Abstract
Purpose
The intestinal microbiota has emerged as a virtual organ with essential functions in human physiology. Antibiotic-induced disruption of the microbiota in critically ill patients may have a negative influence on key energy resources and immunity. We set out to characterize the fecal microbiota composition in critically ill patients both with and without sepsis and to explore the use of microbiota-derived markers for clinical outcome measurements in this setting.
Methods
In this prospective observational cohort study we analyzed the fecal microbiota of 34 patients admitted to the intensive care unit. Fifteen healthy subjects served as controls. The fecal microbiota was phylogenetically characterized by 16S rRNA gene sequencing, and associations with clinical outcome parameters were evaluated.
Results
A marked shift in fecal bacterial composition was seen in all septic and non-septic critically ill patients compared with controls, with extreme interindividual differences. In 13 of the 34 patients, a single bacterial genus made up >50% of the gut microbiota; in 4 patients this was even >75%. A significant decrease in bacterial diversity was observed in half of the patients. No associations were found between microbiota diversity, Firmicutes/Bacteroidetes ratio, or Gram-positive/Gram-negative ratio and outcome measurements such as complications and survival.
Conclusions
We observed highly heterogeneous patterns of intestinal microbiota in both septic and non-septic critically ill patients. Nevertheless, some general patterns were observed, including disappearance of bacterial genera with important functions in host metabolism. More detailed knowledge of the short- and long-term health consequences of these major shifts in intestinal bacterial communities is needed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00134-016-4613-z) contains supplementary material, which is available to authorized users.
Keywords: Gut microbiota, Critically ill, Intensive care unit, Antibiotics, Sepsis
Introduction
While antibiotics are one of the greatest inventions of the twentieth century, their potential side effects are being increasingly recognized. In addition to concerns on the emergence of bacterial resistance, it has been suggested that antibiotic-induced changes to the intestinal flora could have serious and long-lasting consequences for human physiology [1, 2]. Paradoxically, antibiotic treatment can increase susceptibility to opportunistic and nosocomial infections by affecting the resistance of the intestinal microbiota to colonization [3]. A low-diversity community with limited resilience allows pathogens like Clostridium difficile and vancomycin-resistant enterococci to grow unrestrictedly [4]. Additionally, decreased gut microbial diversity has been associated with pathogenic conditions, such as inflammatory bowel disease and increased mortality following allogeneic hematopoietic stem cell transplantation (HSCT) [2, 5]. Physiological functions of the microbiota, such as the metabolism of carbohydrates and production of vitamins, may be lost as well.
A decreased gut microbial diversity could be of particular relevance for critically ill patients, as the vast majority of patients in an intensive care unit (ICU) are treated with antibiotics. A prospective point prevalence study involving 1265 ICUs across the world found that on any given day 75% of patients admitted to these ICUs received antibiotics [6]. In The Netherlands, systemic decontamination of the digestive tract (SDD), consisting of topical application of non-absorbable antibiotics and a short course of systemic cephalosporins, is common practice in ICUs [7, 8]. SDD is a much-debated policy, with reduced infection rates and mortality as potential benefits and increased antimicrobial resistance as potential drawback. While the aim of SDD is to prevent colonization with potential pathogenic microorganisms while preserving anaerobic bacteria, its effect on the composition of the intestinal microbiota as a whole is ill-defined.
Recent advances, such as the implementation of fecal transplantation as therapy for C. difficile infection, suggest that it may be possible to monitor, prevent, or even cure disease by modulating the intestinal microbiota [9–11]. First, however, more knowledge is needed on the composition of the gut microbiota in critically ill patients to better understand the clinical consequences of microbiota disturbances and thus determine which patients might benefit from microbiota-based therapies [9].
Based on previous studies, we hypothesized that microbial diversity would be extremely low in critically ill patients and that low diversity would be associated with worse clinical outcome [5, 12, 13]. In this pilot study we describe the phylogenetic composition of the fecal microbiota in a cohort of critically ill patients using 16S rRNA gene sequence analysis and compare the results in both septic and non-septic critically ill patients with those in healthy subjects. We also explored possibilities to correlate microbiota-derived markers with clinical outcome parameters.
Methods
Subjects
This study was part of a large prospective observational study in critically ill patients admitted to the ICU [Molecular Diagnosis and Risk Stratification of Sepsis (MARS) study; clinicaltrials.gov identifier NCT01905033] [14]. Thirty-four randomly selected adult patients who were admitted to the ICU of the Academic Medical Center (Amsterdam, The Netherlands) between October 2012 and November 2013 were included. Patients who were transferred from other ICUs or had an expected length of ICU stay of <24 h were excluded. All patients met at least two of the following criteria: body temperature of ≤36 or ≥38 °C, tachycardia of >90/min, tachypnea of >20/min or partial pressure of carbon dioxide (pCO2) of < 4.3 kPA, and leukocyte count of <4 × 10E9/L or >12 × 10E9/L [15]. Sepsis was defined when the inclusion criteria were associated with suspected infection within 24 h after ICU admission, with subsequent systemic therapeutic administration of antibiotics to the patient [15]. Sequential Organ Failure Assessment scores were recorded daily. Definitions of acute kidney injury (AKI) or acute respiratory distress syndrome (ARDS) were used exactly as described previously [16, 17]. Data from the municipal personal records database were used to calculate long-term survival. The control group consisted of15 healthy, non-smoking human subjects who had not taken antibiotics during the previous year (ClinicalTrials.gov identifier NCT02127749) [18]. Written informed consent was obtained from all healthy subjects and patients or their legal representative. Ethical approval for both the patient and healthy subject studies was received from the Medical Ethics Committee of the Academic Medical Center in Amsterdam, and all research was conducted in accordance with the declaration of Helsinki.
