [PubMed] [Google Scholar]
- Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat Genet. 3:266–272. [DOI] [PubMed] [Google Scholar]
- Gittins JR, D'Angelo C, Oswald F, Edwards RJ, Wiedenmann J. 2015. Fluorescent protein-mediated colour polymorphism in reef corals: multicopy genes extend the adaptation/acclimatization potential to variable light environments. Mol Ecol. 24:453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JN, Remington SJ. 2005. Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano. Proc Natl Acad Sci U S A. 102:12712–12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59. [DOI] [PubMed] [Google Scholar]
- Johnsen S. 2012. The optics of life. NJ: Princeton University Press. [Google Scholar]
- Kelmanson IV, Matz MV. 2003. Molecular basis and evolutionary origins of color diversity in great star coral Montastraea cavernosa (Scleractinia: Faviida). Mol Biol Evol. 20:1125–1133. [DOI] [PubMed] [Google Scholar]
- Kenkel CD, Traylor MR, Wiedenmann J, Salih A, Matz MV. 2011. Fluorescence of coral larvae predicts their settlement response to crustose coralline algae and reflects stress. Proceedings of the Royal Society B: Biol Sciences 278: 2691–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labas YA, et al. 2002. Diversity and evolution of the green fluorescent protein family. Proc Natl Acad Sci U S A. 99:4256–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger A, et al. 2007. It's cheap to be colorful. FEBS J. 274:2496–2505. [DOI] [PubMed] [Google Scholar]
- Mariano DC, et al. 2015. MapRepeat: an approach for effective assembly of repetitive regions in prokaryotic genomes. Bioinformation 11:276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz MV, et al. 1999. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 17:969–973. [DOI] [PubMed] [Google Scholar]
- Mazel CH. 2003. Contribution of fluorescence to the spectral signature and perceived color of corals. Limnol Oceanogr. 48:390–401. [Google Scholar]
- Moisan TA, Mitchell BG. 2001. UV absorption by mycosporine-like amino acids in Phaeocystis antarctica Karsten induced by photosynthetically available radiation. Mar Biol. 138:217–227. [Google Scholar]
- Oswald F, et al. 2007. Contributions of host and symbiont pigments to the coloration of reef corals. FEBS J. 274:1102–1109. [DOI] [PubMed] [Google Scholar]
- Palmer CV, Modi CK, Mydlarz LD. 2009. Coral fluorescent proteins as antioxidants. PLoS ONE 4:e7298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards ZT, Miller DJ, Wallace CC. 2013. Molecular phylogenetics of geographically restricted Acropora species: implications for threatened species conservation. Mol Phylogenet Evol. 69:837–851. [DOI] [PubMed] [Google Scholar]
- Roth MS, Deheyn DD. 2013. Effects of cold stress and heat stress on coral fluorescence in reef-building corals. Sci Rep 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MS, Fan TY, Deheyn DD. 2013. Life history changes in coral fluorescence and the effects of light intensity on larval physiology and settlement in Seriatopora hystrix. PLoS ONE 8:e59476.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagin DA, et al. 2004. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol. 21:841–850. [DOI] [PubMed] [Google Scholar]
- Shimomura O. 1979. Structure of the chromophore of Aequorea green fluorescent protein. Fed Eur Biochem Soc. 104:220–222. [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476:320–323. [DOI] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Tanaka M, Satoh N. 2012. Fluorescent protein candidate genes in the coral Acropora digitifera genome. Zoolog Sci. 29:260–264. [DOI] [PubMed] [Google Scholar]
- Strader ME, Aglyamova GV, Matz MV. 2016. Red fluorescence in coral larvae is associated with a diapause-like state. Mol Ecol. 25:559–569. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Hayashibara T, Shirayama Y, Fukami H. 2008. Evidence of species-specific habitat selectivity of Acropora corals based on identification of new recruits by two molecular markers. Mar Ecol Prog Ser. 355:149–159. [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729. Epub 2013 Oct 2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veron JEN. 2000. Corals of the world. Townsville, Australia: Australian Institute of Marine Science. [Google Scholar]
- Walsh JB, Stephan W. 2001. Multigene Families: Evolution. In: eLS. John Wiley & Sons, Ltd.
- Weider LJ, et al. 2005. The functional significance of ribosomal (r)DNA variation: impacts on the evolutionary ecology of organisms. Ann Rev Ecol Evol Syst. 36:219–242. [Google Scholar]
- Yue JX, Holland ND, Holland LZ, Deheyn DD. 2016. The evolution of genes encoding for green fluorescent proteins: insights from cephalochordates (amphioxus). Sci Rep. 6:28350.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.