Intestinal microbiota analysis
Fresh stool samples were stored at 4 °C and transferred to −80 °C within 24 h of collection. Samples were processed for 16S rRNA gene sequencing exactly as described by Lankelma et al. [18]. DNA was isolated using a bead-beating protocol [19]. The V1–V2 region of the bacterial 16S rRNA gene was amplified in a two-step protocol using universal primers 27F and 338R and sequenced on the Illumina MiSeq sequencing platform (Illumina, San Diego, CA). Full methods are available in Electronic Supplementary Material (ESM) File 1. The number of reads per sample and taxonomic distribution of the reads are specified in ESM File 2. A canonical correspondence analysis was performed using the Canoco 5 software package (Biometris, Wageningen, The Netherlands).
To investigate whether microbiota diversity was associated with clinical outcome, we divided the cohort into two groups, namely, one with low bacterial diversity (Shannon index < 4.0) and one with normal diversity (Shannon index > 4.0). Previous studies have used similar arbitrary cutoffs [5, 20]; the present cutoff is based on the range of diversity in healthy controls.
Statistical analysis
Continuous nonparametric data were analyzed using a Mann–Whitney U or Kruskal–Wallis test, and the results are presented as the median and interquartile range (IQR). Continuous parametric data, presented as numbers and percentage or as the mean ± standard deviation, were analyzed with a student’s t test or analysis of variance. Categorical data were analyzed using a Chi-square test. All data analyses were performed with R studio built under R version 3.2.3 (R Core Team 2013; R Foundation for Statistical Computing, Vienna, Austria). A p value of <0.05 was considered to be statistically significant. Figures were created using Graphpad Prism (GraphPad Software, San Diego, CA).
Results
Patient characteristics
A single fecal sample was collected from 34 critically ill patients (mean age 64 years; 48% male) between 1 and 21 days after ICU admittance [median 4.5 (IQR 2.8–7) days; ESM Tables S1 and S2]. Of these 34 patients, 25 were admitted because of sepsis and one had a non-infectious diagnosis (ESM Table S1). Fifteen healthy subjects served as controls.
Microbial composition of fecal samples
Compared to healthy subjects, the stool microbiota showed a highly abnormal composition in most patients, based on both the highest bacterial taxonomic ranks (phylum level) and lowest detectable ranks (genus level).
Phylum level
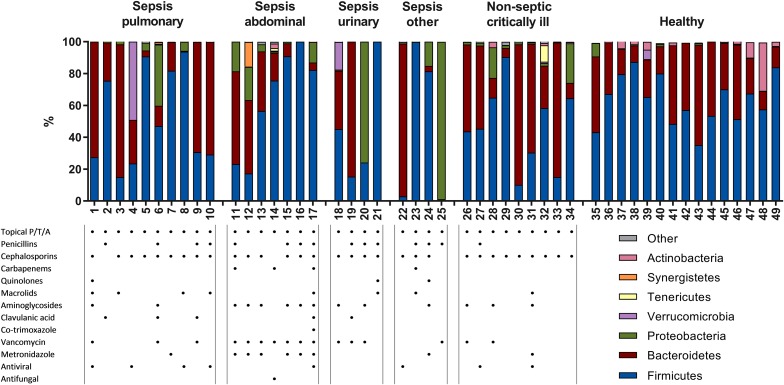
To obtain a global view of the fecal microbiota in critically ill patients, we first investigated microbiota composition at the phylum level (Fig. 1). Firmicutes and Bacteroidetes are the predominant phyla in a stable intestinal microbiota, which was confirmed in our healthy subjects with these two phyla combined constituting 89–98% of all bacteria. However, in 12 of the 34 patients, Firmicutes and Bacteroidetes constituted <89% of all bacteria and even constituted as low as <1% in one sample (p < 0.0001, healthy controls vs. ICU patients). This was the case in both septic and non-septic patients. A relative high abundance of Proteobacteria was observed in approximately one-third of patients (p = 0.02, healthy controls vs. ICU patients). This phylum of Gram-negative bacteria includes many pathogenic and lipopolysaccharide-containing bacteria, such as Escherichia. There was a trend towards an increased ratio of Gram-negative to Gram-positive bacteria in ICU patients (p = 0.0658, healthy vs. ICU patients).
Fig. 1.
High interindividual diversity in fecal microbiota composition at the phylum level in both septic and non-septic critically ill patients. A single fecal sample was collected from septic and non-septic intensive care unit (ICU) patients and healthy subjects. Total bacterial 16S rDNA was isolated and sequenced to investigate the bacterial composition of these samples. Each bar represents the microbiota composition of one individual patient (patient number indicated at bottom of each bar) at the phylum level, which is the highest bacterial taxonomic rank. Data are presented as the percentage of total 16S rDNA reads in each sample; colors indicate different phyla. Patients are grouped according to their main diagnosis. Table shows which classes of antibiotics each patient received during their stay in the intensive care unit prior to fecal sampling. Topical P/T/A Topical application of polymyxin E, tobramycin, and amphotericin B. None of the healthy subjects had received antibiotics for at least 12 months prior to fecal sampling
While it is known that intestinal microbiota is highly personalized, some very striking individual patterns were observed. The fecal sample of patient no. 12 (who had an abdominal Escherichia coli sepsis) showed a high abundance of Synergistetes and those of patient nos. 4 and 18 revealed a high abundance of Verrucomicrobia (discussed in more detail further in the text.
Genus level
Our analysis of bacterial composition at the genus level (ESM Fig. S1) revealed an even greater interpatient diversity than that seen at the phylum level. Septic and non-septic patients did however share some common characteristics when compared to healthy subjects. Most notably, several key bacterial genera that each represent around 5% of the microbiota in healthy subjects had disappeared almost completely, including Faecalibacterium, Blautia, Ruminococcus, Subdoligranulum and Pseudobutyrivibrio (all p < 0.0001, healthy controls vs. ICU patients).
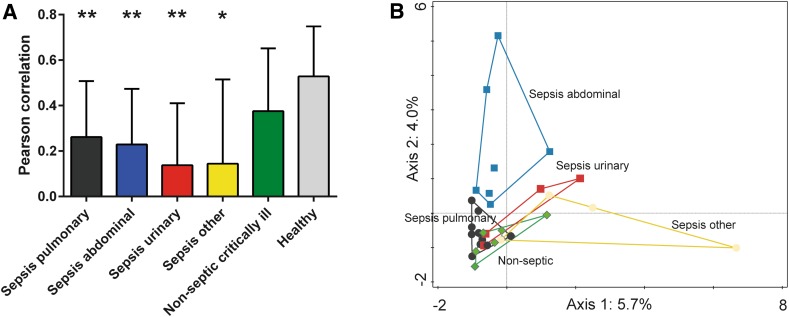
No single bacterial genus colonized >50% of the microbiota in the healthy subjects, whereas this did occur in 13 of the 34 patients, with one genus comprising even >75% of the microbiota in four patients (ESM Fig. S1). These dominating genera included the usual suspects, such as Enterococcus (p = 0.0002 healthy controls vs. ICU patients). Members of Staphylococcus and Escherichia/Shigella were also significantly more abundant in the ICU patients, especially in the abdominal sepsis group (p = 0.042 and p = 0.049, respectively, healthy controls vs. ICU patients; p = 0.0048 and p = 0.001, respectively, healthy controls vs. ICU patients with abdominal sepsis). A canonical correspondence analysis to deduce the major driving forces in the variability within all microbial data indicated that fecal microbiota from abdominal sepsis patients differed the most from that of the other patient groups (Fig. 2).
Fig. 2.
High interindividual diversity in septic critically ill patients with sepsis. a Pearson correlations were calculated to investigate the level of similarity in microbiota composition between patients with a critical illness of different origin (sepsis originating from lung, abdomen, urinary tract, or other location and non-septic critical illness). Patient groups are indicated by colors. Healthy individuals are depicted as additional control group. Data are presented as mean ± standard deviation. *p < 0.05, **p < 0.001 vs. healthy controls; p = 0.0005 for all septic patients vs. healthy controls. b A canonical correspondence analysis to deduce the major driving forces (Axis 1, Axis 2) in the variability within the microbial data of all ICU patients, thereby indicating whether samples are alike (in close proximity to each other) or not (increased distance). The variance is indicated as percentages. Symbols/colors represent patient groups as indicated (see a)
Surprisingly, in some patients bacteria belonging to Lactobacillus spp. (p = 0.051, healthy controls vs. ICU patients with pulmonary sepsis; p = 0.06, healthy controls vs. ICU patients with abdominal sepsis) and Akkermansia spp. (of the phylum Verrucomicrobia; no statistically significant correlation with sepsis) were abundantly present. Notably, A. muciniphila is known for immune stimulation and barrier function improvement [21, 22]. The phylum Synergistetes consists of anaerobic Gram-negative bacteria that have been associated with periodontal disease; those in sample 12 belong to the genus Pyramidobacter. The significance of Pyramidobacter in fecal microbiota is as yet unknown, but the oral isolate P. piscolens is known to produce hydrogen sulphide, which is implied in compromising gut barrier function [22, 23]. Overall, the similarity between samples from septic patients (Pearson correlation) was significantly lower than that of controls (p = 0.0005 healthy controls vs. septic patients; Fig. 2).
In patients undergoing allogeneic HSCT, intestinal domination by vancomycin-resistant enterococci precedes bloodstream infection with these bacteria [24]. We did not observe any correspondence between intestinal abundance and clinical infections (data not shown). Likewise, no cases of C. difficile were recorded.
Antibiotics
All patients received SDD consisting of an oral paste and solution containing non-absorbable polymyxin E, tobramycin, and amphotericin B as well as a third-generation cefalosporin administered intravenously, unless the latter was deemed unnecessary by the treating physician as the patient was already being treated with another broad-spectrum antibiotic (Fig. 1). All septic patients were treated with additional antibiotics, varying from one to ten different classes of antimicrobial agents (Fig. 1; detailed description in ESM Table S2). Septic patients were treated with a significantly higher number of antibiotic classes than were non-septic critically ill patients (p < 0.01), but no differences in microbiota composition were observed between these two groups, both at the phylum and genus levels.
Association between microbiota composition and clinical outcome parameters
To explore whether microbiota-related markers could be related to outcome parameters, we used three parameters that are commonly used to describe the microbiota, namely, bacterial diversity, the Firmicutes/Bacteroidetes ratio (F/B-ratio), and the Gram-positive/Gram-negative bacteria ratio [5, 11, 25, 26].
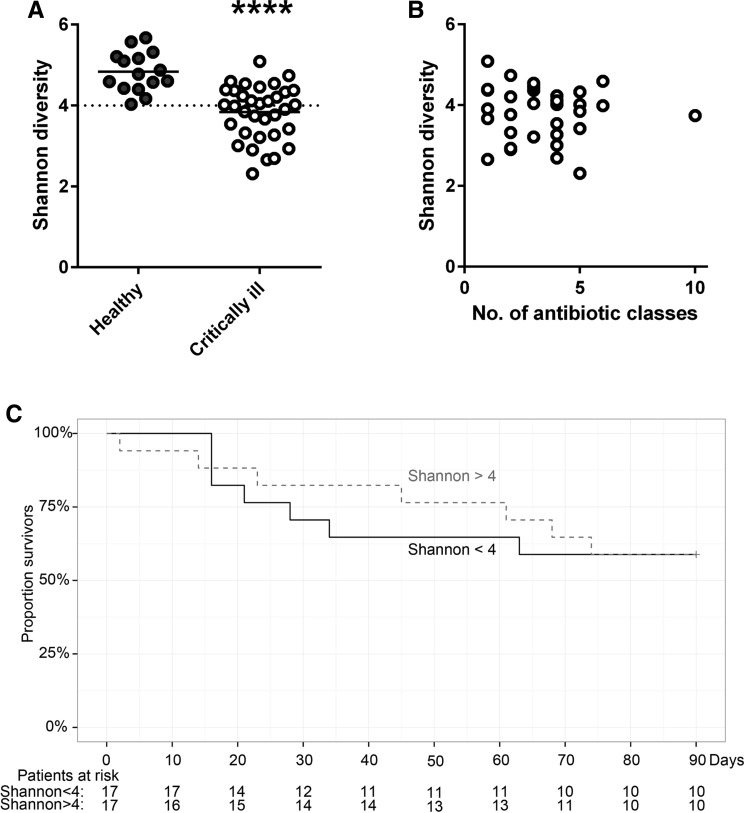
We expected ultra-low bacterial diversity in our patient cohort based on the exposure of these patients to antibiotics and data reported from earlier studies [5, 13]. The critically ill patients in our study had significantly lower bacterial diversity than the control subjects (p < 0.0001; Fig. 3). There was no correlation between the number of antibiotic classes that a patient had received and bacterial diversity (Fig. 3), nor between the number of days between admission and fecal sampling and bacterial diversity (data not shown). When the cohort was divided into two groups with low bacterial diversity (Shannon index < 4.0) and normal diversity (Shannon index > 4.0), there was no intergroup difference in the occurrence of complications such as AKI and ARDS or length of ICU and hospital stay (Fig. 3; ESM Table S3). In accordance with these results, both short- and long-term mortality were not associated with microbiota diversity. Both patient groups were comparable in terms of baseline characteristics, severity of disease, and classes of antibiotics received prior to fecal sampling (Table 1).
Fig. 3.
Decreased intestinal microbiota diversity in critically ill patients is not associated with survival in an exploratory setting. a Microbiota diversity, presented as the Shannon index, was calculated from 15 healthy subjects, as well as from all 34 critically ill patients. Data are presented as dot plot with the mean (solid horizontal line). ****p < 0.0001. b Shannon diversity from all 34 patient samples plotted against the number of different antibiotic classes (categorized as in Fig. 1) that a patient had received prior to fecal sampling. c Based on the range in diversity in healthy subjects, the patient cohort was split into two groups: Shannon index <4 (normal diversity) and Shannon index >4 (high diversity), for which a 90-day Kaplan–Meier survival plot is shown. Numbers below the curve Patients at risk per group
Table 1.
Baseline characteristics of patients with normal or high intestinal microbiota diversity and use of antibiotics prior to fecal sampling
| Baseline characteristics | Normal microbial diversity (Shannon index < 4) | High microbial diversity (Shannon index > 4) | P value |
|---|---|---|---|
| Patients | 17 | 17 | |
| Demographics | |||
| Age (years) | 65.12 ± 13.37 | 62.94 ± 10.96 | |
| Gender (male) | 8 (47.1%) | 8 (47.1%) | |
| Body mass index | 27.09 ± 5.78 | 25.72 ± 4.33 | |
| Race (white) | 15 (88.2%) | 15 (88.2%) | 1 |
| Admission | |||
| Medical admission | 15 (88.2%) | 12 (70.2%) | 0.41 |
| Length of hospital stay prior to ICU admission | 1 [0–2] | 0 [0–2] | 0.70 |
| Admission diagnosis sepsis | 13 (76.5%) | 12 (70.6%) | 0.63 |
| Chronic comorbidity | |||
| None | 5 (29.4%) | 4 (23.5%) | 1 |
| Cardiovascular compromise | 6 (35.3%) | 4 (23.5%) | 0.72 |
| COPD | 2 (11.8%) | 2 (11.8%) | 1 |
| Diabetes mellitus | 3 (17.6%) | 2 (11.8%) | 1 |
| Hypertension | 8 (47.1%) | 6 (35.3%) | 0.74 |
| Malignancy (non-metastatic solid) | 3 (17.6%) | 3 (17.6%) | 1 |
| Renal insufficiency | 3 (17.6%) | 2 (11.8%) | 1 |
| Respiratory insufficiency | 3 (17.6%) | 2 (11.8%) | 1 |
| Modified Charlson Comorbidity Indexa | 4 [3–6] | 4 [3–5] | 0.74 |
| Severity of disease on ICU admission | |||
| APACHE IV score | 71 [59–91] | 95 [79–107] | 0.079 |
| APACHE APS | 62 [47–80] | 82 [64–96] | 0.12 |
| SOFA score | 8 [5.5–10] | 8 [5–10] | 1 |
| Mechanical ventilation | 12 (70.6%) | 15 (88.2%) | 0.40 |
| Organ failure | 15 (88.2%) | 15 (88.2%) | 1 |
| Number of organs failing | |||
| 1 | 4 (23.4%) | 5 (29.4%) | 1 |
| 2 | 7 (41.2%) | 6 (35.3%) | 1 |
| ≥3 | 4 (23.5%) | 4 (23.5%) | 1 |
| Shock | 7 (41.2%) | 9 (52.9%) | 0.71 |
| Antibiotics used prior to sampleb | 0.95 | ||
Data are presented as the median with the interquartile range in square brackets, a number with/without the percentage in parenthesis, or as the mean ± standard deviation, as appropriate
APACHE Acute Physiology and Chronic Health Evaluation, APS acute physiology score, COPD chronic obstructive pulmonary disease, ICU intensive care unit
aModified Charlson Index is calculated including age
bOverall Chi-square test calculated for the number of patients per group that had received a class of antibiotics (as indicated in Fig. 1)
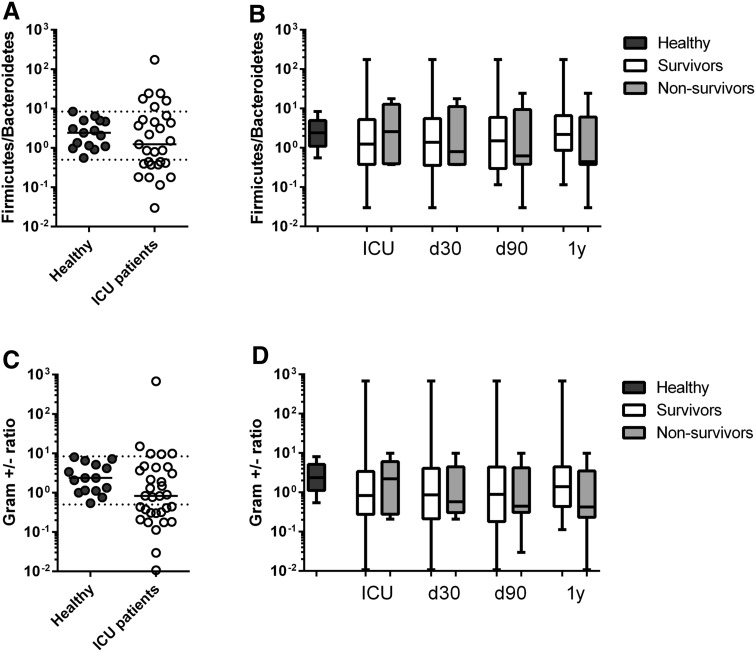
The F/B-ratio was very recently reported to be associated with survival in critically ill patients [12]. The F/B-ratio is known to vary widely in healthy subjects [27], as was the case in our control subjects whose F/B-ratio ranged between 0.5 and 8.4 (median 2.4, IQR 1.1–5.0; Fig. 4). In 11 of our 34 patients the F/B-ratio was <0.5, indicating a relative increase in Bacteroidetes; in six patients, Firmicutes were relatively increased (F/B > 8.4). In five patients, no Bacteroidetes were detected at all, and the F/B-ratio could not be calculated. There was no difference in F/B-ratio between survivors and non-survivors when plotted at different time points (Fig. 4).
Fig. 4.
Firmicutes/Bacteroidetes ratio (F/B-ratio) and Gram-positive/Gram-negative bacteria ratio are not associated with short- and long-term survival in an exploratory setting. a The F/B-ratio was calculated from healthy subjects (dotted horizontal lines range of F/B-ratio in healthy subjects) and from all 34 critically ill patients. and plotted against survival at 4 time points. b Survival data were collected at four time points: during ICU stay (ICU) and at 30 and 90 days and 1 year (d30, d90, 1y, respectively). c The ratio of total Gram-positive/Gram-negative bacteria was calculated from healthy subjects (dotted horizontal lines range of ratio of Gram-positive/Gram-negative bacteria in health subjects. d Survival data collected at same time points as for b. b, d Data are presented as box-and-whisker plots, with median (horizontal line in box), interquartile range (ends of box), and range (whiskers). Black boxes Healthy subjects, white boxes surviving ICU patients, gray boxes non-surviving ICU patients
Similar to our results for the F/B-ratio, the Gram-positive/Gram-negative bacteria ratio varied widely in healthy subjects, ranging between 0.5 and 8.0. In 14 of the 34 patients this ratio was <0.5, indicating a relative increase in Gram-negative bacteria (Fig. 4). In two patients, no Gram-negative bacteria were detected. No differences were found between survivors and non-survivors with respect to the Gram-positive/Gram-negative bacteria ratio (Fig. 4).
Discussion
In this pilot study we performed in-depth fecal microbiota analysis in critically ill patients. We observed marked shifts in fecal bacterial composition of these patients compared to healthy controls. However, a large interindividual diversity was observed at both the phylum and genus levels. No significant differences in microbiota composition were found between patient groups, although patients with an abdominal sepsis showed the strongest changes in relative abundance of Staphylococcus and Escherichia/Shigella. Low bacterial diversity was found in both septic and non-septic patients. In this small cohort, we found no associations between microbiota diversity, F/B-ratio, or Gram-positive/Gram-negative bacteria ratio and outcome parameters, such as the occurrence of complications or mortality.
A common feature in all critically ill patients was the disappearance of genera such as Faecalibacterium, Blautia, Ruminococcus, Subdoligranulum, and Pseudobutyrivibrio. These bacteria degrade complex plant polysaccharides that human cells cannot break down and convert them, for example, into acetate and butyrate, the latter of which is an important energy source for colonic epithelial cells [22]. It is therefore plausible that any microbial disturbance in ICU patients indirectly reduces energy resources. Fecal acetate and butyrate have been shown to be at significantly lower levels in critically ill patients compared to healthy controls [28]. The disappearance of Faecalibacterium prausnitzii, which has known anti-inflammatory properties [25], could potentially promote an unfavorable inflammatory state in the gut. A decreased abundance of Faecalibacterium has been reported in patients with inflammatory bowel disease, psoriasis, and colon cancer [29–31]. SDD has also been reported to affect the numbers of these bacteria [32]. This decrease in beneficial bacteria is in apparent conflict with the potential benefits of SDD in terms of decreased ICU-acquired infections and mortality. As the aim of SDD is to prevent colonization by potential pathogens while preserving anaerobic bacteria, further studies are clearly needed to assess the precise influence of SDD on the microbiota [32]. It will be essential to take these effects into account in the ongoing debate on the use of SDD. In-depth knowledge of the fecal microbiota composition of ICU patients receiving SDD or not will be an important question for follow-up studies.
Knowledge of microbiota changes in critically ill patients has been identified as a key objective for ICU research [33]. A large retrospective cohort study among 10,996 participants of the Health and Retirement Study–Medicare found a strong dose–response relationship between events known to result in microbial disturbance and subsequent hospitalization for severe sepsis [34]. Recent hospitalization for an infection, and especially C. difficile infection, was associated with a higher incidence of rehospitalization for severe sepsis [34]. No microbial analyses were performed in this study. In fact, surprisingly few studies have addressed the microbiota in ICU patients using in-depth sequencing techniques. Two small studies of 12 and 14 ICU patients, respectively, found large interindividual differences in microbiota composition, in line with our findings [12, 13].
A decreased intestinal diversity has been associated with numerous pathogenic conditions, ranging from C. difficile infection, obesity and inflammatory bowel disease to increased mortality following allogeneic HSCT [2, 5, 11]. Our data confirm the expected low bacterial diversity in septic critically ill patients. Strikingly, a similar low diversity was observed in non-septic critically ill, despite great differences in antibiotic treatment. Antibiotics, proton-pump inhibitors, radiation, chemotherapeutics, and poor nutrition may all contribute to the loss of bacterial taxa.
An increased F/B-ratio has been correlated with obesity, asthma, and irritable bowel syndrome [25, 35] and also with ICU mortality in critically ill patients [12]. Our data argue against the usefulness of the F/B-ratio as a microbiota marker in critically ill patients, as both Firmicutes and Bacteroidetes can be strongly but equally decreased. For example, patients no. 6 and 34 in our study had normal F/B-ratios, while still being colonized at 38 and 25%, respectively, with Proteobacteria. We also observed a relative increase in Gram-negative bacteria in 11 patients. A decreased Gram-positive/Gram-negative ratio may be indicative of an increased inflammatory gut status as Gram-negative bacteria produce lipopolysaccharides, some of which are powerful endotoxins.
This study has a number of limitations. As the study started in 2012, sepsis was not defined according to the most recently proposed criteria, including organ failure [36]. However, most patients likely meet these criteria considering that we exclusively studied patients in need of intensive care [36]. It should be emphasized that the sample size is limited and the heterogeneity of critically ill patients substantial; these two limitations make it challenging to establish a potential causal relationship between microbiota composition and prognosis. Furthermore, fecal sampling is trying, as many patients do not defecate. In future studies, rectal swabs could provide a suitable alternative as these have been shown to provide data comparable to stool samples [37]. The single fecal sample studied here should be complemented by longitudinal analyses to provide a dynamic view of the microbiota. It should also be kept in mind that fecal samples do not provide information about the microbiota in the upper gastrointestinal tract. Lastly, the viral and fungal microbiomes were not addressed in this study. This should be a key area of future studies, as these may comprise an important part of the microbiota in ICU patients.
In summary, we found striking abnormalities of the intestinal microbiota in both septic and non-septic critically ill patients, with a large interindividual variation and low bacterial diversity. Numerous questions remain to be answered, including: To what extent does microbial disturbance influence dietary needs, metabolic status, intestinal permeability, and immunity in critically ill patients? Equally important is the question: How might the observed microbiota shifts be leveraged, not only by developing diagnostic and prognostic markers but also therapeutically? Fecal microbiota transplantation is a feasible and effective treatment option for severe C. difficile infection in the ICU [38] and could also become the next big breakthrough in the fight against multi-drug resistant bacteria [3]. Two case reports have recently been published on the successful use of fecal microbiota transplantation in patients with therapy-resistant sepsis and diarrhea [39, 40]. This success raises hope for simple and effective adjunctive therapies based on our expanding knowledge of the gut microbiota that might benefit critically ill patients.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge Maryse A. Wiewel, Friso M. de Beer, Lieuwe D.J. Bos, Gerie J. Glas, Roosmarijn T.M. van Hooijdonk, Michaëla A.M. Huson, Laura R.A. Schouten, Marleen Straat, Esther Witteveen, and Luuk Wieske (Department of Intensive Care, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands) for their participation in data collection. The authors would also like to thank Steven Aalvink (Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands) for his indispensable help in the laboratory work on the fecal samples. This work was supported by the Center for Translational Molecular Medicine [http://www.ctmm.nl; project MARS (Grant 04I-201) and The Netherlands Organization for Scientific Research (ZonMW; Grants 836011024 and 016156475 to WJW; SIAM Gravitation Grant 024.002.002 and Spinoza Award to WMdV)].
Compliance with ethical standards
Conflicts of interest
All authors declare that they have no disclosures.
References
- 1.Blaser MJ. Antibiotic use and its consequences for the normal microbiome. Science. 2016;352:544–545. doi: 10.1126/science.aad9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ianiro G, Tilg H, Gasbarrini A (2016) Antibiotics as deep modulators of gut microbiota: between good and evil. Gut. doi:10.1136/gutjnl-2016-312297 [DOI] [PubMed]
- 3.Pamer EG. Resurrecting the intestinal microbiota to combat antibiotic-resistant pathogens. Science. 2016;352:535–538. doi: 10.1126/science.aad9382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M, Pamer EG. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vincent JL, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K, Investigators EIGo International study of the prevalence and outcomes of infection in intensive care units. JAMA. 2009;302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 7.Kesecioglu J, Eggimann P. What is new in selective decontamination of the digestive tract? Intensive Care Med. 2016;42:1270–1275. doi: 10.1007/s00134-015-4009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Price R, MacLennan G, Glen J, Su DC. Selective digestive or oropharyngeal decontamination and topical oropharyngeal chlorhexidine for prevention of death in general intensive care: systematic review and network meta-analysis. BMJ. 2014;348:g2197. doi: 10.1136/bmj.g2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wischmeyer PE, McDonald D, Knight R. Role of the microbiome, probiotics, and ‘dysbiosis therapy’ in critical illness. Curr Opin Crit Care. 2016;22:347–353. doi: 10.1097/MCC.0000000000000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Debast SB, Bauer MP, Kuijper EJ. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2014;20(Suppl 2):1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 11.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 12.Ojima M, Motooka D, Shimizu K, Gotoh K, Shintani A, Yoshiya K, Nakamura S, Ogura H, Iida T, Shimazu T. Metagenomic analysis reveals dynamic changes of whole gut microbiota in the acute phase of intensive care unit patients. Dig Dis Sci. 2016;61:1628–1634. doi: 10.1007/s10620-015-4011-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, Tirrell M, Tiedje J, Gilbert JA, Zaborina O, Alverdy JC. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. MBio. 2014;5:01314–01361. doi: 10.1128/mBio.01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Vught LA, Klein Klouwenberg PM, Spitoni C, Scicluna BP, Wiewel MA, Horn J, Schultz MJ, Nurnberg P, Bonten MJ, Cremer OL, van der Poll T, MARS Consortium (2016) Incidence, risk factors, and attributable mortality of secondary infections in the intensive care unit after admission for sepsis. JAMA 315:1469–1479 [DOI] [PubMed]
- 15.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 16.Bellomo R, Ronco C, Kellum JA, Mehta RL, Palevsky P, Acute Dialysis Quality Initiative Work group Acute renal failure—definition, outcome measures, animal models, fluid therapy and information technology needs: the Second International Consensus Conference of the Acute Dialysis Quality Initiative (ADQI) Group. Crit Care. 2004;8:R204–R212. doi: 10.1186/cc2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Force ADT, Ranieri VM, Rubenfeld GD, Thompson BT, Ferguson ND, Caldwell E, Fan E, Camporota L, Slutsky AS. Acute respiratory distress syndrome: the Berlin definition. JAMA. 2012;307:2526–2533. doi: 10.1001/jama.2012.5669. [DOI] [PubMed] [Google Scholar]
- 18.Lankelma JM, Cranendonk DR, Belzer C, de Vos AF, de Vos WM, van der Poll T, Wiersinga WJ (2016) Antibiotic-induced gut microbiota disruption during human endotoxemia: a randomised controlled study. Gut. doi: 10.1136/gutjnl-2016-312132 [DOI] [PubMed]
- 19.Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA, Palva A, de Vos WM. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Harris B, Morjaria SM, Littmann ER, Geyer AI, Stover DE, Barker JN, Giralt SA, Taur Y, Pamer EG. Gut microbiota predict pulmonary infiltrates after allogeneic hematopoietic cell transplantation. Am J Respir Crit Care Med. 2016;194:450–463. doi: 10.1164/rccm.201507-1491OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belzer C, de Vos WM. Microbes inside—from diversity to function: the case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Downes J, Vartoukian SR, Dewhirst FE, Izard J, Chen T, Yu WH, Sutcliffe IC, Wade WG. Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int J Syst Evol Microbiol. 2009;59:972–980. doi: 10.1099/ijs.0.000364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MR, Kamboj M, Pamer EG. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev. 2012;70(Suppl 1):S45–S56. doi: 10.1111/j.1753-4887.2012.00505.x. [DOI] [PubMed] [Google Scholar]
- 26.Jalanka J, Salonen A, Salojarvi J, Ritari J, Immonen O, Marciani L, Gowland P, Hoad C, Garsed K, Lam C, Palva A, Spiller RC, de Vos WM. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64:1562–1568. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 27.Human Microbiome Project C Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Keefe SJ, Ou J, Delany JP, Curry S, Zoetendal E, Gaskins HR, Gunn S. Effect of fiber supplementation on the microbiota in critically ill patients. World J Gastrointest Pathophysiol. 2011;2:138–145. doi: 10.4291/wjgp.v2.i6.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eppinga H, Sperna Weiland CJ, Thio HB, van der Woude CJ, Nijsten TE, Peppelenbosch MP, Konstantinov SR (2016) Similar depletion of protective Faecalibacterium prausnitzii in psoriasis and inflammatory bowel disease, but not in hidradenitis suppurativa. J Crohns Colitis 10(9):1067–1075 [DOI] [PubMed]
- 31.Sokol H, Seksik P, Rigottier-Gois L, Lay C, Lepage P, Podglajen I, Marteau P, Dore J. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 32.Benus RF, Harmsen HJ, Welling GW, Spanjersberg R, Zijlstra JG, Degener JE, van der Werf TS. Impact of digestive and oropharyngeal decontamination on the intestinal microbiota in ICU patients. Intensive Care Med. 2010;36:1394–1402. doi: 10.1007/s00134-010-1826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotts JE, Matthay MA. Sepsis: pathophysiology and clinical management. BMJ. 2016;353:i1585. doi: 10.1136/bmj.i1585. [DOI] [PubMed] [Google Scholar]
- 34.Prescott HC, Dickson RP, Rogers MA, Langa KM, Iwashyna TJ. Hospitalization type and subsequent severe sepsis. Am J Respir Crit Care Med. 2015;192:581–588. doi: 10.1164/rccm.201503-0483OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 36.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budding AE, Grasman ME, Eck A, Bogaards JA, Vandenbroucke-Grauls CM, van Bodegraven AA, Savelkoul PH. Rectal swabs for analysis of the intestinal microbiota. PLoS One. 2014;9:e101344. doi: 10.1371/journal.pone.0101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han S, Shannahan S, Pellish R (2016) Fecal microbiota transplant: treatment options for Clostridium difficile infection in the intensive care unit. J Intensive Care Med 31(9):577–786 [DOI] [PubMed]
- 39.Li Q, Wang C, Tang C, He Q, Zhao X, Li N, Li J. Therapeutic modulation and reestablishment of the intestinal microbiota with fecal microbiota transplantation resolves sepsis and diarrhea in a patient. Am J Gastroenterol. 2014;109:1832–1834. doi: 10.1038/ajg.2014.299. [DOI] [PubMed] [Google Scholar]
- 40.Li Q, Wang C, Tang C, He Q, Zhao X, Li N, Li J. Successful treatment of severe sepsis and diarrhea after vagotomy utilizing fecal microbiota transplantation: a case report. Crit Care. 2015;19:37. doi: 10.1186/s13054-015-0738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.